Abstract
Voltage-dependent gating of ion channels is essential for electrical signaling in excitable cells, but the structural basis for voltage sensor function is unknown. We constructed high-resolution structural models of resting, intermediate, and activated states of the voltage-sensing domain of the bacterial sodium channel NaChBac using the Rosetta modeling method, crystal structures of related channels, and experimental data showing state-dependent interactions between the gating charge-carrying arginines in the S4 segment and negatively charged residues in neighboring transmembrane segments. The resulting structural models illustrate a network of ionic and hydrogen-bonding interactions that are made sequentially by the gating charges as they move out under the influence of the electric field. The S4 segment slides 6–8 Å outward through a narrow groove formed by the S1, S2, and S3 segments, rotates ∼30°, and tilts sideways at a pivot point formed by a highly conserved hydrophobic region near the middle of the voltage sensor. The S4 segment has a 310-helical conformation in the narrow inner gating pore, which allows linear movement of the gating charges across the inner one-half of the membrane. Conformational changes of the intracellular one-half of S4 during activation are rigidly coupled to lateral movement of the S4–S5 linker, which could induce movement of the S5 and S6 segments and open the intracellular gate of the pore. We confirmed the validity of these structural models by comparing with a high-resolution structure of a NaChBac homolog and showing predicted molecular interactions of hydrophobic residues in the S4 segment in disulfide-locking studies.
Voltage-gated sodium (NaV) channels are responsible for initiation and propagation of action potentials in nerve, muscle, and endocrine cells (1, 2). They are members of the structurally homologous superfamily of voltage-gated ion channel proteins that also includes voltage-gated potassium (KV), voltage-gated calcium (CaV), and cyclic nucleotide-gated (CNG) channels (3). Mammalian NaV and CaV channels consist of four homologous domains (I through IV), each containing six transmembrane segments (S1 through S6) and a membrane-reentrant pore loop between the S5 and S6 segments (1, 3). Segments S1–S4 of the channel form the voltage-sensing domain (VSD), and segments S5 and S6 and the membrane-reentrant pore loop form the pore. The bacterial NaV channel NaChBac and its relatives consist of tetramers of four identical subunits, which closely resemble one domain of vertebrate NaV and CaV channels, but provide much simpler structures for studying the mechanism of voltage sensing (4, 5). The hallmark feature of the voltage-gated ion channels is the steep voltage dependence of activation, which derives from the voltage-driven outward movement of gating charges in response to the membrane depolarization (6, 7). The S4 transmembrane segment in the VSD has four to seven arginine residues spaced at 3-aa intervals, which serve as gating charges in the voltage-sensing mechanism (8–15). The intracellular S4–S5 linker that connects the VSD to the pore plays a key role in coupling voltage-dependent conformational changes in the VSD to opening and closing of the pore (16). The gating charges are pulled in by the internally negative transmembrane electric field and released to move out on depolarization. Their outward movement must be catalyzed by the voltage sensor to reduce the large thermodynamic barrier to movement of charged amino acid residues across the membrane. The molecular mechanism by which the gating charges are stabilized in the hydrophobic transmembrane environment and the catalytic mechanism through which they are transported across the membrane in response to changes in membrane potential are the subjects of intense research efforts.
Progress has been made in determining high-resolution structures of voltage sensors of KV and NaV channels in activated states (17–20). However, high-resolution structures of resting and intermediate states of voltage sensors are unknown. The majority of evidence supports a sliding helix model of the voltage-dependent gating in which the gating charge-carrying arginines in S4 are proposed to sequentially form ion pairs with negatively charged residues in S1–S3 segments during activation of the channel (9–11, 21). However, the structural basis for stabilization of the gating charges in the membrane and catalysis of their movement through the hydrophobic membrane environment remain uncertain. Here, we have integrated bioinformatics analysis of NaV and KV channel families using the HHPred homology detection server (22–24), high-resolution structural modeling using the Rosetta Membrane (25–27) and Rosetta Symmetry methods (28), the X-ray structures of the Kv1.2-Kv2.1 chimeric channel and NavAb with activated VSDs (19, 20) and the MlotiK1 CNG channel in the resting state (29), and experimental data showing sequential state-dependent interactions between gating charges in S4 and negatively charged residues in S1–S3 (this work and refs. 30–33). Predictions of the resulting voltage-sensing model are confirmed in this work by disulfide-locking studies and mutant cycle analysis of the interactions of hydrophobic residues in the S4 segment. This model reveals structural details of the voltage-dependent conformational changes in the VSD that stabilize and catalyze gating charge movement and are coupled to opening and closing of the intracellular activation gate of the ion-conducting pore.
Results
Modeling the Voltage-Sensing Domain of NaChBac.
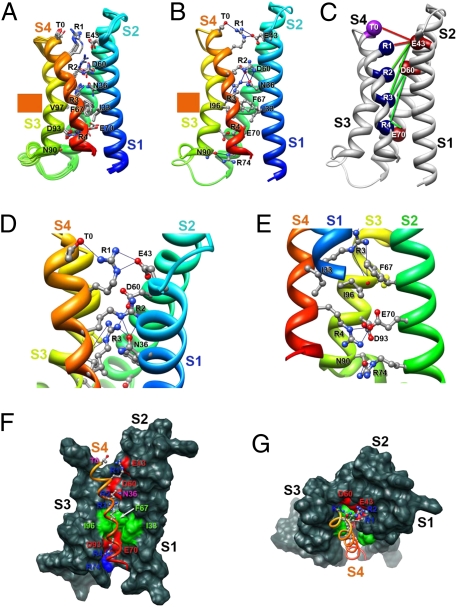
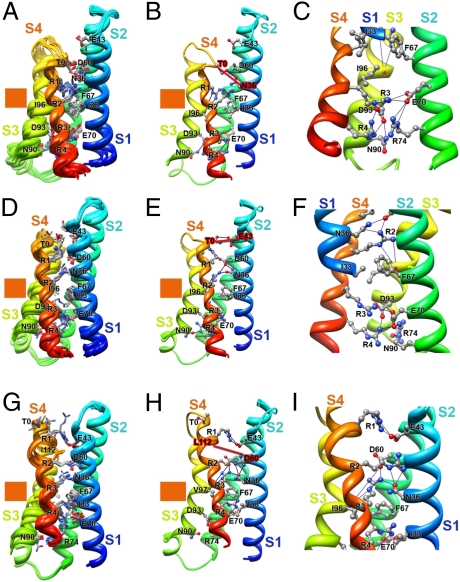
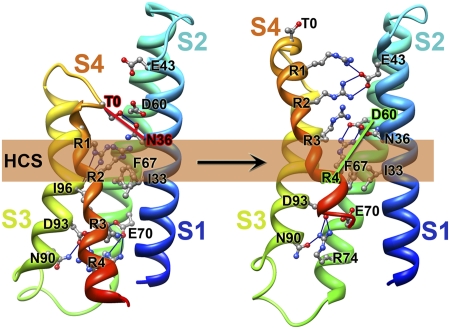
The Rosetta Membrane modeling algorithm has been successful in predicting the conformation of the transmembrane segments of complex multispan membrane proteins (25, 27, 34). We used this method (25–28) to model the structure of NaChBac at high resolution with a database including KV channel structures but no NaV channel structures. Our modeling experiments began with the X-ray structure of the KV1.2-KV2.1 chimera (19) as a template and development of a consensus alignment of the VSDs of NaV and CaV channels (SI Appendix, SI Methods and SI Appendix, Figs. S1–S4). Our final alignment (SI Appendix, Fig. S5) brings many highly conserved residues into register and provides a global template for modeling VSDs (SI Appendix, SI Methods). We used the Rosetta Membrane method to build the poorly conserved S1–S2 and S3–S4 loops de novo and then found the best high-resolution model using the full atom relax protocol that optimizes backbone torsions and repacking of side chain atoms to find the lowest energy conformation. The ensemble of 10 lowest energy full atom models of a single VSD shows close agreement in structure (Fig. 1A). The lowest energy full atom models selected in this unbiased way reveal the voltage sensor in an early activated state (Fig. 1 A–E). As a landmark in our structures, we highlight a highly conserved hydrophobic constriction site (HCS) within the VSD core (35) that is formed by I33 (in S1), F67 (in S2), and I96 and V97 (in S3) in NaChBac (Fig. 1 F and G, green). This hydrophobic cluster lines the narrowest part of the pathway for the gating charge movement through the gating pore of the VSD and likely forms a hydrophobic seal that prevents ion movement through the gating pore (Fig. 1 F and G). It is observed in the crystal structures of KV1.2 and NavAb (18–20).
Fig. 1.
Model of the VSD of NaChBac. (A) Transmembrane view of the ensemble of the 10 lowest energy Rosetta models of the VSD of NaChBac built starting from the KV1.2-KV2.1 chimera channel structure (19). Side chains of the gating charge-carrying arginines in S4 and key residues in S1–S3 segments are shown in stick representation and labeled. Gray, blue, and red atoms are C, N, and O, respectively. The HCS is highlighted by orange bars. (B) The lowest energy Rosetta model of the VSD of NaChBac. Hydrogen bonds between side chains of key residues are shown as blue lines. (C) Transmembrane view of the model shown in B with Cβ-atoms of the gating charge carrying arginines in S4 (colored dark blue), negatively charged residues in S1 and S2 segments (colored red), and T0 in S4 (colored purple) shown in sphere representation and labeled. Disulfide cross-linking–based distance constraints between the gating charge carrying arginines in S4 and negatively charged residues in S1 and S2 are shown by solid red lines for the resting state interactions and solid green lines for the activated state interactions. (D) Close-up view of key interactions in the model shown in B in the extracellular one-half of VSD. (E) Close-up view of key interactions in the model shown in B in the intracellular one-half of VSD. (F) Transmembrane view of the model shown in B with S1–S3 segments shown in surface representation with highly conserved negatively charged and polar residues colored red, positively charged R74 in S2 colored blue, and highly conserved hydrophobic residues in S1–S3 colored green. (G) View of the model shown in F from the extracellular side of the membrane.
In this activated state structure, the first three gating charge-carrying arginines (R1–R3) in S4 are positioned on the extracellular side of the HCS, accessible to the extracellular aqueous vestibule of the VSD (Fig. 1 E and F). A length of the S4 segment in 310-helix conformation starts from the position of R2 and extends beyond R4 to A125, effectively placing the R2–R4 gating charges in a line on one side of the S4 helix (Fig. 1 A–E). The lowest energy models reveal a conserved extracellular negative cluster (ENC) of acidic residues that interacts with gating charges at the extracellular end of the S4 segment, including E43 (in S1) and D60 (in S2) (Fig. 1 A–D). The R1 side chain forms an ion pair (i.e., a salt bridge) with E43 (in S1) and a hydrogen bond with T0 (in S4), and R3 forms an ion pair with D60 (in S2) and hydrogen bonds with N36 (in S1) and the backbone carbonyl of I96 (in S3) in all of the lowest energy models (Fig. 1 D and E). In 7 of 10 lowest energy models, R2 forms hydrogen bonds with N36 (in S1) (Fig. 1 A–D), and in 3 other lowest energy models, it forms ion pairs with E43 (in S1) and D60 (in S2). N36 (in S1) is positioned one helical turn and D60 (in S2) is two helical turns on the extracellular side of the HCS (Fig. 1 A, B, and E). In the KV1.2-KV2.1 chimera structure (19), S176 is at the equivalent position to N36 in NaChBac, and we suggest that the polar side chain at this position is involved in a dynamic hydrogen-bonding network during the transfer of the gating charges through the gating pore.
The backbone carbonyl of I96 in the HCS points to the center of the gating pore, where it is well-positioned to serve as a point of interaction to catalyze the transition of the gating charges through the HCS (Fig. 1E). An intracellular negative cluster (INC) is located on the intracellular side of the HCS, including E70 (in S2) and D93 (in S3). R4 forms an ion pair with E70 (in S2), and R74 (in S2) forms hydrogen bonds with N90 (in S3) in all of the lowest energy models (Fig. 1E). These interactions reveal a network of ionic and hydrogen bonds that stabilize the gating charges on the intracellular side of the HCS, similar to the ENC on the extracellular side. This model and the structures of the HCS, ENC, and INC are in close agreement with a recent high-resolution crystal structure of the NavAb channel (20), a close homolog of NaChBac (SI Appendix, SI Results and SI Appendix, Fig. S6).
Modeling the Voltage Sensor in Activated States.
The voltage sensors of ion channels change conformation under the influence of the transmembrane electric field, move the gating charges outward, and couple those structural changes to opening of the pore. Disulfide cross-linking experiments have defined a series of molecular interactions between pairs of amino acid residues in the S4 segment and the surrounding S1–S3 segments in activated states of the VSD (30–33, 36). NaChBac provides a favorable template for these disulfide-locking experiments, because it is cysteine-free; the disulfide-locking method is ideal for comparison with high-resolution structural data, because the sulfur atoms of a substituted cysteine residue must approach within 2 Å to form a disulfide bond. To give insight into the conformational changes of the voltage sensor in different functional states, we introduced single structural constraints (Fig. 1C, activated state constraints in green) that were derived from these disulfide cross-linking experiments to generate models of the activated states of the voltage sensor of NaChBac (SI Appendix, Figs. S7–S10).
The first activated state of the VSD (Activated State 1) was captured in NaChBac models based on disulfide-locking data showing state-dependent interaction between R2 and E43 (in S1) in an early activated state (33). This constraint is also compatible with the unconstrained structure of the VSD (Fig. 1). R1–R3 are positioned on the extracellular side of the HCS, accessible to the extracellular aqueous vestibule (Fig. 2 A–C), and they interact with the ENC and form a network of hydrogen bonds with additional nearby hydrophilic residues (Fig. 2 A–C). In Activated State 2, the S4 segment moves one helical turn outward based on disulfide-locking data that show state-dependent interaction between R3 and D60 (in S2) (31). Gating charges R1–R3 interact with the ENC on the extracellular side of the HCS, accessible to the extracellular aqueous vestibule (Fig. 2 D–F). Notably, the side chain of R4 flips to the extracellular side of the HCS in a majority of the lowest energy models of Activated State 2 (Fig. 2D) but makes ionic interactions with E70 (in S2) and D93 (in S3) in the INC in the lowest energy model (Fig. 2E). A network of hydrogen bond interactions stabilizes this state (Fig. 2 D–F). The most activated conformation of S4 (Activated State 3) was based on disulfide-locking data showing state-dependent interaction between R4 and D60 (in S2) (32). In this state, the side chains of gating charges R1–R4 interact with the ENC and nearby hydrogen-bonding partners on the extracellular side of the HCS, accessible to the extracellular aqueous vestibule, although the Cα atom of R4 is in the HCS (Fig. 2 G–I). Together, our models of three activated states illustrate progressive outward movement of the S4 gating charges as they exchange hydrogen-bonding and ion pair partners along their path. These interactions catalyze the outward movement of the gating charges from interaction with the INC through the HCS to interaction with the ENC. Because these changes of conformation derive from constraints based on single molecular interactions that occur at progressively more depolarized membrane potentials (33), it is likely that Activated States 1–3 form sequentially during gating of the NaChBac channel. We tested possible models for even more activated states suggested by disulfide-locking results, but they were not stable when the disulfide-locking constraint was relaxed (SI Appendix, SI Results).
Fig. 2.
Models of the VSD of NaChBac in activated states. (A) Transmembrane view of the ensemble of the 10 lowest energy Rosetta models of the VSD of NaChBac in Activated State 1 as presented in Fig. 1A. (B) The lowest energy Rosetta model of the VSD of NaChBac in Activated State 1. (C) Close-up view of key interactions in the model shown in B for the extracellular one-half of the VSD. R1 forms an ion pair with E43 (in S1), R2 forms an ion pair with E43 (in S1) and D60 (in S2), R3 forms an ion pair with D60 (in S2) and a hydrogen bond with N36 (in S1), and R4 forms an ion pair with E70 (in S2; B). (D) Transmembrane view of the ensemble of the 10 lowest energy Rosetta models of the VSD of NaChBac in Activated State 2. (E) The lowest energy Rosetta model of the VSD of NaChBac in Activated State 2. (F) Close-up view of key interactions in the model shown in E in the extracellular one-half of VSD. The lowest energy models predict that R1 forms a hydrogen bond with T0 (in S4), R2 forms an ion pair with E43 (in S1) and D60 (in S2), and R3 forms an ion pair with D60 (in S2). (G) Transmembrane view of the ensemble of the 10 lowest energy Rosetta models of the VSD of NaChBac in Activated State 3. (H) The lowest energy Rosetta model of the VSD of NaChBac in Activated State 3. (I) Close-up view of key interactions in the model shown in H at the extracellular one-half of VSD. The lowest energy models predict that R1 forms an ion pair with E43 (in S1), R2 forms an ion pair with E43 (in S1), R3 forms a hydrogen bond with Y156 (in S5) and makes ionic interactions with D60 (in S2) and E43 (in S1), and R4 forms an ion pair with D60 (in S2).
Disulfide Locking the Voltage Sensor in Resting States.
Because there is no membrane potential in a protein crystal, only the activated conformation that is most stable at 0 mV can be characterized by X-ray crystallography. Therefore, identification of key molecular interactions by disulfide-locking and mutant cycle analysis followed by structural modeling with the Rosetta Membrane method can give unique insights into the structures of VSD resting states. To investigate the molecular interactions of T0 and the outer gating charges R1 and R2 in the S4 segment with D60 in the resting state, we used the disulfide-locking method to analyze the double cysteine mutants D60C:T0C, D60C:R1C, and D60C:R2C. Unfortunately, these double cysteine mutants form disulfide bonds as expected but are not inserted into the cell surface membrane (SI Appendix, SI Results and SI Appendix, Figs. S11 and S12).
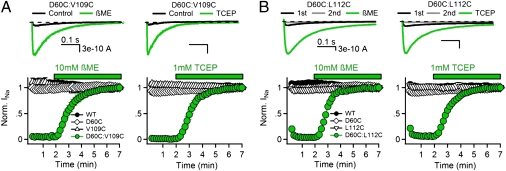
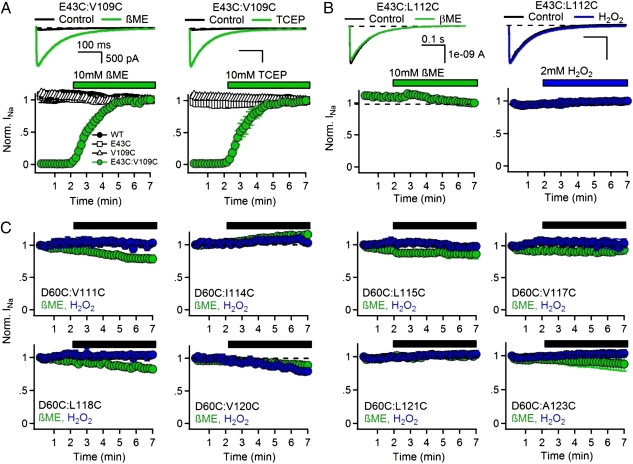
If the gating charges T0, R1, and R2 are able to form disulfide bonds with D60 in the resting state, cysteine residues substituted for the neighboring hydrophobic residues V109 (preceding T0) and L112 (preceding R1) may also be capable of disulfide locking in the resting state, which would be revealed in disulfide-locking experiments by lack of Na+ current (INa) for double cysteine mutants under control conditions and appearance of INa on reduction of disulfide bonds. Repetitive depolarization of WT or the single cysteine mutants from −140 to 0 mV for 500 ms results in inward INa of constant size (Fig. 3 A and B, black symbols), but little or no current was observed for the D60C:V109C or D60C:L112C channels (Fig. 3 A and B, black traces and green symbols). Perfusion of the sulfhydryl reagent β-mercaptoethanol (βME) or the phosphine-reducing agent Tris(2-carboxyethyl)phosphine (TCEP) restored INa within 3 min (Fig. 3 A and B, green). Based on these results, we conclude that the lack of INa for the D60C:V109C and D60C:L112C channels is caused by disulfide locking the voltage sensor in a resting state. Because V109 is one residue extracellular to T0 (T110) and L112 is one residue extracellular to R1 (R113), these results support the conclusion that the V109, T0, L112, R1, and R2 can all interact with D60 in the resting states of the voltage sensor.
Fig. 3.
Disulfide-locking cysteine residues substituted for D60 and the S4 hydrophobic residues V109 and L112. (A, Upper and B, Upper) INa from voltage-clamped tsA-201 cells expressing (A) D60C:V109C and (B) D60C:L112C channels. INa elicited by the first pulse in control conditions (black), the second pulse in control conditions (gray), and the last pulse in the presence of reducing agent (green) during a train of 500-ms depolarizations to 0 mV from a holding potential of −140 mV. (A, Lower and B, Lower) Effects of reducing agents 10 mM βME and 1 mM TCEP (green bars) on the mean normalized peak currents recorded from cells expressing (A) D60C:V109C and (B) D60C:L112C channels (±SEM) recorded during the trains (n = 5–6).
Closer inspection of the experimental results for D60C:L112C reveals a small, but significant Na+ current that is activated in response to the first depolarization without reduction of disulfide bonds (Fig. 3B, Upper, black traces), but it is lost in the second depolarization (Fig. 3B, Upper, gray traces). These results indicate that a small fraction of D60C:L112C channels are in a nondisulfide-locked resting state at the beginning of the experiments, but they are activated and disulfide-locked by the first cycle of depolarization and repolarization. This effect is revealed as a small loss of INa after the first depolarization in the plot of peak INa vs. pulse number (Fig. 3B, green). These results indicate that L112 is in an intermediate position with respect to D60 at the resting membrane potential of our experiments: 88 ± 2% are already disulfide-locked to D60, but the remaining 12 ± 2% are in a more activated position that does not allow disulfide locking with D60 without depolarization/repolarization to induce outward voltage sensor movement followed by complete inward retraction to the resting state. These results indicate molecular proximity between L112 and D60 in an early activated state along the activation pathway.
Mutant Cycle Analysis of Amino Acid Interactions in the Resting State of the Voltage Sensor.
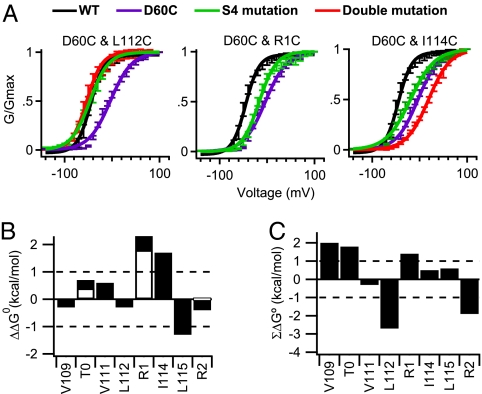
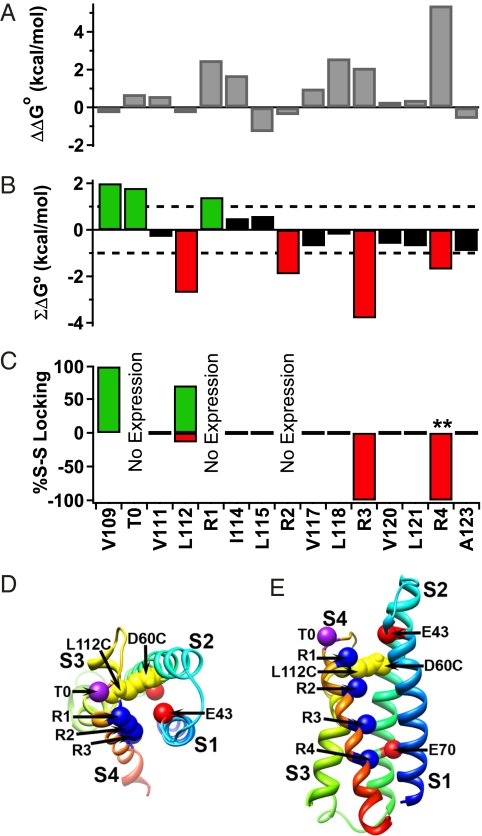
We used mutant cycle analysis to assess the energy of association of these amino acid residues during activation of NaChBac channels. Conductance–voltage profiles were fit to a Boltzmann function (Fig. 4A and SI Appendix, Fig. S13A), and values for Z (slope) and V1/2 (potential of half-maximal activation) were estimated to determine the free energy of activation (ΔG°). Nearly all of our single mutations caused a significant change in V1/2 or Z (SI Appendix, Table S1). The perturbation in free energy (ΔΔG°) and the nonadditive coupling energy (ΣΔG°) were calculated for each mutant (Fig. 4 B and C and SI Appendix, Table S1). If two mutations behave independently, their perturbations in free energy will be additive, yielding little or no coupling energy (ΣΔG° < ±1 kcal/mol) (37) (Fig. 4C, dotted line). Because D60C:V109C channels are disulfide-locked in a resting state, the voltage dependence of activation could only be assessed with βME (SI Appendix, Fig. S13A). We found that D60C:V109C and D60C:L112C channels have significant energies of interaction (2.0 and −2.7 kcal/mol, respectively) (Fig. 4C).
Fig. 4.
Mutant cycle analysis of D60 interactions with the outer gating charges and hydrophobic residues in the resting state. (A) Conductance–voltage relationships are plotted for WT, D60C, the indicated single cysteine S4 mutations, and their double cysteine mutants (n > 9; ±SEM) (SI Appendix, Table S2). (B) Perturbation of free energy (ΔΔG°) caused by single cysteine mutations to cysteine (filled bars) or alanine (open bars). (C) Mutant cycle analysis for double mutants of D60C and the indicated S4 residues mutated to cysteine for hydrophobic residues or alanine for gating charge positions T0, R1, or R2. ΣΔG° is the nonadditive free energy. Because the D60C:V109C channels are disulfide-locked in a resting state, the voltage dependence of activation could only be assessed with βME present.
Because the double cysteine mutants D60C:T0C, D60C:R1C, and D60C:R2C channels are not inserted in the cell membrane, we used mutant cycle analysis with paired substitutions of cysteine and alanine to examine their interactions (Fig. 4 B and C and SI Appendix, Fig. S13B). We found that the ΣΔG° values for the double mutants D60C:T0A, D60C:R1A, and D60C:R2A were 1.8, 1.4, and −1.9 kcal/mol, respectively, indicating that D60 is energetically coupled to T0, R1, and R2 during the gating process (Fig. 4C). The positive ΣΔG° values of the D60C:T0A and D60C:R1A channels suggest that T0 and R1 interact with D60 and stabilize the resting state of the voltage sensor. In contrast, ΣΔG° for D60C:R2A was negative, indicating that R2 and D60 interact and stabilize the activated voltage sensor. Evidently, interactions of D60 with T0 and R1 oppose activation, whereas D60 interactions with R2 favor activation. The interaction of R1 with D60 may represent the transition point at which interactions of more inward gating charges with D60 progressively favor the activated state of the VSD (see below).
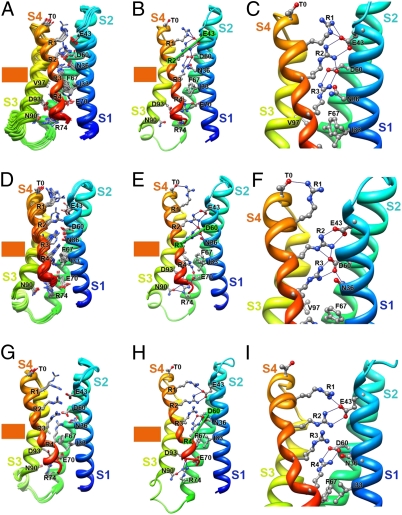
Modeling the Voltage Sensor in Resting States.
The interactions of the gating charges and hydrophobic residues in S4 with E43 and D60 are specific, because the neighboring amino acid residue I59, which points away from the gating pore in our model structures, does not interact with any of the gating charges or hydrophobic residues in the S4 segment in either the resting or activated states (SI Appendix, SI Results and SI Appendix, Fig. S14). Therefore, the interactions detected by disulfide-locking and mutant cycle analysis (Figs. 3 and 4) provide specific molecular constraints with which to develop models of VSD resting states. The deepest stable resting state of the VSD (Resting State 1) was captured in NaChBac models based on disulfide-locking data showing interaction of R1 with I241 (in S1) and I287 (in S2) in the resting state of the Shaker KV channel (30) (SI Appendix, SI Methods). In this state, R3 and R4 are completely on the intracellular side of the HCS, interacting with the INC and exposed to the intracellular aqueous vestibule (Fig. 5 A and B). In contrast, R1 and R2 are positioned within the narrow gating pore region, where they can sense membrane electric field and their side chains can form a barrier to prevent ion leak (Fig. 5 A and B). The gating charges on both sides of the HCS form a network of hydrogen-bonding and ionic interactions with the ENC, INC, and nearby backbone carbonyls and hydrophilic residues (Fig. 5 A–C). These results highlight the increased interaction of the inner gating charges with the INC in the resting states. Deeper resting states, in which the S4 segment is drawn farther inward, were tested and found to be unstable when the disulfide-locking constraint was relaxed (SI Appendix, SI Results).
Fig. 5.
Models of the VSD of NaChBac in resting states. (A) Transmembrane view of the ensemble of the 10 lowest energy Rosetta models of the VSD of NaChBac in Resting State 1 presented as in Fig. 1A. (B) The lowest energy Rosetta model of the VSD of NaChBac in Resting State 1. (C) Close-up view of key interactions in the model shown in B at the intracellular one-half of the VSD. The majority of the lowest energy models predict that R1 forms hydrogen bonds with the backbone carbonyl of I96 (in S3) at the extracellular edge of the HCS. On the intracellular side of the HCS, R3 makes ionic interactions with the amino acid residues of the intracellular negatively charged cluster, including E70 (in S2) and D93 (in S3), and R4 forms an ion pair with D93 (in S3). (D) Transmembrane view of the ensemble of the 10 lowest energy Rosetta models of the VSD of NaChBac in Resting State 2 based on D60-V109 constraint. (E) The lowest energy Rosetta model of the VSD of NaChBac in Resting State 2 based on the D60-V109 constraint. (F) Close-up view of key interactions in the model shown in E in the extracellular one-half of VSD. The majority of the lowest energy models predict that R1 forms an ion pair with D60 (in S2), R2 makes ionic interactions with D60 and hydrogen bonds with N36 (in S1) and the backbone carbonyl of I96 (in S3), R3 forms an ion pair with E70 (in S2), and R4 makes ionic interactions with E70 (in S2) and D93 (in S3). (G) Transmembrane view of the ensemble of the 10 lowest energy Rosetta models of the VSD of NaChBac in Resting State 3 based on D60-L112 constraint. (H) The lowest energy Rosetta model of the VSD of NaChBac in Resting State 3 based on the D60-L112 constraint. (I) Close-up view of key interactions in the model shown in H in the extracellular one-half of VSD. On the extracellular side of the HCS, the majority of the lowest energy models predict that R1 forms an ion pair with E43 (in S1), R2 forms hydrogen bonds with N36 (in S1), and R3 forms an ion pair with D60 and hydrogen bonds with N36 (in S1) and the backbone carbonyl of I96 (in S3). In contrast, R4 forms an ion pair with E70 (in S2), a member of the intracellular negatively charged cluster.
The 310-helix structure of the S4 segment in Resting State 1 starts from R1 and extends to R4 (Fig. 5 A–C). A similar length of 310-helix structure is observed in the X-ray structure of the resting state of the MlotiK1 CNG channel (29). In contrast, the 310-helix structure only extends from R2 to R4 in Activated States 1–3 (Fig. 2). This comparison suggests that the extent of 310-helix conformation of the S4 segment is decreased during activation. This unexpected prediction from our models is confirmed by the disulfide-locking results presented below.
The S4 segment moves out about 2 Å in the model of Resting State 2 (Fig. 5 D–F), which is based on disulfide-locking data showing state-dependent interaction between T0 (in S4) and E43 (in S1) (33). R3 and R4 in S4 are located on the intracellular side of the HCS, interacting with the INC and accessible to the intracellular aqueous vestibule of the VSD. The R2 side chain is positioned within the narrow gating pore region, where it can potentially sense the membrane electric field (Fig. 5 D–F). The last resting state before channel activation (Resting State 3) was captured using disulfide-locking data showing state-dependent proximity of D60 (in S2) and L112 (in S4) (Figs. 3 and 4). Notably, about 88% of D60C-L112C channels were locked in a resting state but ∼12% remained unlocked at the resting membrane potential, suggesting that this state marks a transition between the resting and activated states of the VSD. R1 and R2 are located on the extracellular side of the HCS, accessible to the extracellular aqueous vestibule. The R3 side chain is positioned at the extracellular edge of the HCS, and the R4 side chain is positioned on the intracellular edge of the HCS (Fig. 5 G–I). Both of these gating charges would be partially in the membrane electric field in position to respond to changes in the membrane potential. In both Resting States 2 and 3, an extensive network of ionic and hydrogen-bonding interactions stabilizes the gating charges on both sides of the HCS (Fig. 5 D–I).
The 310-helix structure of S4 in Resting State 3 extends from R2 to R4. Comparison of our Resting States 2 and 3 models (Fig. 5 D–I) suggests that the transition between 310- and α-helix structures in the R1–R2 region of S4 occurs when R3 moves from the intracellular to the extracellular side of the HCS and drives the R1 side chain from interaction with D60 (in S2) to interaction with E43 (in S1). Comparison of the conformations of Resting States 1–3 with Activated States 1–3 gives insights into molecular motions that are potentially associated with pore opening, which is described in Discussion.
Testing Model Predictions by Disulfide-Locking and Mutant Cycle Analysis.
Our structural models of the resting and activated states of the voltage sensor are based on a single molecular constraint from disulfide-locking experiments, usually involving a gating charge (30–33) (Fig. 1C). Therefore, a critical test of these models is to predict the results of disulfide-locking experiments with the hydrophobic residues located between the gating charges in the S4 segment.
V109 precedes the gating charge position T0 and begins the S4 helix in our structural models. L112 precedes the R1 gating charge in the S4 helix. The molecular interactions of these two residues depend on the extent of 310- vs. α-helix at the outer end of S4. In Resting State 2, V109 is predicted to be close enough to E43 (in S1) to form a disulfide bond (Fig. 5). In contrast, L112 is never close enough to E43 to form a disulfide bond in our models (Fig. 5 and SI Appendix, Table S3). We constructed single and double mutants with cysteine residues substituted for E43 and either V109 or L112 and investigated the functional effects of disulfide bond formation (31, 32). If V109 or L112 interacts with E43 in the resting state, we would expect that the double mutant channel would be disulfide-locked and inactivated, but it would be released from disulfide locking by reduction of disulfide bonds and thereby made available for activation by depolarization. Repetitive depolarization of WT or single cysteine mutants results in peak inward sodium currents (INa) of constant size (Fig. 6A, black symbols), but no current was observed for the E43C:V109C channel (Fig. 6A, black traces and green symbols, 0–2 min), suggesting that disulfide bond formation between the substituted cysteine residues locks the VSD in the resting state. Perfusion with the reducing agents βME or TCEP restored INa magnitude within 3 min, confirming that the loss of INa was caused by disulfide bond formation (Fig. 6A, green traces and green symbols, 2–7 min). In contrast, INa from E43C:L112C channels could be elicited over multiple depolarizations, and the magnitude of the peak current did not change (Fig. 6B). To determine if sulfhydryl reagents could affect the E43C:L112C channel, we perfused the oxidizing agent H2O2 or the reducing agent βME but observed no change in peak INa amplitude (Fig. 6B). Based on these results, we conclude that the lack of INa for double mutant E43C:V109C is caused by disulfide locking of the VSD in a resting state, whereas no sulfhydryl interaction occurs in either the resting or activated states for E43C:L112C. These results are consistent with the expected interaction of V109 and E43 in Resting State 3 as well as the lack of interactions of L112 with D60 in all states, and therefore, they provide strong support for our molecular models.
Fig. 6.
Disulfide locking of cysteine residues substituted for amino acid residues in the S4 segment. (A, Upper and B, Upper) INa from the first pulse in control conditions (black) and last pulse in the presence of reducing agent (green) or oxidizing agents (blue) elicited by a 0.1-Hz train of 500-ms depolarizations to 0 mV from a holding potential of −140 mV. (A, Lower and B, Lower) Effects of reducing agents 10 mM βME and 1 mM TCEP (green bars) or 2 mM H2O2 (blue bar) on the mean normalized peak currents (±SEM) recorded during the stimulus trains (n = 5–6). (C) Lack of disulfide locking of cysteine residues substituted for D60 with the indicated hydrophobic residues in the S4 segment. Mean normalized peak currents (±SEM) are plotted from stimulus trains, and the black bar indicates the time when 10 mM βME (green) or 2 mM H2O2 (blue) were perfused (n = 4).
Our observation that the S4 segment must form a 310-helix as it moves through the inner gating pore makes another robust prediction. If the S4 helix moves along a spiral path, as proposed in the sliding helix and helical screw models (9–11), the gating charges and some of the hydrophobic residues between them would be available for disulfide locking to the same amino acid residues (SI Appendix, Fig. S15). However, if the S4 segment moves through the narrow gating pore in a 310-conformation, the gating charges would be positioned in nearly a straight line on one side of the helix, and the hydrophobic residues would be constrained to the opposite sides of the 310-helix and would make different molecular interactions (SI Appendix, Fig. S15). We used the disulfide-locking method to test this prediction. Single and double cysteine mutants of D60 (in S2) and every residue in the S4 segment were constructed and analyzed (Fig. 6C). We found that none of the hydrophobic residues between R1 and R4 were capable of disulfide locking with D60, although R112 immediately preceding R1 and the R1–R4 gating charges themselves are all capable of disulfide bond formation with D60C in either the resting or activated state (Figs. 4 and 6).
We conducted mutant cycle analysis to determine the energy of interaction of these pairs of amino acid residues (Fig. 7 and SI Appendix, Fig. S16). We found that most single mutations in the S4 segment increased the energy required to activate the channels (ΔΔG), with especially large effects for neutralization of the gating charges R1, R3, and R4 (Fig. 7A). Calculation of the interaction energies (ΣΔG) revealed no significant energy of interaction for any of the hydrophobic amino acid residues between R1 and R4 (Fig. 7B, black bars), which is in sharp contrast to the substantial interaction energies of V109, T0, and R1 with D60 in the resting state (Fig. 7B, green bars) and R2, R3, and R4 with D60 in the activated state (Fig. 7B, red bars). The results of our disulfide-locking experiments on all of the residues in the S4 segment are summarized as a bar graph in Fig. 7C, with percent disulfide locking in the resting state illustrated by green bars and in the activated state illustrated by red bars. Together with the mutant cycle analysis, these results show that T0 and V109, which precedes it, both interact with D60 in the resting state and stabilize the resting state (Fig. 7 B and C, green bars). Similarly, R2, R3, and R4 all interact with D60 in the activated state and stabilize the activated state (Fig. 7 B and C, red bars). R2 and L112, which precedes it, define a transition zone with respect to state dependence of D60 interactions—both interact with D60, but L112 preferentially interacts with the resting state, whereas R1 preferentially stabilizes the activated state (Fig. 7 B and C, red and green bars). In contrast, the hydrophobic residues between R1 and R4 are all unable to interact with D60, as assessed by disulfide-locking and mutant cycle analysis (Fig. 7 B and C, black bars). This complex pattern of interactions of hydrophobic amino acid residues is exactly as predicted from our molecular models, in which the S4 segment is in an α-helical conformation from V109 to R2 but in a 310-conformation from R2 to R4 during activation of the voltage sensor, thereby allowing the inner gating charges to move outward in a nearly linear path past their ion pair partners (Fig. 7D).
Fig. 7.
Mutant cycle analysis of energy coupling between D60 and interacting residues in the S4 segment. (A) The perturbation of free energy (ΔΔG°) from single cysteine mutations to the S4 of the NaChBac channel. (B) Summary of results of mutant cycle analysis (SI Appendix, Table S2) of double cysteine mutations of S4 residues in combination with D60C. Green bars indicate energy coupling favoring deactivation, red bars indicate energy coupling favoring activation, and black bars indicate no energy coupling. All of the indicated S4 residues are mutated to cysteine except T0A, R1A, and R2A. (C) Summary of disulfide interactions of S4 amino acid residues with D60C in the resting (green) and activated (red) states as assessed by disulfide locking. (D) Low-resolution Rosetta model of the gating pore viewed from the extracellular side. Note the linear array of S4 gating charges opposite D60 and E43. (E) Low-resolution Rosetta model of the gating pore viewed from the membrane.
We considered the predictions for interactions of amino acid residues in the S4 segment with D60 in the S2 segment in more detail in SI Appendix, Tables S3–S5, which present the minimum predicted distance of the Cβ-atom of each S4 residue from the Cβ-atom of D60 in all of the states modeled here together with the interactions observed by disulfide locking (+ or −). A maximum Cβ distance of ≤9–10 Å was set as the criterion for disulfide bond formation, which is based on a requirement for 5–6 Å to form disulfides in rigid structures and up to 2 Å movement for each side chain within the VSD based on molecular dynamics simulations (38, 39). For our models presented in Figs. 2 and 5, which have a combination of α-helical conformation at the extracellular end of S4 and 310-helical conformation at the intracellular end, all predictions are verified as noted by green highlights in SI Appendix, Table S3. However, a model with α-helical conformation throughout the length of S4 fits the data for the extracellular end but not the intracellular end, which is noted by red highlights in SI Appendix, Table S5, whereas a model with 310-helical conformation throughout S4 fits the data for the intracellular end but not the extracellular end, which is noted by the red highlights in SI Appendix, Table S4. This analysis shows that the models of Figs. 2 and 5, which have a combination of α- and 310-helix, fit the data on protein interactions substantially better than an α- or a 310-helix over the length of the S4 segment. It is particularly significant that the intracellular part of S4, which moves through the narrow HCS in the gating pore, is in 310-helix conformation, because it allows the R1–R4 gating charges to interact specifically with D60 and shields the intervening hydrophobic residues from that charge interaction. These models and the significance of the extended 310-helix are considered further in SI Appendix, SI Discussion.
Discussion
Comparison of Voltage-Sensing Models with Experimental Data.
Our structure prediction methods are still significantly less accurate than crystallographic structure determination. Therefore, we have highest confidence in the prediction of overall secondary structure and the spatial relationships between amino acid residues in different interacting transmembrane segments within the VSD. This aspect of our structures also derives direct support from disulfide-locking experiments. We have less confidence in the exact ion pair and hydrogen-bonding partners within the interacting networks derived from these high-resolution structural models, because these interactions are likely to be dynamic and interchanging as the S4 segment moves through the gating pore. Nevertheless, we believe that the hydrogen-bonding networks that we present here are representative of the molecular interactions that stabilize and catalyze gating charge movement in voltage sensors. Our structural models for NaChBac are very similar to the Rosetta models of KV1.2 channels derived previously (26) and are compatible with the vast majority of current experimental data on the structure and function of the VSD (SI Appendix, Table S6). These numerous points of agreement between our structural models and prior experimental data support the accuracy of these models and the mechanism of voltage sensor function derived from them. Moreover, our models made predictions about the structure of the extracellular end of the S4 segment and the 310-helix component of the S4 segment that were unique and fully confirmed in our experiments with disulfide-locking and mutant cycle analysis (Figs. 6 and 7 and SI Appendix, Tables S3–S5).
Conformational Changes in the Voltage-Sensing Domain During Gating.
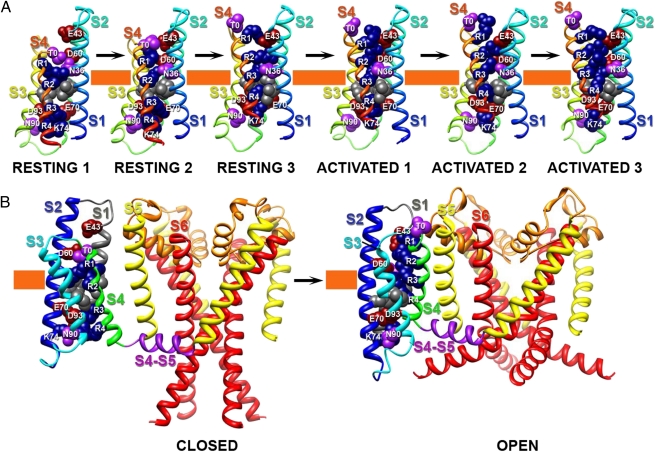
Comparison of our structural models of resting and activated states of NaChBac suggests several probable features of the structural mechanism for catalysis of gating charge movement and its coupling to pore opening (Fig. 8 and Movies S1, S2, and S3). These molecular motions are most easily appreciated from Movies S1, S2, and S3.
Fig. 8.
Model of conformational changes in NaChBac during gating. (A) Transmembrane view of the ribbon representation of Rosetta models of three resting and three activated states of the VSD of NaChBac. Segments S1–S4 are colored individually and labeled. Side chain atoms of the gating charge-carrying arginines in S4 (colored dark blue), negatively charged residues in S1–S3 segments (colored red), polar residues in S1, S3, and S4 (colored purple), and key hydrophobic residues in S1–S3 (colored gray) are shown in sphere representation and labeled. The HCS is highlighted by orange bars. (B) Transmembrane view of the ribbon representation of Rosetta models of Resting State 1 and Activated State 3 of NaChBac VSD and pore domain. Only a single VSD is shown attached to a tetramer of the pore domain for clarity. Transmembrane segments S1–S6 are colored individually and labeled. The S4–S5 linker is colored purple. Side chains atoms of key residues in VSD are represented as in A. The narrow region of focused membrane electrical field at the hydrophobic constriction site is highlighted by orange bars.
i) A highly conserved cluster of the large hydrophobic side chains of I33 (in S1), F67 (in S2), and I96 (in S3) creates the HCS, a tightly packed structural element of ∼5 Å thickness (Figs. 1 F and G and 8, orange band). This cluster of hydrophobic residues allows movement of R1 through R4 from the intracellular to the extracellular vestibule without ion leak; however, ionic leak current through VSDs is observed when the arginine gating charges or hydrophobic residues in S1 and S2 are mutated to smaller amino acids (30, 40–43). In our structural models, the transmembrane electrical potential would drop across a short stretch of the gating pore, which was predicted from studies showing a focused electric field near the center of the VSD (44–48). We consider the focused electrical field to drop primarily across the HCS, which forms a hydrophobic seal of 5–6 Å thickness just above the center of the VSD (Fig. 8, orange band). This structure is in close agreement with continuum electrostatic calculations (47), fluorometric measurements (49), estimates based on tethered charges of different length (46), and molecular dynamics simulations (39). Comparison of our Resting State 1 and Activated State 3 models shows that at least three arginine gating charges move completely through the HCS (Fig. 8), which is in close agreement with experimental observations suggesting that 3–4e are transferred across the membrane electric field per voltage sensor (50–52).
ii) S4 moves outward, rotates, and tilts as it passes through the HCS (Fig. 8). Its outward movement is about 6–8 Å relative to the S1 and S2 segments, which is in close agreement with previous studies (46, 53, 54). S4 also moves outward relative to S3 (Fig. 8) (36), and this conformational change is allowed by a bend of the extracellular one-half of S3 at a conserved position that contains only amino acid residues with small side chains analogous to G100 in NaChBac (SI Appendix, Figs. S2 and S3). S4 rotates ∼30° around its own axis with respect to S1 and S2. Finally, S4 also tilts relative to the S4–S5 linker from 100° in Resting State 1 to ∼60° in Activated State 3. An even larger tilting movement was proposed based on studies of the KAT1 channel by genetic complementation experiments in yeast (55). Our results give two distinct views of the conformation and rotation of the S4 segment. An α-helical conformation of the outer gating charges from T0 to R2 precisely accommodates the pattern of disulfide locking that we have observed. However, the pattern of disulfide locking observed for the hydrophobic residues from R1 to R4 implies that they are in a 310-helical conformation and move linearly through the gating pore without rotation. These two seemingly incompatible aspects of our results are accommodated by our model, in which the part of the S4 segment that is interacting with D60 and E70 in the gating pore is in a 310-helical conformation. Our estimated rotation of S4 is within the range measured for the Shaker channel (30–60°) from FRET and Luminescence Resonance Energy Transfer (LRET) data, suggesting that the proposed rotation in our model is accurate and likely to be a conserved motion in VSDs (56–59).
iii) On the extracellular side of the VSD, the combination of sideways and transmembrane movements of S4 moves the S3–S4 loop from close proximity to the S1–S2 loop in the resting states to a position in which there is a substantial cleft between the S1–S2 and S3–S4 loops in the activated states (Fig. 8). Structural proximity between the extracellular ends of S1–S4 segments in the resting state is observed in the MlotiK1 CNG channel structure (29), and separation between these two extracellular loops is observed in the structure of KV1.2-KV2.1 and also is implied by data showing that the wedge-shaped β-scorpion toxins bind between the S1–S2 and S3–S4 loops of sodium channels in the activated state (60).
iv) On the intracellular side of the VSD, the combination of the outward, rotating, and tilting motions of S4 imposes a sideways gating movement of the S4–S5 linker that is nearly parallel (within 3–5 Å) to the plane of the inner surface of the membrane (Fig. 8 and Movie S1). This movement may push the intracellular end of S4 and the adjacent S4–S5 linker in direction of S6 and may cause the entire VSD to rotate relative to the axis of the pore during pore opening (Movies S2 and S3). A similar gating movement parallel to the plane of the membrane can be inferred from comparison of the structures of the resting state of MlotiK1 (29) with the activated state of the KV1.2-KV2.1 (19), and this motion is seen more definitively from comparison of the recent crystal structure of NavAb in a preopen state (20) with the structure of the KV1.2/KV2.1 chimera in the open state (19).
v) Within the VSD, the S4 segment contains a section of 310-helix from R1 to R4 in Resting States 1 and 2, which is shortened to extend only from R2 to R4 in Activated States 1–3. This change in secondary structure allows the R1–R3 region of S4 to move nearly linearly through a narrow groove formed by the S1–S3 segments (Figs. 1 F and G and 8). Because of this 310-helix structure, the magnitude of S4 rotational movement is significantly less than suggested by the original versions of the sliding helix and helical screw models (11, 26, 61). This mechanism may also allow the S4 segment to store the energy derived from its movement through the electric field in its high-energy 310-conformation and then release it to drive pore opening by converting the extracellular end to α-helical conformation (SI Appendix, SI Discussion). This mode of movement through the gating pore is consistent with disulfide-locking experiments showing that only the S4 gating charges themselves, and not the intervening hydrophobic residues, interact with D60 in the ENC (Figs. 6C and 8). The gating charges R1–R4 form an extensive network of ionic and hydrogen-bonding interactions with negatively charged and polar residues in the intracellular and extracellular halves of the VSD and the backbone carbonyl of I96 in the HCS in the middle of S3. This network of hydrophilic interactions catalyzes movement of the gating charges from interactions with the INC in the resting state to interactions with the ENC in the activated state. The extracellular and intracellular vestibules within the VSD enable exposure of the gating charges to the aqueous environment in the activated and resting states, respectively.
Overall, our structural models of resting and activated states of the NaChBac VSD fit both previous and current experimental data surprisingly well and explain many unexpected features of these experimental results. These models will serve as valuable templates for modeling and structure function studies of the more complex mammalian voltage-gated sodium and calcium channels. These models require additional testing and validation, but our present results show that structural modeling with the Rosetta Membrane Symmetry method using available crystal structures and experimental data can be a useful approach for structure prediction of multiple functional states of ion channels and other symmetric membrane proteins at high resolution.
Methods
Structural modeling was carried out with the Rosetta Membrane Program (25, 27, 28). Disulfide-locking (31, 32) and mutant cycle analysis (31, 32) were carried out as described previously. Please see SI Appendix, SI Methods for details.
Supplementary Material
Acknowledgments
We thank Jian Payandeh, Frank DiMaio, Stanislav Sokolov, Gilbert Martinez, and Tamer Gamaleldin for helpful discussions; Eduardo Perozo for the structural model of the VSD of NaChBac; Robert Guy for structural models of NaChBac; and Keith Laidig and Darwin Alonso for excellent administration of computational resources. This work was supported by National Institutes of Health Grants P20GM076222 (to D.B.) and R01NS015751 (to W.A.C.).
Footnotes
The authors declare no conflict of interest.
See Author Summary on page 367.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1118434109/-/DCSupplemental.
References
- 1.Hille B. Ion Channels of Excitable Membranes. 3rd Ed. Sunderland, MA: Sinauer; 2001. [Google Scholar]
- 2.Catterall WA. From ionic currents to molecular mechanisms: The structure and function of voltage-gated sodium channels. Neuron. 2000;26:13–25. doi: 10.1016/s0896-6273(00)81133-2. [DOI] [PubMed] [Google Scholar]
- 3.Yu FH, Catterall WA. The VGL-chanome: A protein superfamily specialized for electrical signaling and ionic homeostasis. Sci STKE. 2004;2004:re15. doi: 10.1126/stke.2532004re15. [DOI] [PubMed] [Google Scholar]
- 4.Ren D, et al. A prokaryotic voltage-gated sodium channel. Science. 2001;294:2372–2375. doi: 10.1126/science.1065635. [DOI] [PubMed] [Google Scholar]
- 5.Koishi R, et al. A superfamily of voltage-gated sodium channels in bacteria. J Biol Chem. 2004;279:9532–9538. doi: 10.1074/jbc.M313100200. [DOI] [PubMed] [Google Scholar]
- 6.Hodgkin AL, Huxley AF. A quantitative description of membrane current and its application to conduction and excitation in nerve. J Physiol. 1952;117:500–544. doi: 10.1113/jphysiol.1952.sp004764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Armstrong CM. Sodium channels and gating currents. Physiol Rev. 1981;61:644–683. doi: 10.1152/physrev.1981.61.3.644. [DOI] [PubMed] [Google Scholar]
- 8.Bezanilla F. The voltage sensor in voltage-dependent ion channels. Physiol Rev. 2000;80:555–592. doi: 10.1152/physrev.2000.80.2.555. [DOI] [PubMed] [Google Scholar]
- 9.Catterall WA. Voltage-dependent gating of sodium channels: Correlating structure and function. Trends Neurosci. 1986;9:7–10. [Google Scholar]
- 10.Catterall WA. Molecular properties of voltage-sensitive sodium channels. Annu Rev Biochem. 1986;55:953–985. doi: 10.1146/annurev.bi.55.070186.004513. [DOI] [PubMed] [Google Scholar]
- 11.Guy HR, Seetharamulu P. Molecular model of the action potential sodium channel. Proc Natl Acad Sci USA. 1986;83:508–512. doi: 10.1073/pnas.83.2.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sigworth FJ. Voltage gating of ion channels. Q Rev Biophys. 1994;27:1–40. doi: 10.1017/s0033583500002894. [DOI] [PubMed] [Google Scholar]
- 13.Kontis KJ, Rounaghi A, Goldin AL. Sodium channel activation gating is affected by substitutions of voltage sensor positive charges in all four domains. J Gen Physiol. 1997;110:391–401. doi: 10.1085/jgp.110.4.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Papazian DM, Timpe LC, Jan YN, Jan LY. Alteration of voltage-dependence of Shaker potassium channel by mutations in the S4 sequence. Nature. 1991;349:305–310. doi: 10.1038/349305a0. [DOI] [PubMed] [Google Scholar]
- 15.Stühmer W, et al. Structural parts involved in activation and inactivation of the sodium channel. Nature. 1989;339:597–603. doi: 10.1038/339597a0. [DOI] [PubMed] [Google Scholar]
- 16.Long SB, Campbell EB, Mackinnon R. Voltage sensor of Kv1.2: Structural basis of electromechanical coupling. Science. 2005;309:903–908. doi: 10.1126/science.1116270. [DOI] [PubMed] [Google Scholar]
- 17.Jiang Y, et al. X-ray structure of a voltage-dependent K+ channel. Nature. 2003;423:33–41. doi: 10.1038/nature01580. [DOI] [PubMed] [Google Scholar]
- 18.Long SB, Campbell EB, Mackinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 19.Long SB, Tao X, Campbell EB, MacKinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007;450:376–382. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- 20.Payandeh J, Scheuer T, Zheng N, Catterall WA. The crystal structure of a voltage-gated sodium channel. Nature. 2011;475:353–358. doi: 10.1038/nature10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Catterall WA. Ion channel voltage sensors: Structure, function, and pathophysiology. Neuron. 2010;67:915–928. doi: 10.1016/j.neuron.2010.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Söding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 23.Söding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hildebrand A, Remmert M, Biegert A, Söding J. Fast and accurate automatic structure prediction with HHpred. Proteins. 2009;77(Suppl 9):128–132. doi: 10.1002/prot.22499. [DOI] [PubMed] [Google Scholar]
- 25.Barth P, Schonbrun J, Baker D. Toward high-resolution prediction and design of transmembrane helical protein structures. Proc Natl Acad Sci USA. 2007;104:15682–15687. doi: 10.1073/pnas.0702515104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yarov-Yarovoy V, Baker D, Catterall WA. Voltage sensor conformations in the open and closed states in ROSETTA structural models of K+ channels. Proc Natl Acad Sci USA. 2006;103:7292–7297. doi: 10.1073/pnas.0602350103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yarov-Yarovoy V, Schonbrun J, Baker D. Multipass membrane protein structure prediction using Rosetta. Proteins. 2006;62:1010–1025. doi: 10.1002/prot.20817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.André I, Bradley P, Wang C, Baker D. Prediction of the structure of symmetrical protein assemblies. Proc Natl Acad Sci USA. 2007;104:17656–17661. doi: 10.1073/pnas.0702626104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clayton GM, Altieri S, Heginbotham L, Unger VM, Morais-Cabral JH. Structure of the transmembrane regions of a bacterial cyclic nucleotide-regulated channel. Proc Natl Acad Sci USA. 2008;105:1511–1515. doi: 10.1073/pnas.0711533105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Campos FV, Chanda B, Roux B, Bezanilla F. Two atomic constraints unambiguously position the S4 segment relative to S1 and S2 segments in the closed state of Shaker K channel. Proc Natl Acad Sci USA. 2007;104:7904–7909. doi: 10.1073/pnas.0702638104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DeCaen PG, Yarov-Yarovoy V, Zhao Y, Scheuer T, Catterall WA. Disulfide locking a sodium channel voltage sensor reveals ion pair formation during activation. Proc Natl Acad Sci USA. 2008;105:15142–15147. doi: 10.1073/pnas.0806486105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DeCaen PG, Yarov-Yarovoy V, Sharp EM, Scheuer T, Catterall WA. Sequential formation of ion pairs during activation of a sodium channel voltage sensor. Proc Natl Acad Sci USA. 2009;106:22498–22503. doi: 10.1073/pnas.0912307106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Decaen PG, Yarov-Yarovoy V, Scheuer T, Catterall WA. Gating charge interactions with the S1 segment during activation of a Na+ channel voltage sensor. Proc Natl Acad Sci USA. 2011;108:18825–18830. doi: 10.1073/pnas.1116449108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barth P, Wallner B, Baker D. Prediction of membrane protein structures with complex topologies using limited constraints. Proc Natl Acad Sci USA. 2009;106:1409–1414. doi: 10.1073/pnas.0808323106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen X, Wang Q, Ni F, Ma J. Structure of the full-length Shaker potassium channel Kv1.2 by normal-mode-based X-ray crystallographic refinement. Proc Natl Acad Sci USA. 2010;107:11352–11357. doi: 10.1073/pnas.1000142107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Broomand A, Elinder F. Large-scale movement within the voltage-sensor paddle of a potassium channel-support for a helical-screw motion. Neuron. 2008;59:770–777. doi: 10.1016/j.neuron.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 37.Yifrach O, MacKinnon R. Energetics of pore opening in a voltage-gated K+ channel. Cell. 2002;111:231–239. doi: 10.1016/s0092-8674(02)01013-9. [DOI] [PubMed] [Google Scholar]
- 38.Careaga CL, Falke JJ. Thermal motions of surface alpha-helices in the D-galactose chemosensory receptor. Detection by disulfide trapping. J Mol Biol. 1992;226:1219–1235. doi: 10.1016/0022-2836(92)91063-u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jogini V, Roux B. Dynamics of the Kv1.2 voltage-gated K+ channel in a membrane environment. Biophys J. 2007;93:3070–3082. doi: 10.1529/biophysj.107.112540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sokolov S, Scheuer T, Catterall WA. Ion permeation through a voltage- sensitive gating pore in brain sodium channels having voltage sensor mutations. Neuron. 2005;47:183–189. doi: 10.1016/j.neuron.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 41.Sokolov S, Scheuer T, Catterall WA. Ion permeation and block of the gating pore in the voltage sensor of NaV1.4 channels with hypokalemic periodic paralysis mutations. J Gen Physiol. 2010;136:225–236. doi: 10.1085/jgp.201010414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tombola F, Pathak MM, Gorostiza P, Isacoff EY. The twisted ion-permeation pathway of a resting voltage-sensing domain. Nature. 2007;445:546–549. doi: 10.1038/nature05396. [DOI] [PubMed] [Google Scholar]
- 43.Tombola F, Pathak MM, Isacoff EY. Voltage-sensing arginines in a potassium channel permeate and occlude cation-selective pores. Neuron. 2005;45:379–388. doi: 10.1016/j.neuron.2004.12.047. [DOI] [PubMed] [Google Scholar]
- 44.Yang N, George AL, Jr., Horn R. Molecular basis of charge movement in voltage-gated sodium channels. Neuron. 1996;16:113–122. doi: 10.1016/s0896-6273(00)80028-8. [DOI] [PubMed] [Google Scholar]
- 45.Yang N, George AL, Jr, Horn R. Probing the outer vestibule of a sodium channel voltage sensor. Biophys J. 1997;73:2260–2268. doi: 10.1016/S0006-3495(97)78258-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ahern CA, Horn R. Focused electric field across the voltage sensor of potassium channels. Neuron. 2005;48:25–29. doi: 10.1016/j.neuron.2005.08.020. [DOI] [PubMed] [Google Scholar]
- 47.Islas LD, Sigworth FJ. Electrostatics and the gating pore of Shaker potassium channels. J Gen Physiol. 2001;117:69–89. doi: 10.1085/jgp.117.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Larsson HP, Baker OS, Dhillon DS, Isacoff EY. Transmembrane movement of the shaker K+ channel S4. Neuron. 1996;16:387–397. doi: 10.1016/s0896-6273(00)80056-2. [DOI] [PubMed] [Google Scholar]
- 49.Asamoah OK, Wuskell JP, Loew LM, Bezanilla F. A fluorometric approach to local electric field measurements in a voltage-gated ion channel. Neuron. 2003;37:85–97. doi: 10.1016/s0896-6273(02)01126-1. [DOI] [PubMed] [Google Scholar]
- 50.Aggarwal SK, MacKinnon R. Contribution of the S4 segment to gating charge in the Shaker K+ channel. Neuron. 1996;16:1169–1177. doi: 10.1016/s0896-6273(00)80143-9. [DOI] [PubMed] [Google Scholar]
- 51.Kuzmenkin A, Bezanilla F, Correa AM. Gating of the bacterial sodium channel, NaChBac: Voltage-dependent charge movement and gating currents. J Gen Physiol. 2004;124:349–356. doi: 10.1085/jgp.200409139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seoh SA, Sigg D, Papazian DM, Bezanilla F. Voltage-sensing residues in the S2 and S4 segments of the Shaker K+ channel. Neuron. 1996;16:1159–1167. doi: 10.1016/s0896-6273(00)80142-7. [DOI] [PubMed] [Google Scholar]
- 53.Pathak MM, et al. Closing in on the resting state of the Shaker K+ channel. Neuron. 2007;56:124–140. doi: 10.1016/j.neuron.2007.09.023. [DOI] [PubMed] [Google Scholar]
- 54.Posson DJ, Selvin PR. Extent of voltage sensor movement during gating of shaker K+ channels. Neuron. 2008;59:98–109. doi: 10.1016/j.neuron.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grabe M, Lai HC, Jain M, Jan YN, Jan LY. Structure prediction for the down state of a potassium channel voltage sensor. Nature. 2007;445:550–553. doi: 10.1038/nature05494. [DOI] [PubMed] [Google Scholar]
- 56.Mannuzzu LM, Moronne MM, Isacoff EY. Direct physical measure of conformational rearrangement underlying potassium channel gating. Science. 1996;271:213–216. doi: 10.1126/science.271.5246.213. [DOI] [PubMed] [Google Scholar]
- 57.Glauner KS, Mannuzzu LM, Gandhi CS, Isacoff EY. Spectroscopic mapping of voltage sensor movement in the Shaker potassium channel. Nature. 1999;402:813–817. doi: 10.1038/45561. [DOI] [PubMed] [Google Scholar]
- 58.Cha A, Snyder GE, Selvin PR, Bezanilla F. Atomic scale movement of the voltage-sensing region in a potassium channel measured via spectroscopy. Nature. 1999;402:809–813. doi: 10.1038/45552. [DOI] [PubMed] [Google Scholar]
- 59.Cha A, Bezanilla F. Structural implications of fluorescence quenching in the Shaker K+ channel. J Gen Physiol. 1998;112:391–408. doi: 10.1085/jgp.112.4.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cestèle S, et al. Structure and function of the voltage sensor of sodium channels probed by a beta-scorpion toxin. J Biol Chem. 2006;281:21332–21344. doi: 10.1074/jbc.M603814200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shafrir Y, Durell SR, Guy HR. Models of voltage-dependent conformational changes in NaChBac channels. Biophys J. 2008;95:3663–3676. doi: 10.1529/biophysj.108.135335. [DOI] [PMC free article] [PubMed] [Google Scholar]