Abstract
Flower production and crop yields are highly influenced by the architectures of inflorescences. In the compound inflorescences of tomato and related nightshades (Solanaceae), new lateral inflorescence branches develop on the flanks of older branches that have terminated in flowers through a program of plant growth known as “sympodial.” Variability in the number and organization of sympodial branches produces a remarkable array of inflorescence architectures, but little is known about the mechanisms underlying sympodial growth and branching diversity. One hypothesis is that the rate of termination modulates branching. By performing deep sequencing of transcriptomes, we have captured gene expression dynamics from individual shoot meristems in tomato as they gradually transition from a vegetative state to a terminal flower. Surprisingly, we find thousands of age-dependent expression changes, even when there is little change in meristem morphology. From these data, we reveal that meristem maturation is an extremely gradual process defined molecularly by a “meristem maturation clock.” Using hundreds of stage-enriched marker genes that compose this clock, we show that extreme branching, conditioned by loss of expression of the COMPOUND INFLORESCENCE gene, is driven by delaying the maturation of both apical and lateral meristems. In contrast, we find that wild tomato species display a delayed maturation only in apical meristems, which leads to modest branching. Our systems genetics approach reveals that the program for inflorescence branching is initiated surprisingly early during meristem maturation and that evolutionary diversity in inflorescence architecture is modulated by heterochronic shifts in the acquisition of floral fate.
Keywords: flowering, development, cyme, evolution, transcriptomics
Inflorescences develop from small groups of pluripotent cells in growing tips called shoot apical meristems (SAM). SAMs first give rise to leaves before transitioning to inflorescence meristems (IM), which can produce lateral (axillary) meristems that either transition into flower-bearing shoots or differentiate directly into flowers (1–3). Diversity in inflorescence architecture is based on two major growth habits. In “monopodial” plants, such as Arabidopsis and other annual species that transition to flowering only once in a lifetime, IMs grow continuously and initiate axillary flowers laterally, which results in a narrow range of inflorescence architectures. In contrast, in “sympodial” plants, such as trees and myriad other perennial species that experience multiple flowering transitions throughout life, IMs terminate in flowers and growth continues from a variable number of new axillary (sympodial) IMs that repeat this process to form compound inflorescence shoots (4). The alternation between meristem termination and renewal in sympodial plants provides the foundation for a wide range of inflorescence types showing extensive variation in branch and flower number (5, 6), but the basis for sympodial growth and branching diversity is poorly understood.
Plants in the nightshade (Solanaceae) family (7), particularly tomato (Solanum lycopersicum), are models for sympodial growth, and inflorescence architectures range from a solitary flower, as in petunia (8), to progressively more complex structures in which the meristem proliferates into multiple branches with dozens of flowers, as in several wild species of tomato (9). Most cultivated tomatoes produce inflorescences with a few flowers arranged in a zigzag branching pattern (Fig. 1 A–E), but we previously showed that variants with highly branched inflorescences bearing hundreds of flowers arose more than a century ago due to loss-of-function mutations in a homeobox transcription factor encoded by the COMPOUND INFLORESCENCE (S) gene (Fig. 1G) (10). Because meristem ontogenies between s mutants and normal plants are nearly identical, we proposed that s inflorescences might be highly branched due to a molecular delay in meristem maturation and termination that results in an extended period of indeterminacy, during which additional sympodial meristems, and therefore branches, develop. We further hypothesized that, because wild tomato species with branched inflorescences resemble s inflorescences, branching diversity in the tomato clade might similarly be based on delaying meristem termination (Fig. 1 F–H) (10); however, reproductive barriers between branched and unbranched tomato species prohibit a genetic dissection of branching variation (9, 11). Therefore, to decipher the basis for evolutionary diversity in tomato inflorescence architecture, we took a systems genetics approach and used quantitative transcriptomics to characterize and compare meristem maturation, termination, and branching in the domesticated tomato, the highly branched s mutant, and the modestly branched wild species, Solanum peruvianum.
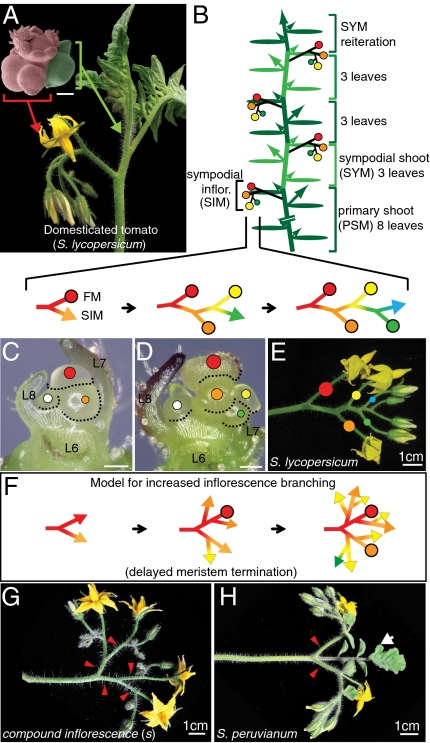
Fig. 1.
Tomato sympodial growth and inflorescence branching diversity. (A) A typical tomato plant and pseudocolored scanning electron microscope image showing the meristems giving rise to a multiflowered compound inflorescence (red) and vegetative shoot with leaves (green). (Scale bar, bracketed image, 100 μm.) (B) Schematic diagram depicting sympodial growth of tomato (cv. M82), in which meristems terminate in flowers and growth continues from specialized axillary (sympodial) meristems. The primary shoot meristem (PSM) produces eight leaves and terminates in a multiflowered inflorescence. Upright growth continues from the reiteration of sympodial shoot meristems (SYM), where each successive SYM produces three leaves, a terminal inflorescence, and the next SYM, resulting in a compound vegetative shoot. Dark and light-green ovals depict leaves associated with successive sympodial units. Small arrows represent canonical axillary shoots, which go on to produce their own SYMs and SIMs similar to the PSM, leading to a bushy plant with many branches and inflorescences. (C–E) Sequential stereoscope images (C and D) of the earliest stages of inflorescence development showing the zigzag reiteration of sympodial inflorescence meristems (SIM). Each SIM produces one new lateral SIM before terminating in a flower meristem (FM), which becomes a flower (E). Corresponding colored lines and dots in schematics and images reflect SIM initiation (lines) and FM termination (dots). The white dot indicates the first SYM in the axil of the last formed leaf (L8) on the primary shoot. (Scale bars in C and D, 100 μm.) (F) Proposed model for sympodial inflorescence branching in which meristem maturation and termination is delayed molecularly, thereby enabling additional SIMs, and therefore branches, to develop (10). (G and H) Diversity in tomato inflorescence branching ranging from highly branched inflorescences of s mutants (G) to the modestly branched “forked” inflorescences of the wild species, S. peruvianum (H). Red arrowheads point to branches. Most wild tomatoes produce modified leaves (bracts) in their inflorescences (white arrow in H), which manifest only after several flowers have formed.
Results
Tomato Sympodial Growth and Transcriptome Dynamics of Meristem Maturation.
Tomato plants develop from distinct types of shoot meristems (12, 13), each of which is large and easily dissected from surrounding leaves (Fig. 1 A–E). The primary shoot meristem (PSM) originates from the embryo and produces between 7 and 12 leaves before switching to reproductive growth and terminating in an inflorescence (SI Appendix, Fig. S1). Upright growth continues from the formation of a specialized axillary meristem called a sympodial shoot meristem (SYM), which develops in the axil of the last leaf on the PSM and terminates after producing only three leaves. A new SYM subsequently forms in the axil of the last leaf produced by the previous SYM, and this process reiterates such that all subsequent SYMs develop three leaves, a terminal inflorescence, and the next SYM to produce a compound vegetative shoot (Fig. 1B). Inflorescences are also compound shoots, resulting from the zigzag reiteration of sympodial inflorescence meristems (SIM), each of which gives rise to another SIM before terminating in a flower meristem (FM) (Fig. 1 C–E). Each new SIM develops perpendicular to the one formed previously with no intervening leaf development, resulting in the familiar zigzag pattern of tomato inflorescences. Thus, all aspects of tomato sympodial growth can be reduced to the sequential maturation and termination of individual shoot meristems.
To study the role of meristem maturation and termination in sympodial inflorescence branching, we first captured gene expression profiles underlying the gradual transition of the PSM from a vegetative state to a terminal flower. From a wild-type (WT) unbranched tomato (S. lycopersicum) cultivar known as M82 that consistently initiates eight leaves before flowering, we measured the plastochron index (14) and morphological characters to a timescale based on “days after germination” (DAG) (SI Appendix, Fig. S1). We defined five distinct stages during PSM maturation: the early, middle, and late vegetative meristems (EVM: fifth leaf initiated; MVM: sixth leaf initiated; LVM: seventh leaf initiated), the transition meristem (TM: eighth leaf initiated), and the FM (Fig. 2A and SI Appendix). We profiled the transcriptome of each stage by isolating mRNA and subjecting cDNA libraries to Illumina sequencing (SI Appendix) (15). In total, 30–40 million high-quality sequence reads were generated for each stage, and reads were mapped to the protein-coding sequences of the tomato genome annotation and quantified (SI Appendix, Fig. S2 and Dataset S1) (16). Depending on the meristem stage, between 17,882 and 18,084 genes are detected, and a total of 19,100 genes (54% of the annotated transcriptome) are detected when summed across all stages, of which 16,958 are common to all five meristems.
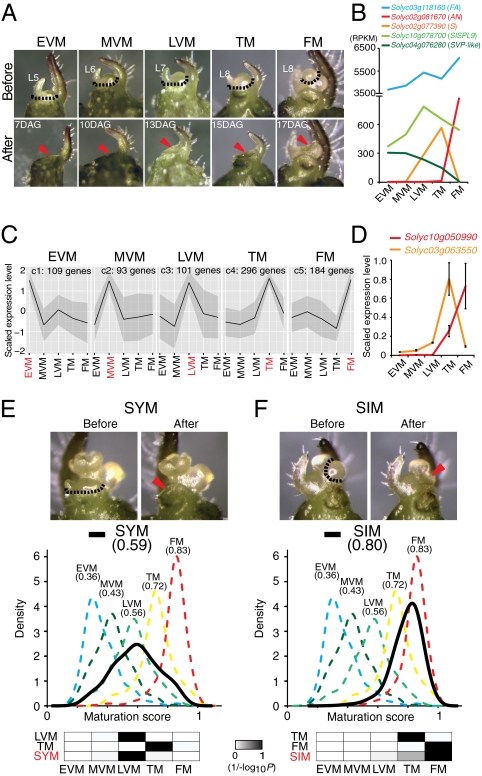
Fig. 2.
Characterization of primary shoot meristem maturation and underlying gene expression dynamics. (A) Microdissection of the five meristem stages of PSM for transcriptome profiling. Dashed lines indicate dissected tissue line. Red arrowheads indicate dissected meristem. DAG, days after germination; L, leaf number from first leaf. (B) Dynamic expression of the tomato inflorescence branching genes S, FALSIFLORA (FA), and ANANTHA (AN), and the vegetative-to-reproductive phase transition marker genes SlSPL9 (ortholog of Arabidopsis SPL9) and SVP-like (homolog of Arabidopsis SVP) through five successive stages of PSM maturation ending in flower meristem (FM) termination. Mean RPKM values are shown on the y axis. (C) Clusters of stage-enriched marker genes derived from K-means clustering of 3,919 dynamically expressed genes during PSM maturation. Five clusters are shown to illustrate PSM stage-enriched expression patterns from 20 clusters (SI Appendix, Fig. S4). Cluster name (e.g., C4) and number of genes are indicated on top. (D) Quantitative RT-PCR validation of two newly discovered lowly expressed marker genes of unknown function, which peak in the TM (Solyc03g063550) and FM (Solyc10g050990), respectively. (E and F) Molecular quantification of sympodial meristem maturation states. Dissection of the SYM (E, top) and SIM (F, top) were used for mRNA-seq as in A. DDI quantifications of meristem maturation scores for the SYM (E, bottom) and SIM (F, bottom) relative to five PSM stages used to develop a meristem maturation clock based on 841 DDI-selected marker genes. Black curves, which are closest to the LVM and the TM/FM, respectively, show the DDI-estimated maturation state of the SYM (E, bottom) and SIM (F, bottom). Numbers in parentheses indicate mean maturation scores. Scaled 1/(−log10P) values are shown as grayscale heat maps, where darker color indicates greater similarity in maturation state. P values are derived from Student's t tests between estimated maturation scores of calibrations and query samples based on the marker genes derived by the DDI algorithm (Datasets S3 and S4).
Our transcriptome profiling captured gene expression dynamics at a high temporal resolution. For example, our data revealed distinct age-dependent increases in expression during PSM maturation for the S gene and two additional tomato inflorescence branching genes, FALSIFLORA and ANANTHA [orthologs of Arabidopsis LEAFY and UNUSUAL FLORAL ORGANS, respectively (Fig. 2B)], which other detection techniques (e.g., in situ hybridization) could not readily expose (10, 17). More broadly, our data revealed previously unknown and surprisingly dynamic expression changes through the vegetative-to-reproductive transition for genes encoding MADS-box transcription factors related to the Arabidopsis flowering genes SHORT VEGETATIVE PHASE (SVP), APETALA1 (AP1), and SEPALLATA (SEP). For example, transcripts from the tomato ortholog of AP1 (SlAP1/MC: Solyc05g056620) are not restricted to the flower meristem, but accumulate gradually beginning already in the LVM. Other genes with roles in the flowering transition, such as the microRNA-regulated SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) transcription factors, are also dynamically expressed as the PSM matures from a vegetative to a reproductive state (Fig. 2B and SI Appendix, Fig. S3) (18).
To explore our data more deeply and further characterize and define meristem maturation and termination molecularly beyond the limited number of genes with known roles in flowering, we identified 3,919 differentially expressed genes [False Discovery Rate (FDR): twofold change and P ≤ 0.05] among the five PSM stages (Dataset S2) and grouped them according to expression dynamics using the K-means clustering algorithm (SI Appendix). Five clusters showed a stage-enriched peak of expression, revealing modules of coexpressed genes that precisely define each stage of maturation (Fig. 2C). Fifteen additional clusters resolved into two major groups: gradients of increasing or decreasing expression or peaks spanning consecutive vegetative or reproductive stages, revealing coexpressed genes showing waves of expression changes that further define PSM maturation (SI Appendix, Fig. S4). All clusters are enriched for several functional categories of genes as determined by MapMan classifications (SI Appendix, Fig. S5 and Dataset S2). Particularly noteworthy are genes encoding transcription factors, which, despite typically showing low transcript abundance, are readily detected in our data, compose the most-enriched functional category among all clusters, and show diverse age-dependent expression dynamics during meristem maturation (SI Appendix, Fig. S6). Importantly, however, our data expose thousands of additional genes from diverse functional categories, as well as 375 genes of unknown function that are also expressed dynamically in an age-dependent manner, and we validated the dynamics of 16 genes by quantitative RT-PCR (Fig. 2D and SI Appendix, Fig. S7). Thus, in addition to generating the first high-resolution gene expression atlas for meristem maturation, our transcriptomics profiling of the tomato PSM reveals that thousands of genes with dynamic, age-dependent expression changes define the gradual maturation of a single meristem from a vegetative state to a terminal flower. To visualize expression levels during meristem maturation, we developed a tomato electronic fluorescent pictograph (eFP) browser (http://tomatolab.cshl.edu/efp/cgi-bin/efpWeb.cgi) (19).
Establishment of a “Meristem Maturation Clock.”
The transcriptional hallmarks defining PSM maturation provide a rich source of molecular markers for evaluating maturation states of other types of meristems. The SYM and SIM of tomato originate from the uppermost axillary meristems on the primary shoot and terminate after producing only three leaves or immediately in a flower, respectively, suggesting that these meristems are initiated in differing maturation states. To test this idea, we profiled the transcriptomes of the SYM (second leaf initiated) and the SIM (Fig. 2 E and F and Dataset S1) and compared their maturation states against the stages of PSM maturation by establishing a meristem maturation clock with the digital differentiation index (DDI) algorithm (20). DDI, originally developed to study leaf ontogeny, molecularly quantifies the maturation states of known tissue samples (calibration stages) by identifying marker genes that show an expression peak at each calibration stage and then comparing the expression of these stage-enriched marker genes from an unknown sample to quantify the relative maturation state (see details in SI Appendix). We first tested whether DDI can be used to quantify meristem maturation by using one replicate of transcriptome data for calibration and predicting the “maturation scores” of a second biological replicate, treated blindly as unknowns (SI Appendix, Fig. S8A). All meristems were accurately and precisely assigned from 841 DDI-selected marker genes (Dataset S3), revealing that DDI is an extremely robust tool to quantify the meristem maturation state and that the relative maturation state of each meristem can be captured solely on the basis of gene expression profiles. We next used the five stages of PSM maturation as calibration stages and measured the maturation scores of the SYM and the SIM. DDI revealed that the SYM is closest to the LVM, consistent with the SYM terminating after only three leaves (Fig. 2E and Dataset S4). The maturation score of the SIM, on the other hand, lies between the TM and FM (Fig. 2F and Dataset S4), consistent with the SIM behaving like a PSM fast approaching termination. These findings suggest that sympodial growth is based on termination of the primary shoot causing the uppermost axillary meristems to adopt advanced states of maturation and terminate after brief (SYM) or no (SIM) vegetative growth. The termination of each new side shoot in a flower leads subsequent uppermost axillary meristems to adopt the same developmental fate as their predecessors, thereby establishing a self-perpetuating mechanism for sympodial meristem reiteration (13).
Extreme Branching in compound inflorescence Is Based on Sequential Delays in the Maturation of Two Different Meristems.
The finding that the SIM of domesticated tomato is initiated in an advanced state of maturation suggests that inflorescences are unbranched because each successive SIM-to-FM transition is rapid, providing only a brief developmental window for a single axillary meristem (i.e., the next SIM) to form before flower termination. We previously proposed that inflorescence branching in s mutants is based on each SIM giving rise to multiple axillary SIMs due to sequential delays in SIM termination (10). To test this prediction, we carefully compared s PSM maturation and reproductive meristems with WT (SI Appendix, Fig. S9 and Dataset S5). Like WT, the FM and SIM of s manifest as an apical dome and a lateral dome, respectively, following the TM stage. Meristem morphology remains indistinguishable between the two genotypes until sepals begin developing on the WT FM, whereas the s FM remains a uniform dome. This suggests that the PSM of s might take longer to reach terminal fate compared with WT (SI Appendix, Fig. S9). We profiled transcriptomes from the PSM stages of s and also incorporated the SIM, positioned between the TM and FM based on its intermediate maturation state (Fig. 2F). Specifically, we profiled s meristems before (MVM), during (LVM, TM, SIM), and after (FM) the accumulation of S transcripts and compared them with their corresponding stages in WT. Importantly, that the developmental progression (ontogenies) of the s and WT PSM and SIM are so similar suggests that expression differences should reflect primarily changes in the underlying meristem maturation program, as opposed to expression differences being driven by morphological change (Fig. 3 A–C and SI Appendix, Fig. S9).
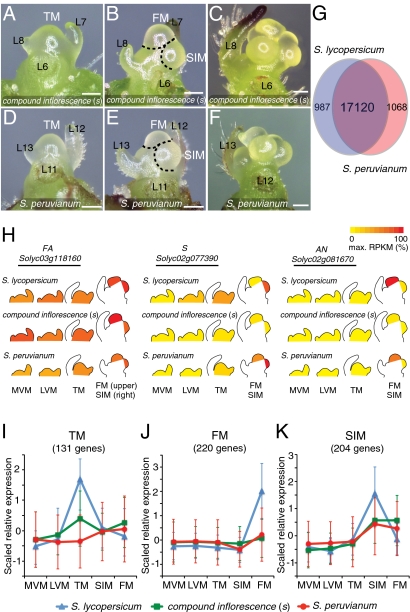
Fig. 3.
Disrupted expression dynamics in s mutants and the branched wild species S. peruvianum point to an early delay in meristem maturation and termination. (A–F) Stereoscope images from the primary shoot apex comparing the TM, SIM, and FM stages of s (A and B) with the wild tomato species, S. peruvianum (D and E). Meristem ontogenies are indistinguishable until well after branching has begun (compare B and E with C and F) (SI Appendix, Fig. S9). (G) Overlap of detected genes in meristem transcriptomes of S. lycopersicum and S. peruvianum. (H) eFP browser views of temporal expression changes during meristem maturation for the tomato inflorescence branching genes FA, S, and AN in S. lycopersicum, s, and S. peruvianum. (I–K) Altered expression dynamics in s and S. peruvianum within three S. lycopersicum stage-specific clusters. TM (I), FM (J), and SIM (K) clusters are shown (comparisons of all 20 clusters from three genotypes are shown in SI Appendix, Fig. S12). Scaled RPKM expression values of genes in the TM (I), FM (J), and SIM (K) clusters were plotted for S. lycopersicum (blue triangles and lines), s (green squares and lines), and S. peruvianum (red circles and lines) across five meristem stages. Error bars indicate SDs.
We first noted that S expression is strongly reduced in s mutants, revealing that this allele is based on a transcriptional defect (Fig. 3H) (10). We also found that AN is no longer expressed in the FM of s, suggesting that s FMs might not be terminating as rapidly as wild type. FA, on the other hand, showed little change, indicating that some elements of the meristem maturation program remain intact in s mutants (Fig. 3H). To further evaluate if and how meristem maturation is changed in s, we compared corresponding stages between s and WT and identified 4,192 differentially expressed genes (FDR: twofold change and P ≤ 0.05) representing diverse functional categories, although changes among transcription factors are most prominent (Dataset S6 and SI Appendix, Fig. S10). Hundreds of genes are differentially expressed in the SIM, where S normally peaks; however, widespread changes are also detected in the MVM, LVM, TM, and FM, even though S is normally much less transcribed in these stages. One explanation for these findings is that S also functions throughout PSM maturation and therefore loss of S transcription alters meristem maturation early on, leading to expression changes that reflect delayed termination not only of the SIM, but also of the PSM. Consistent with this idea, reproductive transition and flower identity marker genes, such as SPL and SEP, are increased (SPL) and reduced (SEP) in expression in the TM (SPL) and FM (SEP) of s mutants, respectively (SI Appendix, Fig. S11). Moreover, many of the clusters of dynamically expressed genes are altered in s in a direction suggesting a delay in both meristems (SI Appendix, Fig. S12). In particular, transcripts of TM and FM stage-enriched marker genes no longer accumulate in corresponding stages of s, and genes that normally peak in the SIM are greatly reduced in s mutants and become more expressed in s FMs (Fig. 3 I–K).
To quantify the changes in PSM and SIM maturation in s, we again used the meristem maturation clock and expanded our original DDI analysis to include the SIM as an additional calibration stage between the TM and FM, which enabled direct comparisons of the WT and mutant maturation states for each stage (Fig. 2F and SI Appendix, Fig. S8B). DDI revealed that the TM of s is markedly less mature than its corresponding stages in wild type (Fig. 4A and Dataset S4) and that the FM of s has the hallmarks of a wild-type SIM (Fig. 4B and Dataset S4). These results indicate that s mutants experience a remarkably early and extended delay in PSM maturation and that the s FM adopts the branching potential of a SIM. We further found that the SIM of s is most similar to the earlier maturation state of the TM of wild type, indicating that maturation of s SIMs is also delayed (Fig. 4C and Dataset S4). Thus, the extreme branching of s inflorescences is based on a surprisingly early and prolonged maturation of two different meristems: first, multiple SIMs develop on the primary shoot due to a delay in PSM termination, and second, branching continues through delays in SIM termination (Fig. 4G).
Fig. 4.
A delay in primary shoot meristem maturation drives inflorescence branching in wild tomatoes. (A–F) DDI quantifications of meristem maturation states for the TM, FM, and SIM stages of s (A–C) and S. peruvianum (D–F). The first replicate of S. lycopersicum was used for calibration (Dataset S3), and dashed color curves are DDI predictions for a second biological replicate (MVM, LVM, TM, SIM, FM), as in Fig. 2. Black curves represent DDI predictions for each s and S. peruvianum meristem stage based on 685 DDI-selected marker genes. Numbers in parentheses indicate mean maturation scores. Heat maps of scaled 1/(−log10P) values are shown below each graph (Dataset S4). (G) Schematic diagram representing the program of inflorescence branching in tomato. The molecular maturation underlying ontogeny for the PSM is depicted by the color gradient in the bar, and the red dot indicates floral termination. The rate of floral termination in S. lycopersicum allows for only one SIM to develop, whereas, in s mutants, floral termination of the PSM is delayed (extended green and yellow), leading to the initiation of additional SIMs (yellow dots), which are themselves delayed. This enables SIMs to elaborate indefinitely. In contrast, in S. peruvianum, the PSM is much more delayed compared with the SIM, leading to modest branching. Cartoons of meristems below the bars show when meristems were collected for all three genotoypes, as determined by shared, and nearly identical, morphological parameters used to define corresponding stages of meristem maturation (SI Appendix, Fig. S9).
Delay in the Maturation of the Primary Shoot Meristem Drives Inflorescence Branching in Wild Tomatoes.
The discovery of a two-tiered branching program for s led us to hypothesize that inflorescence branching in wild tomato species might be based on a similar mechanism. Several tomato species produce branched inflorescences (9), and, in our growth conditions, the inflorescences of Solanum chilense, Solanum habrochaites, and S. peruvianum consistently develop two to three branches often referred to as “forked” inflorescences (Fig. 1H and SI Appendix, Fig. S13). Because wild species branch only a few times compared with s mutants, we speculated that forked branching might be based on delaying only PSM termination because also delaying SIM termination should lead to continuous branching. We tested this idea using S. peruvianum, which transitions to flowering 7 d after domesticated tomato does and develops large meristems (SI Appendix). We observed that meristem ontogenies of S. peruvianum and s are indistinguishable until well after branching has begun, suggesting that meristem morphology fails to distinguish branching programs (Fig. 3 A–F). We therefore used equivalent criteria as s for defining meristem stages (SI Appendix, Fig. S9 and Dataset S5) and profiled transcriptomes of the S. peruvianum MVM, LVM, TM, SIM, and FM stages (Dataset S1). We first assembled a S. peruvianum transcriptome de novo, which revealed an average protein-coding sequence divergence from domesticated tomato of only 0.8% (SI Appendix). This high sequence similarity, combined with 95% overlap in the genes that are detected in S. lycopersicum (Fig. 3G), permitted mapping and quantification of S. peruvianum reads against the WT reference transcriptome and a direct comparison of expression profiles (SI Appendix, Fig. S14).
In a stage-by-stage comparison, we found 7,729 differentially expressed genes between S. peruvianum and domesticated tomato, which is nearly twice the number detected when comparing s to domesticated tomato (FDR: twofold change and P < 0.05) (Dataset S7). One explanation for this difference is that species divergence has resulted in broad gene expression changes unrelated to inflorescence branching programs, which we showed by comparing the combined transcriptomes from each genotype in a principal component analysis (PCA) (SI Appendix, Fig. S15). To determine if inflorescence branching in S. peruvianum is caused by delaying maturation of the PSM, the SIM, or both, we focused on comparing dynamically expressed genes between S. peruvianum, domesticated tomato, and s mutants. We first noted that the temporal expression dynamic of the S gene is delayed beginning in the TM of S. peruvianum and that AN expression is reduced in the FM, consistent with PSM maturation being delayed (Fig. 3H). Beyond these two markers, we observed that the clusters of dynamically expressed genes are drastically altered in S. peruvianum and follow trajectories of change that resemble s, particularly for the TM, FM, and SIM clusters (Fig. 3 I–K). To quantitatively determine how maturation states for the TM, FM, and SIM of wild species differ from domesticated tomato, we applied DDI and found that the TM of S. peruvianum is significantly less mature than the TM of both domesticated tomato and s (Fig. 4D and Dataset S4). Furthermore, the FM of S. peruvianum has the hallmarks of both a TM and a SIM (Fig. 4E and Dataset S4). These results indicate that, like s, the branching program of S. peruvianum is initiated through a delay in the maturation and termination of the PSM. Significantly, the maturation state of the SIM of S. peruvianum lies in between the SIM of domesticated tomato and s, suggesting that S. peruvianum gives rise to SIMs with less branching potential than s SIMs, which are therefore more similar to domesticated SIMs (Fig. 4F and Dataset S4). This likely explains why only two to three branches develop on each inflorescence in S. peruvianum. Thus, the modestly branched forked inflorescences of S. peruvianum are based predominantly on a delay in PSM termination (Fig. 4G).
Discussion
Mathematical modeling has suggested that evolutionary partitioning of inflorescences into distinct types such as racemes (e.g., Arabidopsis), panicles (e.g., rice), and cymes (e.g., tomato) might be based on differences in the relative rates of termination in apical versus lateral meristems, modulated by a theoretical parameter called vegetativeness (veg) (21). Using deep transcriptomic sequencing, we captured gene expression dynamics underlying the process of meristem maturation and termination from tomato genotypes representing a range of inflorescence complexity and experimentally quantified a reflection of veg for the first time. This systems genetics approach (22) revealed that the evolutionary switch to branched cymes is associated with a surprisingly early delay in primary meristem maturation, well before lateral inflorescence meristems are initiated. This delay may permit the formation of additional, often cryptic, lateral organs on a nearly terminated apex with each organ hosting a new inflorescence meristem in its axil, even though there is no ontogenetic change reflecting continued meristem activity for the PSM or SIMs in s mutants or S. peruvianum. Importantly, modified leaves known as bracts frequently manifest in mutants with highly branched inflorescences, such as s (10), and in branched wild tomato species (9). Remarkably, our data reveal that the transcriptome profile of the TM not only indicates the speed of PSM maturation, but also provides an early predictor of whether an inflorescence will be unbranched or branched. Thus, whereas delaying maturation promotes branching, faster PSM maturation early in ontogeny might lead to a swift adoption of floral fate, perhaps explaining single-flower inflorescences of solanaceous plants like petunia and tobacco (Nicotiana benthamiana). Although differential expression of S might contribute to branching diversity in tomato (Fig. 3H, Center), our data suggest that many other factors could be responsible for the evolution of inflorescence branching in tomato and related Solanaceae and that such factors may further differ between species. In this respect, it is telling that petunia plants mutated in the ortholog of S not only fail to branch, but also mostly lack flowers altogether (23). Because meristem termination in sympodial plants is synonymous with flowering (13), another potential modulator of branching is the mobile flowering inducer and termination signal, florigen, encoded in tomato by the SINGLE FLOWER TRUSS (SFT) gene (24). Importantly, overexpression of SFT generates single-flower inflorescences when the related gene and floral repressor, SELF PRUNING, is absent (13), and we find that a high percentage of primary inflorescences in loss-of-function sft mutants are branched (SI Appendix, Fig. S16) (25). Intriguingly, unlike in Arabidopsis, our RNA-seq data reveal that both SFT and SP transcripts accumulate during tomato meristem maturation and that SFT peaks in the TM and SIM, perhaps functioning to boost leaf-derived SFT protein levels and termination activity over the higher levels of veg conferred by the activity of SP (SI Appendix, Fig. S16).
Beyond the Solanaceae, much evidence suggests that not all genes dictating monopodial and sympodial inflorescence architecture are shared; however, all players seem to converge on a universal task: to establish and regulate the rate of meristem maturation. Indeed, strong parallels are evident between the regulation of SIM maturation through S and the regulation of branch meristem and spikelet meristem determinacy in maize (26) through the RAMOSA and INDETERMINATE SPIKELET 1 genes, respectively (27). Advancing the systems genetics approach presented here to maturing meristems in Solanaceae species representing a continuum of inflorescence complexity, along with corresponding data from nonsympodial plants, such as maize and other grasses, can expose to what extent general mechanistic principles underlie the rate of meristem maturation and inflorescence development. Findings from such studies will provide a deeper understanding of inflorescence evolution and have widespread implications for the manipulation of inflorescence architecture, flower production, and crop yields.
Materials and Methods
Plant Material, Tissue Collection, and Library Preparation.
The domesticated tomato (S. lycopersicum) cultivar M82 was used in this study, and the isogenic mutant compound inflorescence (s) (10), isolated in the M82 background, was backcrossed four times. The self-incompatible wild species, S. peruvianum, was sib-mated to increase seed, and seedlings for all three genotypes were grown in greenhouses for meristem imaging and tissue collection (SI Appendix). Meristems were imaged and dissected using a stereoscope, and tissue was processed for RNA stabilization using an acetone fixation technique (SI Appendix). RNA was extracted using the PicoPure RNA Extraction kit (Arcturus). More than 70 meristems were collected for each stage for each genotype, yielding 1–3 μg RNA, which was enriched for mRNA and processed into cDNA libraries according to standard protocols for Illumina mRNA sequencing. cDNA libraries of 250–350 bp were quantified using a Bioanalyzer 2100 (Agilent), and paired-end 50-base sequencing was performed on the Illumina GAII platform (Dataset S1). Samples were randomized across sequencing flow cells and lanes within flow cells, and two biological replicates were used for all library constructions.
Read Mapping and Statistical Analysis.
All reads were aligned against predicted protein coding sequences from tomato iTAG version 2.3 (http://solgenomics.net/organism/solanum_lycopersicum/genome) (16) using the short read-mapping software Bowtie with custom parameters (28). Low evolutionary divergence between S. peruvianum and S. lycopersicum estimated on the basis of read-mapping statistics, de novo assembly, and PCA analysis of transcriptomes enabled use of the same quantification pipeline. All gene expression level estimations [reads per kilobase of transcript per million of sequenced reads (RPKM) calculation], differential expression tests, and comparative clustering were conducted in edgeR with custom R scripts (29). The DDI algorithm (20) was applied with threefold change to select marker genes and predict s and S. peruvianum meristem maturation states using five stages of S. lycopersicum meristems (MVM, LVM, TM, SIM, and FM) as calibrations (SI Appendix). Quantitative and semiquantitative RT-PCR validation of DDI-selected marker genes and other dynamically expressed genes was performed according to standard protocols (SI Appendix).
Supplementary Material
Acknowledgments
We thank M. Strahl, A. Goldshmidt, and M. Regulski for assistance with Illumina sequencing; I. Efroni for assistance with DDI; M. Hammell for statistical advice; M. Paradesi for developing the eFP browser; the International Tomato Genome Sequencing Consortium for prepublication use of the tomato genome annotation; and J. Giovannoni and F. Zhangjun for transcription factor annotation. We thank D. Jackson, N. Ori, and Y. Eshed for comments on the manuscript. We also thank the Tomato Genetics Research Center at the University of California, Davis, for providing wild species seed stocks and T. Mulligan and P. Hanlon for plant care. This research was supported by a National Science Foundation Plant Genome Research Program Grant (DBI-0922442) to Z.B.L. and a Career Development Award from the International Human Frontier Science Program Organization to Z.B.L.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the Solanaceae Genomics Network, http://solgenomics.net/.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1114963109/-/DCSupplemental.
References
- 1.Benlloch R, Berbel A, Serrano-Mislata A, Madueño F. Floral initiation and inflorescence architecture: A comparative view. Ann Bot (Lond) 2007;100:659–676. doi: 10.1093/aob/mcm146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kwiatkowska D. Flowering and apical meristem growth dynamics. J Exp Bot. 2008;59:187–201. doi: 10.1093/jxb/erm290. [DOI] [PubMed] [Google Scholar]
- 3.Turck F, Fornara F, Coupland G. Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu Rev Plant Biol. 2008;59:573–594. doi: 10.1146/annurev.arplant.59.032607.092755. [DOI] [PubMed] [Google Scholar]
- 4.Bell AD. Plant Form: An Illustrated Guide to Flowering Plant Morphology. Oxford: Oxford University Press; 1993. [Google Scholar]
- 5.Halle F, Oldeman RAA, Tomlinson PB. Tropical Trees and Forests: An Architectural Analysis. Berlin: Springer-Verlag; 1978. [Google Scholar]
- 6.Rickett HW. The classification of inflorescences. Bot Rev. 1944;10:187–231. [Google Scholar]
- 7.Knapp S, Bohs L, Nee M, Spooner DM. Solanaceae: A model for linking genomics with biodiversity. Comp Funct Genomics. 2004;5:285–291. doi: 10.1002/cfg.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Napoli CA, Ruele J. New mutations affecting meristem growth and potential in Petunia hybrida Vilm. J Hered. 1996;87:371–377. [Google Scholar]
- 9.Peralta IE, Spooner DM. Morphological characterization and relationships of wild tomatoes (Solanum L. Section Lycopersicon) Monogr Syst Bot Missouri Bot Gard. 2005;104:227–257. [Google Scholar]
- 10.Lippman ZB, et al. The making of a compound inflorescence in tomato and related nightshades. PLoS Biol. 2008;6:e288. doi: 10.1371/journal.pbio.0060288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rick CM. The tomato. Sci Am. 1978;239(2):76–87. [Google Scholar]
- 12.Pnueli L, et al. The SELF-PRUNING gene of tomato regulates vegetative to reproductive switching of sympodial meristems and is the ortholog of CEN and TFL1. Development. 1998;125:1979–1989. doi: 10.1242/dev.125.11.1979. [DOI] [PubMed] [Google Scholar]
- 13.Shalit A, et al. The flowering hormone florigen functions as a general systemic regulator of growth and termination. Proc Natl Acad Sci USA. 2009;106:8392–8397. doi: 10.1073/pnas.0810810106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erickson RO, Michelini FJ. The plastochron index. Am J Bot. 1957;44:297–305. [Google Scholar]
- 15.Wang Z, Gerstein M, Snyder M. RNA-Seq: A revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.The International Tomato Genome Sequencing Consortium The official annotation for the tomato genome is provided by the International Tomato Annotation Group (ITAG), a multinational consortium, funded in part by the EU-SOL project. 2011 Available at http://solgenomics.net/organism/Solanum_lycopersicum/genome#itag23_annotation_release. [Google Scholar]
- 17.Molinero-Rosales N, et al. FALSIFLORA, the tomato orthologue of FLORICAULA and LEAFY, controls flowering time and floral meristem identity. Plant J. 1999;20:685–693. doi: 10.1046/j.1365-313x.1999.00641.x. [DOI] [PubMed] [Google Scholar]
- 18.Schmid M, et al. Dissection of floral induction pathways using global expression analysis. Development. 2003;130:6001–6012. doi: 10.1242/dev.00842. [DOI] [PubMed] [Google Scholar]
- 19.Winter D, et al. An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE. 2007;2:e718. doi: 10.1371/journal.pone.0000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Efroni I, Blum E, Goldshmidt A, Eshed Y. A protracted and dynamic maturation schedule underlies Arabidopsis leaf development. Plant Cell. 2008;20:2293–2306. doi: 10.1105/tpc.107.057521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prusinkiewicz P, Erasmus Y, Lane B, Harder LD, Coen E. Evolution and development of inflorescence architectures. Science. 2007;316:1452–1456. doi: 10.1126/science.1140429. [DOI] [PubMed] [Google Scholar]
- 22.Nadeau JH, Dudley AM. Genetics: Systems genetics. Science. 2011;331:1015–1016. doi: 10.1126/science.1203869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rebocho AB, et al. Role of EVERGREEN in the development of the cymose petunia inflorescence. Dev Cell. 2008;15:437–447. doi: 10.1016/j.devcel.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 24.Lifschitz E, Eshed Y. Universal florigenic signals triggered by FT homologues regulate growth and flowering cycles in perennial day-neutral tomato. J Exp Bot. 2006;57:3405–3414. doi: 10.1093/jxb/erl106. [DOI] [PubMed] [Google Scholar]
- 25.Quinet M, et al. Characterization of tomato (Solanum lycopersicum L.) mutants affected in their flowering time and in the morphogenesis of their reproductive structure. J Exp Bot. 2006;57:1381–1390. doi: 10.1093/jxb/erj117. [DOI] [PubMed] [Google Scholar]
- 26.Irish EE, Nelson TM. Identification of multiple stages in the conversion of maize meristems from vegetative to floral development. Development. 1991;112:891–898. [Google Scholar]
- 27.Vollbrecht E, Springer PS, Goh L, Buckler ES, IV, Martienssen R. Architecture of floral branch systems in maize and related grasses. Nature. 2005;436:1119–1126. doi: 10.1038/nature03892. [DOI] [PubMed] [Google Scholar]
- 28.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Team RDC. R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; 2011. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.