Abstract
Systemic lupus erythematosus (SLE), the prototypic systemic autoimmune disease, is a debilitating multisystem autoimmune disorder characterized by chronic inflammation and extensive immune dysregulation in multiple organ systems, resulting in significant morbidity and mortality. Here, we present a multidisciplinary approach resulting in the identification of neutrophil cytosolic factor 2 (NCF2) as an important risk factor for SLE and the detailed characterization of its causal variant. We show that NCF2 is strongly associated with increased SLE risk in two independent populations: childhood-onset SLE and adult-onset SLE. The association between NCF2 and SLE can be attributed to a single nonsynonymous coding mutation in exon 12, the effect of which is the substitution of histidine-389 with glutamine (H389Q) in the PB1 domain of the NCF2 protein, with glutamine being the risk allele. Computational modeling suggests that the NCF2 H389Q mutation reduces the binding efficiency of NCF2 with the guanine nucleotide exchange factor Vav1. The model predicts that NCF2/H389 residue interacts with Vav1 residues E509, N510, E556, and G559 in the ZF domain of Vav1. Furthermore, replacing H389 with Q results in 1.5 kcal/mol weaker binding. To examine the effect of the NCF2 H389Q mutation on NADPH oxidase function, site-specific mutations at the 389 position in NCF2 were tested. Results show that an H389Q mutation causes a twofold decrease in reactive oxygen species production induced by the activation of the Vav-dependent Fcγ receptor-elicited NADPH oxidase activity. Our study completes the chain of evidence from genetic association to specific molecular function.
Keywords: autoimmunity, NOX2
Fine localization of the polymorphisms responsible for genotype-phenotype correlations is emerging as a difficult hurdle in the implementation and interpretation of genetic association studies. Candidate gene studies and, more recently, genome-wide association studies (GWAS), have begun to elucidate the complex genetic profile of systemic lupus erythematosus (SLE) with identification of ∼30 risk loci (1–3). However, for almost all these identified loci, the causal polymorphism that leads to lupus susceptibility has not been discovered. GWAS have been praised for representing an “agnostic” approach that is unbiased by prior assumptions regarding genetic association with the disease. However, such an approach typically ignores all valuable prior information collected over decades about the pathogenesis and genetic basis of diseases that have been previously studied. This inevitably leads to the inclusion of regions (and additional SNPs) that have little to no possibility of being associated with a disease, increasing the number of tests. More tests mean a more stringent multiple testing correction and a reduction of power or a greater number of subjects to overcome the reduction of power. To avoid this reduction of power, we have developed a two-step bioinformatics-driven design that increases the power of gene association studies using a partial Bayesian approach. The first step uses a family-based study to identify “noteworthy” genes (having a multitest-corrected probability of being associated with SLE of less than 0.5) from a larger panel of genes selected on the basis of increased prior likelihood of association because of their known function or genomic location (4, 5). The second step follows up these noteworthy genes in a targeted investigation.
The hypothesis that neutrophil cytosolic factor 2 (NCF2) is a candidate gene for SLE was derived from testing our bioinformatics-driven approach in a moderately sized family-based linkage study using the family-based transmission disequilibrium test (TDT). In that study, SNP rs2274065 showed likely association with SLE [χ2 = 15.7, P = 7.28 × 10−5, false discovery rate (FDR) = 0.15]. In the present study, we demonstrate highly significant association of NCF2 with SLE by genotyping additional SNPs in two independent case–control populations, identify a causal mutation, and characterize the consequences of the causal mutation on the function of the NADPH oxidase complex.
Results and Discussion
Childhood-onset SLE presents a unique subgroup of patients for genetic studies because of the likelihood of a higher genetic load or higher degree of penetrance driving an earlier disease onset, more severe disease course, greater frequency of family history of SLE, and lesser effect of sex hormones in disease development (6, 7). Therefore, in stage 1, we genotyped an independent cohort of 769 subjects with childhood-onset SLE. In stage 2, we genotyped an additional population of 5,139 North American subjects with adult-onset SLE. Because children can develop SLE later in life, to ensure that our controls are SLE-free, we used healthy adults as controls for both stages. A total of 5,163 controls were genotyped. A number of subjects with SLE and controls were excluded as a result of principal component analysis (PCA), relatedness, or genotyping failure. After applying these quality control and adjustment measures, 4,578 subjects with adult-onset SLE, 663 subjects with childhood-onset SLE, and independent cohorts of 879 controls for the childhood-onset group and 3,910 controls for the adult-onset group were used for analyses. The ethnic background of patients with SLE and controls are detailed in Table S1, and clinical characteristics of the patients with SLE are given in Table S2.
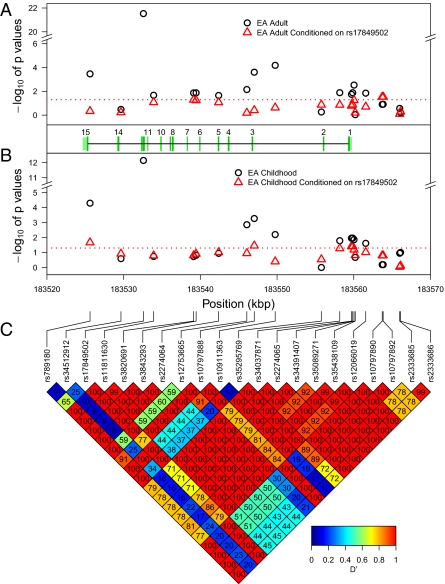
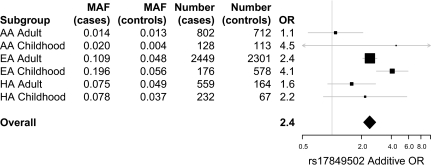
A total of 25 SNPs were genotyped; SNPs were chosen to provide uniform mapping of the gene with the minor allele frequency (MAF) in at least one previously genotyped population exceeding 5%. When appropriate, preference was given to nonsynonymous SNPs. An additional 256 imputed SNPs were included in the analysis. Imputation was performed using IMPUTE2 (8) and the 2010/08/04 release of the 1000 Genomes Project (9) European cohort. As shown in Fig. 1 and Table S3, the typed SNP with the highest association, rs17849502, showed an unparalleled association with P values for European Americans (EA; subjects of European ancestry) of 2.95 × 10−22 in adult-onset SLE and 7.13 × 10−13 in childhood-onset SLE, corresponding to multitest-corrected FDRs of 4.62 × 10−20 and 5.97 × 10−10, respectively. Fig. 2 shows a forest plot depicting the contribution of different ethnicities to the overall odds ratio (OR) and significance of rs17849502. Significant association of rs17849502 was observed only for the EA subgroup. The lack of association in the case of Asian Americans (AsA) and African Americans (AA) is attributable to the extremely low MAF, 0.00, for all cohorts in the case of AsA and an MAF varying from 0.00 to 0.02 in the case of AA. Thus, we have no evidence of the association of rs17849502 with SLE in the AsA and AA populations. The association of this SNP in Hispanic Americans (HA) is inconclusive because the number of subjects was too low to have enough power, given the low MAF of this SNP in the HA population.
Fig. 1.
Association of NCF2 SNPs with SLE. Data are plotted with their P values (shown as −log10 values). Association of NCF2 SNPs with SLE in adult-onset (A) and childhood-onset (B) SLE cases in the EA subgroup with or without conditioning on rs17849502. The positions of exons (green rectangles) and introns (connecting lines) are indicated in the middle plot. The pale green rectangles at the two ends correspond to the UTRs. The dashed horizontal red lines correspond to P = 0.05. (C) LD heat map shows D′ between each of the typed SNPs in the region plotted, with D′ × 100 reported in each diamond. Chromosomal positions are according to National Center for Biotechnology Information build 37.1.
Fig. 2.
Relative contribution of different ethnicities to the overall OR for SNP rs17849502. Forest plot of additive OR ± 95% CI in indicated subgroups and overall OR (Mantel–Haenszel). AsA are not shown because the risk allele of rs17849502 was not found in this population. Size of rectangles is proportional to the weighting of the study in the meta-analysis, the top and bottom of diamond are the overall OR, the left and right whiskers (or left and right diamond edges) are the ±95% CI of the corresponding OR.
In the EA subgroup, 9 SNPs exhibited significant association with SLE in either the childhood-onset or adult-onset group (Fig. 1 and Table S3). Of the 256 imputed SNPs, 16 SNPs showed strong association with SLE after FDR correction (FDR < 10−3; Fig. S1 and Table S4). Conditional regression shows that the significant association is completely explained by rs17849502 (Fig. 1 A and B and Fig. S1). This demonstrates that SNP rs17849502 is the sole independently significant SNP. Therefore, the previously reported association of rs10911363 with SLE (10) was attributable exclusively to linkage disequilibrium (LD) with rs17849502, with a D′ of 1 (Fig. 1C). SNP rs17849502 is a nonsynonymous coding polymorphism in exon 12 of the NCF2 gene, the effect of which is a substitution of histidine (His) 389 with glutamine (Gln) 389 in the PB1 domain of the NCF2 protein. A comparison of a His/His genotype with a Gln/Gln genotype shows that the latter is associated with SLE, with an OR of 12.37 [95% confidence interval (CI): 8.34, 18.37] in adult-onset disease and 33.50 (95% CI: 15.72, 71.39) in childhood-onset disease (Table 1), indicating that individuals with Gln/Gln are far more likely to develop SLE than those with His/His, a finding with significant clinical relevance.
Table 1.
Comparison of genotypes in position 389 of the NCF2 protein
| Adult-onset SLE | Childhood-onset SLE | OR | |||||
| Genotype | Case | Control | Case | Control | Transition | Adult onset | Childhood onset |
| H/H | 2,000 | 2,088 | 123 | 515 | H/H→H/Q | 1.85 (1.69, 2.03) | 2.53 (2.01, 3.2) |
| Q/H | 366 | 206 | 37 | 61 | H/H→Q/Q | 12.37 (8.34, 18.37) | 33.50 (15.72, 71.39) |
| Q/Q | 83 | 7 | 16 | 2 | H/Q→Q/Q | 6.67 (4.46, 10.99) | 13.19 (6.06, 28.73) |
Sample counts of genotypes in position 389 are shown in subjects who have SLE (adult/childhood onset) and controls (EA). ORs for the transition between H/H, H/Q, and Q/Q are shown with 95% CIs in parentheses.
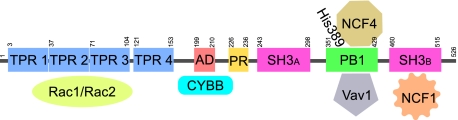
NCF2 is a component of the leukocyte NADPH oxidase complex that produces superoxide (11). The NADPH oxidase complex is composed of multiple subunits, two of which (CYBA and CYBB, also known as NOX2) are localized in cellular membranes, constituting the flavocytochrome b558. On activation, the three cytosolic subunits (NCF1, NCF2, and NCF4) colocalize and translocate to the membrane, where they, along with the membrane subunits and the small GTPase (Rac1 in monocytes/macrophages and Rac2 in neutrophils), form the active NOX2 complex (11, 12). Genetic defects in subunits of NADPH oxidase in humans cause chronic granulomatous disease (CGD), which is associated with life-threatening bacterial and fungal infections (13).
To add further complexity, there is evidence for both positive and negative regulation of the NADPH oxidase complex. In resting cells, Rac-GDP is present as a complex with Rho-GDP disassociation inhibitor, a negative regulator of Rho family GTPases, but this complex rapidly dissociates in stimulated cells, forming Rac-GTP (12). This process is facilitated by activation of guanine nucleotide exchange factors (GEFs) and is accompanied by translocation of Rac to the plasma membrane (11, 12). Vav1 is such a GEF found in the cytoplasm of hematopoietic lineage cells. On phosphorylation, Vav1 has GEF activity for the Rac GTPase, facilitating Rac's transition from an inactive GDP-bound state to an active GTP-bound state (14). Recent work demonstrated a direct interaction of Vav1 with the C terminus of NCF2 that enhances the GEF activity of Vav1 and causes a positive feedback loop for amplifying Rac activation (15).
We next asked whether the H389Q mutation in the PB1 domain of NCF2 affects any of its multiple protein-protein interactions. The binding sites on NCF2 to Rac1/2, CYBB, and NCF1 are clearly outside the PB1 domain (11, 12). Although the PB1 domain of NCF2 interacts with the PB1 domain of NCF4, the structure of this heterodimeric complex (16) shows that residue H389 makes no contact with NCF4 and is highly exposed in the complex; hence, the mutation H389Q is unlikely to affect the NCF2/NCF4 interaction. The structure of the NCF2–Vav1 complex is not resolved. To test if the mutation H389Q is likely to affect the binding with Vav1, we constructed a model of interaction using a protein modeling method developed by one of the authors (M.E.), which has been demonstrated to be highly accurate in predicting other protein-protein interactions (17).
The modeling consisted of several stages, most of which used only the structures of the molecules and their physical properties, and did not require that H389 be at the binding interface. First, His binding sites on the entire surface of Vav1 (His anchoring spots) were mapped using ANCHORSmap, a computational procedure developed by one of the authors (M.E.) (18). The existence of low ΔG His anchoring spots indicated that an interface can be formed in which an exposed His residue from a binding partner acts as an anchoring hot spot. To test if NCF2 is the relevant binding partner, we evaluated the physicochemical surface complementarity between NCF2 and Vav1 utilizing a multistep approach. Most of the steps used only the structures of the molecules and their physical properties, and did not require that H389 be at the binding interface. In the first step, protein-protein docking between NCF2 and Vav1 was conducted using the geometric-electrostatic-hydrophobic (GEH) version of the MolFit program (19). Because experimental data regarding the interaction interface between NCF2 and Vav1 are lacking, the docking scans were based only on the physicochemical properties of the molecules calculated from their coordinates. Hence, in the docking scans, the molecules were not guided to interact via NCF2/H389 or to avoid the NCF2/NCF4 interface. The MolFit program produced an ensemble of 12,256 docking models evaluated by GEH score. These models underwent postscan filtering in which additional properties of the interface, desolvation energy and statistical propensity measures, were tested and rescored (20). This postscan filtering was passed by 366 docking models. In the next step, a condition was added that the residue NCF2/H389 should be located at the interface; this condition was fulfilled only in 12 cases. In the last step, the 12 docking models were matched with the anchoring spots map, searching for docking solutions in which H389 is positioned near a His anchor with binding ΔG less than −4 kcal/mol. The distances between the Cβ and Cε1 atoms of H389 and the corresponding atoms in nearby anchors were calculated and averaged. In three cases, H389 was located within 3 Å of an anchor; in the other cases, the distance was significantly larger (5 Å or more) or there was no nearby anchoring site. Remarkably, the highest scoring docking model was the one that used an anchoring site with highest preference of His over Gln, by 1.5 kcal/mol. Two additional tests were used to validate the model. First, it was verified that the model allowed simultaneous binding of the PB1 domain of NCF2 to Vav1 and NCF4 without clash. Second, in agreement with experimental results, the best model of simultaneous binding of Vav1 and NCF4 to NCF2 involved the Rac-binding conformation of Vav1.
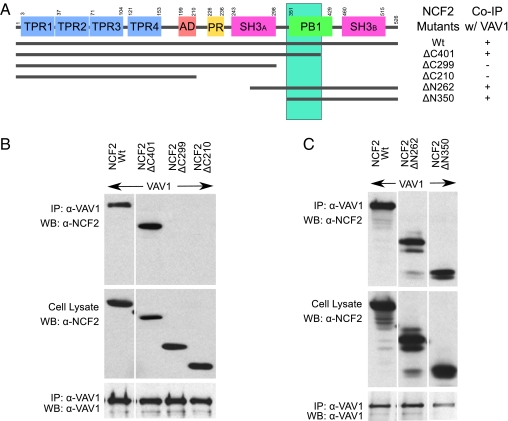
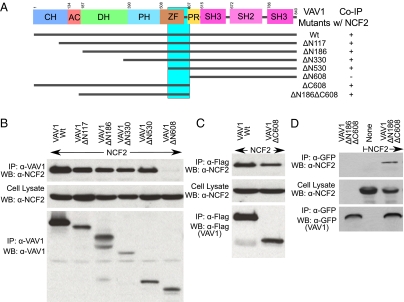
The NCF2/Vav1 interaction model thus obtained was supported by the available information and led to the formation of the hypothesis that the H389Q mutation will affect the interaction of NCF2 with Vav1, and consequently alter the function of the NADPH oxidase complex. To test the first part of this hypothesis, we conducted coimmunoprecipitation (Co-IP) experiments showing that NCF2 binding to Vav1 involves their respective PB1 and ZF domains. A series of Co-IP experiments of deletion and truncation constructs of NCF2 (Fig. 3) and Vav1 (Fig. 4) provides strong evidence that the PB1 domain of NCF2 is directly involved in binding to the ZF domain (also known as the C1 domain) of Vav1.
Fig. 3.
(A) Summary of interaction of truncated NCF2 derivatives with Vav1 as determined by Co-IP experiments and proposed binding region (vertical green bar) for Vav1 based on these experiments. (B and C) Immunoprecipitation (IP) of full-length or truncated NCF2 mutants with full-length Vav from lysates prepared from COS7 cells transiently transfected with corresponding expression constructs. Lysates were immunoprecipitated with a rabbit polyclonal antibody against Vav1 (Santa Cruz Biotechnology), and immunoprecipitates and lysates were analyzed by immunoblotting with the indicated antibodies. To detect NCF2, either a rabbit polyclonal antibody that reacts with the N terminus of NCF2 (Santa Cruz Biotechnology) or a mouse monoclonal antibody directed against the C terminus of NCF2 (BD Transduction Laboratories) was used in B and C, respectively. Samples shown in each panel were run on the same gel from the same experiment, but irrelevant lanes were excised for simplicity and clarity of presentation. Upon C-terminal truncation of NCF2, the removal of the PB1 domain led to a failure to coimmunoprecipitate with Vav1 (B, lanes ΔC299 and ΔC210), in contrast to an NCF2 derivative that included only the C-terminal PB1 and SH3 domains (C, lanes ΔN262 and ΔN350). WB, Western blot; Wt, wild type.
Fig. 4.
(A) Summary of interactions of truncated Vav1 derivatives with NCF2 as determined by Co-IP experiments and proposed binding region (vertical cyan bar) for NCF2. (B–D) Immunoprecipitation of full-length or truncated Vav1 mutants with full-length NCF2 from lysates prepared from COS7 cells transiently transfected with corresponding expression constructs. Lysates were immunoprecipitated with antibodies directed against Vav1 or against epitope tags present on truncated Vav1 derivatives as indicated, and immunoprecipitates and lysates were analyzed by immunoblotting with the indicated antibodies. A Vav1 derivative lacking the ZF domain failed to coimmunoprecipitate with NCF2 (B, lane ΔN608), in contrast to full-length Vav1 or other truncated Vav1 proteins that included the ZF domain (B–D, all lanes except ΔN608). IP, immunoprecipitation; WB, Western blot.
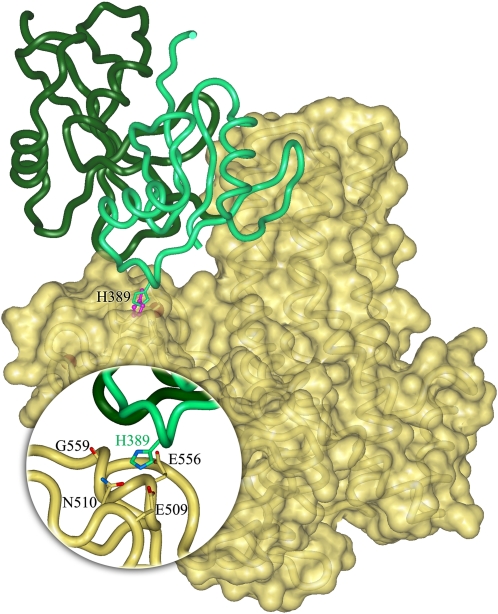
The docking model presented in Fig. 5 fits all the available experimental information. Accordingly, our model suggests that the NCF2 H389 residue sits in a pocket on the surface of the Vav1 ZF domain, surrounded by Vav1 residues E509, N510, E556, and G559. In addition, an adjacent loop of NCF2 interacts with the C-terminal part of domain DH of Vav1 (residues 341–361) and stabilizes a conformation adequate for Rac binding. Previous studies have suggested that a R394Q substitution in mouse NCF2 (21) and a R395W substitution in human NCF2 (22) affected NCF2 interaction with NCF4 and resulted in CGD. Indeed, the experimental structure of the NCF2/NCF4 complex and our model support the findings that NCF2 amino acids 394 and 395 are involved in NCF2 binding to NCF4 but not in the binding with Vav1.
Fig. 5.
Model of the interaction of NCF2 with Vav1 via His389 in the presence of NCF4. Starting coordinates of Vav1 and NCF2 were taken from the PDB. The structure of the NCF2 PB1 domain was extracted from the complex with the PB1 domain of NCF4 [chain A in PDB entry 1oey (16)], and the structure of the DH-PH-ZF domains of Vav1 was taken from the complex with full-length Rac1 [chain B in PDB entry 2vrw (42)]. Coordinates of missing residues in the Vav1 domain DH were completed by superimposing the corresponding domain in the autoinhibited structure of Vav1 [PDB entry 3ky9 (43)]. The solvent-accessible surface of Vav1 is shown in yellow; NCF2 is shown as a ribbon diagram in green with H389 indicated. The nearby His anchoring spot is shown in magenta. NCF4 is shown in dark green. (Inset) Interaction site of H389 detected in the anchoring spots mapping is highlighted. The surface of Vav1 was made transparent to show the side chains of residues that are within 3 Å and interact with H389 in the docking model.
Most importantly, the model predicts a significantly reduced binding of NCF2 to Vav1 as a consequence of an H389Q mutation. Gln binds 1.5 kcal/mol weaker than His at the predicted position. Considering that the GTP-bound form of Rac is the physiological mediator of oxidase activation in the NOX2 oxidase (11, 12), decreased binding of Vav1 to NCF2 should decrease amplification of Rac activation, which, in turn, should decrease production of reactive oxygen species (ROS).
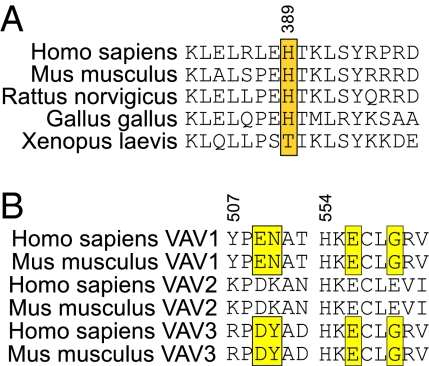
NCF2 His-389 is conserved across species (Fig. 6), suggesting that there is selective pressure to maintain this residue. Functional activation of neutrophils and macrophages via integrins or Fcγ receptors (FcγR) is markedly impaired in mice lacking both Vav1 and Vav3 (23–25), consistent with the finding that these are the main Vav isoforms expressed by these cells (23, 24). Vav1 residues E509, N510, E556, and G559 in the ZF domain, predicted to interact with NCF2 His-389 (Fig. 5), are either identical in Vav3 or have conservative substitutions capable of mediating interactions with NCF2 His-389 (Fig. 6).
Fig. 6.
Conservation of H389 in NCF2 and its target amino acids in the binding pocket in the ZF domain of Vav. (A) H389 (boxed in yellow) is conserved from Gallus gallus (chicken) to human. (B) Sequence alignment of amino acid residues in the binding pocket in the ZF domain of the three Vav isoforms in the human and mouse are shown. Each human Vav isoform is identical to its relevant mouse homolog. The target amino acids (509, 510, 556, and 559) in Vav1 (yellow) are identical or have conservative substitution in Vav3 (yellow). Replacement of E by D in position 509 is considered a conserved charge; position 510 is less conserved, but the model suggests that NCF2 H389 binds Y510 similar to N510. The conservation of P508 and the general similarity of the character of the residues indicate that this fragment has a similar fold in the three Vav isoforms. The second fragment is even better conserved. E556 is identical in all three variants, and G559 is conserved in Vav1 and Vav3.
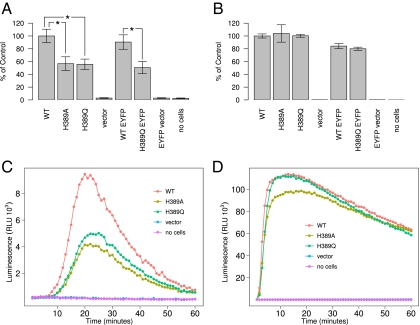
To test the prediction that NCF2 H389Q would reduce the function of NADPH oxidase, site-specific mutations at the 389 position in NCF2 were tested experimentally. Experiments were conducted in human K562 myeloid cells expressing endogenous CYBA, CYBB, and NCF1 as stable transgenes and transiently expressing NCF2 and NCF4. We compared WT NCF2 (with His at position 389) and NCF2 with either Gln or alanine substitutions at position 389. We also compared WT NCF2 and H389Q NCF2 derivatives tagged at the C terminus with enhanced yellow fluorescent protein (EYFP). NADPH oxidase was activated either by cross-linking FcγR with IgG-opsonized latex beads or by phorbol 12-myristate 13-acetate (PMA). ROS production was measured by a luminescence-based assay system. As shown in Fig. 7, IgG bead-elicited ROS responses for either the Q389 or A389 mutation were reduced by approximately twofold compared with WT NCF2, whereas there was no effect on PMA-elicited ROS release. The differential effect of NCF2 H389 substitutions on PMA- vs. IgG bead-elicited ROS release is especially relevant, because it is consistent with the lack of participation of any Vav isoform in PMA-elicited NADPH oxidase activity, whereas Vav1 and Vav3 are essential for FcγR-elicited NADPH oxidase activity (25). Furthermore, these results are consistent with previous results in which an R395W mutation reduced the PMA-induced (Vav-independent) NADPH oxidase activity and caused CGD (16), supporting the notion that amino acid 395 is involved in the binding to NCF4 rather than to Vav1.
Fig. 7.
NCF2 H389Q mutation leads to decreased ROS release in K562 cells. K562 cells stably transfected with CYBB and NCF1 were cotransfected with plasmids for expression of NCF4 and NCF2 WT, NCF2 H389A mutant, or NCF2 H389Q mutant with the Amaxa Nucleofector kit V, under the T-16 program. Derivatives of NCF2 tagged at the C terminus with EYFP were also tested. According to our model, a Q in position 389 binds 1.5 kcal/mol weaker than H, whereas an A binds 2.0 kcal/mol weaker than H in this position. (A) ROS production in K562 cells in response to hIgG beads in the presence of luminol and HRP (n = 4; mean ± SEM; *P < 0.05). (B) ROS production in K562 cells in response to PMA in the presence of isoluminol and HRP (n = 4; mean ± SEM; In A and B, the total relative light unit (RLU) values over 60 min, measured at 1-min intervals, are expressed as the percentage of the RLU output of NCF2 WT-transfected cells in each experiment. Data were normalized to correct for interexperiment variability and log-transformed to stabilize variance; they were tested for significance using the Student t test. Representative kinetic curves are taken from one of four independent experiments for ROS production in K562 cells in response to hIgG beads (C) or PMA (D).
It is noteworthy that a variant NCF1 allele identified by positional cloning based on its association with the severity of pristane-induced arthritis in a susceptible rat strain results in a reduction in neutrophil oxidase activity of a similar magnitude (26). Hence, a twofold reduction in NADPH oxidase activity is highly likely to be biologically significant.
That a CGD phenotype is not seen in our SLE subjects even in the presence of a homozygous 389Q genotype indicates that the ability of phagocytes to generate a respiratory burst sufficient for antimicrobial activity is not impaired by reduced NCF2 function. However, in other immune cells (e.g., antigen-presenting cells), ROS production is much more limited (26–28) and the role of ROS is not microbicidal. NADPH oxidase activity regulates phagosomal pH, and derivative ROS can function as signaling molecules within and between neighboring immune cells; reduced levels of ROS in these cells may thus influence antigen processing, immunoregulation, control of cell activation, and differentiation (28, 29).
Our results corroborate more recent findings suggesting that ROS produced by the NOX2 complex are used in fine-tuning inflammatory responses, depending on when, where, and at what level the ROS are produced (29, 30). A point mutation in NCF1 results in decreased ROS production (comparable to those seen in our H389Q experiments), leading to increased susceptibility to autoimmune arthritis in rodents (31, 32). Furthermore, both patients who have CGD and carriers have a higher risk for developing autoimmune diseases (33). Finally, patients who have CGD and mothers carrying X-linked CYBB mutations have a 5–10% incidence of cutaneous and/or mucosal lesions characteristic of those seen in SLE (34–36).
The present study demonstrates the advantages of our study design, where the utilization of prior information coupled with a two-step experimental approach allows for more efficient identification of disease-associated genes. Our findings demonstrate that a single identified mutation in NCF2 leads to decreased ROS production and plays a role in predisposition to SLE. This finding provides further support for the emerging paradigm shift that ROS are not solely proinflammatory byproducts of cellular responses to infectious or inflammatory stimuli but may have a more nuanced function in immunoregulation and inflammation-limiting processes, with notable consequences for autoimmune disease development.
Materials and Methods
Recruitment and Biological Sample Collection.
Subjects were enrolled in the Lupus Genetic Study Groups at the University of Southern California, Oklahoma Medical Research Foundation, University of Alabama at Birmingham, and Imperial College London using identical protocols. All patients met the revised 1997 American College of Rheumatology criteria for the classification of SLE (37). Controls were collected in parallel with the SLE cases in the same institutions. Ethnicity was self-reported and verified by parental and grandparental ethnicity when known. Controls were defined as adults with self-reported absence of SLE or any other autoimmune disease in the subjects or their first-degree relatives. Blood samples were collected from each participant, and genomic DNA was isolated and stored using standard methods. Cases were defined as childhood onset according to the criterion that the diagnosis of SLE was made before the age of 13 y by at least one pediatric rheumatologist participating in the study. Previously typed trios from our published TDT studies (5) were not regenotyped, and they were not used in the present study. The University of Southern California Institutional Review Board for research on human subjects approved this study. The study was also approved by human subject institutional review board at each institution where subjects were recruited. Informed consent was obtained from all subjects.
Genotyping.
Genotyping was performed using Illumina iSelect Infinium II Assays on the BeadStation 500GX system (Illumina). For analysis, only genotype data from SNPs with a call frequency greater than 90% in the samples tested and an Illumina GenTrain score greater than 0.7 were used. GenTrain scores measure the reliability of SNP detection based on the distribution of genotypic classes. The average SNP call rate for all samples was 97.18%. To minimize sample misidentification, data from 91 SNPs that had been previously genotyped on 42.12% of the samples were used to verify sample identity. In addition, at least one sample previously genotyped was randomly placed on each Illumina Infinium BeadChip and used to track samples throughout the genotyping process.
Stratification Analyses.
To account for potential confounding substructure or admixture in these samples, PCAs were performed (38) for all samples using a large set of SNPs (18,446, which were genotyped on these subjects as part of a larger effort). Four principal components were identified that explained a total of ∼60% of the observed genetic variation. These were used to identify individuals who were genetically distant from other samples in the same ethnic subset, and thus capable of introducing admixture bias. Increasing the number of principal components to 10 did not significantly change the inflation factor. We then performed genomic control analysis to calculate the inflation factor λ using the same set of SNPs. This yielded a λ of 1.13 in EA samples, 1.03 in HA samples, 1.08 in AA samples, and 1.04 in AsA samples. Q/Q plots were performed to verify a lack of departure from unity and possible substructure (Fig. S2). The Q/Q plots show no meaningful systematic departure from expectation (i.e., line of unity), suggesting that population structure is not a biasing influence in these results.
Statistical Analyses.
Testing for association in the case–control studies was completed using the freely available programs SNPGWA (http://www.phs.wfubmc.edu/web/public_bios/sec_gene/downloads.cfm) and PLINK (39). For each SNP, missing data proportions for cases and controls, MAF, and exact test results for departures from Hardy–Weinberg expectations were calculated. In addition to an allelic test of association, the additive genetic model was used as the primary hypothesis of statistical inference. The R module genetics (available from http://cran.rproject.org/web/packages/genetics/index.html) were used to estimate the LD between markers and haplotype structures in different ethnicities.
To have separate and independent control groups for both adult-onset and childhood-onset cases, the control cohort was split into two control groups at random to achieve a ratio of childhood to adult controls the same as the ratio of childhood to adult cases in all subgroups, except the EA subgroup, where the ratio was threefold the ratio of childhood to adult cases to maximize the power of the childhood stage while maintaining the power of the adult stage.
FDR estimates using q values were calculated for different ethnicities using the q value package (available from http://cran.r-project.org), which implements the q value extension of FDR (40). The FD for combined results were estimated using the Benjamini and Hochberg procedure (41), because the proportion of correctly rejected null hypotheses was possibly overestimated when using the q value extension and the Benjamini and Hochberg procedure provides a more conservative estimation of FDR (with less power). The FDR corresponds to the proportion of false-positive results among the total results. Thus, an estimate of FDR less than 0.05 signifies that less than 5% of the results accepted as true are false-positive results and is taken as a measure of significance.
Molecular Modeling of the NCF2/NCF4/Vav1/Rac1 Complex.
Coordinates of the molecules.
Starting coordinates of Vav1 and NCF2 were taken from the Protein Data Bank (PDB). The structure of the NCF2 PB1 domain was extracted from the complex with the PB1 domain of NCF4 [chain A in PDB entry 1oey (16)], and the structure of Vav1 was taken from the complex with Rac [chain B in PDB entry 2vrw (42)]. Coordinates of missing residues in Vav1 domain DH were completed by superposing the corresponding domain in the autoinhibited structure of Vav1 [PDB entry 3ky9 (43)].
Computational anchoring spots mapping.
ANCHORSmap (developed by M.E.) identifies preferred binding sites of amino acid side chains (anchors) on the surface of a protein, taking into consideration that this amino acid is part of a hypothetical protein (18). The procedure detects small cavities and subcavities on the surface of a protein, which are adequate for binding single amino acid side chains. Next, thousands of amino acid probes are scattered near the cavities, and optimal probe positions are determined through iterative energy minimization and spatial clustering. The binding ΔGs of the optimally posed probes are calculated by means of a scoring function that includes van der Waals, electrostatic, and solvation energy terms. The electrostatic energy is corrected for the dielectric shielding exerted by the approaching protein. Finally, the anchoring spots are clustered to produce an ensemble of mean anchoring spots, which were found to be very useful for detecting interacting surfaces. The standard parameters described by Ben-Shimon and Eisenstein (18) were used in the current study. Only anchoring spots with ΔG less than −4 kcal/mol were considered in subsequent analyses, because such low-energy anchoring spots often correspond to experimentally detected hot spots (18).
Rigid body docking with MolFit.
Protein-protein docking was executed using the docking program MolFit4. MolFit performs a comprehensive stepwise rotation/translation docking scan. It uses grid representations of the molecules and fast Fourier transformations to evaluate the surface complementarity for each pose (44). In this study, we used the GEH version of MolFit, which tests the shape complementarity of the contacting surfaces, together with the electrostatic (45) and hydrophobic (19) complementarity. Standard translation and rotation grid intervals, 1.05 Å and 12°, respectively, were used. The resulting lists of docking models were intersected to produce a list of models evaluated by a GEH score (19). Higher scores indicate better shape, electrostatic, and hydrophobic complementarities of the interacting surfaces.
Propensity and solvation postscan filter.
The comprehensive docking scan was followed by an efficient postscan filtering and rescoring procedure that tests additional descriptors of the interfaces (20). The procedure determines the interface core (46) for each docking model and calculates several interface core residue and residue-residue propensities and a measure of the spatial clustering of the interface core atoms. These descriptors, together with the whole interface solvation energy, are used in class-specific filters that retain models for which any M of the N descriptors exceed their calibrated thresholds. These models are reevaluated by class-specific scoring functions that combine the same interface descriptors with the GEH score.
Normal modes analysis.
The analysis was performed using the ElNemo Web server, which computes the low-frequency normal modes of a protein (http://igs-server.cnrs-mrs.fr/elnemo) (47). The low-frequency normal modes (excluding the 6 translational and rotational modes) represent gross deformations of the molecule, such as domain movements.
Plasmid Constructs.
Constructs for mouse Vav1 and human NCF2 cDNAs were subcloned into pcDNA3.1 (Invitrogen), NCF2 in pEYFP-N1 (BD Biosciences Clontech), and human NCF4 in pRK5 (BD Biosciences) as previously described (15, 48). Site-directed mutagenesis was performed in NCF2 in pcDNA3.1 using the QuikChange site-directed mutagenesis kit (Stratagene) to generate NCF2 H389A. An NCF2 H389Q cDNA was synthesized by Genscript and cloned into pcDNA3.1 and pEYFP-N1. The constructs were confirmed by sequencing.
Truncated derivatives of human NCF2 were generated using internal restriction sites or PCR, except for the fragments N1-210 and N1-299, which were gifts from D. Lambeth (Emory University, Atlanta, GA) and cloned into pCDNA3.1. Truncated derivatives of mouse Vav1 generated in a similar fashion were also cloned into pcDNA3.1 or pCMV-Tag3c (Myc tagged; Stratagene). Some derivatives were based on a truncated derivative of Vav1 tagged at the N-terminus with green fluorescence protein using pEGFP-C3 (BD Biosciences Clontech), or a full-length Vav1 tagged at the N-terminus with a Flag epitope tag (gift of D. Billadeau, Mayo Clinic, Rochester MN). Details of constructs are available on request.
Analysis of Interactions Between NCF2 and Vav1.
Immunoprecipitation experiments to analyze interactions between NCF2 and Vav1 derivatives expressed in COS7 cells were performed as previously described (15). COS7 cells were transiently transfected with expression constructs for either full-length Vav1 in combination with full-length or truncated NCF2 derivatives or full-length human NCF2 and truncated Vav1 derivatives. Lysates were immunoprecipitated with antibodies against either Vav1, NCF2, Myc (9E10; Upstate Biotechnology), or Flag (F-3165; Sigma–Aldrich) as appropriate, and lysates and immunoprecipitates were analyzed by immunoblotting (15).
Preparation of Transfected Lines for NADPH Oxidase Activity Studies.
K562 myeloid cells (American Type Culture Collection) were grown in RPMI 1640 with 10% (vol/vol) FCS and 1% penicillin/streptomycin at 37 °C in 5% CO2 as previously described (48). To generate K562 cells stably expressing human CYBB and NCF1, K562 cells were transected with the CYBB cDNA in pEF-PGKpac and NCF2 cDNA in pEF-PGKhygro (49) using an Amaxa Nucleofector (Lonza) and Amaxa program T-16. Cells were selected in the presence of 250 μg/mL hygromycin B and 2 μg/mL puromycin. For analysis of NADPH oxidase activity, the Amaxa Nucleofector kit V (Lonza) was used to transfect 2 × 106 CYBB/NCF1-expressing K562 cells transiently with 2 μg of pRK5-pNCF4 and 2 μg of pcDNA3-pNCF2 WT or mutant (H389Q or H389A). After 24 h, transfected K562 cells were collected for NADPH oxidase assays and for immunoblot analysis. NCF1, NCF2, and NCF4 expression was evaluated by immunoblotting as described (48) using anti-NCF1 monoclonal antibody (BD Biosciences Clontech), anti-NCF2 monoclonal antibody (BD Biosciences Clontech), and anti-NCF4 polyclonal antibody (Millipore), respectively. Representative blots are shown in Fig. S3.
NADPH Oxidase Activation in K562 Cells.
NADPH oxidase activity was assayed using chemiluminescence enhanced by luminol or isoluminol, which is membrane-impermeable as described (48); both compounds detect superoxide in a peroxidase-dependent reaction (48). In our hands, the chemiluminescence signal for luminol is greater than for isoluminol; thus, we used the former to assay human IgG (hIgG) latex bead-elicited ROS release more sensitively, which is less than that elicited by PMA. Note that although the K562 cells used in these studies bind hIgG beads, they are not ingested. For measurement of ROS release, PMA (300 ng/mL) or hIgG-latex beads (6.25 × 105 hIgG-latex particles) were used to activate 2 × 105 K562-CYBB/NCF1 cells, which were cotransfected the previous day with plasmids for expression of NCF4 and WT or mutant NCF2, in the presence of 20 μM isoluminol (for PMA stimulation) or 20 μM luminol (for hIgG-latex bead stimulation) and 20 units/mL HRP, without or with superoxide dismutase (SOD; final concentration of 75 μg/mL). A Spectromax L two-channel microplate luminometer (Molecular Devices) was used to record luminescence every 1 min at 37 °C for a total of 60 readings. Chemiluminescence was not detected in the presence of SOD. Activity was normalized to NCF2 expression determined by densitometry.
Supplementary Material
Acknowledgments
We thank Y.X. Wu for technical assistance, C. Marchal and M. Stefanovic for assistance with generating some of the NCF2 expression constructs, J. Matute and X. Li for generating the transgenic K562 cell line, and Dr. XingPing Cui for fruitful discussions. Support for this study was provided by National Institutes of Health Grants AR043815 (to C.O.J.); HL45635 (to M.C.D.); AR62277 (to K.L.M.); AI08394, AI024717, AR042460 (to J.B.H.); AR049084 and AR33062 (to R.P.K.); and RR020143 and AI063274 (to P.M.G.). Additional support was provided by the Alliance for Lupus Research (C.O.J., J.B.H., and R.Z.), Riley Children's Foundation (M.C.D.), Children's Discovery Institute (M.C.D.), and US Department of Veterans Affairs Medical Center, Cincinnati (J.B.H.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
See Author Summary on page 359.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1113251108/-/DCSupplemental.
References
- 1.Tan W, et al. BIOLUPUS Network. GENLES Network Association of PPP2CA polymorphisms with systemic lupus erythematosus susceptibility in multiple ethnic groups. Arthritis Rheum. 2011;63:2755–2763. doi: 10.1002/art.30452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deng Y, Tsao BP. Genetic susceptibility to systemic lupus erythematosus in the genomic era. Nat Rev Rheumatol. 2010;6:683–692. doi: 10.1038/nrrheum.2010.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harley JB, et al. International Consortium for Systemic Lupus Erythematosus Genetics (SLEGEN) Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40:204–210. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Armstrong DL, Jacob CO, Zidovetzki R. Function2Gene: A gene selection tool to increase the power of genetic association studies by utilizing public databases and expert knowledge. BMC Bioinformatics. 2008;9:311. doi: 10.1186/1471-2105-9-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jacob CO, et al. Identification of novel susceptibility genes in childhood-onset systemic lupus erythematosus using a uniquely designed candidate gene pathway platform. Arthritis Rheum. 2007;56:4164–4173. doi: 10.1002/art.23060. [DOI] [PubMed] [Google Scholar]
- 6.Cassidy J, Petty R. Textbook of Pediatric Rheumatology. Philadelphia: Elsevier Saunders; 1996. [Google Scholar]
- 7.Lehman T. Dubois’ lupus erythematosus. In: Wallace D, Hahn B, editors. Dubois’ Lupus Erythematosus. Philadelphia: Lippincott Williams & Wilkins; 2002. pp. 863–884. [Google Scholar]
- 8.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anonymous , 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gateva V, et al. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat Genet. 2009;41:1228–1233. doi: 10.1038/ng.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nauseef WM. Assembly of the phagocyte NADPH oxidase. Histochem Cell Biol. 2004;122:277–291. doi: 10.1007/s00418-004-0679-8. [DOI] [PubMed] [Google Scholar]
- 12.Groemping Y, Rittinger K. Activation and assembly of the NADPH oxidase:A structural perspective. Biochem J. 2005;386:401–416. doi: 10.1042/BJ20041835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stasia MJ, Li XJ. Genetics and immunopathology of chronic granulomatous disease. Semin Immunopathol. 2008;30:209–235. doi: 10.1007/s00281-008-0121-8. [DOI] [PubMed] [Google Scholar]
- 14.Katzav S. Vav1: A hematopoietic signal transduction molecule involved in human malignancies. Int J Biochem Cell Biol. 2009;41:1245–1248. doi: 10.1016/j.biocel.2008.11.006. [DOI] [PubMed] [Google Scholar]
- 15.Ming W, Li S, Billadeau DD, Quilliam LA, Dinauer MC. The Rac effector p67phox regulates phagocyte NADPH oxidase by stimulating Vav1 guanine nucleotide exchange activity. Mol Cell Biol. 2007;27:312–323. doi: 10.1128/MCB.00985-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilson MI, Gill DJ, Perisic O, Quinn MT, Williams RL. PB1 domain-mediated heterodimerization in NADPH oxidase and signaling complexes of atypical protein kinase C with Par6 and p62. Mol Cell. 2003;12:39–50. doi: 10.1016/s1097-2765(03)00246-6. [DOI] [PubMed] [Google Scholar]
- 17.Eisenstein M, Ben-Shimon A, Frankenstein Z, Kowalsman N. CAPRI targets T29-T42: Proving ground for new docking procedures. Proteins. 2010;78:3174–3181. doi: 10.1002/prot.22793. [DOI] [PubMed] [Google Scholar]
- 18.Ben-Shimon A, Eisenstein M. Computational mapping of anchoring spots on protein surfaces. J Mol Biol. 2010;402:259–277. doi: 10.1016/j.jmb.2010.07.021. [DOI] [PubMed] [Google Scholar]
- 19.Berchanski A, Shapira B, Eisenstein M. Hydrophobic complementarity in protein-protein docking. Proteins. 2004;56:130–142. doi: 10.1002/prot.20145. [DOI] [PubMed] [Google Scholar]
- 20.Kowalsman N, Eisenstein M. Combining interface core and whole interface descriptors in postscan processing of protein-protein docking models. Proteins. 2009;77:297–318. doi: 10.1002/prot.22436. [DOI] [PubMed] [Google Scholar]
- 21.Sancho-Shimizu V, Malo D. Sequencing, expression, and functional analyses support the candidacy of Ncf2 in susceptibility to Salmonella typhimurium infection in wild-derived mice. J Immunol. 2006;176:6954–6961. doi: 10.4049/jimmunol.176.11.6954. [DOI] [PubMed] [Google Scholar]
- 22.Patiño PJ, et al. Molecular characterization of autosomal recessive chronic granulomatous disease caused by a defect of the nicotinamide adenine dinucleotide phosphate (reduced form) oxidase component p67-phox. Blood. 1999;94:2505–2514. [PubMed] [Google Scholar]
- 23.Gakidis MAM, et al. Vav GEFs are required for β2 integrin-dependent functions of neutrophils. J Cell Biol. 2004;166:273–282. doi: 10.1083/jcb.200404166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Graham DB, et al. Neutrophil-mediated oxidative burst and host defense are controlled by a Vav-PLCgamma2 signaling axis in mice. J Clin Invest. 2007;117:3445–3452. doi: 10.1172/JCI32729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Utomo A, Cullere X, Glogauer M, Swat W, Mayadas TN. Vav proteins in neutrophils are required for FcgammaR-mediated signaling to Rac GTPases and nicotinamide adenine dinucleotide phosphate oxidase component p40(phox) J Immunol. 2006;177:6388–6397. doi: 10.4049/jimmunol.177.9.6388. [DOI] [PubMed] [Google Scholar]
- 26.Paclet M-H, Coleman AW, Burritt J, Morel F. NADPH oxidase of Epstein-Barr-virus immortalized B lymphocytes. Effect of cytochrome b(558) glycosylation. Eur J Biochem. 2001;268:5197–5208. doi: 10.1046/j.0014-2956.2001.02455.x. [DOI] [PubMed] [Google Scholar]
- 27.Savina A, et al. NOX2 controls phagosomal pH to regulate antigen processing during crosspresentation by dendritic cells. Cell. 2006;126:205–218. doi: 10.1016/j.cell.2006.05.035. [DOI] [PubMed] [Google Scholar]
- 28.Mantegazza AR, et al. NADPH oxidase controls phagosomal pH and antigen cross-presentation in human dendritic cells. Blood. 2008;112:4712–4722. doi: 10.1182/blood-2008-01-134791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hultqvist M, Olsson LM, Gelderman KA, Holmdahl R. The protective role of ROS in autoimmune disease. Trends Immunol. 2009;30:201–208. doi: 10.1016/j.it.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 30.Schäppi MG, Jaquet V, Belli DC, Krause K-H. Hyperinflammation in chronic granulomatous disease and anti-inflammatory role of the phagocyte NADPH oxidase. Semin Immunopathol. 2008;30:255–271. doi: 10.1007/s00281-008-0119-2. [DOI] [PubMed] [Google Scholar]
- 31.Olofsson P, et al. Positional identification of Ncf1 as a gene that regulates arthritis severity in rats. Nat Genet. 2003;33:25–32. doi: 10.1038/ng1058. [DOI] [PubMed] [Google Scholar]
- 32.Hultqvist M, et al. Enhanced autoimmunity, arthritis, and encephalomyelitis in mice with a reduced oxidative burst due to a mutation in the Ncf1 gene. Proc Natl Acad Sci USA. 2004;101:12646–12651. doi: 10.1073/pnas.0403831101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Ravin SS, et al. Chronic granulomatous disease as a risk factor for autoimmune disease. J Allergy Clin Immunol. 2008;122:1097–1103. doi: 10.1016/j.jaci.2008.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Winkelstein JA, et al. Chronic granulomatous disease. Report on a national registry of 368 patients. Medicine (Baltimore) 2000;79:155–169. doi: 10.1097/00005792-200005000-00003. [DOI] [PubMed] [Google Scholar]
- 35.Cale CM, Morton L, Goldblatt D. Cutaneous and other lupus-like symptoms in carriers of X-linked chronic granulomatous disease: Incidence and autoimmune serology. Clin Exp Immunol. 2007;148:79–84. doi: 10.1111/j.1365-2249.2007.03321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van den Berg JM, et al. Chronic granulomatous disease: The European experience. PLoS ONE. 2009;4:e5234. doi: 10.1371/journal.pone.0005234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997;40:1725. doi: 10.1002/art.1780400928. [DOI] [PubMed] [Google Scholar]
- 38.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 39.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Series B. 1995;57:289–300. [Google Scholar]
- 42.Rapley J, Tybulewicz VLJ, Rittinger K. Crucial structural role for the PH and C1 domains of the Vav1 exchange factor. EMBO Rep. 2008;9:655–661. doi: 10.1038/embor.2008.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu B, et al. Structural and energetic mechanisms of cooperative autoinhibition and activation of Vav1. Cell. 2010;140:246–256. doi: 10.1016/j.cell.2009.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Katchalski-Katzir E, et al. Molecular surface recognition: Determination of geometric fit between proteins and their ligands by correlation techniques. Proc Natl Acad Sci USA. 1992;89:2195–2199. doi: 10.1073/pnas.89.6.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heifetz A, Katchalski-Katzir E, Eisenstein M. Electrostatics in protein-protein docking. Protein Sci. 2002;11:571–587. doi: 10.1110/ps.26002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chakrabarti P, Janin J. Dissecting protein-protein recognition sites. Proteins. 2002;47:334–343. doi: 10.1002/prot.10085. [DOI] [PubMed] [Google Scholar]
- 47.Suhre K, Sanejouand Y-H. ElNemo: A normal mode web server for protein movement analysis and the generation of templates for molecular replacement. Nucleic Acids Res. 2004;32(Web Server issue) Suppl 2:W610–W614. doi: 10.1093/nar/gkh368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li XJ, Marchal CC, Stull ND, Stahelin RV, Dinauer MC. p47phox Phox homology domain regulates plasma membrane but not phagosome neutrophil NADPH oxidase activation. J Biol Chem. 2010;285:35169–35179. doi: 10.1074/jbc.M110.164475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Price MO, et al. Creation of a genetic system for analysis of the phagocyte respiratory burst: High-level reconstitution of the NADPH oxidase in a nonhematopoietic system. Blood. 2002;99:2653–2661. doi: 10.1182/blood.v99.8.2653. [DOI] [PubMed] [Google Scholar]