Abstract
Aims
The sinus venous myocardium, comprising the sinoatrial node (SAN) and sinus horns (SH), is a region subject to congenital malformations and cardiac arrhythmias. It differentiates from symmetric bilateral mesenchymal precursors, but morphological, molecular, and functional left/right differences are progressively established through development. The role of the laterality gene Pitx2 in this process is unknown. We aimed to elucidate the molecular events driving left/right patterning in the sinus venosus (SV) myocardium by using a myocardial Pitx2 knockout mouse.
Methods and results
We generated a myocardial specific Pitx2 knockout model (cTP mice). cTP embryos present several features of Pitx2 null, including right atrial isomerism with bilateral SANs and symmetric atrial entrance of the systemic veins. By in situ hybridization and optical mapping analysis, we compared throughout development the molecular and functional properties of the SV myocardium in wt and mutant embryos. We observed that Pitx2 prevents the expansion of the left-SAN primordium at the onset of its differentiation into myocardium; Pitx2 promotes expansion of the left SH through development; Pitx2 dose-dependently represses the autorhythmic properties of the left SV myocardium at mid-gestation (E14.5); Pitx2 modulates late foetal gene expression at the left SH-derived superior caval vein.
Conclusion
Pitx2 drives left/right patterning of the SV myocardium through multiple developmental steps. Overall, Pitx2 plays a crucial functional role by negatively modulating a nodal-type programme in the left SV myocardium.
Keywords: Pitx2, Sinus venosus myocardium, Optical mapping, Mouse cardiac development
1. Introduction
In human and mouse embryos, the sinus venosus (SV) myocardium includes the sinoatrial node (SAN) and the sinus horns (SH), from which the myocardial walls of the left superior caval vein (LSCV) and right superior caval vein (RSCV) will differentiate.1,2 SV-derived structures generate great medical interest, since various congenital malformations3 and arrhythmias4,5 have their origin in this region. The molecular mechanisms underlying SV genetic origin and the molecular pathways driving its cellular differentiation have been extensively investigated.6
The SV myocardium is originated by a single genetic cell lineage, derived from Tbx18-positive/Nkx2.5-negative mesenchymal precursors, originally (E8.25 in mouse) located at the lateral rims of the splanchnic mesoderm.7 The molecular mechanisms driving progressive reorganization from bilateral SV mesenchymal precursors into a lateralized SAN and an asymmetrically structured SH myocardium have not yet been clarified.
At early developmental stages, the entire SV region presents pacemaker properties.8–11 Starting at mid-foetal stages, the SH myocardium, but not the SAN, progressively matures to obtain a molecular phenotype comparable to the atrial working myocardium.7 Eventually, in the adult heart, pacemaker activity is confined to the SAN. Mistakes in developmental modulation of this functional maturation programme may be the leading cause for adult SV-originated arrhythmias; however, the genetic mechanisms regulating this process are still not understood.
The homeobox transcription factor Pitx2 has been indicated as a susceptibility gene for atrial arrhythmias in humans12 and in mice.13–15 Pitx2 mediates early signalling events into left cardiac morphogenesis,16 as Pitx2-null embryos present severe cardiac defects including right atrial isomerism (RAI).17–20 This phenotype was recapitulated by conditional deletion of the gene in second heart field (SHF) progenitors,21 but not in the developing myocardium;22 therefore, left cardiac identity is due to Pitx2 action in cardiogenic precursors or at early cardiogenesis. To address this question, we have conditionally inactivated Pitx2 from the onset of cardiomyogenesis. We show here that Pitx2 is required from early cardiomyogenesis to confer left identity to the entire sinoatrial region, including the SV myocardium. Within the SV, left morphological, molecular, and functional identity is achieved through multiple developmental steps, corresponding to distinct actions of Pitx2 on SV cardiomyocytes. Our results highlight the myocardial role of Pitx2 in preventing the onset and maintenance of a nodal-type programme in left SV cardiomyocytes.
2. Methods
2.1. Mouse lines
Pitx2 floxed, Pitx2 constitutive mutant, cTnT Cre, and R26R transgenic mouse lines have previously been described;17,22–24 mice were kept on a C57Bl6/J background. DNA for PCR screening was extracted with DNeasy Blood and Tissue Kit (Qiagen) from tails of anaesthetized mice (Zoletil, 30 mg/kg, ip) and from amniotic sac of embryos isolated after sacrifice of the anaesthetized mother by cervical dislocation. A PCR amplification protocol for Pitx2 and Cre has been described.22
This study was performed conforming to Guide for the Care and Use of Laboratory Animals described by Directive 2010/63/EU of the European Parliament. Animal work was approved by the Ethics Committee for Animal Experiments of the University of Padua, in compliance with NIH, and carried out in compliance with Italian government guidelines.
2.2. Histology
Stage E8.5, E10.5, and E14.5 embryos were fixed overnight with 4% paraformaldehyde, dehydrated through graded ethanol series, and embedded in paraffin. Sections were cut at 12 μm and processed for haematoxylin and eosin staining.
2.3. In situ hybridization
Non-radioactive in situ hybridization (ISH) on sections was performed as described previously.22 RNA probes complementary to mouse Pitx2, Tbx3, Shox2, Hcn4, Tbx18, NKX2.5, Cx40, and the ATP-binding site of myosin heavy chain (MHC) were generated using standard protocols. Images were taken using a Leica DC300 digital camera.
2.4. Real-time PCR
E14.5 hearts were dissected from freshly isolated embryos and stored in liquid nitrogen. Total RNA was extracted using Trizol® (Invitrogen); then cDNA was synthesized using SuperScript III Reverse Transcriptase and random primers (Invitrogen). Amplifications were performed on three samples for each genotype using an iQ5 Real-time machine (Bio-Rad); Pitx2 expression values were normalized with the housekeeping genes GAPDH and β-actin. Primer sequences are reported in the Supplementary material online.
2.5. Magnetic resonance imaging analysis
Magnetic resonance imaging was performed on a horizontal 9.4 T/21 cm VNMRS Direct Drive MR system (Varian Inc., Palo Alto, CA, USA) on E14.5 embryos, as described previously.25
2.6. Optical mapping
E14.5 embryos were dissected on ice, their heart removed and stained for 10min with di-4-ANEPPS (Invitrogen) at 4°C. Then, they were pinned, their dorsal part up, on the bottom of a silicone-lined copper dish filled with oxygenated Tyrode-HEPES solution (pH 7.4), with Blebbistatin added, positioned on a temperature-controlled stage (TH 60, 37°C) of an upright epifluorescence microscope (Leica DML-FS). To accommodate the entire heart, 4× and 10× water-dipping objectives and 0.63 photo tubes were used. Recordings were performed in the dorsal view, with the posterior atrial wall facing the optical apparatus. See also the Supplementary material online.
2.7. Statistical analysis
Atrial activation and propagation patterns could be grouped into three categories, each having a minimum of 10 hearts; differences between groups were analysed using Pearson's χ2-test. Atrial activation times were calculated as an average between 4× and 10× recordings of each sample when these numbers did not differ more than 2 ms. Data are presented as mean ± standard deviations; comparisons between wt, cTP het, and cTP ko values were performed using a two-tailed Student's t-test; P-values <0.05 were considered significant.
3. Results
3.1. Characterization of the cTnT Cre-Pitx2 mouse line
We investigated the myocardial role of Pitx2 with a conditional ko approach by crossing Pitx2 floxed mice (Pitx2loxP/loxP)14 with Troponin T (TnT) Cre deletor mice, which are active from early cardiomyogenesis23 (Figure 1A).
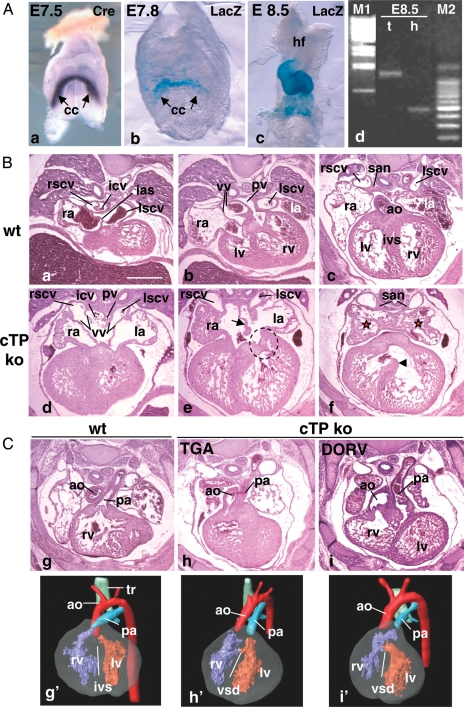
Figure 1.
cTP mouse line characterization. (A) TnT Cre line characterization. Onset of Cre mRNA expression visualized by whole-mount ISH (Cre) (a) and onset of Cre activity (b), assessed by crossing TnT Cre with R26R mice (LacZ); (c) by E8.5 Cre activity is visible in the entire heart; (d) Pitx2 PCR on genomic DNA from trunk (t) and isolated heart (h) of a E8.5 cTP ko embryo to assess cardiac-specific recombination at the Pitx2 locus. M1: Lambda phage DNA, BstEII digested; M2: 100 bp ladder. From the embryonic trunk only the floxed allele is amplified (1232 bp band). In the corresponding heart only the 500 bp band is visible, indicating complete Cre-driven recombination.22 (B and C) H/E staining and MRI analysis of E14.5 wt and cTP ko hearts. Note that the sinoatrial region of the ko hearts presents the morphological features of RAI. Red stars in (f) show symmetrical pectinate muscles arrangement in atria. (e) Arrow indicates ASD, dotted circle indicates abnormal shape of the left atrioventricular valve; (f) arrowhead indicates VSD. (C) (h and i) Ventriculo-arterial alignment defects in cTP ko and (h′ and i′) their corresponding 3D MRI reconstructions. Cc, cardiac crescent; hf, headfolds; ias, interatrial septum; la, ra, left and right atrium; ivs, interventricular septum; san, sinoatrial node; lscv, rscv, left and right superior caval veins; vv, venous valves; icv, inferior caval vein; pv, pulmonary veins; lv, rv, left and right ventricle; ao, aorta; pa, pulmonary artery; tr, trachea. Scale bar: 0.5 mm.
Myocardial deletion of one loxP allele (TnT Cre; Pitx2loxP/wt = Pitx2 hetmyo, from here onwards referred as cTP het) resulted in viable and fertile offspring. We then crossed cTP hets with Pitx2loxP/loxP mice to generate cTP ko mice. No cTP ko pups were identified at post-natal day (P)3; however, their embryonic distribution at E17.5 was according to the Mendelian ratio (data not shown); we concluded that the cTP ko phenotype is not viable.
Quantification of Pitx2 mRNA in E14.5 wt and mutant hearts indicated a dose-dependent reduction in Pitx2 transcript levels (see Supplementary material online, Figure S1). Histological and MRI analysis of E14.5 cTP embryos did not reveal obvious morphological defects in cTP hets (see Supplementary material online, Figure S2A; Table 1), whereas the ko hearts presented a complex phenotype (Figure 1B and C, and Table 1): the left auricle was identical in shape and orientation to the right one (Figure 1B, d–f) and the atrial septum was reduced or totally absent (ASD) (Figure 1B, e); additionally, we detected bilateral SAN and venous valves, bilateral caval vein (CV) entrance into the atria, and drainage of the inferior caval vein (ICV) and pulmonary vein (PV) into the medial part of the common atrium (Figure 1B, d–f). Overall, these are typical features of RAI; therefore, we concluded that early myocardial Pitx2 action is required to confer left atrial (LA) identity.
Table 1.
Characterization of cardiac defects in wt and cTP mutants by MRI analysis
| wt (n = 7) | het (n = 5) | ko (n = 5) | |
|---|---|---|---|
| Heart position | |||
| Left-sided | 6 | 5 | 0 |
| Right-sided | 0 | 0 | 5 |
| Midline | 1 | 0 | 0 |
| Apex direction | |||
| Left | 6 | 0 | 0 |
| Right | 0 | 5 | 4 |
| Down | 1 | 0 | 1 |
| Atrial shape | |||
| No ASD, no RAI | 7 | 5 | 0 |
| ASD plus RAI | 0 | 0 | 5 |
| ICV drainage | |||
| Into RA | 7 | 5 | 0 |
| Into common atrium | 0 | 0 | 5 |
| PV drainage | |||
| Into LA | 7 | 5 | 0 |
| Into common atrium | 0 | 0 | 5 |
| AV junction | |||
| Normal | 7 | 5 | 0 |
| Abnormal | 0 | 0 | 5 |
| Ao exit | |||
| From LV | 7 | 5 | 0 |
| From RV | 0 | 0 | 5 |
| PA exit | |||
| From RV | 7 | 5 | 1 |
| From LV | 0 | 0 | 4 |
| TGA | 0 | 0 | 4 |
| DORV | 0 | 0 | 1 |
Additionally, in the cTP ko embryos, we detected abnormal atrioventricular junction with malformed left atrioventricular valves (Figure 1B, e), ventricular septal defects (VSD), and abnormal ventriculoarterial alignment, resulting in transposition of the great arteries (TGA) and double outlet right ventricle (DORV) (Figure 1C and Table 1).
To get insights into the myocardial role of the gene, we histologically compared Pitx2 constitutive mutants (Pitx2wt/− and Pitx2−/−). E14.5 Pitx2wt/− embryos were normal, except one sample that presented mild VSD (see Supplementary material online, Figure S2B and Table S1). This indicates a differential sensitivity to Pitx2 gene dosage within the developing heart, the ventricles being more sensitive than the atria. In line with previous data,18 E14.5 Pitx2−/− displayed RAI (see Supplementary material online, Figure S2D) and strong impairment in atrioventricular canal (AVC) and ventricular remodelling, resulting in common AVC (see Supplementary material online, Figure S2E), atrioventricular valve defects, severe VSD, and ventriculoarterial alignment defects (see Supplementary material online, Figure S2F). Additionally, the ventricular compact wall was thinner (see Supplementary material online, Figure S2F). Altogether, cTP and Pitx2 mutant hearts present some morphological differences outside the sinoatrial region (see Supplementary material online, Table S1); therefore, myocardial Pitx2 action is sufficient to confer the LA identity but not to drive complete cardiac morphogenesis.
3.2. Pitx2 prevents the expansion of left-SAN precursors cells as they differentiate in myocardium
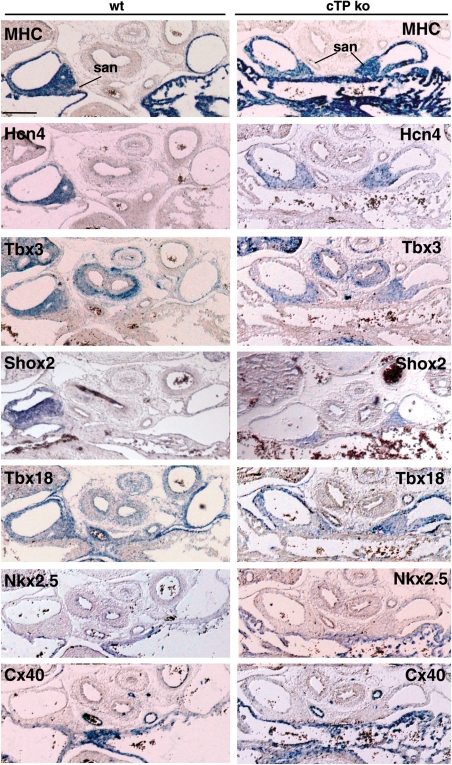
ISH analysis (Figure 2) showed that in cTP ko embryos, both SANs were myocardialized and correctly expressed the nodal differentiation and functional markers Tbx18, Tbx3, Shox2, and Hcn4, but not the chamber myocardium markers Nkx2.5 and CX40.7,26 Thus, we decided to investigate the role of Pitx2 in the earliest events of asymmetric right-SAN (R-SAN) formation.
Figure 2.
The L-SAN of the cTP ko presents correct molecular pattern. ISH analysis of E14.5 wt and cTP ko embryos to assess L-SAN molecular signature. Note SAN expression of MHC, Hcn4, Tbx3, Shox2, Tbx18, and the negative staining with Nkx2.5 and CX40 antisense probes. Scale bar: 0.2 mm.
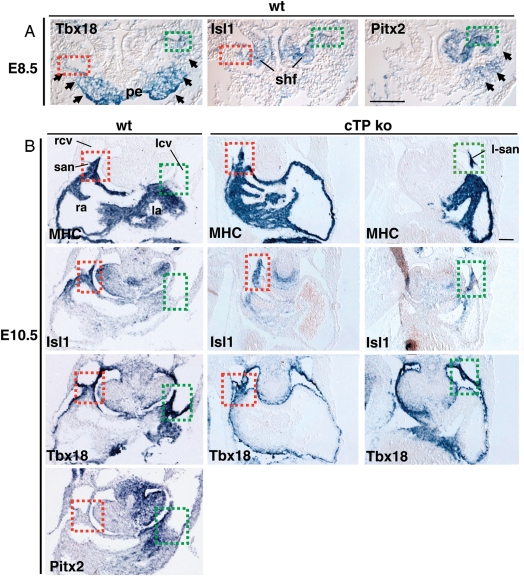
SAN cardiomyocytes constitute a subpopulation of the SV myocardium derived from Tbx18 mesenchymal precursors, which at E8.5 are symmetrically located at the lateral rims of the splanchnic mesoderm,26 flanking the proepicardial organ (Figure 3A, arrows). At the most lateral borders, SAN precursors can be identified by the co-expression of the SHF marker Isl1;27 the left portion of the SV mesenchyme, including left-SAN (L-SAN) precursors, additionally co-expresses Pitx2 (Figure 3A). In E10.5 wt embryos, the embryonic SAN structure is right-sided and can be identified as a thickening of Isl1- and Tbx18-positive myocardial cells at the border between the RSCV and the RA7 (Figure 3B), while the corresponding region on the left side is not myocardialized (Figure 3B). On the contrary, in E10.5 cTP ko embryos, the borders between the CVs and atria were both myocardialized and co-expressing Isl1 and Tbx18, thus indicating the presence of a second L-SAN, which presented a correct molecular pattern (data not shown).
Figure 3.
L-SAN mesenchymal precursors differentiate and expand into the left ectopic SAN of the cTP ko. (A) ISH of E8.5 wt embryos shows Tbx18 expression in SV mesenchymal precursors (arrows) and proepicardial organ (pe); Isl1 is bilaterally expressed in the second heart field (shf) overlapping Tbx18 at the SAN progenitors (dotted squares: green, right; red, left). Pitx2 expression delineates the left SV domain (arrows), including L-SAN progenitors (green dotted square). (B) At E10.5, cTP ko embryos present bilateral MHC, Isl1- and Tbx18-positive regions at the borders between left and right cardinal veins (lcv, rcv) and atria, identifying the early differentiated bilateral SAN. Scale bar: 0.1 mm.
The presence of an ectopic myocardial structure in the cTP ko embryos indicates that early left Isl1/Tbx18-positive SAN cardiomyocytes have expanded bilaterally. We conclude that Pitx2 prevents the expansion of the left Isl1/Tbx18-positive SAN precursors as they differentiate in cardiomyocytes.
3.3. Pitx2 modulates the developmental programme of the LSCV
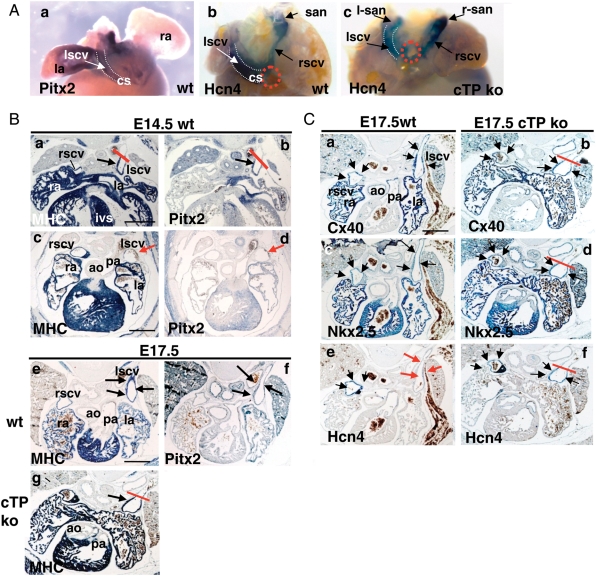
We then investigated the role of Pitx2 in the left SH-derived CV myocardium. In E14.5 wt embryos, ICV entrance is positioned on the right (Figure 1B, a, and dotted circles in Figure 4A, b), at the site of RSCV confluence with the coronary sinus. Conversely, in the cTP ko, ICV enters medially into the common atrium (Figure 1B, d), the coronary sinus is absent, and CVs run symmetrically (Figure 4A, b and c); this has been highlighted by ISH with the hyperpolarization-activated channel Hcn4, which at this stage presents SV-wide expression. We conclude that symmetric organization of the CVs is due to the lack of Pitx2-dependent early left cardiomyocyte expansion into the coronary sinus,2,28 which is missing in the cTP ko.
Figure 4.
Pitx2 modulates the LSCV developmental programme. (A) Whole-mount ISH of E14.5 wt hearts (dorsal view) showing expression of Pitx2 in LA and LSCV (a) and of Hcn4 in the entire SV myocardium, including the SAN (b). Red dotted circles (b and c) indicate ICV entrance. Note the absence of coronary sinus (cs) and symmetric CVs arrangement in the cTP ko (c). (B) Correlation between Pitx2 expression and MHC extension in the LSCV. In E14.5 wt hearts, note Pitx2 and MHC co-expression in the LSCV in four-chamber view sections (a and b) but not more distally (c and d); red bar indicates the border of probe detection. In E17.5 wt, MHC and Pitx2 expression is visible more distally (e and f), but in the cTP ko distal MHC extension is impaired (red bar in g). (C) The atrialization programme is partially impaired in the cTP ko. E17.5 wt embryos bilaterally express Cx40 (a) and Nkx2.5 (c) in the CVs, while Hcn4 expression is strongly down-regulated only in the LSCV (red arrows, e). In the cTP ko, Cx40 and Nkx2.5 signals are still present (b and d) in the reduced LSCV myocardial domain. Note the absence of Hcn4 down-regulation in the LSCV (black arrows in f). Scale bar: 0.5 mm.
The SH myocardium will progressively expand and form the proximal myocardial cuff of CVs by E12.5;1 we wondered if Pitx2 could affect LSCV cardiomyocyte expansion at later stages. In E14.5 wt embryos, LSCV myocardium is restricted to its most proximal portion; these SV cardiomyocytes are Pitx2-positive (Figure 4B, a–d), and at E17.5, they extend more distally (Figure 4B, e and f). This was not observed in the cTP ko (Figure 4B, g); therefore, we concluded that Pitx2 promotes LSCV cardiomyocyte expansion both at early and late developmental stages.
We then tested whether Pitx2 could modulate LSCV transcriptional properties. A progressive shift towards an atrial-type gene expression programme is started in the CV myocardium at E14.5 and is clearly visible by E17.5:7 Nkx2.5 is up-regulated in the SAN and CVs, whereas Cx40 is up-regulated exclusively in CVs, without obvious left–right differences (Figure 4C, a and c); concomitantly, Hcn4 expression is down-regulated exclusively in the LSCV (red arrows in Figure 4C, e). Since Hcn4 is responsible for the generation of pacemaker potentials,29 this indicates that the late foetal RSCV myocardium retains a more nodal-type phenotype than the LSCV. In the cTP ko, the LSCV atrial gene programme was restricted to the most proximal, myocardialized region (red bars in Figure 4C, b, d, and f); moreover, it was incomplete since Nkx2.5 and Cx40 expression was unaffected, while Hcn4 expression was not down-regulated (Figure 4C, f). As a result, in the cTP ko, the nodal-type molecular profile was retained bilaterally.
3.4. Pitx2 represses left pacemaker activity in the SV-derived myocardium of E14.5 embryos
The functional properties of mid-foetal mouse hearts are as yet quite elusive. We approached this problem and investigated the atrial electrophysiology of E14.5 wt and cTP mutant hearts by optical mapping.
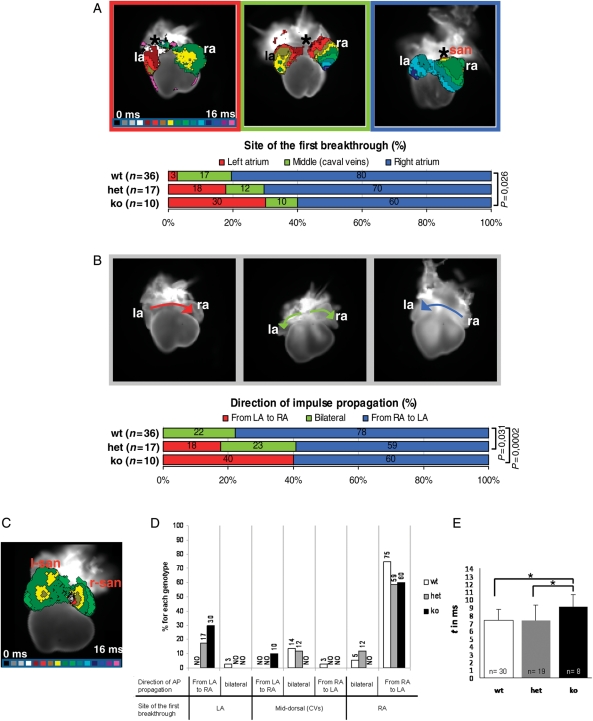
In wt hearts (n = 36), the site of first activated region was predominantly detected in the RA (n = 29; 80%) mainly around the R-SAN region; in a smaller group (n = 6; 17%), it mapped medially, in correspondence to the CVs, while in a single sample, impulse origin was left-sided (n = 1; 3%) (Figure 5A). The direction of action potential (AP) propagation was mostly sequential from RA to LA (n = 28; 78%), but synchronous bilateral propagation (≈ 1–2 ms difference between LA and RA activation times) was also recorded (n = 8; 22%) (Figure 5B). Therefore, in wt embryos, both the site of first atrial breakthrough and the AP direction of propagation presented a variable distribution within the SV region with a pronounced right-sided dominance.
Figure 5.
Pitx2 represses left pacemaker activity within the SV myocardium of E14.5 embryos. (A) Site of first breakthrough. Top: prototypical maps (dorsal view) showing the three main atrial activation patterns recorded in isolated E14.5 hearts by optical mapping. The earliest activated region is indicated with an asterisk. Isochronal lines, delimiting regions activated within the same time frame, are spaced at 1 ms intervals; colour progression visualizes the advancing activation wavefront. Below: diagram showing the distribution of the activation patterns in the three genotypes. n = classified samples. (B) Direction of impulse propagation. Top: three representative examples of observed impulse propagation patterns; arrows depict the direction of electrical activation spread. Below: diagram illustrating pattern distribution within the genotypes. P-values <0.05 are indicated. (C) The L-SAN is functional. Activation map of a cTP ko heart showing the almost synchronous activation of the two SANs. (D and E) Atrial conduction properties in E14.5 wt and cTP mutants. (D) Correlation between the site of first atrial breakthrough and direction of impulse propagation. Columns indicate the combined distribution of atrial activation and propagation patterns within genotypes. Data are expressed as percentages; NO, never observed. (E) Atrial activation times. Data are presented as averages ± standard deviations; *P < 0.05.
In the cTP ko hearts (n = 10), we found a reduced percentage of samples presenting right-sided (n = 6; 60%) or mid-dorsal (n = 1; 10%) impulse initiation, and higher incidence of left-originating breakthroughs (n = 3; 30%). Bilateral AP propagation was not detected, whereas a new left-to-right direction of impulse spread was present (n = 4; 40%). In a single ko sample, a double concomitant impulse firing was observed, with the two earliest activated sites located in the R- and L-SAN regions (Figure 5C), thus indicating that the L-SAN is functional. The cTP het samples (n = 17), although morphologically normal, presented an intermediate behaviour both in pacemaker location distribution and in the impulse propagation profile (Figure 5A and B). Heart rate did not significantly differ among genotypes (wt: 102 ± 24, het: 91 ± 36, ko: 104 ± 42; P> 0.05). Movies of representative activation and propagation patterns are presented as Supplementary material online.
In conclusion, our optical mapping analyses have revealed some functional plasticity in E14.5 hearts, which display pacemaker activity at both the R-SAN and CVs. In wt hearts, SV-wide capacity to generate the first electrical activity is mainly restricted to the right side where the SAN is located; when Pitx2 gene dosage is reduced (cTP het) or its action is lacking (cTP ko), a left pacemaker potential is progressively uncovered. Therefore, Pitx2 prevents the occurrence of left pacemaker activity in the SV myocardium in a dose-dependent way, thus restricting impulse generation to its right side.
Moreover, correlation between pacemaker location and AP spread direction (Figure 5D) demonstrated that given a site of impulse initiation, atrial propagation patterns are different in the three genotypes, indicating differences in their conduction properties. In line with this observation, we found that time of atrial impulse propagation (Figure 5E) is significantly higher in cTP ko hearts compared with wt and het (wt: 7.3 ms; het: 7.3 ms; ko: 9.0 ms).
4. Discussion
4.1. The myocardial role of Pitx2
Here, we have presented the outcome of Pitx2 deletion from the onset of cardiomyogenesis. At the morphological level, we found that the cTP ko partially recapitulates the cardiac phenotype of Pitx2 constitutive nulls. In particular, RAI was detected in both models, thus indicating that Pitx2 action within the myocardium is sufficient to confer LA identity.
Additionally, the comparative analysis of cTP and constitutive mutant embryos has shown that unlike the sinoatrial region, the AVC and ventricles are more severely affected in Pitx2 constitutive mutants, indicating that morphogenesis of these regions is extremely sensitive to Pitx2 dosage. Moreover, it has uncovered that Pitx2 function is additionally required in precardiac cells of the venous and arterial poles contributing to those regions and/or in early cardiomyocytes before E8.5, when cTnT Cre-driven recombination is complete (Figure 1A).
We have also shown that the SV myocardium is molecularly left/right patterned through multiple developmental steps. A later myocardial deletion of Pitx2 with α-MHC Cre driver mice had resulted in molecular atrial isomerism in the absence of any morphological alterations;22 at the light of this previous result, we propose that the molecular features of the cTP hearts are not the indirect consequence of the earlier Pitx2 deletion, but correspond to a specific and distinct action of Pitx2 on SV cardiomyocytes.
4.2. Opposite effects of Pitx2 within the left SV myocardium: a regional-specific differential transcriptional modulation?
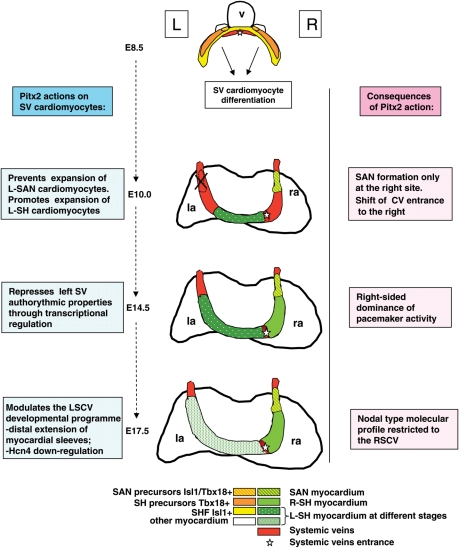
We have shown here that myocardial loss of Pitx2 leads to symmetrical morphological organization of the CVs, absence of the coronary sinus, and reduced expansion of LSCV myocardium. These results suggest that Pitx2 promotes the higher proliferation and/or migration rate in Tbx18-derived cardiomyocytes of the left SH from early to late foetal stages (Figure 6). On the other hand, myocardial loss of Pitx2 leads to the presence of a novel L-SAN, visible from its onset; therefore, we conclude that Pitx2 prevents the expansion of the left Isl1/Tbx18 + SAN precursors at the onset of their differentiation into nodal cells. Thus, Pitx2 seems to exert an opposite role on SH and SAN cardiomyocytes (Figure 6).
Figure 6.
Proposed model for Pitx2 action in left SV cardiomyocytes; hearts are shown in the dorsal view. Left columns summarize the temporal actions of Pitx2 in the developing SV region; right columns summarize the resulting morphogenetic and functional effects; v, embryonic ventricle; la, ra, left and right atrium.
SH differentiate through progressive recruitment of Tbx18-positive mesenchymal precursors, their myocardial differentiation, and subsequent proliferation.1 SAN mesenchymal precursors additionally co-express Isl1,27 thus having features of both SHF (Isl1+) and SV (Tbx18+) progenitors. Isl1 promotes the proliferation of cardiogenic precursors;30 since its expression is selectively retained in the SAN7,31 through early development, it might exert a similar function also in nodal cardiomyocytes. It is possible that Pitx2 could differentially modulate Tbx18 and Isl1 transcriptional action in the left SV-derived cardiomyocytes; however, the characterization of its molecular mechanisms of action still requires additional analysis.
4.3. Pitx2 restricts nodal conductive properties to the right SV region: implications for adult heart disease
In the adult heart, pacemaker activity is restricted to the SAN as shown by Hcn4 expression,32 crucial for this function.29 Conversely, broader Hcn4 expression in the embryo7 suggests the existence of wider areas with pacemaker potential, in line with several functional studies performed in chick8,10,11 and in mouse pre-early somite embryos.9 Functional characterization of mid-foetal mouse hearts has so far been elusive.
Our optical mapping analyses have shown that mid-foetal hearts present a heterogeneous profile of pacemaker activity originating within the SV, thereby revealing some functional plasticity, which can be modulated by Pitx2. In this respect, the intermediate functional properties of the cTP hets, morphologically normal, must be solely due to a Pitx2 dose-dependent modulation of SV molecular properties (Figure 6). The capacity of Pitx2 to inhibit the expression of transcripts crucial for the SAN programme in developing and adult atria has been shown previously.13 Our results additionally highlight the importance of a correct Pitx2 gene dosage for repressing the LSCV autorhythmic potential, in order to restrict pacemaker activity to the SAN.
Critical for confinement of pacemaker activity to the adult SAN is the onset, at mid-foetal stages, of a novel genetic programme in CV myocardium, which progressively acquires a molecular phenotype comparable to the atrial working myocardium.7 Crucial for this developmental programme is the transcriptional left down-regulation of pacemaker channel Hcn4 which, as we have shown here, is Pitx2-dependent.
Atrial arrhythmias are devastating diseases of the adult heart caused by ageing, acquired diseases, or genetic defects. Arrhythmogenic foci are mostly located at the PVs33 or in left SV-derived structures, such as the coronary sinus or LSCV.4,5 Ectopic pacemaker foci in the left SV myocardium might occur if the developmental programme repressing nodal properties of the left SV structures does not occur completely, or alternatively, if an embryonic ‘left’ programme is reinitiated there. Our results suggest that proper regulation of Pitx2 dosage in the left SV is crucial to prevent this process. An additional role of Pitx2 in other left arrhythmogenic areas, such as the PV, cannot be ruled out and is currently being explored.
Genome-wide study populations in humans12 have indicated PITX2 as a candidate susceptibility gene for atrial arrhythmias, later confirmed by functional studies in adult Pitx2 heterozygous mice.13–15 Parallel microarray analysis has additionally identified a wide range of Pitx2 transcriptionally modulated left targets, possibly mediating its action.13,14 Future studies will be required to precisely refine their sites of expression and effective role, thereby delineating a Pitx2-dependent anti-arrhythmogenic road map.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Funding
This work was supported by grants from European Community's Sixth Framework Programme (I.P Project ‘Heart repair’, n. LSHM-CT-2005-018630) and Telethon Grant n. GGP08112 to M.C.; by Purkinje Fellowship and AV0Z50110509 of the Academy of Sciences of the Czech Republic, Ministry of Education VZ 0021620806 and Grant Agency of the Czech Republic 304/08/0615 to D.S. (David Sedmera). Funding to pay the Open Access publication charges for this article was provided by Telethon.
Supplementary Material
Acknowledgements
We thank Kai Jiao for providing cTnT Cre mice, Marta Martin and Giulia Rainato for initial contribution with experiments, and Anne Picard and Sandra Furlan for support with real-time PCR analysis.
Conflict of interest: none declared.
References
- 1.Christoffels VM, Mommersteeg MT, Trowe MO, Prall OW, de Gier-de Vries C, Soufan AT, et al. Formation of the venous pole of the heart from an Nkx2-5-negative precursor population requires Tbx18. Circ Res. 2006;98:1555–1563. doi: 10.1161/01.RES.0000227571.84189.65. [DOI] [PubMed] [Google Scholar]
- 2.Sizarov A, Anderson RH, Christoffels VM, Moorman AF. Three-dimensional and molecular analysis of the venous pole of the developing human heart. Circulation. 2010;122:798–807. doi: 10.1161/CIRCULATIONAHA.110.953844. [DOI] [PubMed] [Google Scholar]
- 3.Attenhofer Jost CH, Connolly HM, Danielson GK, Bailey KR, Schaff HV, Shen WK, et al. Sinus venosus atrial septal defect: long-term postoperative outcome for 115 patients. Circulation. 2005;112:1953–1958. doi: 10.1161/CIRCULATIONAHA.104.493775. [DOI] [PubMed] [Google Scholar]
- 4.Lin WS, Tai CT, Hsieh MH, Tsai CF, Lin YK, Tsao HM, et al. Catheter ablation of paroxysmal atrial fibrillation initiated by non-pulmonary vein ectopy. Circulation. 2003;107:3176–3183. doi: 10.1161/01.CIR.0000074206.52056.2D. [DOI] [PubMed] [Google Scholar]
- 5.Katritsis DG, Giazitzoglou E, Korovesis S, Karvouni E, Anagnostopoulos CE, Camm AJ. Conduction patterns in the cardiac veins: electrophysiologic characteristics of the connections between left atrial and coronary sinus musculature. J Interv Card Electrophysiol. 2004;10:51–58. doi: 10.1023/B:JICE.0000011485.98197.df. [DOI] [PubMed] [Google Scholar]
- 6.Christoffels VM, Smits GJ, Kispert A, Moorman AF. Development of the pacemaker tissues of the heart. Circ Res. 2010;106:240–254. doi: 10.1161/CIRCRESAHA.109.205419. [DOI] [PubMed] [Google Scholar]
- 7.Mommersteeg MT, Hoogaars WM, Prall OW, de Gier-de Vries C, Wiese C, Clout DE, et al. Molecular pathway for the localized formation of the sinoatrial node. Circ Res. 2007;100:354–362. doi: 10.1161/01.RES.0000258019.74591.b3. [DOI] [PubMed] [Google Scholar]
- 8.Van Mierop LH. Location of pacemaker in chick embryo heart at the time of initiation of heartbeat. Am J Physiol. 1967;212:407–415. doi: 10.1152/ajplegacy.1967.212.2.407. [DOI] [PubMed] [Google Scholar]
- 9.Kamino K, Hirota A, Fujii S. Localization of pacemaking activity in early embryonic heart monitored using voltage-sensitive dye. Nature. 1981;290:595–597. doi: 10.1038/290595a0. [DOI] [PubMed] [Google Scholar]
- 10.Sedmera D, Wessels A, Trusk TC, Thompson RP, Hewett KW, Gourdie RG. Changes in activation sequence of embryonic chick atria correlate with developing myocardial architecture. Am J Physiol Heart Circ Physiol. 2006;291:H1646–H1652. doi: 10.1152/ajpheart.01007.2005. [DOI] [PubMed] [Google Scholar]
- 11.Vicente-Steijn R, Kolditz DP, Mahtab EA, Askar SF, Bax NA, Van Der Graaf LM, et al. Electrical activation of sinus venosus myocardium and expression patterns of RhoA and Isl-1 in the chick embryo. J Cardiovasc Electrophysiol. 2010;21:1284–1292. doi: 10.1111/j.1540-8167.2010.01790.x. [DOI] [PubMed] [Google Scholar]
- 12.Gudbjartsson DF, Arnar DO, Helgadottir A, Gretarsdottir S, Holm H, Sigurdsson A, et al. Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature. 2007;448:353–357. doi: 10.1038/nature06007. [DOI] [PubMed] [Google Scholar]
- 13.Wang J, Klysik E, Sood S, Johnson RL, Wehrens XH, Martin JF. Pitx2 prevents susceptibility to atrial arrhythmias by inhibiting left-sided pacemaker specification. Proc Natl Acad Sci USA. 2010;107:9753–9758. doi: 10.1073/pnas.0912585107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kirchhof P, Kahr PC, Kaese S, Piccini I, Vokshi I, Scheld HH, et al. PITX2c is expressed in the adult left atrium, and reducing Pitx2c expression promotes atrial fibrillation inducibility and complex changes in gene expression. Circ Cardiovasc Genet. 2011;4:123–133. doi: 10.1161/CIRCGENETICS.110.958058. [DOI] [PubMed] [Google Scholar]
- 15.Chinchilla A, Daimi H, Lozano-Velasco E, Dominguez JN, Caballero R, Delpón E, et al. PITX2 insufficiency leads to atrial electrical and structural remodeling linked to arrhythmogenesis. Circ Cardiovasc Genet. 2011;4:269–279. doi: 10.1161/CIRCGENETICS.110.958116. [DOI] [PubMed] [Google Scholar]
- 16.Campione M, Steinbeisser H, Schweichert A, Deissler K, van Bebber F, Lowe LA, et al. The homeobox gene Pitx2: mediator of asymmetric left-right signaling in vertebrate heart and gut looping. Development. 1999;126:1225–1234. doi: 10.1242/dev.126.6.1225. [DOI] [PubMed] [Google Scholar]
- 17.Gage PJ, Suh H, Camper SA. Dosage requirement of Pitx2 for development of multiple organs. Development. 1999;126:4643–4651. doi: 10.1242/dev.126.20.4643. [DOI] [PubMed] [Google Scholar]
- 18.Liu C, Liu W, Lu MF, Brown NA, Martin JF. Regulation of left-right asymmetry by thresholds of Pitx2c activity. Development. 2001;128:2039–2048. doi: 10.1242/dev.128.11.2039. [DOI] [PubMed] [Google Scholar]
- 19.Liu C, Liu W, Palie J, Lu MF, Brown NA, Martin JF. Pitx2c patterns anterior myocardium and aortic arch vessels and is required for local cell movement into atrioventricular cushions. Development. 2002;129:5081–5091. doi: 10.1242/dev.129.21.5081. [DOI] [PubMed] [Google Scholar]
- 20.Lu MF, Pressman C, Dyer R, Johnson RL, Martin JF. Function of Rieger syndrome gene in left-right asymmetry and craniofacial development. Nature. 1999;401:276–278. doi: 10.1038/45797. [DOI] [PubMed] [Google Scholar]
- 21.Ai D, Liu W, Ma L, Dong F, Lu MF, Wang D, et al. Pitx2 regulates cardiac left-right asymmetry by patterning second cardiac lineage-derived myocardium. Dev Biol. 2006;296:437–449. doi: 10.1016/j.ydbio.2006.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tessari A, Pietrobon M, Notte A, Cifelli G, Gage PJ, Schneider MD, et al. Myocardial Pitx2 differentially regulates the left atrial identity and ventricular asymmetric remodeling programs. Circ Res. 2008;102:813–822. doi: 10.1161/CIRCRESAHA.107.163188. [DOI] [PubMed] [Google Scholar]
- 23.Jiao K, Kulessa H, Tompkins K, Zhou Y, Batts L, Baldwin HS, et al. An essential role of Bmp4 in the atrioventricular septation of the mouse heart. Genes Dev. 2003;17:2362–2367. doi: 10.1101/gad.1124803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:532–537. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 25.Schneider JE, Böse J, Bamforth SD, Gruber AD, Broadbent C, Clarke K, et al. Identification of cardiac malformations in mice lacking Ptdsr using a novel high-throughput magnetic resonance imaging technique. BMC Dev Biol. 2004;22:4–16. doi: 10.1186/1471-213X-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blaschke RJ, Hahurij ND, Kuijper S, Just S, Wisse LJ, Deissler K, et al. Targeted mutation reveals essential functions of the homeodomain transcription factor Shox2 in sinoatrial and pacemaking development. Circulation. 2007;115:1830–1838. doi: 10.1161/CIRCULATIONAHA.106.637819. [DOI] [PubMed] [Google Scholar]
- 27.Mommersteeg MT, Domínguez JN, Wiese C, Norden J, de Gier-de Vries C, Burch JB, et al. The sinus venosus progenitors separate and diversify from the first and second heart fields early in development. Cardiovasc Res. 2010;87:92–101. doi: 10.1093/cvr/cvq033. [DOI] [PubMed] [Google Scholar]
- 28.Soufan AT, van den Hoff MJ, Ruijter JM, de Boer PA, Hagoort J, Webb S, et al. Reconstruction of the patterns of gene expression in the developing mouse heart reveals an architectural arrangement that facilitates the understanding of atrial malformations and arrhythmias. Circ Res. 2004;95:1207–1215. doi: 10.1161/01.RES.0000150852.04747.e1. [DOI] [PubMed] [Google Scholar]
- 29.Stieber J, Herrmann S, Feil S, Löster J, Feil R, Biel M, et al. The hyperpolarization-activated channel HCN4 is required for the generation of pacemaker action potentials in the embryonic heart. Proc Natl Acad Sci USA. 2003;100:15235–15240. doi: 10.1073/pnas.2434235100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cai CL, Liang X, Shi Y, Chu PH, Pfaff SL, Chen J, et al. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell. 2003;5:877–889. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun Y, Liang X, Najafi N, Cass M, Lin L, Cai CL, et al. Islet 1 is expressed in distinct cardiovascular lineages, including pacemaker and coronary vascular cells. Dev Biol. 2007;304:286–296. doi: 10.1016/j.ydbio.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu J, Dobrzynski H, Yanni J, Boyett MR, Lei M. Organisation of the mouse sinoatrial node: structure and expression of HCN channels. Cardiovasc Res. 2007;73:729–738. doi: 10.1016/j.cardiores.2006.11.016. [DOI] [PubMed] [Google Scholar]
- 33.Haïssaguerre M, Jaïs P, Shah DC, Takahashi A, Hocini M, Quiniou G, et al. Spontaneous initiation of atrial fibrillation by ectopic beats originating in the pulmonary veins. N Engl J Med. 1998;339:659–666. doi: 10.1056/NEJM199809033391003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.