Abstract
Reactive oxygen and nitrogen species change cellular responses through diverse mechanisms that are now being defined. At low levels, they are signalling molecules, and at high levels, they damage organelles, particularly the mitochondria. Oxidative damage and the associated mitochondrial dysfunction may result in energy depletion, accumulation of cytotoxic mediators and cell death. Understanding the interface between stress adaptation and cell death then is important for understanding redox biology and disease pathogenesis. Recent studies have found that one major sensor of redox signalling at this switch in cellular responses is autophagy. Autophagic activities are mediated by a complex molecular machinery including more than 30 Atg (AuTophaGy-related) proteins and 50 lysosomal hydrolases. Autophagosomes form membrane structures, sequester damaged, oxidized or dysfunctional intracellular components and organelles, and direct them to the lysosomes for degradation. This autophagic process is the sole known mechanism for mitochondrial turnover. It has been speculated that dysfunction of autophagy may result in abnormal mitochondrial function and oxidative or nitrative stress. Emerging investigations have provided new understanding of how autophagy of mitochondria (also known as mitophagy) is controlled, and the impact of autophagic dysfunction on cellular oxidative stress. The present review highlights recent studies on redox signalling in the regulation of autophagy, in the context of the basic mechanisms of mitophagy. Furthermore, we discuss the impact of autophagy on mitochondrial function and accumulation of reactive species. This is particularly relevant to degenerative diseases in which oxidative stress occurs over time, and dysfunction in both the mitochondrial and autophagic pathways play a role.
Keywords: autophagy, mitochondrion, neurodegeneration, nitrative stress, oxidative stress, redox signalling
Abbreviations: ALS, amyotrophic lateral sclerosis; AMPK, 5′-AMP-activated protein kinase; ATG, AuTophaGy-related; BAG, Bcl-2-associated athanogene; BNIP, Bcl-2/adenovirus E18 19-kDa-interacting protein; BNIP3L, BNIP3-like; Drp1, dynamin-related protein 1; ECH, enoyl-CoA hydratase; EM, electron microscopy; ER, endoplasmic reticulum; FIP200, focal adhesion kinase family-interacting protein of 200 kDa; GABARAP, GABAA (γ-aminobutyric acid type A)-receptor-associated protein; GFP, green fluorescent protein; HIF-1, hypoxia-inducible factor 1; HNE, 4-hydroxynonenal; IκB, inhibitor of nuclear factor κB; IKKβ, IκB kinase β; IP3, inositol 1,4,5-trisphosphate; JNK1, c-Jun N-terminal kinase 1; Keap1, Kelch-like ECH-associated protein 1; LAMP, lysosome-associated membrane protein; LC3, light chain 3; LRRK2, leucine-rich repeat kinase 2; 3-MA, 3-methyladenine; mETC, mitochondrial electron-transport chain; mtDNA, mitochondrial DNA; mTOR, mammalian target of rapamycin; NAC, N-acetyl-L-cysteine; NBR1, neighbour of BRCA1 (breast cancer early-onset 1); NF-κB, nuclear factor κB; NGF, nerve growth factor; NOS, nitric oxide synthase; NOX, NADPH oxidase; Nrf2, nuclear factor-erythroid 2-related factor 2; PE, phosphatidylethanolamine; PI3K, phosphoinositide 3-kinase; PI3P, phosphatidylinositol 3-phosphate; PINK1, PTEN (phosphatase and tensin homologue deleted on chromosome 10)-induced kinase 1; RFP, red fluorescent protein; RLS, reactive lipid species; RNS, reactive nitrogen species; ROS, reactive oxygen species; Rubicon, RUN domain- and cysteine-rich domain-containing beclin-1-interacting protein; siRNA, small interfering RNA; SOD, superoxide dismutase; TAC, transverse aortic constriction; tfLC3, tandem fluorescently tagged LC3; TIGAR, TP53 (tumour protein 53)-induced glycolysis and apoptosis regulator; TNFα, tumour necrosis factor α; TOR, target of rapamycin; Tzb, trastuzumab; UCP, uncoupling protein; ULK, unc (unco-ordinated family member)-51-like kinase; VDAC, voltage-dependent anion channel; Vps34, vacuolar protein sorting 34
INTRODUCTION
Autophagy (or self-eating) was first described by Christian de Duve in 1963 as a lysosome-mediated degradation process for non-essential or damaged cellular constituents [1,2]. Physiologically, it serves to preserve the balance between organelle biogenesis, protein synthesis and their clearance. Autophagy is emerging as an important mediator of pathological responses and engages in cross-talk with ROS (reactive oxygen species) and RNS (reactive nitrogen species) in both cell signalling and protein damage.
Impaired mitochondrial function, oxidative stress, accumulation of protein aggregates and autophagic stress are common in many pathologies, including neurodegenerative diseases [3,4]. Oxidative stress can lead to the non-specific post-translational modifications of proteins and contributes to protein aggregation. Since the brain uses 20% of the inspired oxygen and 90% of the consumed oxygen to produce energy during oxidative phosphorylation, it is not surprising that neuronal cells are particularly sensitive to oxidative stress. During oxidative phosphorylation, neurons in the brain are vulnerable to oxidative damage because of their high metabolic activity, low antioxidant capacity and non-replicative nature.
The highly abundant mitochondria in brain cells are a major site of generation and action of ROS/RNS. Specific forms of ROS and RNS include hydrogen peroxide (H2O2), superoxide (O2•−), nitric oxide (NO), peroxynitrite (ONOO−) and RLS (reactive lipid species). Lipid peroxidation is a consistent feature of neurodegenerative diseases and biologically active RLS, such as HNE (4-hydroxynonenal), accumulates in brains of individuals with Parkinson's or Alzheimer's disease [5–10]. Other mechanisms of protein modification are NO-dependent. For example, NO reacts with O2•− and generates ONOO−, which is capable of initiating further protein oxidation and nitration [11,12]. The nitrogen dioxide radical, formed biologically from the reaction of NO with oxygen or decomposition from ONOO−, reacts with tyrosine residues, resulting in 3-nitrotyrosine formation [13]. The addition of NO to thiol groups on proteins, S-nitrosation (also referred to as S-nitrosylation), has also been reported in neurodegenerative diseases. This adduct has been detected in a broad range of pathologies, including Parkinson's disease, which is associated with both nitrated α-synuclein and S-nitrosated parkin [14]. Likewise, ONOO−-dependent modifications of proteins are widespread in brains of individuals with Alzheimer's disease [15].
Oxidative stress is inseparably linked to mitochondrial dysfunction, as mitochondria are both generators of and targets for reactive species [16]. Mitochondrial turnover is dependent on autophagy, which declines with age and is frequently dysfunctional in neurodegenerative diseases [3]. The cross-talk between autophagy, redox signalling and mitochondrial dysfunction is not well understood. Some possibilities include the accumulation of toxic proteins and the decrease in mitochondrial function, leading to further oxidative stress when the autophagic process is disrupted [17–19]. Dysregulated redox signalling or mitochondrial dysfunction can also influence autophagic activities. How particular species of modified proteins participate in the neurodegenerative process remains unclear. The present review highlights recent studies on redox signalling in regulation of autophagy, as well as mechanisms of mitophagy. Furthermore, we discuss the impact of autophagy on mitochondrial function, accumulation of ROS and relevance to chronic pathologies, with a particular emphasis on neurodegenerative diseases.
AUTOPHAGY MECHANISMS
Autophagy machinery
Three classes of autophagy have been described, based on how protein substrates for degradation reach the lumen of the lysosome.
(i) Macroautophagy (generally referred to as autophagy) is a multi-step process, involving the formation of double-membrane vesicles known as autophagosomes. Autophagosomes mature and fuse with lysosomes to degrade their contents in the acidic environment mediated by acidic hydrolases. More than 30 Atg (AuTophaGy-related) proteins conserved from yeast to mammals participate in autophagy at different steps throughout the process [20,21].
(ii) Microautophagy is a process in which lysosomes directly wrap around cytosolic constituents and ingest cargo by membrane involution [22–25].
(iii) Chaperone-mediated autophagy targets chaperones to proteins that contain a motif biochemically related to the pentapeptide KFERQ. The chaperone–KFERQ-containing protein complex then binds LAMP (lysosome-associated membrane protein)-2A receptors on the lysosomal membrane, and translocates the target proteins into the lysosomes for degradation [26].
Macroautophagy is the focus of the present review. Selective macroautophagy and autophagy of specific organelles are subsets of macroautophagy that depend on selective combination of Atg proteins [20], and are specifically discussed whenever appropriate. Figure 1 is a simplified diagram describing these complex processes and regulation. Many of these key steps have been shown to be modulated by ROS/RNS or their metabolites, including lipid peroxidation products [20,24,26].
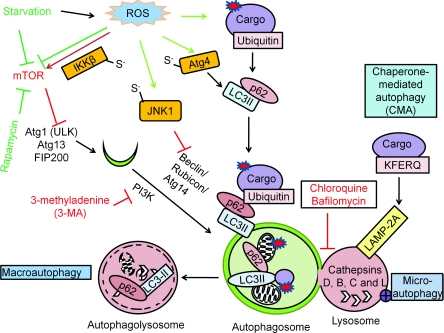
Figure 1. Autophagy mechanisms and regulation.
There are three different mechanisms of autophagy: (i) macroautophagy, the bulk degradation of cargo; (ii) microautophagy, involution of the lysosome around cargo; and (iii) chaperone-mediated autophagy (CMA), the degradation of the tagged cargo. The autophagosome and the lysosome fuse to degrade the cargo by lysosomal acidic hydrolases, including cathepsins D, B and L. Macroautophagy is regulated by starvation through the Atg1–Atg13 complex, which is inhibited by mTOR activation. Rapamycin or nutrient starvation causes mTOR inactivation, subsequent Atg1–Atg13 activation and activates macroautophagy. The beclin-1–PI3K complex is involved in autophagosomal expansion. 3-MA inhibits PI3K activity. Atg8–pro-LC3 is cleaved by Atg4, modified by PE to become LC3-II and inserted into the autophagosomes. The puncta formed by LC3-II and the electrophoretic separation of LC3-I and LC3-II are used as a marker for autophagosomal accumulation. RFP–GFP–LC3 (tfLC3) form puncta with both red and green fluorescence in the autophagosomes, and only red puncta when the autophagosomes fuse with lysosomes where GFP is inactivated, therefore tfLC3 is used to monitor autophagic flux. The degradation rate of long-lived proteins is another method for measurement of autophagic flux. p62 binds both LC3 and ubiquitin and therefore plays a role in targeting ubiquitinated proteins to the autophagosomes. Red arrows are indicative of negative regulators of autophagy, and green arrows are indicative of positive regulators of autophagy. Atg4 Cys81 thiol modification is involved in starvation and H2O2-induced autophagy. S-nitrosation of IKKβ and JNK1 are involved in NO-induced inhibition of autophagy.
To understand the impact of ROS/RNS on autophagy, we briefly outline key elements of the molecular mechanisms and their control. In mammalian cells, when the signalling protein TOR (target of rapamycin) is inhibited in response to starvation, dephosphorylation of Atg13 occurs. Dephosphorylated Atg13 associates with and activates Atg1 homologues {ULK [unc (unco-ordinated family member)-51-like kinase] 1 and 2}, leading to the recruitment of FIP200 (focal adhesion kinase family-interacting protein of 200 kDa), an orthologue of yeast Atg17. The ULKs–Atg13–FIP200 complex participates in elongation of the isolation membrane and progression towards a complete autophagosomal structure [20,21,27–29] (Figure 1).
Generation of the pre-autophagosomal structure requires the beclin-1–class III PI3K (phosphoinositide 3-kinase) complex, as well as generation and insertion of LC3 (light chain 3)-II into the autophagosomal membrane [21]. Atg8/LC3 is a ubiquitin-like protein and it undergoes post-translational modifications when LC3-I becomes LC3-II [30–32]. The C-terminal flanking region of nascent LC3 (proLC3) is cleaved by Atg4, a protease, to become LC3-I, which then has an exposed C-terminal glycine residue. LC3-I is modified with PE (phosphatidylethanolamine) and is now referred to as LC3-II; this step involves a ubiquitination-like reaction mediated by Atg7 (E1-like activating enzyme), Atg3 (E2-like conjugating enzyme) and the Atg16L complex (E3-like ligase enzyme). First, Atg7 plays a role in the conjugation of Atg10 to Atg12, and conjugation of Atg3 to LC3-I. Then, E2-like enzymes Atg3 and Atg10 catalyse the conjugation of Atg5 to Atg12, and the conjugation of LC3-I to PE to convert LC3-I into LC3-II. These conjugation processes play an essential role in the expansion of the autophagosomes [30–32]. Atg7, Atg3 and Atg10 use cysteine residues in their catalytic sites to catalyse ubiquitin transfer. These proteins may then be sensitive to redox signalling, since the thiol groups on cysteine residues are particularly susceptible to oxidative modifications by ROS/RNS [33]. Lipidation of LC3-I by PE enables it to be inserted into the autophagosomal membrane. The LC3-related molecules GABARAP [GABAA (γ-aminobutyric acid type A)-receptor-associated protein] and GABARAPL1 (GABARAP-like 1 protein) may play similar roles in the autophagosomal expansion process [34]. LC3 homologues form many functional interactions in the autophagy network [35] (Figure 1).
Fusion of autophagosomes to lysosomes may be mediated by Rab7 and the lysosomal transmembrane protein LAMP-2 [36–39]. Degradation of autophagosomal contents requires lysosomal proteases, such as cathepsins D, B and L [40–42]. The cathepsins are a particularly interesting class of proteases which have different abilities to degrade modified proteins. Many members of the cathepsin family that are important in regulation of autophagy have a cysteine at the active group, which may be susceptible to oxidative modification by metals and redox regulation [43]. For example, cathepsin B can be inhibited by oxidized low-density lipoprotein which may then contribute to the accumulation of oxidized lipoproteins in atherosclerotic lesions [44] (Figure 1).
Tremendous progress has been made in the field of autophagy in terms of identification of the molecular components involved in the autophagy process, especially in model systems such as yeast. Whereas the overall mechanics of the autophagy–lysosome pathway are now clear, how these vary in different cell types needs to be established. Major gaps in our knowledge in the field now being addressed are how the physical and functional interactions between the autophagosomes are controlled to achieve assembly and the subsequent structural changes needed to engulf cargos and fuse with lysosomes. It is also likely that the response to different ROS/RNS will place distinct demands on the autophagy system, particularly under conditions of inflammation, which involves the concerted action of multiple cell types.
Regulation of autophagy
As mentioned above, mTOR (mammalian TOR) is activated in the presence of growth factors and abundant cellular nutrients such as amino acids. mTOR functions as an inhibitor of the initiation step of macroautophagy. Under nutrient-rich conditions, mTOR is active and phosphorylates Atg13, preventing its association with ULK and FIP200 recruitment [27,28]. During starvation, mTOR is inhibited, resulting in the activation of phosphatases and partial dephosphorylation of Atg13 leading to autophagosomal formation (Figure 1). A possible mechanism of mTOR regulation could involve the targeting of mTOR to cellular compartments, resulting in interactions with different populations of protein substrates [45]. For example, amino acid availability stimulates the translocation of mTORC1 (mTOR complex 1) to the lysosomal surface, where it associates with its co-activator Rheb and activates mTOR activity [45,46]. Conversely, during starvation, mTOR co-localizes with LC3 after autolysosome formation, suggesting that it may regulate autophagosome and lysosome reformation directly [45]. This regulatory step may be subject to ROS-dependent regulation, as mTOR oxidation results in the inhibition of its activity [47]. The activity of the beclin-1–class III PI3K complex is regulated by the interaction of a series of cofactors, including Atg14/Barkor (beclin-1-associated autophagy-related key regulator), UVRAG (UV radiation resistance-associated gene), Bif-1 (Bax-interacting factor 1), Ambra1 (autophagy/beclin-1 regulator 1) and Rubicon (RUN domain- and cysteine-rich domain-containing beclin-1-interacting protein) (Figure 1). Beclin is regulated by the transcription factor NF-κB (nuclear factor κB) [48] and K63-linked ubiquitination [49].
The beclin-1–class III PI3K complex activity is cysteine-rich, but the potential regulation by ROS/RNS has not been investigated. One positive regulator of the beclin–Bcl-2 interaction in the ER (endoplasmic reticulum) is NAF-1 (nutrient-deprivation autophagy factor-1), a component of the IP3 (inositol 1,4,5-trisphosphate) receptor complex, which contains a redox-sensitive CDGSH iron–sulfur-binding domain [50]. Another regulator of the complex is Rubicon, a negative regulator with a candidate redox-sensing cysteine-rich domain [51]. The multiprotein beclin-1–class III PI3K complex must be formed for the allosteric activation of the class III PI3K Vps34 (vacuolar protein sorting 34) to generate PI3P (phosphatidylinositol 3-phosphate). PI3P recruits effectors such as DFCP1 (double FYVE domain-containing protein 1) [52–55] and WIPI (WD-repeat protein interacting with phosphoinositides) family proteins to mediate the vesicle nucleation and autophagosome formation [56,57]. Additional molecules involved in macroautophagy regulation include AMPK (5′-AMP-activated protein kinase) [58], DAPK (death-associated protein kinase) [59] and IP3R (IP3 receptor) [60]. Exactly how these molecules respond to starvation, proteasomal dysfunction and accumulation of damaged proteins and/or organelles is unclear, although redox regulation may be a key component.
The selectivity of macroautophagy can occur at different levels. ‘Adaptor’ molecules, including p62/SQSTM1, have been shown to bind to ubiquitinated proteins and target them to LC3-II, to promote selective uptake and degradation [61,62] (Figure 1). The LRS (LC3-recognition sequence) is in an uncharacterized linker region between the zinc finger and UBA (ubiquitin-associated) domains in murine p62. As zinc fingers are particularly susceptible to oxidative and nitrative stress, p62 may be regulated by redox signalling [43]. NBR1 [neighbour of BRCA1 (breast cancer early-onset 1)], is another ‘adaptor’ molecule with both a LC3-binding domain and a ubiquitin-binding domain [63]. These binding domains may work in concert or separately to target mitochondria to the autophagic pathway. p62 is not required for autophagosomal formation and overall long-lived protein degradation, but plays a role as a specific substrate and a target-recognition molecule for autophagy [64].
The understanding of how nutrient regulation of autophagy regulation occurs has advanced in the last few decades, especially at the junction of the mTOR/Atg1/Atg13 cascades. How ROS/RNS signals and mitochondrial dysfunction regulate autophagy has not been fully understood. Since mitochondria are both a source and target of ROS/RNS, a major effort is needed to determine how the organelle communicates with the lysosome to combat protein oxidation, mitochondrial damage and the maintenance of mitochondrial networks through fission and fusion.
Measurement of autophagy
Defining the stages of autophagy and its progress in a tissue or cell is important for understanding its function and regulation. Ultrastructural analysis by EM (electron microscopy) of autophagosomes at different stages of maturation has been the classic approach for confirming autophagic activities. EM can distinguish early autophagosomes or phagopores, and amphisomes, from autolysosomes generated by fusion of amphisomes or autophagosomes fusing lysosomes [2]. Measuring the long-lived protein degradation rate by pulse–chase methods is also a commonly used complementary approach [65]. These two methods have helped to determine autophagic pathways and time lapses during autophagy.
Another widely used tool for measuring autophagy is based on Atg8/LC3. As mentioned above, LC3-I undergoes post-translational modifications by PE to become LC3-II [30–32]. LC3-I and LC3-II can be discriminated by their difference in mobility on gel electrophoresis [30–32]. LC3-II insertion into the autophagosomal membrane is a consistent key step in autophagosomal formation, and its level reflects the relative amount of autophagosomes in the cell. LC3-II protein can also be visualized by immunocytochemistry as vesicular puncta when associated with autophagosomes [30–32].
The fate of individual LC3-II molecules depends on their localization. During recycling, LC3-II that is located on the outer face of the autophagosome is delipidated and converted back into the LC3-I form in cytosol; this population of LC3 is presumably reused for another round of conjugation by PE [30–32]. In contrast, the LC3-II that localizes to the inner autophagosome is degraded by lysosomal proteases, after the fusion of the autophagosome with the lysosome, as part of the normal maturation process [66]. When autophagy–lysosome activities are highly efficient, the transient increase in LC3-II can be overlooked. The additional use of a lysosome and autophagosome fusion inhibitor, such as chloroquine, to block the formation/degradation of the autophagolysosome is necessary in these cases to reveal the increase in LC3-II [67].
Currently available antibodies recognize both LC3-I and LC3-II. Under conditions of autophagosomal accumulation, immunohistochemistry and immunocytochemistry signals increase and appear punctate due to increased LC3-II which is localized to autophagic structures. Expression of exogenously introduced GFP (green fluorescent protein)-fused LC3 has provided a convenient method for measuring autophagy both qualitatively and quantitatively (by measuring the punctate green fluorescence in cells [32]. Transgenic mice expressing GFP–LC3 have been used to aid autophagy studies in vivo [68]. Interpretation of these studies needs caution, since under specific conditions, LC3 may form protein aggregates independently of autophagy [69], bind to the smooth ER [70] or form puncta in response to detergents [71].
The accumulation of autophagosomes may be due to increased autophagic initiation or decreased autophagic completion. In the latter scenario, when autophagic flux is blocked, the degradation of the inner membrane-localized LC3-II cannot occur; and the number of LC3-positive puncta increases as a result of the accumulation of autophagosomes. To distinguish whether the accumulation of autophagosomes is due to increased autophagosomal formation or decreased fusion of autophagosomes with lysosomes, the tfLC3 (tandem fluorescently tagged LC3) method has been developed [72]. This assay is based on the sensitivity of the fluorescent signal of GFP to the lysosomal acidic/proteolytic environment, whereas RFP (red fluorescent protein) is resistant to the acidic environment. LC3 tandemly tagged with GFP and RFP exhibit overlapping GFP and RFP signals in the autophagosomes before fusion with the lysosomes, but once the maturation to an autolysosome occurs, it exhibits only the RFP signal. Therefore the appearance of the puncta exhibiting only an RFP signal indicates the normal autophagic maturation process and flux activities. This allows for a live imaging of autophagy, unlike EM, which is in fixed tissue. Transgenic mice expressing tfLC3 have been generated to study autophagic flux in the heart, and other transgenic tfLC3 models will be extremely important for studying many diseases [73].
Currently, multiple approaches for the measurement of autophagy are necessary to draw conclusions regarding the status of the autophagy process, whether activated or repressed, in response to ROS/RNS signalling. Live-cell imaging will be very helpful to simultaneously follow autophagosomes as they form, move and fuse with lysosomes. This will help to determine changes in autophagic flux and to determine in real time how ROS/RNS affect autophagic activity. The availability of domain-specific redox sensors offers an interesting opportunity to map the progression of autophagy in the cell with oxidative stress [74].
AUTOPHAGY REGULATION BY REDOX SIGNALLING
It has long been known that the conditions that regulate the activity of the autophagic process are also associated with changes in the production of ROS/RNS in cells. As shown in Figure 2, ROS/RNS are a range of oxygen-derived molecules formed by the incomplete reduction of oxygen during oxidative metabolism and have both specific mechanisms of production and intracellular targets. The most important biologically are O2•− and H2O2, since both can be formed by controlled mechanisms in cells and are cell signalling molecules [75]. O2•− and H2O2 can interact with NO to generate nitrating species, such as ONOO−, and oxidized lipids to produce RLS [12,76]. A major endogenous source of both O2•− and H2O2 is the mETC (mitochondrial electron-transport chain), where continuous electron leakage to O2 occurs during aerobic respiration [16]. In addition to the mETC, low levels of ROS are produced by membrane-localized NOXs (NADPH oxidases) [77], peroxisomes [78] and the cytochrome p450 system [79]. It is important to recognize that the ROS cannot be considered as discrete or independent redox messengers since they can be interconverted.
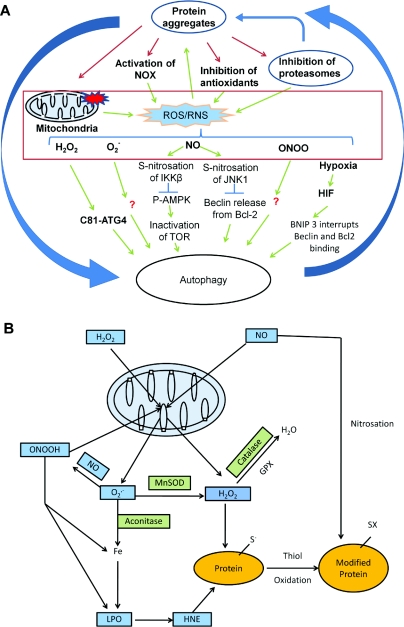
Figure 2. ROS/RNS production and signalling.
(A) ROS/RNS production and signalling of autophagy. ROS and RNS production can be induced by protein aggregates, generated by the mitochondrial respiratory activities and by other cellular oxidases. These ROS/RNS species can modify signalling molecules to stimulate or inhibit autophagy. Atg4 Cys81 thiol modification is involved in starvation and H2O2-induced autophagy. S-nitrosation of IKKβ and JNK1 are involved in NO-induced inhibition of autophagy. Although many ROS/RNS have effects on autophagy, besides a handful of known ROS/RNS-targeted autophagy regulators, the exact mechanisms for the effect of ROS/RNS on autophagy are largely unknown. (B) Mitochondrial production of ROS/RNS. Regardless of their sources, H2O2 and NO can target to the mitochondria. Mitochondrially produced superoxide (O2•−) can react with NO to produce peroxynitrite (ONOOH). O2•−, H2O2 and ONOOH can further damage the mitochondria in combination with iron and produce lipid peroxidation (LPO) products, including HNE. MnSOD, aconitase, GPX (glutathione peroxidase) and catalase are involved in generation and reduction of these products. HNE, NO and H2O2 can modify proteins to initiate signalling mechanisms or damage. The most sensitive moieties to oxidative modification are the thiols on cysteine residues. Redox reactions in protein modification are involved in signalling of autophagy.
For example, starvation not only increases ROS levels in the cell, but also stimulates autophagy [80,81]. Both O2•− and H2O2 have been shown to mediate autophagy induction [80,82]. For example, H2O2 can modify mitochondrial proteins damaging the electron-transfer process, so that they now generate intra-mitochondrial O2•−. In a further example, oxidative modification of the NOSs (nitric oxide synthases) can cause disruption of electron transfer within the enzymes, inhibiting the formation of nitric oxide and generating O2•− [83,84]. In a biological setting, it is then likely that the cell is responding to the combined effects of ROS/RNS acting at different sites within the autophagic process.
Mitochondria generate ROS from a number of different redox centres in the respiratory chain and other metabolic pathways [16]. The primary ROS generated from mitochondria is O2•−, which can then be converted into other ROS such as H2O2 or ONOO− (Figure 2). At low levels, it is thought that mitochondrial ROS play a role in cell signalling, but, at higher levels, mitochondrial proteins are susceptible to damage because of the concentration of both oxidizable lipids and abundant redox-active proteins which can amplify oxidative damage [16,85]. It is then essential to regulate autophagy at multiple levels, including removal of defective proteins generating uncontrolled ROS and the entire organelle by mitophagy. Not surprisingly, the mitochondrion and mitochondrially produced ROS are emerging as important players in autophagy and mitophagy [80,86].
The reduction of superoxide and generation of H2O2 is controlled by the SODs (superoxide dismutases). The mitochondrial form is located in the matrix and contains a manganese prosthetic group and is known as SOD2 or MnSOD. Mitochondrial MnSOD-deficient cells exhibit decreased oxygen consumption and increased O2•− production, suggesting a key role in the response to pathological stressors [87,88]. For example, in response to acute alcohol binge in mice, MnSOD overexpression prevents, and MnSOD deficiency exacerbates, NOS expression, plasma nitrites/nitrates, nitration of complex I and V proteins, and mtDNA (mitochondrial DNA) depletion [89]. In a transgenic Alzheimer's disease mouse, MnSOD overexpression led to increased catalase protein levels, decreased total oxidized proteins and amyloid burden, and improved spatial memory [90]. Mutations of SOD1 or Cu,ZnSOD, the cytoplasmic form, have been found to be involved in ALS (amyotrophic lateral sclerosis), which is a pathology associated with oxidative stress, mitochondrial dysfunction and intracellular protein aggregation in degenerating motor neurons [91–95]. Accumulation of autophagosomes and p62 also occurs in motor neurons in ALS patients' spinal cords [96], as well as in experimental animal models [97].
Mitochondrial ROS production and oxidation of mitochondrial lipids have been shown to play a role in autophagy. In yeast, rapamycin induces ROS, fatty acid modification, autophagy and mitophagy [98]. Resveratrol and, to a lesser extent, the soluble antioxidant NAC (N-acetyl-L-cysteine) inhibit these effects [98]. In mammalian cells, starvation-induced autophagy is associated with increased oxidative stress, and both H2O2 and O2•− have been shown to induce autophagy [80,82]. In addition to mitochondrially generated ROS, NOX activities have also been shown to play an important role in antibacterial phagosome autophagy, neutrophil autophagy and ER stress autophagy [99]. Below, we review the current understanding of how different ROS/RNS are sensed by the autophagy machinery, which is also summarized in Table 1.
Table 1. Redox modification of autophagy proteins and impact of autophagy deficits on oxidative stress.
Summary of the current literature on the impact of alterations of autophagy gene levels on mitochondrial function and cellular oxidative stress. Many of these observations have been mentioned in the text. Furthermore, specific examples of protein modifications of autophagy regulatory molecules in response to ROS/RNS alterations are also listed.
| Autophagy gene | Impact on mitochondrial function | Impact on oxidative stress | Redox modification |
|---|---|---|---|
| ULK1 | ULK1 knockout results in decreased mitochondrial clearance during reticulocyte maturation [27] | ||
| FIP200 | The livers from FIP200 knockouts exhibit significant increase in mitochondrial mass [139] | The livers from FIP200 knockouts exhibit significant increase in ROS [139] | |
| mTOR | Rapamycin treatment, or siRNA knockdown of TSC2 (tuberous sclerosis complex 2), S6K1 (ribosomal S6 kinase 1), raptor (regulatory associated protein of mTOR) or rictor (rapamycin-insensitive companion of mTOR), lowered the mitochondrial membrane potential and oxygen consumption [278] | NO-induced S-nitrosation of IKKβ inhibits its activity and prevents the inactivation of TOR [110] | |
| Atg3 | Atg3−/− T-cells exhibited reduced autophagy and expanded mitochondria [279] | Atg3−/− T-cells exhibit increased ROS production [279] | |
| Atg4 | Atg4b knockout results in decreased proteolytic processing of LC3A, LC3B, GABARAP and Atg8L [280]. Atg4d has an affinity for damaged mitochondria in cells treated with H2O2 [281] | Starvation and H2O2 induce Atg4 thiol modification, which is important for autophagy [80] | |
| Atg5 | Atg5−/− cells have deformed and dysfunctional mitochondria [282,283]. Mitochondrial gene regulation is significantly altered in Atg5−/− T-cells [284]. Mitochondrial content and cross-sectional mitochondrial surface area are increased in Atg5−/− T-cells [285] | Atg5−/− cells exhibit accumulation of ubiquitinated proteins and accumulation of ROS [80,142,143,283]. Reduction of Atg5 and Atg10 further increases ROS in response to starvation [81]. Genes involved in ROS generation are significantly altered in Atg5−/− T-cells [284] | |
| Atg7 | Reticulocyte maturation is diminished, but not abolished in Atg7−/− mice [286]. Atg7−/− erythrocytes accumulate damaged mitochondria [287]. Mitochondrial content and cross-sectional mitochondrial surface area are increased in Atg7−/− T-cells [285]. Haemopoietic stem cells that are Atg7-deficient accumulate mitochondria with high membrane potential [288]. Atg7−/− MEFs (mouse embryonic fibroblasts) exhibit dysfunctional mitochondrial respiration [142] | Atg7−/− cells have increased ROS [285]. Haemopoietic stem cells with Atg7 deficiency exhibit enhanced mitochondrial superoxide accumulation [288]. p62 and ROS are increased in Atg7−/− cells [142] | |
| Beclin/Atg6 | Apoptosis-deficient Bcl-2-overexpressing iBMK (immortalized baby mouse kidney) cells undergo autophagy in response to stress. Abnormal mitochondria accumulate in either Beclin+/− (Beclin haploinsufficiency) or Atg5−/− animals [270] | ROS accumulate in either Beclin+/− or Atg5−/− animals [270] | NO-induced S-nitrosation of JNK1 inhibits its activity and prevents the release of beclin from binding to Bcl-2 [110] |
| Atg8 (LC3B) | In response to LPS (lipopolysaccharide), LC3B−/− macrophages had more swollen mitochondria with severely disrupted cristae [289] | LC3B−/− macrophages generate more superoxide [289] | |
| Nix | Decreased clearance of mitochondria in erythrocyte maturation [290] | ||
| Parkin | Parkin-deficient fibroblasts in Nix−/− mice exhibit altered mitochondrial morphology and activities [240] | Parkin can be inhibited by S-nitrosation [238] | |
| PINK1 | PINK1−/− mice exhibit impaired mitochondrial respiratory activities [241]. PINK1-deficient dopaminergic neurons accumulate fragmented mitochondria [242]. Down-regulation of PINK1 in neuroblastoma SH-SY5Y cells induced mitochondrial fragmentation [243] | PINK1−/− mice exhibit increased sensitivity to oxidative stress [241]. PINK1-deficient dopaminergic neurons accumulate mitochondrial superoxide [242] | |
| DJ-1 | Knockout of DJ-1 in fruitflies reduced the RCR (respiratory control ratio) and mitochondrial DNA/nuclear DNA ratio without changing membrane potential or complex I subunit NDUFS3 [NADH dehydrogenase (ubiquinone) 1β subcomplex 8] level [244]. DJ-1−/− mice exhibit reduced skeletal muscle ATP [244], down-regulated UCP4 and UCP5 [245–247], and reduced mitochondrial length and fusion rate [248]. Stable knockdown of DJ-1 in neuroblastoma cells led to a reduction in mitochondrial membrane potential, mitochondrial fragmentation and accumulation of LC3 around mitochondria. Overexpression of DJ-1 protects against rotenone-induced mitochondrial fragmentation; this function is independent of PINK1 [248,250] | DJ-1−/− mice exhibit increased sensitivity to oxidative stress, and increased H2O2 [245–247]. Addition of glutathione or NAC attenuates DJ-1-deficiency-induced phenotypes [248–250]. |
Autophagy regulation by H2O2
H2O2 plays an important role in autophagy in response to nutrient starvation, rapamycin, TNFα (tumour necrosis factor α) [100] and NGF (nerve growth factor) deprivation [101,102]. NGF deprivation induces both early burst and late-phase ROS accumulation [103], accompanied by autophagosome accumulation [102]. In this study, NAC decreased both cellular ROS production and autophagy, implicating redox thiol signalling as an important regulator of autophagy [102]. In Ewing sarcoma cells, TNFα treatment in the presence of IκB (inhibitor of NF-κB), led to increased ROS, protein oxidation and beclin-1 levels; these effects are mimicked by exogenous H2O2 and suppressed by the lipid radical scavenger BHA (butylated hydroxyanisole) or BNP (N-butyl-α-phenylnitrone), or NF-κB activation of mTOR [100,104]. These findings are consistent with H2O2 effects being mediated through the production of RLS, which are well known to modify proteins and change protein function.
Although the mechanisms remain unidentified, redox modification of transcription factors by electrophilic lipid oxidation products may mediate the regulation of the levels of autophagy genes, as has been shown with other redox signalling pathways [105]. In support of specific mechanisms mediated by redox signalling proteins, starvation-induced ROS formation has been shown to play a key role in autophagic induction through thiol modification of the Cys81 site of Atg4 [80]. The thiol reduced form of Atg4 enables the conversion of LC3-I into LC3-II, lipidation and insertion into the autophagosome, and then recycles LC3-II after the autophagosome fuses with the lysosome [80]. In vitro, Atg4 can be modified by H2O2 [80]. In vivo, it is known that exogenous exposure to H2O2 induces autophagy, but it is unknown whether autophagy is solely mediated by H2O2's primary effects on Atg4 or also secondary effects via an increase in mitochondrial O2•− through oxidative modification of the mETC [16]. The thiol modification of Atg4 is one of the few known mechanisms of redox control of autophagy.
Autophagy regulation by mitochondrial O2•−
Although exogenous exposure to H2O2 induces autophagy, some evidence suggests that endogenously produced O2•− may also play an important role in the induction of autophagy. For example, autophagy in response to starvation in HeLa cells requires a mitochondria-dependent regulation of O2•−, since the overexpression of MnSOD decreases O2•− and decreases autophagy [82]. Consistent with a role of mitochondrial O2•−, siRNA (small interfering RNA) knockdown of MnSOD increased autophagy, while raising levels of O2•− and decreasing H2O2. These studies indicated that O2•− plays a pivotal role, perhaps through its interactions with aconitase which can result in iron release and promotion of lipid peroxidation [82] (Figure 2B). In support of this idea, the lipid peroxidation product HNE induces accumulation of LC3-II in vascular smooth muscle cells [106]. The mechanisms stimulating mitochondrial O2•− are not clear, but they must be independent of class III PI3K, since the inhibitors 3-methyladenine and wortmannin did not change O2•− or H2O2 levels [82]. Neither do the mechanisms seem to involve beclin, since siRNA knockdown of beclin does not change O2•− levels. In glioma, paraquat and selenite induced O2•− production and autophagic cell death. Selenite-induced autophagy and cell death has been found to be inhibited by overexpression of MnSOD and Cu,ZnSOD, but not by catalase.
Autophagy regulation by nitric oxide (NO)
NO has a variety of effects on autophagy depending upon the cell type. In glioma cells, when combined with hypothermia, NO donors SNP (sodium nitroprusside), S-nitrosoglutathione or PAPA-NONOate (propylamine propylamine NONOate) inhibit the completion of autophagy, as is evident by LC3-II accumulation [107], whereas in primary neurons, NO induces Drp1 (dynamin-related protein 1)-mediated mitochondrial fission, which further causes an increase in mitophagy [108]. Furthermore, in a heart disease model, homocysteine increased mitochondrial O2•− and ONOO− levels as detected by MitoSOX and dihydrorhodamine-123. Functionally, this resulted in a decreased oxygen consumption rate and increased accumulation of LC3-II [109].
Molecular studies in primary cortical neurons, HeLa and HEK (human embryonic kidney)-293 cells have shown that NO-induced S-nitrosation of IKKβ (IκB kinase β) and JNK1 (c-Jun N-terminal kinase 1) inhibits their activities, and the inhibition of IKKβ and JNK1 in turn prevents the inactivation of TOR and the release of Beclin from binding to Bcl-2. Overexpression of NOS impairs autophagosome synthesis, therefore, in these systems, NO appears to play an inhibitory role in autophagy [110]. L-NAME (NG-nitro-L-arginine methyl ester), a nitric oxide synthase inhibitor, prevents NO inhibition of autophagy, and, in this case, induction of autophagy is independent of effects on TOR inhibition or Bcl-2 by NO [110].
Autophagy regulation and cross-talk with transcriptional regulation of cellular antioxidants by redox signalling
Some of the post-translational protein modifications by ROS and RNS have a profound biological impact on the cell, by regulation of transcription factor activities that lead to global gene expression changes, including those that regulate autophagy and antioxidant defence in the cell. A prominent redox signalling pathway responsive to ROS/RNS/RLS is the Nrf2 (nuclear factor-erythroid 2-related factor 2)/Keap1 [Kelch-like ECH (enoyl-CoA hydratase)-associated protein 1] pathway [111]. Nrf2 can bind to the ARE (antioxidant-response element) [also called ERE (electrophile-response element)] sequence to activate transcription of antioxidant and cellular detoxification genes [112–115]. Interestingly, it also activates autophagy by increasing p62, thereby enhancing the cells' ability to process damaged proteins, as well as scavenge reactive species which cause protein damage. Nrf2-deficient cells exhibit a deficient stress response [116]. Nrf2 is negatively regulated by the cytoplasmic redox-sensor protein Keap1 via its Neh2 (Nrf2/ECH homology 2) domain [117]. Cysteine residue modifications of Keap1 release Nrf2 from binding to Keap1. These modifications inhibit the proteasomal degradation of Nrf2 and cause the translocation of Nrf2 to the nucleus to activate the transcription of antioxidant genes, such as NQO1 [NAD(P)H:quinone acceptor oxidoreductase 1] and GSTs (glutathione transferases) [118], and the autophagy genes such as p62 [119,120]. Therefore, indirectly, autophagy can be regulated by transcriptional mechanisms in response to ROS/RNS.
Another transcriptional sensor of ROS, which is intimately linked to autophagy, is p53, which is elevated in response to ROS [121,122]. It also regulates target genes that either promote or decrease ROS [123–125]. p53 regulation of autophagy is complex. Downstream of p53, both autophagy-attenuating and autophagy-promoting genes have been found. One p53 target gene is the TIGAR [TP53 (tumour protein 53)-induced glycolysis and apoptosis regulator] gene which has a fructose-2,6-bisphosphatase activity. TIGAR decreases glycolysis, increases NADPH, increases the GSH/GSSG ratio and reduces p53-induced intracellular ROS [126]. TIGAR overexpression attenuates starvation-induced and hypoxia/glucose deprivation-induced autophagy [81]. Another p53-regulated gene, DRAM (damage-regulated autophagy modulator) promotes autophagy [127]. p53-regulated sestrin proteins enhance autophagy via activation of AMPK which inhibits mTOR [128], although evidence for whether sestrin 2 acts as a reductase of sulfinylated peroxidredoxins or has other functions is being investigated [129,130].
AUTOPHAGY REGULATION OF ROS AND RNS
Dysregulation of autophagy leads to increases in oxidative stress, as shown by many pharmacological inhibitor and knockout studies (summarized in Table 1). Autophagy inhibition by the lysosomotropic agent chloroquine or the cathepsin D inhibitor pepstatin A increases the formation of reactive species [131–135]. Cathepsin D-knockout mice exhibit increased iNOS (inducible NOS) and NO production [136]. Many lysosomal diseases exhibit autophagosomal accumulation, as indicated by an increase in LC3-II, and an accumulation of p62, a known substrate of autophagic degradation. Among the lysosomal diseases, there are various changes in mitochondrial morphology and mitochondrial function, in both tissues and isolated mitochondria [137]. Furthermore, disruption of the autophagy pathway at earlier steps also leads to accumulation of ubiquitinated proteins, increased ROS and dysfunctional mitochondria. Genetically engineered mouse models have provided abundant evidence for the important role of autophagy in mitochondrial integrity, intracellular protein clearance and ROS control. For example, AMPK- and ULK1- (Atg1 homologue) knockout liver exhibits p62 accumulation and defective mitophagy [138]. Downstream of Atg1, FIP200- (Atg17 homologue) knockout livers exhibit significant increases in mitochondrial mass and ROS [139]. Atg5- and Atg7-knockout mice or cells accumulate ubiquitinated proteins and ROS [80,140–143]. Decreased levels of Atg5 and Atg10 further increase ROS in response to starvation [81].
Autophagic degradation of proteins
Under normal conditions, protein degradation is mediated by both the autophagy–lysosome pathway and the ubiquitin–proteasome system. The ubiquitin–proteasome system is thought to be primarily responsible for the degradation of short-lived signalling molecules or misfolded proteins tagged with ubiquitin [144]. The proteasome contains a 19S regulatory subunit which docks on the α-subunit rings of the catalytically active 20S core. The 19S regulatory particle contains ~18 subunits, including a protein that recognizes ubiquitinated substrates and positions them for proteasomal degradation [145]. Numerous studies have shown that the ubiquitin–proteasome system is one pathway through which oxidatively modified proteins can be degraded [146–152]. Under severe stress, however, this ubiquitin–proteasome pathway can be overwhelmed, and the autophagy–lysosome pathway is then required to increase its activity to compensate for the increased protein damage.
Autophagic degradation can target selective groups of proteins, lipids and organelles, depending on not only cellular ATP, but also the type, distribution and level of oxidative stress [29]. The pioneer studies in yeast in identification of autophagic process and molecular components of the autophagy–lysosome (vacuolar) degradation machinery laid the foundation for studying autophagic degradation of proteins and organelles in mammalian cells. Yeast vacuolar (equivalent of mammalian lysosomes) proteases are responsible for up to 70% of cellular protein turnover under nutrient-deprivation conditions in response to the sporulation signal [153]. ROS regulation of yeast autophagy is evident by an up-regulation of yeast lysosomal cathepsin D homologue (PEP4) in response to H2O2. PEP4-deficient cells exhibit reduced protein degradation, accumulation of oxidized proteins and shortened lifespan [154]. A role of autophagy in protection against accumulation of oxidized proteins is a conserved process through evolution. Cathepsin D-deficient sheep, mice and humans exhibit accumulation of ubiquitinated proteins and α-synuclein [41,42], indicating insufficient protein degradation.
The autophagy–lysosome pathway also degrades ubiquitinated proteins. p62, NBR1 and NDP52 (nuclear domain 10 protein) recognize ubiquitin moieties and target ubiquitinated proteins or ubiquitin-coated bacteria to the autophagosomes via binding to Atg8/LC3 [62,63,155–157]. A functional homologue of p62, Alfy (autophagy-linked FYVE protein) is highly expressed in the brain and has been found to recognize protein aggregates and bring them to p62, Atg5 and Vps34 [53,158,159], supporting cell-type- and tissue-specific autophagy regulation.
When cells are subject to starvation, stress or pathological damage, autophagy may degrade proteasomes [160]. Alternatively, autophagy can compensate for deficiencies in proteasomal degradation [161,162]. Deficits in the autophagy pathway lead to accumulation of ubiquitinated proteins [119,140,141,163]. The accumulation of ubiquitinated proteins in these scenarios can be attributed to the combined consequence of lack of autophagic flux and p62 accumulation, which also inhibits protein targeting to the proteasome [119,140,141,163]. Conversely, proteasomal activities also play a role in regulating autophagic activities. A recent study reported that, because of its ubiquitin-like domain and its N-terminal domain, LC3 can be degraded by the proteasome in vitro [164]. When p62 is available to bind to LC3, this degradation of LC3 by the proteasome can be prevented, thus making LC3 engagement to the autophagosomes possible [164].
Another transcription factor, FOXO3 (forkhead box O3), is activated in response to oxidative stress, and regulates the transcription of target genes involved in both proteasomal and autophagic activities [165,166]. Ubiquitin ligases, parkin and the CHIP [C-terminus of Hsc70 (heat-shock cognate 70)-interacting protein] have been shown to contribute to both proteasomal and autophagosomal targeting of proteins and organelles [167–170]. The relative levels of co-chaperone, BAG (Bcl-2-associated athanogene) 1 and BAG3 proteins balance the cellular dependence on either proteasomal or autophagic activities [171–173].
It is well established that dysfunction of autophagic activities results in accumulation of protein aggregates. However, it is not clear how protein aggregates are recognized and targeted to the autophagosomes.
Autophagic degradation of damaged mitochondria
Since mitochondria lack a compact chromatin structure and have an abbreviated replication and repair system compared with nuclear DNA, mtDNA is 10–20 times more likely to have mutations and incurs more damage with age [174]. As mentioned above, autophagy plays an important role in the regulation of mitochondrial function. In normal mammalian brains, it has been shown that mitochondria have an average half-life of 10–25 days [175]. During starvation, mitochondrial turnover can be accelerated [160] by an autophagic process specifically called mitophagy [176].
Mitochondrial membrane depolarization has been shown to precede the induction of autophagy by nutrient deprivation in hepatocytes, and the depolarized mitochondria co-localize with autophagosomes [177]. Further studies have shown that mitochondrial fission also co-ordinates with mitophagy [178–180]. Mitochondria will fuse every 5–20 min to reduce mitochondrial depolarization in both COS7 and INS1 cells [181]. Mitochondrial remodelling through fission, fusion or mitophagy is important for mitochondrial homoeostasis. In cultured liver hepatocytes, although mitochondria undergo fission, fusion or mitophagy, the overall mtDNA/nuclear DNA ratio remains unchanged, implying that areas of the mitochondria which contain the mitochondrial genome are not degraded. This is indicative of the specificity of fission, fusion and mitophagy [178]. Mitophagy may also be important in attenuating apoptosis or necrosis, by clearance of damaged mitochondria. This could then prevent the inadvertent release of cytochrome c, AIF (apoptosis-inducing factor) and other apoptotic factors.
Mitophagy is controlled either in conjunction with general macroautophagy or selectively through specific mitophagy genes. AMPK is activated in response to decreased intracellular ATP and then phosphorylates ULK1 and ULK2 (two Atg1 homologues) to activate both general macroautophagy and mitophagy [138]. AMPK- or ULK1-deficient hepatocytes exhibit accumulation of p62, ubiquitin aggregates and abnormal mitochondria [138]. The selectivity of mitophagy is controlled by a variety of proteins, including PINK1 [PTEN (phosphatase and tensin homologue deleted on chromosome 10)-induced kinase 1], parkin, BNIP (Bcl-2/adenovirus E18 19-kDa-interacting protein) and Atg32, as well as the processes of mitochondrial fission and fusion [168,182–185]. The yeast mitophagy-specific protein Atg32 localizes to the mitochondrial outer membrane, where it interacts with Atg11. Atg11 then interacts with Atg8 (LC3) and incorporates the mitochondria into the autophagosome [182,183]. Mitophagy regulation by Atg32 is controlled by the phosphorylation of Ser114 [186]. The mammalian sequence homologue of Atg32 has not been identified, but putative functional homologues include Nix protein, a gene disruption of which results in defects in clearance of mitochondria in erythrocyte maturation [187], and BNIP. Autophagy induction by BNIP in Bax/Bak double-knockout cells is independent of UCPs (uncoupling proteins) or the mtDNA/nuclear DNA ratio, and is protective against cell death. BNIP overexpression in Bax/Bak double-knockout cells increases mitochondrial protease activities, decreases mitochondrial complex III subunit 2, complex IV subunits I and IV, complex V α-subunit, complex II subunit 30 kDa and complex I subunit NDUFB8 [NADH dehydrogenase (ubiquinone) 1β subcomplex 8], but has no effects on MnSOD or Tom20 [188]. It is clear that mitophagy is highly regulated, although the exact mechanisms have not been defined. Figure 3 outlines the important events and the roles of some of the key proteins in mitophagy.
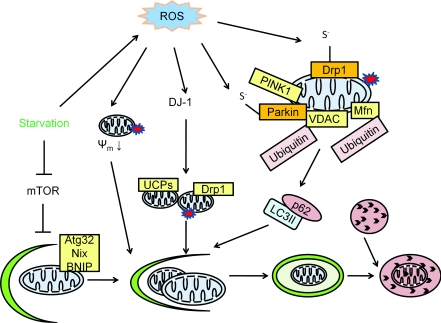
Figure 3. Mitophagy.
Mitophagy is the specific degradation of the mitochondria in response to global signals, including starvation and oxidative stress, or specific signals including mitochondrial targeting of signalling proteins or modification of mitochondrial proteins. In yeast, Atg32 is specific for mitophagy by targeting the mitochondria to the autophagosome. In mammalian cells, Nix is involved in mitochondrial clearance during erythrocyte maturation. In response to reduced cellular ATP, AMPK is activated and phosphorylates ULK1 and ULK2 (two Atg1 homologues) to activate both general macroautophagy and mitophagy. Parkinson's disease genes encoding α-synuclein, parkin, PINK1 and DJ-1 are all involved in mitophagy. A decrease in mitochondrial membrane potential (ψm) can be induced by ROS, and by targeting α-synuclein to the mitochondria. This decrease in mitochondrial membrane potential serves as a signal for mitophagy. In addition to a decrease in mitochondrial membrane potential, mitochondrial fission is another signal for mitophagy. PINK1 facilitates parkin targeting to the mitochondria, and ubiquitinates the mitochondrial outer membrane protein VDAC. Ubiquitinated VDAC can be recognized by p62 to initiate mitophagy. DJ-1 senses oxidative stress and serves a parallel pathway to maintain mitochondrial membrane potential and preserve mitochondria from fragmentation. Many of the regulators of mitophagy can be regulated by ROS. For example, α-synuclein is nitrated and, as a consequence, increases its aggregation propensity. Parkin can be sulfonated and S-nitrosated. Drp1 S-nitrosation is also involved in regulation of mitochondrial fission and associated induction of mitophagy.
Mitochondrial depolarization, fission and VDAC (voltage-dependent anion channel) ubiquitination are all candidates and are associated with mitophagy, but how these potential signals are integrated is not clear. In addition to genetic screening for mutants that fail to perform mitophagy, future efforts will need to clarify the molecular signals for mitophagy.
AUTOPHAGY IN REDOX SIGNALLING, OXIDATIVE AND NITRATIVE STRESS IN THE CONTEXT OF DISEASE PATHOGENESIS
One of the major questions in autophagy, redox signalling, oxidative and nitrative stress is whether autophagy regulation of ROS/RNS levels, intracellular location and effects play different roles in distinct cell types or tissues and in different chronic pathologies. Likewise, it is also unclear whether different cells and tissues have different redox signalling mechanisms for regulation of autophagy. Below, we discuss briefly what is known in the field of neurodegenerative and heart diseases, atherosclerosis and cancer.
Parkinson's disease
Parkinson's disease is a movement disorder manifested by neurodegeneration of specific areas of the brain, including the pronounced loss of substantia nigra pars compacta dopaminergic neurons, resulting in dopamine depletion [189]. Several lines of evidence have implicated the protein α-synuclein and its modification as having important roles in Parkinson's pathogenesis. Genetic mutations and gene amplifications of α-synuclein are the cause of a subset of familial Parkinson's disease [189a]. α-Synuclein is a major component of Lewy bodies, which accumulate in many brain regions of both familial and the majority of sporadic Parkinson's disease patients depending upon the stage of disease progression. How α-synuclein plays a role in Parkinson's disease pathogenesis is still under debate. Much evidence suggests an involvement of α-synuclein in autophagy, mitochondrial function and ROS-induced cellular damage. α-Synuclein can be modified by nitrative stress, which increases its propensity to aggregate [190–192]. α-Synuclein also targets to mitochondria [193–197], where it causes a decrease in complex I activity and/or mitochondrial damage [196,198]. This mitochondrial damage causes an increase in mitophagy, presumably as an attempt to clear damaged mitochondria [199]. Mutations of another autosomal dominant Parkinson's disease gene, LRRK2 (leucine-rich repeat kinase 2), has been shown to induce or inhibit autophagy depending on the cell type [200–202]. LRRK2 can increase α-synuclein levels [203] and interact with α-synuclein [204]. Furthermore, LRRK2 overexpression in transgenic mice carrying the A53T α-synuclein transgene exhibit accelerated neurodegeneration and α-synuclein accumulation [205]. LRRK2-knockout mice exhibit normal substantia nigra dopaminergic neuron histology, whereas kidneys from these mice exhibit cell loss, accumulation and aggregation of α-synuclein and ubiquitinated proteins, and accumulation of lipofuscin, LC3-II and p62 [206].
That a redox imbalance is a prominent feature in brains of individuals with Parkinson's disease is supported by a number of findings. Post-mortem studies have observed increased iron levels [207–210], an early and profound loss of the antioxidant GSH [210–212], increased lipid, protein and DNA oxidation and/or nitration, and increased SOD2 [213–216]. Lipid peroxidation products, such as HNE, accumulate in brains of individuals with Parkinson's disease and are known to promote autophagy [5–10,106,217]. Oxyradical-mediated DNA damage [specifically the production of 8-OHdG (8-hydroxydeoxyguanosine)] occurs to a greater extent in Parkinson's disease patients than in age-matched controls [218]. The activity of complex I, a major component of the mETC, is decreased in the substantia nigra [219,220] and frontal cortex [221] in patients with Parkinson's disease. Mitochondrial damage or deficiencies can contribute to cell death and neurodegeneration, as is evident in a variety of cell and animal models [222–228].
Brains of individuals with Parkinson's disease also accumulate autophagosome-like structures, indicating autophagic stress [229–231]. Supporting a link to pathology, it has been reported that recessively inherited forms of Parkinson's disease can be caused by loss-of-function mutations in genes encoding proteins that target to the mitochondria and mediate autophagy, including parkin, PINK1 and DJ-1 [232,233]. Parkin is an E3 ubiquitin ligase [168]. Some of the important parkin substrates include VDAC [234], mitofusin assembly regulatory factor [235,236], mitofusin 1 [237] and mitofusin 2 [237]. Parkin can be inhibited by S-nitrosation, which provides an important link to the generation of RNS prevalent in neurodegenerative disease [238]. MPP+ (1-methyl-4-phenylpyridinium ion) treatment of neuroblastoma SH-SY5Y cells induces parkin sulfonation, decreases parkin's E3 ligase activity and promotes subsequent self-aggregation [239]. Proteasome activities also seem to be involved in parkin-induced mitophagy [169]. p62 has been shown to bind VDAC and both mitofusin 1 and 2 on the mitochondria, where they are then ubiquitinated in a parkin-mediated process, and finally destined for mitophagic degradation [234,237]. Parkin-deficient fibroblasts exhibit altered mitochondrial morphology and activities [240]. Thus parkin may be both a target of RNS signalling and a modulator of mitochondrial ROS generation.
PINK1 is present in most mitochondrial membranes, but is usually degraded. When mitochondrial damage occurs, PINK1 is not degraded and it recruits and phosphorylates parkin, which initiates mitophagy [168]. PINK1-knockout mice exhibit impaired mitochondrial respiratory activities and increased sensitivity to oxidative stress [241]. PINK1-deficient dopaminergic neurons accumulate fragmented mitochondria and increased mitochondrial O2•− [242]. Down-regulation of PINK1 in neuroblastoma SH-SY5Y cells induced mitochondrial fragmentation and autophagy, which can be rescued by a dominant-negative Drp1 mutant [243]. PINK1/parkin-induced mitophagy may be mediated by VDAC and it is hypothesized that p62 plays an important role in the eventual clearance of the mitophagosome [234]. These observations indicate that PINK1 functions in both mitophagy and in regulating mitochondrial generation of ROS. Whether PINK1 is a target of redox modification is currently unknown.
DJ-1 also plays an important role in mitochondrial integrity. Mutant DJ-1 proteins that are associated with Parkinson's disease are stabilized by binding to parkin. This interaction is enhanced further by oxidative stress [170]. DJ-1-knockout animal models exhibit a variety of mitochondrial deficits. Knockout of DJ-1 in fruitflies reduced the RCR (respiratory control ratio) and mtDNA/nuclear DNA ratio, without changing membrane potential or complex I subunit NDUFS3 [NADH dehydrogenase (ubiquinone) Fe–S protein 3] level [244]. Mice with a DJ-1 knockout exhibit decreased skeletal muscle ATP [244], increased sensitivity to oxidative stress, increased H2O2, down-regulated UCP4 and UCP5 [245–247], and decreased mitochondrial length and fusion rate [248]. These changes are also associated with enhanced autophagic flux [248,249]. Although mitochondrial membrane potential appears to be preserved in knockout fruitflies [244], stable knockdown of DJ-1 in neuroblastoma cells led to a decrease in mitochondrial membrane potential, increase in mitochondrial fragmentation and accumulation of LC3 around mitochondria. Overexpression of DJ-1 protects neuronal cells against rotenone-induced mitochondrial fragmentation; this function is independent of PINK1, suggesting that both PINK1 and DJ-1 can independently regulate mitochondrial fragmentation [250]. Addition of glutathione or NAC, or overexpression of parkin and PINK1, attenuates DJ-1 deficiency-induced phenotypes [248–250]. This observation leads to the hypothesis that DJ-1 is regulated by oxidative stress, although the target of ROS is unknown [249]. Figure 3 provides a model summarizing the involvement of parkin, PINK1 and DJ-1 in mitophagy.
Although parkin, PINK1 and DJ-1 were initially identified as familial Parkinson's disease genes, their involvement in mitophagy extends beyond Parkinson's disease. Parkin overexpression attenuates accumulation of Aβ and deficient mitochondria in a 3×Tg Alzheimer's disease mouse model [251]. Also involved in mitophagy and being a potential parkin substrate, mitochondrial fission protein Drp1 is S-nitrosated in response also to amyloid β-peptide and is involved in mitochondrial fragmentation [252]. Mitochondrial dynamics play an important role in Alzheimer's disease. In post-mortem brains from individuals with Alzheimer's disease, changes in the fusion and fission protein levels by transcriptional regulation of Drp1, Fis1, Opa1, Mfn1 and Mfn2 have been observed [253,254]. Autophagy and mitophagy regulation are also involved in heart diseases and cancer. We discuss these topics further in the next section.
Cardiovascular disease
As in neurodegenerative diseases, autophagic stress occurs in hypertension and heart failure. Basal levels of autophagy are required for normal heart function. For example in Danon disease, where there is a deficiency of LAMP-2, a protein involved in autophagosome–lysosome fusion, there is cardioskeletal myopathy [255–257]. In disease models, it has been shown that autophagy can play both a protective and a detrimental response to these pathological conditions depending on specific experimental scenarios. Cardiac myocytes exhibit autophagy in response to coronary occlusion [258]. In TAC (transverse aortic constriction) models, Atg5 deficiency has been shown to exacerbate cell death [259], whereas Beclin deficiency protected against deterioration of cardiac function in response to TAC [258]. To interpret these observations properly, it is important to keep in mind that some of the autophagy inhibitors have off-target effects and some Atg genes can have activities not directly related to autophagy. For example, Beclin interacts with Bcl-2 and therefore may inhibit Bcl-2 anti-apoptotic function [260]. Thus decreases in Beclin can result in increased free Bcl-2 and therefore increase the anti-apoptotic effects of Bcl-2 and cause increases in cell viability.
Hypoxia in rabbit hearts triggers immediate accumulation of autophagosomes near swollen and fragmented mitochondria [261]. Although parkin was initially identified as a familial Parkinson's disease gene, it has also been shown to play a role in mitophagy in the heart. Parkin and p62 interaction plays a key role in pre-conditioning mitophagy in the heart [262]. Whether hypoxia increases or decreases ROS/RNS is currently under investigation [263]. Nonetheless, loss of MnSOD decreased mTOR signalling and increased the LC3-II/LC3-I ratio [88]. In response to ischaemia/reperfusion, rapamycin treatment or overexpression of Beclin, both induce autophagy and have been shown to play a protective role [264]. Wortmannin (an inhibitor of PI3K and autophagy), as well as Atg7 deficiency have been shown to exacerbate cell death [264]. HIF-1 (hypoxia-inducible factor 1) transcription regulation of antioxidant proteins is important for signalling from ROS/RNS to the autophagy pathway. Upon moderate hypoxia in mouse embryonic fibroblasts and a variety of human cancer cells (1–3% oxygen), HIF-1 activates the transcription of BNIP3 and BNIP3L (BNIP3-like) (Nix) and enhances cell survival [265]. BNIP3L is present at the outer surface of mitochondria, possesses a WXXL motif that binds to LC3 and its homologue GABARAP [266] and thus may be responsible for targeting mitochondria for autophagic clearance [267]. Whether BNIP-mediated mitophagy is protective or detrimental to cells may depend on cell type and age, because of the existence of contradictory findings that have not been resolved; for example, in an in vivo study, BNIP3 has been shown to promote ischaemia/reperfusion-induced cardiomyocyte cell death, and BNIP−/− mice exhibit decreased cell death [268].
Cancer
Autophagic activities participate in the tumorigenesis process at several levels. First, a lack of autophagic clearance of damaged proteins, organelles and DNA can increase mutagenesis and lead to tumorigenesis [269]. Secondly, tumour cells probably suffer from nutrient deprivation and hypoxia due to their highly proliferative nature which rapidly exhausts nutrient and oxygen supply in solid tumours. Autophagy up-regulation can provide tumour cells with several survival advantages. It has been shown that autophagy-deficient mice are more likely to develop tumours [143,260,270–272]. Furthermore, in response to cancer therapeutic agents, autophagy up-regulation may help to clear the drug-induced damage and result in cancer cells resistant to chemotherapy. Autophagy allows metabolites and nutrients to be recycled and prevents the cancer cells from accumulating dysfunctional mitochondria [273]. Autophagy inhibition in drug-resistant cancers has been used in conjunction with other cancer treatments and has caused tumour regression. Chloroquine potentiated Bcr/Abl tyrosine kinase inhibitor-induced cell death in chronic myeloid leukaemia primary cells and p210BCR/ABL-expressing myeloid precursor cells [274]. The PI3K inhibitor 3-MA (3-methyladenine) or LC3 siRNA resensitized breast cancer cells that gained resistance to the anti-HER2 antibody Tzb (trastuzumab) to growth inhibition by Tzb [275]. siRNA against Beclin-1, Atg3, Atg4, Atg5 or Atg12 augmented the effectiveness of irradiation in breast, lung and cervical squamous cell carcinomas [276]. In these studies, it is hypothesized that cancer cells which are autophagy-dependent are easier to treat, since removal of autophagy will remove the cell survival mechanisms.
Conversely, excessive autophagy can induce cell death and can also be used as a therapeutic strategy. Autophagy has been shown to induce cell senescence, which is able to stop cancer progression [277]. Therefore how autophagy–mitochondria function and ROS/RNS stress can be cross-regulated will continue to be an important topic for future investigation of mechanisms of tumorigenesis and cancer therapy.
FUTURE DIRECTIONS AND CONCLUDING REMARKS
Autophagy is an integral biological process, important in cellular and organismic health. Much progress has been made in understanding the molecules involved in this complex process, and how it is regulated. As the present review shows, a newer hypothesis which is gaining support is that autophagic activities are sensitive to the ROS/RNS levels, species, and intracellular and extracellular locations, although it is still unclear whether specific ROS/RNS elicit specific autophagic responses.
As we have discussed, defining these mechanisms is intimately related to the issue of how and why autophagy is protective or detrimental to cellular health. This question resonates with current controversies in the redox biology field. The concept of oxidative stress simply implied that ROS/RNS are toxic species because of their highly reactive nature. Similarly, it has been suggested that autophagy, or ‘self-eating’, is bad for the cell since it is an unhealthy cell which has multiple vesicles. However, depending on the particular situation, ‘self-cleansing’ can be good for the cell. Rapid reaction of ROS/RNS can elicit rapid responses to adapt to environmental situations. Resolving the issue of whether autophagy damages or protects the cells, especially in response to redox signals, will also be an area of focus for the field.
In summary, ROS/RNS formation is part of normal cellular physiology. Excessive or abnormal free radical production and accumulation result in oxidative stress which is a significant pathology in many diseases, including neurodegenerative diseases. Although the general inclination is to categorize autophagy as the activity that decreases ROS/RNS damage, experimental proof is still lacking regarding whether this is true for different species of ROS/RNS produced endogenously at different intracellular locations. Autophagy plays an important role in both sensing oxidative stress and removing oxidatively damaged proteins and organelles, as well as the cellular machineries responsible for excessive ROS/RNS production. Investigations into the specific molecular targets of ROS in the autophagy pathway and the specific signalling mechanisms will be important for our understanding of biology and diseases.
ACKNOWLEDGMENT
We thank Dr Victor Darley-Usmar for discussion and reading this review before submission.
FUNDING
This work was supported by the National Institutes of Health [grant number NIHR01-NS064090] and a Veterans Affairs merit award (to J.Z.).
References
- 1.de Duve C. The lysosome. Sci. Am. 1963;208:64–72. doi: 10.1038/scientificamerican0563-64. [DOI] [PubMed] [Google Scholar]
- 2.de Duve C., Wattiaux R. Functions of lysosomes. Annu. Rev. Physiol. 1966;28:435–492. doi: 10.1146/annurev.ph.28.030166.002251. [DOI] [PubMed] [Google Scholar]
- 3.Shacka J. J., Roth K. A., Zhang J. The autophagy–lysosomal degradation pathway: role in neurodegenerative disease and therapy. Front. Biosci. 2008;13:718–736. doi: 10.2741/2714. [DOI] [PubMed] [Google Scholar]
- 4.Schapira A. H., Gegg M. Mitochondrial contribution to Parkinson's disease pathogenesis. Parkinsons Dis. 2011;2011:159160. doi: 10.4061/2011/159160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crifo C., Capuozzo E., Siems W., Salerno C. Inhibition of ion transport ATPases by HNE. Biofactors. 2005;24:137–140. doi: 10.1002/biof.5520240116. [DOI] [PubMed] [Google Scholar]
- 6.Crifo C., Siems W., Soro S., Salerno C. Inhibition of defective adenylosuccinate lyase by HNE: a neurological disease that may be affected by oxidative stress. Biofactors. 2005;24:131–136. doi: 10.1002/biof.5520240115. [DOI] [PubMed] [Google Scholar]
- 7.Siems W., Grune T., Sommerburg O., Flohé L., Cadenas E. HNE and further lipid peroxidation products. Biofactors. 2005;24:1–4. doi: 10.1002/biof.5520240101. [DOI] [PubMed] [Google Scholar]
- 8.Castellani R. J., Perry G., Siedlak S. L., Nunomura A., Shimohama S., Zhang J., Montine T., Sayre L. M., Smith M. A. Hydroxynonenal adducts indicate a role for lipid peroxidation in neocortical and brainstem Lewy bodies in humans. Neurosci. Lett. 2002;319:25–28. doi: 10.1016/s0304-3940(01)02514-9. [DOI] [PubMed] [Google Scholar]
- 9.Sayre L. M., Zelasko D. A., Harris P. L., Perry G., Salomon R. G., Smith M. A. 4-Hydroxynonenal-derived advanced lipid peroxidation end products are increased in Alzheimer's disease. J. Neurochem. 1997;68:2092–2097. doi: 10.1046/j.1471-4159.1997.68052092.x. [DOI] [PubMed] [Google Scholar]
- 10.Dalfo E., Ferrer I. Early α-synuclein lipoxidation in neocortex in Lewy body diseases. Neurobiol. Aging. 2008;29:408–417. doi: 10.1016/j.neurobiolaging.2006.10.022. [DOI] [PubMed] [Google Scholar]
- 11.Pacher P., Beckman J. S., Liaudet L. Nitric oxide and peroxynitrite in health and disease. Physiol. Rev. 2007;87:315–424. doi: 10.1152/physrev.00029.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beckman J. S. Protein tyrosine nitration and peroxynitrite. FASEB J. 2002;16:1144. doi: 10.1096/fj.02-0133lte. [DOI] [PubMed] [Google Scholar]
- 13.Hill B. G., Dranka B. P., Bailey S. M., Lancaster J. R., Jr, Darley-Usmar V. M. What part of NO don't you understand? Some answers to the cardinal questions in nitric oxide biology. J. Biol. Chem. 2010;285:19699–19704. doi: 10.1074/jbc.R110.101618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malkus K. A., Tsika E., Ischiropoulos H. Oxidative modifications, mitochondrial dysfunction, and impaired protein degradation in Parkinson's disease: how neurons are lost in the Bermuda triangle. Mol. Neurodegener. 2009;4:24. doi: 10.1186/1750-1326-4-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith M. A., Richey Harris P. L., Sayre L. M., Beckman J. S., Perry G. Widespread peroxynitrite-mediated damage in Alzheimer's disease. J. Neurosci. 1997;17:2653–2657. doi: 10.1523/JNEUROSCI.17-08-02653.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murphy M. P. How mitochondria produce reactive oxygen species. Biochem. J. 2009;417:1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hashimoto M., Rockenstein E., Crews L., Masliah E. Role of protein aggregation in mitochondrial dysfunction and neurodegeneration in Alzheimer's and Parkinson's diseases. NeuroMol. Med. 2003;4:21–36. doi: 10.1385/NMM:4:1-2:21. [DOI] [PubMed] [Google Scholar]
- 18.Gottlieb R. A., Carreira R. S. Autophagy in health and disease. 5. Mitophagy as a way of life. Am. J. Physiol. Cell Physiol. 2010;299:C203–C210. doi: 10.1152/ajpcell.00097.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schneider L., Zhang J. Lysosomal function in macromolecular homeostasis and bioenergetics in Parkinson's disease. Mol. Neurodegener. 2010;5:14. doi: 10.1186/1750-1326-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He C., Klionsky D. J. Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Y., Klionsky D. J. The regulation of autophagy: unanswered questions. J. Cell Sci. 2011;124:161–170. doi: 10.1242/jcs.064576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mortimore G. E., Hutson N. J., Surmacz C. A. Quantitative correlation between proteolysis and macro- and microautophagy in mouse hepatocytes during starvation and refeeding. Proc. Natl. Acad. Sci. U.S.A. 1983;80:2179–2183. doi: 10.1073/pnas.80.8.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mortimore G. E., Lardeux B. R., Adams C. E. Regulation of microautophagy and basal protein turnover in rat liver: effects of short-term starvation. J. Biol. Chem. 1988;263:2506–2512. [PubMed] [Google Scholar]
- 24.Mijaljica D., Prescott M., Devenish R. J. Microautophagy in mammalian cells: revisiting a 40-year-old conundrum. Autophagy. 2011;7:673–682. doi: 10.4161/auto.7.7.14733. [DOI] [PubMed] [Google Scholar]
- 25.Sahu R., Kaushik S., Clement C. C., Cannizzo E. S., Scharf B., Follenzi A., Potolicchio I., Nieves E., Cuervo A. M., Santambrogio L. Microautophagy of cytosolic proteins by late endosomes. Dev. Cell. 2011;20:131–139. doi: 10.1016/j.devcel.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bejarano E., Cuervo A. M. Chaperone-mediated autophagy. Proc. Am. Thorac. Soc. 2010;7:29–39. doi: 10.1513/pats.200909-102JS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kundu M. ULK1, mammalian target of rapamycin, and mitochondria: linking nutrient availability and autophagy. Antioxid. Redox Signaling. 2011;14:1953–1958. doi: 10.1089/ars.2010.3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chan E. Y., Longatti A., McKnight N. C., Tooze S. A. Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domains using an Atg13-independent mechanism. Mol. Cell. Biol. 2009;29:157–171. doi: 10.1128/MCB.01082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weidberg H., Shvets E., Elazar Z. Biogenesis and cargo selectivity of autophagosomes. Annu. Rev. Biochem. 2011;80:125–156. doi: 10.1146/annurev-biochem-052709-094552. [DOI] [PubMed] [Google Scholar]
- 30.Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kabeya Y., Mizushima N., Yamamoto A., Oshitani-Okamoto S., Ohsumi Y., Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J. Cell Sci. 2004;117:2805–2812. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- 32.Tanida I., Ueno T., Kominami E. LC3 conjugation system in mammalian autophagy. Int. J. Biochem. Cell Biol. 2004;36:2503–2518. doi: 10.1016/j.biocel.2004.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cooper C. E., Patel R. P., Brookes P. S., Darley-Usmar V. M. Nanotransducers in cellular redox signaling: modification of thiols by reactive oxygen and nitrogen species. Trends Biochem. Sci. 2002;27:489–492. doi: 10.1016/s0968-0004(02)02191-6. [DOI] [PubMed] [Google Scholar]
- 34.Le Grand J. N., Chakrama F. Z., Seguin-Py S., Fraichard A., age-Mourroux R., Jouvenot M., Boyer-Guittaut M. GABARAPL1 (GEC1): original or copycat? Autophagy. 2011;7:1098–1107. doi: 10.4161/auto.7.10.15904. [DOI] [PubMed] [Google Scholar]
- 35.Behrends C., Sowa M. E., Gygi S. P., Harper J. W. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saftig P., Beertsen W., Eskelinen E. L. LAMP-2: a control step for phagosome and autophagosome maturation. Autophagy. 2008;4:510–512. doi: 10.4161/auto.5724. [DOI] [PubMed] [Google Scholar]
- 37.Jager S., Bucci C., Tanida I., Ueno T., Kominami E., Saftig P., Eskelinen E. L. Role for Rab7 in maturation of late autophagic vacuoles. J. Cell Sci. 2004;117:4837–4848. doi: 10.1242/jcs.01370. [DOI] [PubMed] [Google Scholar]
- 38.Gutierrez M. G., Munafo D. B., Beron W., Colombo M. I. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J. Cell Sci. 2004;117:2687–2697. doi: 10.1242/jcs.01114. [DOI] [PubMed] [Google Scholar]
- 39.Ganley I. G., Wong P. M., Gammoh N., Jiang X. Distinct autophagosomal–lysosomal fusion mechanism revealed by thapsigargin-induced autophagy arrest. Mol. Cell. 2011;42:731–743. doi: 10.1016/j.molcel.2011.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koike M., Shibata M., Waguri S., Yoshimura K., Tanida I., Kominami E., Gotow T., Peters C., von Figura K., Mizushima N., et al. Participation of autophagy in storage of lysosomes in neurons from mouse models of neuronal ceroid-lipofuscinoses (Batten disease) Am. J. Pathol. 2005;167:1713–1728. doi: 10.1016/S0002-9440(10)61253-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qiao L., Hamamichi S., Caldwell K. A., Caldwell G. A., Yacoubian T. A., Wilson S., Xie Z. L., Speake L. D., Parks R., Crabtree D., et al. Lysosomal enzyme cathepsin D protects against α-synuclein aggregation and toxicity. Mol. Brain. 2008;1:17. doi: 10.1186/1756-6606-1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cullen V., Lindfors M., Ng J., Paetau A., Swinton E., Kolodziej P., Boston H., Saftig P., Woulfe J., Feany M. B., et al. Cathepsin D expression level affects α-synuclein processing, aggregation, and toxicity in vivo. Mol. Brain. 2009;2:5. doi: 10.1186/1756-6606-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Giles N. M., Gutowski N. J., Giles G. I., Jacob C. Redox catalysts as sensitisers towards oxidative stress. FEBS Lett. 2003;535:179–182. doi: 10.1016/s0014-5793(02)03890-5. [DOI] [PubMed] [Google Scholar]
- 44.O'Neil J., Hoppe G., Sayre L. M., Hoff H. F. Inactivation of cathepsin B by oxidized LDL involves complex formation induced by binding of putative reactive sites exposed at low pH to thiols on the enzyme. Free Radical Biol. Med. 1997;23:215–225. doi: 10.1016/s0891-5849(96)00612-0. [DOI] [PubMed] [Google Scholar]
- 45.Yu L., McPhee C. K., Zheng L., Mardones G. A., Rong Y., Peng J., Mi N., Zhao Y., Liu Z., Wan F., et al. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature. 2010;465:942–946. doi: 10.1038/nature09076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sancak Y., Bar-Peled L., Zoncu R., Markhard A. L., Nada S., Sabatini D. M. Ragulator–Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dames S. A., Mulet J. M., Rathgeb-Szabo K., Hall M. N., Grzesiek S. The solution structure of the FATC domain of the protein kinase target of rapamycin suggests a role for redox-dependent structural and cellular stability. J. Biol. Chem. 2005;280:20558–20564. doi: 10.1074/jbc.M501116200. [DOI] [PubMed] [Google Scholar]
- 48.Copetti T., Bertoli C., Dalla E., Demarchi F., Schneider C. p65/RelA modulates BECN1 transcription and autophagy. Mol. Cell. Biol. 2009;29:2594–2608. doi: 10.1128/MCB.01396-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shi C. S., Kehrl J. H. TRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagy. Sci. Signaling. 2010;3:ra42. doi: 10.1126/scisignal.2000751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chang N. C., Nguyen M., Germain M., Shore G. C. Antagonism of Beclin 1-dependent autophagy by BCL-2 at the endoplasmic reticulum requires NAF-1. EMBO J. 2010;29:606–618. doi: 10.1038/emboj.2009.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhong Y., Wang Q. J., Li X., Yan Y., Backer J. M., Chait B. T., Heintz N., Yue Z. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1–phosphatidylinositol-3-kinase complex. Nat. Cell Biol. 2009;11:468–476. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Burd C. G., Emr S. D. Phosphatidylinositol(3)-phosphate signaling mediated by specific binding to RING FYVE domains. Mol. Cell. 1998;2:157–162. doi: 10.1016/s1097-2765(00)80125-2. [DOI] [PubMed] [Google Scholar]
- 53.Simonsen A., Birkeland H. C., Gillooly D. J., Mizushima N., Kuma A., Yoshimori T., Slagsvold T., Brech A., Stenmark H. Alfy, a novel FYVE-domain-containing protein associated with protein granules and autophagic membranes. J. Cell Sci. 2004;117:4239–4251. doi: 10.1242/jcs.01287. [DOI] [PubMed] [Google Scholar]
- 54.Chen W., Li N., Chen T., Han Y., Li C., Wang Y., He W., Zhang L., Wan T., Cao X. The lysosome-associated apoptosis-inducing protein containing the pleckstrin homology (PH) and FYVE domains (LAPF), representative of a novel family of PH and FYVE domain-containing proteins, induces caspase-independent apoptosis via the lysosomal–mitochondrial pathway. J. Biol. Chem. 2005;280:40985–40995. doi: 10.1074/jbc.M502190200. [DOI] [PubMed] [Google Scholar]
- 55.Matsunaga K., Morita E., Saitoh T., Akira S., Ktistakis N. T., Izumi T., Noda T., Yoshimori T. Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via Atg14L. J. Cell Biol. 2010;190:511–521. doi: 10.1083/jcb.200911141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Proikas-Cezanne T., Waddell S., Gaugel A., Frickey T., Lupas A., Nordheim A. WIPI-1α (WIPI49), a member of the novel 7-bladed WIPI protein family, is aberrantly expressed in human cancer and is linked to starvation-induced autophagy. Oncogene. 2004;23:9314–9325. doi: 10.1038/sj.onc.1208331. [DOI] [PubMed] [Google Scholar]
- 57.Polson H. E., de Lartique J., Rigden D. J., Reedijk M., Urbé S., Clague M. J., Tooze S. A. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy. 2010;6:506–522. doi: 10.4161/auto.6.4.11863. [DOI] [PubMed] [Google Scholar]
- 58.Zhao M., Klionsky D. J. AMPK-dependent phosphorylation of ULK1 induces autophagy. Cell Metab. 2011;13:119–120. doi: 10.1016/j.cmet.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Harrison B., Kraus M., Burch L., Stevens C., Craig A., Gordon-Weeks P., Hupp T. R. DAPK-1 binding to a linear peptide motif in MAP1B stimulates autophagy and membrane blebbing. J. Biol. Chem. 2008;283:9999–10014. doi: 10.1074/jbc.M706040200. [DOI] [PubMed] [Google Scholar]
- 60.Sarkar S., Rubinsztein D. C. Inositol and IP3 levels regulate autophagy: biology and therapeutic speculations. Autophagy. 2006;2:132–134. doi: 10.4161/auto.2387. [DOI] [PubMed] [Google Scholar]
- 61.Vadlamudi R. K., Joung I., Strominger J. L., Shin J. p62, a phosphotyrosine-independent ligand of the SH2 domain of p56lck, belongs to a new class of ubiquitin-binding proteins. J. Biol. Chem. 1996;271:20235–20237. doi: 10.1074/jbc.271.34.20235. [DOI] [PubMed] [Google Scholar]
- 62.Bjørkøy G., Lamark T., Brech A., Outzen H., Perander M., Overvatn A., Stenmark H., Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kirkin V., Lamark T., Sou Y. S., Bjørkøy G., Nunn J. L., Bruun J. A., Shvets E., McEwan D. G., Clausen T. H., Wild P., et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell. 2009;33:505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 64.Komatsu M., Waguri S., Koike M., Sou Y. S., Ueno T., Hara T., Mizushima N., Iwata J. I., Ezaki J., Murata S., et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 65.Klionsky D. J., Abeliovich H., Agostinis P., Agrawal D. K., Aliev G., Askew D. S., Baba M., Baehrecke E. H., Bahr B. A., Ballabio A., et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kimura S., Fujita N., Noda T., Yoshimori T. Monitoring autophagy in mammalian cultured cells through the dynamics of LC3. Methods Enzymol. 2009;452:1–12. doi: 10.1016/S0076-6879(08)03601-X. [DOI] [PubMed] [Google Scholar]
- 67.Ni H. M., Bockus A., Wozniak A. L., Jones K., Weinman S., Yin X. M., Ding W. X. Dissecting the dynamic turnover of GFP–LC3 in the autolysosome. Autophagy. 2011;7:188–204. doi: 10.4161/auto.7.2.14181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mizushima N. Methods for monitoring autophagy using GFP-LC3 transgenic mice. Methods Enzymol. 2009;452:13–23. doi: 10.1016/S0076-6879(08)03602-1. [DOI] [PubMed] [Google Scholar]
- 69.Kuma A., Matsui M., Mizushima N. LC3, an autophagosome marker, can be incorporated into protein aggregates independent of autophagy: caution in the interpretation of LC3 localization. Autophagy. 2007;3:323–328. doi: 10.4161/auto.4012. [DOI] [PubMed] [Google Scholar]
- 70.Korkhov V. M. GFP–LC3 labels organised smooth endoplasmic reticulum membranes independently of autophagy. J. Cell. Biochem. 2009;107:86–95. doi: 10.1002/jcb.22103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ciechomska I. A., Tolkovsky A. M. Non-autophagic GFP–LC3 puncta induced by saponin and other detergents. Autophagy. 2007;3:586–590. doi: 10.4161/auto.4843. [DOI] [PubMed] [Google Scholar]
- 72.Kimura S., Noda T., Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 73.Hariharan N., Zhai P., Sadoshima J. Oxidative stress stimulates autophagic flux during ischemia/reperfusion. Antioxid. Redox Signaling. 2011;14:2179–2190. doi: 10.1089/ars.2010.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Waypa G. B., Schumacker P. T. Hypoxia-induced changes in pulmonary and systemic vascular resistance: where is the O2 sensor? Respir. Physiol. Neurobiol. 2010;174:201–211. doi: 10.1016/j.resp.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Murphy M. P. Induction of mitochondrial ROS production by electrophilic lipids: a new pathway of redox signaling? Am. J. Physiol. Heart Circ. Physiol. 2006;290:H1754–H1755. doi: 10.1152/ajpheart.00040.2006. [DOI] [PubMed] [Google Scholar]
- 76.Beckman J. S., Beckman T. W., Chen J., Marshall P. A., Freeman B. A. Apparent hydroxyl radical production by peroxynitrite: implications for endothelial injury from nitric oxide and superoxide. Proc. Natl. Acad. Sci. U.S.A. 1990;87:1620–1624. doi: 10.1073/pnas.87.4.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lambeth J. D. Nox enzymes, ROS, and chronic disease: an example of antagonistic pleiotropy. Free Radical Biol. Med. 2007;43:332–347. doi: 10.1016/j.freeradbiomed.2007.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schrader M., Fahimi H. D. Peroxisomes and oxidative stress. Biochim. Biophys. Acta. 2006;1763:1755–1766. doi: 10.1016/j.bbamcr.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 79.Hanukoglu I. Antioxidant protective mechanisms against reactive oxygen species (ROS) generated by mitochondrial P450 systems in steroidogenic cells. Drug Metab. Rev. 2006;38:171–196. doi: 10.1080/03602530600570040. [DOI] [PubMed] [Google Scholar]
- 80.Scherz-Shouval R., Shvets E., Fass E., Shorer H., Gil L., Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007;26:1749–1760. doi: 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bensaad K., Cheung E. C., Vousden K. H. Modulation of intracellular ROS levels by TIGAR controls autophagy. EMBO J. 2009;28:3015–3026. doi: 10.1038/emboj.2009.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen Y., Azad M. B., Gibson S. B. Superoxide is the major reactive oxygen species regulating autophagy. Cell Death Differ. 2009;16:1040–1052. doi: 10.1038/cdd.2009.49. [DOI] [PubMed] [Google Scholar]
- 83.Forstermann U. Janus-faced role of endothelial NO synthase in vascular disease: uncoupling of oxygen reduction from NO synthesis and its pharmacological reversal. Biol. Chem. 2006;387:1521–1533. doi: 10.1515/BC.2006.190. [DOI] [PubMed] [Google Scholar]
- 84.Velayutham M., Hemann C., Zweier J. L. Removal of HO and generation of superoxide radical: role of cytochrome c and NADH. Free Radical Biol. Med. 2011;51:160–170. doi: 10.1016/j.freeradbiomed.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ballinger S. W., Patterson C., Yan C. N., Doan R., Burow D. L., Young C. G., Yakes F. M., Van Houten B., Ballinger C. A., Freeman B. A., Runge M. S. Hydrogen peroxide- and peroxynitrite-induced mitochondrial DNA damage and dysfunction in vascular endothelial and smooth muscle cells. Circ. Res. 2000;86:960–966. doi: 10.1161/01.res.86.9.960. [DOI] [PubMed] [Google Scholar]
- 86.Scherz-Shouval R., Elazar Z. Regulation of autophagy by ROS: physiology and pathology. Trends Biochem. Sci. 2011;36:30–38. doi: 10.1016/j.tibs.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 87.Williams M. D., Van Remmen H., Conrad C. C., Huang T. T., Epstein C. J., Richardson A. Increased oxidative damage is correlated to altered mitochondrial function in heterozygous manganese superoxide dismutase knockout mice. J. Biol. Chem. 1998;273:28510–28515. doi: 10.1074/jbc.273.43.28510. [DOI] [PubMed] [Google Scholar]
- 88.Zhang Y., Zhang H. M., Shi Y., Lustgarten M., Li Y., Qi W., Zhang B. X., Van Remmen H. Loss of manganese superoxide dismutase leads to abnormal growth and signal transduction in mouse embryonic fibroblasts. Free Radical Biol. Med. 2010;49:1255–1262. doi: 10.1016/j.freeradbiomed.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Larosche I., Letteron P., Berson A., Fromenty B., Huang T. T., Moreau R., Pessayre D., Mansouri A. Hepatic mitochondrial DNA depletion after an alcohol binge in mice: probable role of peroxynitrite and modulation by manganese superoxide dismutase. J. Pharmacol. Exp. Ther. 2010;332:886–897. doi: 10.1124/jpet.109.160879. [DOI] [PubMed] [Google Scholar]
- 90.Dumont M., Wille E., Stack C., Calingasan N. Y., Beal M. F., Lin M. T. Reduction of oxidative stress, amyloid deposition, and memory deficit by manganese superoxide dismutase overexpression in a transgenic mouse model of Alzheimer's disease. FASEB J. 2009;23:2459–2466. doi: 10.1096/fj.09-132928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Beckman J. S., Carson M., Smith C. D., Koppenol W. H. ALS, SOD and peroxynitrite. Nature. 1993;364:584. doi: 10.1038/364584a0. [DOI] [PubMed] [Google Scholar]
- 92.Deng H. X., Hentati A., Tainer J. A., Iqbal Z., Cayabyab A., Hung W. Y., Getzoff E. D., Hu P., Herzfeldt B., Roos R. P., et al. Amyotrophic lateral sclerosis and structural defects in Cu,Zn superoxide dismutase. Science. 1993;261:1047–1051. doi: 10.1126/science.8351519. [DOI] [PubMed] [Google Scholar]
- 93.Rosen D. R., Siddique T., Patterson D., Figlewicz D. A., Sapp P., Hentati A., Donaldson D., Goto J., O'Regan J. P., Deng H. X., et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 94.Beckman J. S., Chen J., Crow J. P., Ye Y. Z. Reactions of nitric oxide, superoxide and peroxynitrite with superoxide dismutase in neurodegeneration. Prog. Brain Res. 1994;103:371–380. doi: 10.1016/s0079-6123(08)61151-6. [DOI] [PubMed] [Google Scholar]
- 95.Gurney M. E., Pu H., Chiu A. Y., Dal Canto M. C., Polchow C. Y., Alexander D. D., Caliendo J., Hentati A., Kwon Y. W., Deng H. X., et al. Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science. 1994;264:1772–1775. doi: 10.1126/science.8209258. [DOI] [PubMed] [Google Scholar]
- 96.Sasaki S. Autophagy in spinal cord motor neurons in sporadic amyotrophic lateral sclerosis. J. Neuropathol. Exp. Neurol. 2011;70:349–359. doi: 10.1097/NEN.0b013e3182160690. [DOI] [PubMed] [Google Scholar]
- 97.Zhang X., Li L., Chen S., Yang D., Wang Y., Zhang X., Wang Z., Le W. Rapamycin treatment augments motor neuron degeneration in SOD1(G93A) mouse model of amyotrophic lateral sclerosis. Autophagy. 2011;7:412–425. doi: 10.4161/auto.7.4.14541. [DOI] [PubMed] [Google Scholar]
- 98.Kissova I., Deffieu M., Samokhvalov V., Velours G., Bessoule J. J., Manon S., Camougrand N. Lipid oxidation and autophagy in yeast. Free Radical Biol. Med. 2006;41:1655–1661. doi: 10.1016/j.freeradbiomed.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 99.Huang J., Brumell J. H. NADPH oxidases contribute to autophagy regulation. Autophagy. 2009;5:887–889. doi: 10.4161/auto.9125. [DOI] [PubMed] [Google Scholar]
- 100.Djavaheri-Mergny M., Amelotti M., Mathieu J., Besancon F., Bauvy C., Souquere S., Pierron G., Codogno P. NF-κB activation represses tumor necrosis factor-α-induced autophagy. J. Biol. Chem. 2006;281:30373–30382. doi: 10.1074/jbc.M602097200. [DOI] [PubMed] [Google Scholar]
- 101.Kirkland R. A., Saavedra G. M., Franklin J. L. Rapid activation of antioxidant defenses by nerve growth factor suppresses reactive oxygen species during neuronal apoptosis: evidence for a role in cytochrome c redistribution. J. Neurosci. 2007;27:11315–11326. doi: 10.1523/JNEUROSCI.3590-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kirkland R. A., Adibhatla R. M., Hatcher J. F., Franklin J. L. Loss of cardiolipin and mitochondria during programmed neuronal death: evidence of a role for lipid peroxidation and autophagy. Neuroscience. 2002;115:587–602. doi: 10.1016/s0306-4522(02)00512-2. [DOI] [PubMed] [Google Scholar]
- 103.Kirkland R. A., Franklin J. L. Evidence for redox regulation of cytochrome c release during programmed neuronal death: antioxidant effects of protein synthesis and caspase inhibition. J. Neurosci. 2001;21:1949–1963. doi: 10.1523/JNEUROSCI.21-06-01949.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Djavaheri-Mergny M., Javelaud D., Wietzerbin J., Besancon F. NF-κB activation prevents apoptotic oxidative stress via an increase of both thioredoxin and MnSOD levels in TNFα-treated Ewing sarcoma cells. FEBS Lett. 2004;578:111–115. doi: 10.1016/j.febslet.2004.10.082. [DOI] [PubMed] [Google Scholar]
- 105.Levonen A. L., Landar A., Ramachandran A., Ceaser E. K., Dickinson D. A., Zanoni G., Morrow J. D., Darley-Usmar V. M. Cellular mechanisms of redox cell signalling: role of cysteine modification in controlling antioxidant defences in response to electrophilic lipid oxidation products. Biochem. J. 2004;378:373–382. doi: 10.1042/BJ20031049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hill B. G., Haberzettl P., Ahmed Y., Srivastava S., Bhatnagar A. Unsaturated lipid peroxidation-derived aldehydes activate autophagy in vascular smooth-muscle cells. Biochem. J. 2008;410:525–534. doi: 10.1042/BJ20071063. [DOI] [PubMed] [Google Scholar]
- 107.Janjetovic K., Misirkic M., Vucicevic L., Harhaji L., Trajkovic V. Synergistic antiglioma action of hyperthermia and nitric oxide. Eur. J. Pharmacol. 2008;583:1–10. doi: 10.1016/j.ejphar.2007.12.028. [DOI] [PubMed] [Google Scholar]
- 108.Barsoum M. J., Yuan H., Gerencser A. A., Liot G., Kushnareva Y., Graber S., Kovacs I., Lee W. D., Waggoner J., Cui J., et al. Nitric oxide-induced mitochondrial fission is regulated by dynamin-related GTPases in neurons. EMBO J. 2006;25:3900–3911. doi: 10.1038/sj.emboj.7601253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tyagi N., Vacek J. C., Givvimani S., Sen U., Tyagi S. C. Cardiac specific deletion of N-methyl-D-aspartate receptor 1 ameliorates mtMMP-9 mediated autophagy/mitophagy in hyperhomocysteinemia. J. Recept. Signal Transduction Res. 2010;30:78–87. doi: 10.3109/10799891003614808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sarkar S., Korolchuk V. I., Renna M., Imarisio S., Fleming A., Williams A., Garcia-Arencibia M., Rose C., Luo S., Underwood B. R., et al. Complex inhibitory effects of nitric oxide on autophagy. Mol. Cell. 2011;43:19–32. doi: 10.1016/j.molcel.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Itoh K., Chiba T., Takahashi S., Ishii T., Igarashi K., Katoh Y., Oyake T., Hayashi N., Satoh K., Hatayama I., et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 112.Friling R. S., Bensimon A., Tichauer Y., Daniel V. Xenobiotic-inducible expression of murine glutathione S-transferase Ya subunit gene is controlled by an electrophile-responsive element. Proc. Natl. Acad. Sci. U.S.A. 1990;87:6258–6262. doi: 10.1073/pnas.87.16.6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rushmore T. H., Pickett C. B. Transcriptional regulation of the rat glutathione S-transferase Ya subunit gene: characterization of a xenobiotic-responsive element controlling inducible expression by phenolic antioxidants. J. Biol. Chem. 1990;265:14648–14653. [PubMed] [Google Scholar]
- 114.Malhotra D., Portales-Casamar E., Singh A., Srivastava S., Arenillas D., Happel C., Shyr C., Wakabayashi N., Kensler T. W., Wasserman W. W., Biswal S. Global mapping of binding sites for Nrf2 identifies novel targets in cell survival response through ChIP-Seq profiling and network analysis. Nucleic Acids Res. 2010;38:5718–5734. doi: 10.1093/nar/gkq212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wasserman W. W., Fahl W. E. Functional antioxidant responsive elements. Proc. Natl. Acad. Sci. U.S.A. 1997;94:5361–5366. doi: 10.1073/pnas.94.10.5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ishii T., Itoh K., Yamamoto M. Roles of Nrf2 in activation of antioxidant enzyme genes via antioxidant responsive elements. Methods Enzymol. 2002;348:182–190. doi: 10.1016/s0076-6879(02)48637-5. [DOI] [PubMed] [Google Scholar]
- 117.Itoh K., Wakabayashi N., Katoh Y., Ishii T., Igarashi K., Engel J. D., Yamamoto M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Talalay P., De Long M. J., Prochaska H. J. Identification of a common chemical signal regulating the induction of enzymes that protect against chemical carcinogenesis. Proc. Natl. Acad. Sci. U.S.A. 1988;85:8261–8265. doi: 10.1073/pnas.85.21.8261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Riley B. E., Kaiser S. E., Shaler T. A., Ng A. C., Hara T., Hipp M. S., Lage K., Xavier R. J., Ryu K. Y., Taguchi K., et al. Ubiquitin accumulation in autophagy-deficient mice is dependent on the Nrf2-mediated stress response pathway: a potential role for protein aggregation in autophagic substrate selection. J. Cell Biol. 2010;191:537–552. doi: 10.1083/jcb.201005012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Fujita K., Maeda D., Xiao Q., Srinivasula S. M. Nrf2-mediated induction of p62 controls Toll-like receptor-4-driven aggresome-like induced structure formation and autophagic degradation. Proc. Natl. Acad. Sci. U.S.A. 2011;108:1427–1432. doi: 10.1073/pnas.1014156108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chen K., Albano A., Ho A., Keaney J. F., Jr Activation of p53 by oxidative stress involves platelet-derived growth factor-β receptor-mediated ataxia telangiectasia mutated (ATM) kinase activation. J. Biol. Chem. 2003;278:39527–39533. doi: 10.1074/jbc.M304423200. [DOI] [PubMed] [Google Scholar]
- 122.Martindale J. L., Holbrook N. J. Cellular response to oxidative stress: signaling for suicide and survival. J. Cell. Physiol. 2002;192:1–15. doi: 10.1002/jcp.10119. [DOI] [PubMed] [Google Scholar]
- 123.Johnson T. M., Yu Z. X., Ferrans V. J., Lowenstein R. A., Finkel T. Reactive oxygen species are downstream mediators of p53-dependent apoptosis. Proc. Natl. Acad. Sci. U.S.A. 1996;93:11848–11852. doi: 10.1073/pnas.93.21.11848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Li P. F., Dietz R., von Harsdorf R. p53 regulates mitochondrial membrane potential through reactive oxygen species and induces cytochrome c-independent apoptosis blocked by Bcl-2. EMBO J. 1999;18:6027–6036. doi: 10.1093/emboj/18.21.6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Polyak K., Waldman T., He T. C., Kinzler K. W., Vogelstein B. Genetic determinants of p53-induced apoptosis and growth arrest. Genes Dev. 1996;10:1945–1952. doi: 10.1101/gad.10.15.1945. [DOI] [PubMed] [Google Scholar]
- 126.Bensaad K., Tsuruta A., Selak M. A., Vidal M. N., Nakano K., Bartrons R., Gottlieb E., Vousden K. H. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 127.Crighton D., Wilkinson S., O'Prey J., Syed N., Smith P., Harrison P. R., Gasco M., Garrone O., Crook T., Ryan K. M. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–134. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 128.Budanov A. V., Karin M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell. 2008;134:451–460. doi: 10.1016/j.cell.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Budanov A. V., Sablina A. A., Feinstein E., Koonin E. V., Chumakov P. M. Regeneration of peroxiredoxins by p53-regulated sestrins, homologs of bacterial AhpD. Science. 2004;304:596–600. doi: 10.1126/science.1095569. [DOI] [PubMed] [Google Scholar]
- 130.Woo H. A., Bae S. H., Park S., Rhee S. G. Sestrin 2 is not a reductase for cysteine sulfinic acid of peroxiredoxins. Antioxid. Redox Signaling. 2009;11:739–745. doi: 10.1089/ars.2008.2360. [DOI] [PubMed] [Google Scholar]
- 131.Rouschop K. M., Ramaekers C. H., Schaaf M. B., Keulers T. G., Savelkouls K. G., Lambin P., Koritzinsky M., Wouters B. G. Autophagy is required during cycling hypoxia to lower production of reactive oxygen species. Radiother. Oncol. 2009;92:411–416. doi: 10.1016/j.radonc.2009.06.029. [DOI] [PubMed] [Google Scholar]
- 132.Park B. C., Park S. H., Paek S. H., Park S. Y., Kwak M. K., Choi H. G., Yong C. S., Yoo B. K., Kim J. A. Chloroquine-induced nitric oxide increase and cell death is dependent on cellular GSH depletion in A172 human glioblastoma cells. Toxicol. Lett. 2008;178:52–60. doi: 10.1016/j.toxlet.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 133.Farombi E. O. Genotoxicity of chloroquine in rat liver cells: protective role of free radical scavengers. Cell Biol. Toxicol. 2006;22:159–167. doi: 10.1007/s10565-006-0173-2. [DOI] [PubMed] [Google Scholar]
- 134.Park J., Choi K., Jeong E., Kwon D., Benveniste E. N., Choi C. Reactive oxygen species mediate chloroquine-induced expression of chemokines by human astroglial cells. Glia. 2004;47:9–20. doi: 10.1002/glia.20017. [DOI] [PubMed] [Google Scholar]
- 135.Yamasaki R., Zhang J., Koshiishi I., Sastradipura Suniarti D. F., Wu Z., Peters C., Schwake M., Uchiyama Y., Kira J., Saftig P., et al. Involvement of lysosomal storage-induced p38 MAP kinase activation in the overproduction of nitric oxide by microglia in cathepsin D-deficient mice. Mol. Cell. Neurosci. 2007;35:573–584. doi: 10.1016/j.mcn.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 136.Nakanishi H., Zhang J., Koike M., Nishioku T., Okamoto Y., Kominami E., von Figura K., Peters C., Yamamoto K., Saftig P., Uchiyama Y. Involvement of nitric oxide released from microglia–macrophages in pathological changes of cathepsin D-deficient mice. J. Neurosci. 2001;21:7526–7533. doi: 10.1523/JNEUROSCI.21-19-07526.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Jolly R. D., Brown S., Das A. M., Walkley S. U. Mitochondrial dysfunction in the neuronal ceroid-lipofuscinoses (Batten disease) Neurochem. Int. 2002;40:565–571. doi: 10.1016/s0197-0186(01)00128-0. [DOI] [PubMed] [Google Scholar]
- 138.Egan D. F., Shackelford D. B., Mihaylova M. M., Gelino S., Kohnz R. A., Mair W., Vasquez D. S., Joshi A., Gwinn D. M., Taylor R., et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Liu F., Lee J. Y., Wei H., Tanabe O., Engel J. D., Morrison S. J., Guan J. L. FIP200 is required for the cell-autonomous maintenance of fetal hematopoietic stem cells. Blood. 2010;116:4806–4814. doi: 10.1182/blood-2010-06-288589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hara T., Nakamura K., Matsui M., Yamamoto A., Nakahara Y., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H., Mizushima N. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 141.Komatsu M., Waguri S., Chiba T., Murata S., Iwata J. I., Tanida I., Ueno T., Koike M., Uchiyama Y., Kominami E., Tanaka K. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 142.Wu J. J., Quijano C., Chen E., Liu H., Cao L., Fergusson M. M., Rovira I. I., Gutkind S., Daniels M. P., Komatsu M., Finkel T. Mitochondrial dysfunction and oxidative stress mediate the physiological impairment induced by the disruption of autophagy. Aging. 2009;1:425–437. doi: 10.18632/aging.100038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Takamura A., Komatsu M., Hara T., Sakamoto A., Kishi C., Waguri S., Eishi Y., Hino O., Tanaka K., Mizushima N. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25:795–800. doi: 10.1101/gad.2016211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Varshavsky A. The ubiquitin system. Trends Biochem. Sci. 1997;22:383–387. doi: 10.1016/s0968-0004(97)01122-5. [DOI] [PubMed] [Google Scholar]
- 145.Deveraux Q., Ustrell V., Pickart C., Rechsteiner M. A 26 S protease subunit that binds ubiquitin conjugates. J. Biol. Chem. 1994;269:7059–7061. [PubMed] [Google Scholar]
- 146.Grune T., Reinheckel T., Davies K. J. Degradation of oxidized proteins in mammalian cells. FASEB J. 1997;11:526–534. [PubMed] [Google Scholar]
- 147.Shringarpure R., Grune T., Davies K. J. Protein oxidation and 20S proteasome-dependent proteolysis in mammalian cells. Cell. Mol. Life Sci. 2001;58:1442–1450. doi: 10.1007/PL00000787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Grune T., Davies K. J. The proteasomal system and HNE-modified proteins. Mol. Aspects Med. 2003;24:195–204. doi: 10.1016/s0098-2997(03)00014-1. [DOI] [PubMed] [Google Scholar]
- 149.Grune T., Merker K., Sandig G., Davies K. J. Selective degradation of oxidatively modified protein substrates by the proteasome. Biochem. Biophys. Res. Commun. 2003;305:709–718. doi: 10.1016/s0006-291x(03)00809-x. [DOI] [PubMed] [Google Scholar]
- 150.Jung T., Grune T. The proteasome and its role in the degradation of oxidized proteins. IUBMB Life. 2008;60:743–752. doi: 10.1002/iub.114. [DOI] [PubMed] [Google Scholar]
- 151.Pickering A. M., Koop A. L., Teoh C. Y., Ermak G., Grune T., Davies K. J. The immunoproteasome, the 20S proteasome and the PA28αβ proteasome regulator are oxidative-stress-adaptive proteolytic complexes. Biochem. J. 2010;432:585–594. doi: 10.1042/BJ20100878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Grune T., Catalgol B., Licht A., Ermak G., Pickering A., Ngo J. K., Davies K. J. HSP70 mediates dissociation and reassociation of the 26S proteasome during adaptation to oxidative stress. Free Radical Biol. Med. 2011;51:1355–1364. doi: 10.1016/j.freeradbiomed.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zubenko G. S., Jones E. W. Protein degradation, meiosis and sporulation in proteinase-deficient mutants of Saccharomyces cerevisiae. Genetics. 1981;97:45–64. doi: 10.1093/genetics/97.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Marques M., Mojzita D., Amorim M. A., Almeida T., Hohmann S., Moradas-Ferreira P., Costa V. The Pep4p vacuolar proteinase contributes to the turnover of oxidized proteins but PEP4 overexpression is not sufficient to increase chronological lifespan in Saccharomyces cerevisiae. Microbiology. 2006;152:3595–3605. doi: 10.1099/mic.0.29040-0. [DOI] [PubMed] [Google Scholar]
- 155.Pankiv S., Clausen T. H., Lamark T., Brech A., Bruun J. A., Outzen H., Overvatn A., Bjørkøy G., Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 156.Kirkin V., Lamark T., Johansen T., Dikic I. NBR1 cooperates with p62 in selective autophagy of ubiquitinated targets. Autophagy. 2009;5:732–733. doi: 10.4161/auto.5.5.8566. [DOI] [PubMed] [Google Scholar]
- 157.Thurston T. L., Ryzhakov G., Bloor S., von Muhlinen N., Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat. Immunol. 2009;10:1215–1221. doi: 10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- 158.Clausen T. H., Lamark T., Isakson P., Finley K., Larsen K. B., Brech A., Overvatn A., Stenmark H., Bjørkøy G., Simonsen A., Johansen T. p62/SQSTM1 and ALFY interact to facilitate the formation of p62 bodies/ALIS and their degradation by autophagy. Autophagy. 2010;6:330–344. doi: 10.4161/auto.6.3.11226. [DOI] [PubMed] [Google Scholar]
- 159.Filimonenko M., Isakson P., Finley K. D., Anderson M., Jeong H., Melia T. J., Bartlett B. J., Myers K. M., Birkeland H. C., Lamark T., et al. The selective macroautophagic degradation of aggregated proteins requires the PI3P-binding protein Alfy. Mol. Cell. 2010;38:265–279. doi: 10.1016/j.molcel.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Dengjel J., Kristensen A. R., Andersen J. S. Ordered bulk degradation via autophagy. Autophagy. 2008;4:1057–1059. doi: 10.4161/auto.6824. [DOI] [PubMed] [Google Scholar]
- 161.Pandey U. B., Nie Z., Batlevi Y., McCray B. A., Ritson G. P., Nedelsky N. B., Schwartz S. L., DiProspero N. A., Knight M. A., Schuldiner O., et al. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447:859–863. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- 162.Rubinsztein D. C. Autophagy induction rescues toxicity mediated by proteasome inhibition. Neuron. 2007;54:854–856. doi: 10.1016/j.neuron.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 163.Korolchuk V. I., Menzies F. M., Rubinsztein D. C. A novel link between autophagy and the ubiquitin–proteasome system. Autophagy. 2009;5:862–863. doi: 10.4161/auto.8840. [DOI] [PubMed] [Google Scholar]
- 164.Gao Z., Gammoh N., Wong P. M., Erdjument-Bromage H., Tempst P., Jiang X. Processing of autophagic protein LC3 by the 20S proteasome. Autophagy. 2010;6:126–137. doi: 10.4161/auto.6.1.10928. [DOI] [PubMed] [Google Scholar]
- 165.Zhao J., Brault J. J., Schild A., Cao P., Sandri M., Schiaffino S., Lecker S. H., Goldberg A. L. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6:472–483. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 166.Jegga A. G., Schneider L., Ouyang X., Zhang J. Systems biology of the autophagy–lysosomal pathway. Autophagy. 2011;7:477–489. doi: 10.4161/auto.7.5.14811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Olzmann J. A., Li L., Chudaev M. V., Chen J., Perez F. A., Palmiter R. D., Chin L. S. Parkin-mediated K63-linked polyubiquitination targets misfolded DJ-1 to aggresomes via binding to HDAC6. J. Cell Biol. 2007;178:1025–1038. doi: 10.1083/jcb.200611128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Narendra D., Tanaka A., Suen D. F., Youle R. J. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Tanaka A., Cleland M. M., Xu S., Narendra D. P., Suen D. F., Karbowski M., Youle R. J. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J. Cell Biol. 2010;191:1367–1380. doi: 10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Moore D. J., Zhang L., Troncoso J., Lee M. K., Hattori N., Mizuno Y., Dawson T. M., Dawson V. L. Association of DJ-1 and parkin mediated by pathogenic DJ-1 mutations and oxidative stress. Hum. Mol. Genet. 2005;14:71–84. doi: 10.1093/hmg/ddi007. [DOI] [PubMed] [Google Scholar]
- 171.Gamerdinger M., Hajieva P., Kaya A. M., Wolfrum U., Hartl F. U., Behl C. Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. EMBO J. 2009;28:889–901. doi: 10.1038/emboj.2009.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Arndt V., Dick N., Tawo R., Dreiseidler M., Wenzel D., Hesse M., Furst D. O., Saftig P., Saint R., Fleischmann B. K., et al. Chaperone-assisted selective autophagy is essential for muscle maintenance. Curr. Biol. 2010;20:143–148. doi: 10.1016/j.cub.2009.11.022. [DOI] [PubMed] [Google Scholar]
- 173.Carra S., Seguin S. J., Lambert H., Landry J. HspB8 chaperone activity toward poly(Q)-containing proteins depends on its association with Bag3, a stimulator of macroautophagy. J. Biol. Chem. 2008;283:1437–1444. doi: 10.1074/jbc.M706304200. [DOI] [PubMed] [Google Scholar]
- 174.Yakes F. M., Van Houten B. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc. Natl. Acad. Sci. U.S.A. 1997;94:514–519. doi: 10.1073/pnas.94.2.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Menzies R. A., Gold P. H. The turnover of mitochondria in a variety of tissues of young adult and aged rats. J. Biol. Chem. 1971;246:2425–2429. [PubMed] [Google Scholar]
- 176.Lemasters J. J. Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuvenation Res. 2005;8:3–5. doi: 10.1089/rej.2005.8.3. [DOI] [PubMed] [Google Scholar]
- 177.Elmore S. P., Qian T., Grissom S. F., Lemasters J. J. The mitochondrial permeability transition initiates autophagy in rat hepatocytes. FASEB J. 2001;15:2286–2287. doi: 10.1096/fj.01-0206fje. [DOI] [PubMed] [Google Scholar]
- 178.Rodríguez-Enríquez S., Kim I., Currin R. T., Lemasters J. J. Tracker dyes to probe mitochondrial autophagy (mitophagy) in rat hepatocytes. Autophagy. 2006;2:39–46. doi: 10.4161/auto.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Kim I., Rodríguez-Enríquez S., Lemasters J. J. Selective degradation of mitochondria by mitophagy. Arch. Biochem. Biophys. 2007;462:245–253. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Kim I., Lemasters J. J. Mitochondrial degradation by autophagy (mitophagy) in GFP-LC3 transgenic hepatocytes during nutrient deprivation. Am. J. Physiol. Cell Physiol. 2011;300:C308–C317. doi: 10.1152/ajpcell.00056.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Twig G., Elorza A., Molina A. J., Mohamed H., Wikstrom J. D., Walzer G., Stiles L., Haigh S. E., Katz S., Las G., et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Kanki T., Wang K., Cao Y., Baba M., Klionsky D. J. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev. Cell. 2009;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Okamoto K., Kondo-Okamoto N., Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev. Cell. 2009;17:87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 184.Schweers R. L., Zhang J., Randall M. S., Loyd M. R., Li W., Dorsey F. C., Kundu M., Opferman J. T., Cleveland J. L., Miller J. L., Ney P. A. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc. Natl. Acad. Sci. U.S.A. 2007;104:19500–19505. doi: 10.1073/pnas.0708818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Tracy K., Dibling B. C., Spike B. T., Knabb J. R., Schumacker P., Macleod K. F. BNIP3 is an RB/E2F target gene required for hypoxia-induced autophagy. Mol. Cell. Biol. 2007;27:6229–6242. doi: 10.1128/MCB.02246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Aoki Y., Kanki T., Hirota Y., Kurihara Y., Saigusa T., Uchiumi T., Kang D. Phosphorylation of Ser114 on Atg32 mediates mitophagy. Mol. Biol. Cell. 2011;22:3206–3217. doi: 10.1091/mbc.E11-02-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Kanki T. Nix, a receptor protein for mitophagy in mammals. Autophagy. 2010;6:433–435. doi: 10.4161/auto.6.3.11420. [DOI] [PubMed] [Google Scholar]
- 188.Rikka S., Quinsay M. N., Thomas R. L., Kubli D. A., Zhang X., Murphy A. N., Gustafsson A. B. Bnip3 impairs mitochondrial bioenergetics and stimulates mitochondrial turnover. Cell Death Differ. 2011;18:721–731. doi: 10.1038/cdd.2010.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Thomas B., Beal M. F. Parkinson's disease. Hum. Mol. Genet. 2007;16(Suppl. R2):R183–R194. doi: 10.1093/hmg/ddm159. [DOI] [PubMed] [Google Scholar]
- 189a.Martin I., Dawson V. L., Dawson T. M. Recent advances in the genetics of Parkinson's disease. Annu. Rev. Genomics Hum. Genet. 2011;12:301–325. doi: 10.1146/annurev-genom-082410-101440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Takahashi T., Yamashita H., Nakamura T., Nagano Y., Nakamura S. Tyrosine 125 of α-synuclein plays a critical role for dimerization following nitrative stress. Brain Res. 2002;938:73–80. doi: 10.1016/s0006-8993(02)02498-8. [DOI] [PubMed] [Google Scholar]
- 191.Paxinou E., Chen Q., Weisse M., Giasson B. I., Norris E. H., Rueter S. M., Trojanowski J. Q., Lee V. M., Ischiropoulos H. Induction of α-synuclein aggregation by intracellular nitrative insult. J. Neurosci. 2001;21:8053–8061. doi: 10.1523/JNEUROSCI.21-20-08053.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Souza J. M., Giasson B. I., Chen Q., Lee V. M., Ischiropoulos H. Dityrosine cross-linking promotes formation of stable α-synuclein polymers: implication of nitrative and oxidative stress in the pathogenesis of neurodegenerative synucleinopathies. J. Biol. Chem. 2000;275:18344–18349. doi: 10.1074/jbc.M000206200. [DOI] [PubMed] [Google Scholar]
- 193.Li W. W., Yang R., Guo J. C., Ren H. M., Zha X. L., Cheng J. S., Cai D. F. Localization of α-synuclein to mitochondria within midbrain of mice. NeuroReport. 2007;18:1543–1546. doi: 10.1097/WNR.0b013e3282f03db4. [DOI] [PubMed] [Google Scholar]
- 194.Cole N. B., Dieuliis D., Leo P., Mitchell D. C., Nussbaum R. L. Mitochondrial translocation of α-synuclein is promoted by intracellular acidification. Exp. Cell Res. 2008;314:2076–2089. doi: 10.1016/j.yexcr.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Parihar M. S., Parihar A., Fujita M., Hashimoto M., Ghafourifar P. Mitochondrial association of α-synuclein causes oxidative stress. Cell. Mol. Life Sci. 2008;65:1272–1284. doi: 10.1007/s00018-008-7589-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Shavali S., Brown-Borg H. M., Ebadi M., Porter J. Mitochondrial localization of α-synuclein protein in α-synuclein overexpressing cells. Neurosci. Lett. 2008;439:125–128. doi: 10.1016/j.neulet.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Parihar M. S., Parihar A., Fujita M., Hashimoto M., Ghafourifar P. α-Synuclein overexpression and aggregation exacerbates impairment of mitochondrial functions by augmenting oxidative stress in human neuroblastoma cells. Int. J. Biochem. Cell Biol. 2009;41:2015–2024. doi: 10.1016/j.biocel.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 198.Devi L., Raghavendran V., Prabhu B. M., Avadhani N. G., Anandatheerthavarada H. K. Mitochondrial import and accumulation of α-synuclein impair complex I in human dopaminergic neuronal cultures and Parkinson disease brain. J. Biol. Chem. 2008;283:9089–9100. doi: 10.1074/jbc.M710012200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Chinta S. J., Mallajosyula J. K., Rane A., Andersen J. K. Mitochondrial α-synuclein accumulation impairs complex I function in dopaminergic neurons and results in increased mitophagy in vivo. Neurosci. Lett. 2010;486:235–239. doi: 10.1016/j.neulet.2010.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Plowey E. D., Cherra S. J., III, Liu Y. J., Chu C. T. Role of autophagy in G2019S-LRRK2-associated neurite shortening in differentiated SH-SY5Y cells. J. Neurochem. 2008;105:1048–1056. doi: 10.1111/j.1471-4159.2008.05217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Cherra S. J., III, Kulich S. M., Uechi G., Balasubramani M., Mountzouris J., Day B. W., Chu C. T. Regulation of the autophagy protein LC3 by phosphorylation. J. Cell Biol. 2010;190:533–539. doi: 10.1083/jcb.201002108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Alegre-Abarrategui J., Christian H., Lufino M. M., Mutihac R., Venda L. L., Ansorge O., Wade-Martins R. LRRK2 regulates autophagic activity and localizes to specific membrane microdomains in a novel human genomic reporter cellular model. Hum. Mol. Genet. 2009;18:4022–4034. doi: 10.1093/hmg/ddp346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Carballo-Carbajal I., Weber-Endress S., Rovelli G., Chan D., Wolozin B., Klein C. L., Patenge N., Gasser T., Kahle P. J. Leucine-rich repeat kinase 2 induces α-synuclein expression via the extracellular signal-regulated kinase pathway. Cell. Signalling. 2010;22:821–827. doi: 10.1016/j.cellsig.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Qing H., Zhang Y., Deng Y., McGeer E. G., McGeer P. L. Lrrk2 interaction with α-synuclein in diffuse Lewy body disease. Biochem. Biophys. Res. Commun. 2009;390:1229–1234. doi: 10.1016/j.bbrc.2009.10.126. [DOI] [PubMed] [Google Scholar]
- 205.Lin X., Parisiadou L., Gu X. L., Wang L., Shim H., Sun L., Xie C., Long C. X., Yang W. J., Ding J., et al. Leucine-rich repeat kinase 2 regulates the progression of neuropathology induced by Parkinson's-disease-related mutant α-synuclein. Neuron. 2009;64:807–827. doi: 10.1016/j.neuron.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Tong Y., Yamaguchi H., Giaime E., Boyle S., Kopan R., Kelleher R. J., III, Shen J. Loss of leucine-rich repeat kinase 2 causes impairment of protein degradation pathways, accumulation of α-synuclein, and apoptotic cell death in aged mice. Proc. Natl. Acad. Sci. U.S.A. 2010;107:9879–9884. doi: 10.1073/pnas.1004676107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Dexter D. T., Carter C. J., Wells F. R., Javoy-Agid F., Agid Y., Lees A., Jenner P., Marsden C. D. Basal lipid peroxidation in substantia nigra is increased in Parkinson's disease. J. Neurochem. 1989;52:381–389. doi: 10.1111/j.1471-4159.1989.tb09133.x. [DOI] [PubMed] [Google Scholar]
- 208.Riederer P., Sofic E., Rausch W. D., Schmidt B., Reynolds G. P., Jellinger K., Youdim M. B. Transition metals, ferritin, glutathione, and ascorbic acid in parkinsonian brains. J. Neurochem. 1989;52:515–520. doi: 10.1111/j.1471-4159.1989.tb09150.x. [DOI] [PubMed] [Google Scholar]
- 209.Sofic E., Sapcanin A., Tahirovic I., Gavrankapetanovic I., Jellinger K., Reynolds G. P., Tatschner T., Riederer P. Antioxidant capacity in postmortem brain tissues of Parkinson's and Alzheimer's diseases. J. Neural Transm. Suppl. 2006:39–43. doi: 10.1007/978-3-211-33328-0_5. [DOI] [PubMed] [Google Scholar]
- 210.Sofic E., Lange K. W., Jellinger K., Riederer P. Reduced and oxidized glutathione in the substantia nigra of patients with Parkinson's disease. Neurosci. Lett. 1992;142:128–130. doi: 10.1016/0304-3940(92)90355-b. [DOI] [PubMed] [Google Scholar]
- 211.Sian J., Dexter D. T., Lees A. J., Daniel S., Agid Y., Javoy-Agid F., Jenner P., Marsden C. D. Alterations in glutathione levels in Parkinson's disease and other neurodegenerative disorders affecting basal ganglia. Ann. Neurol. 1994;36:348–355. doi: 10.1002/ana.410360305. [DOI] [PubMed] [Google Scholar]
- 212.Perry T. L., Yong V. W. Idiopathic Parkinson's disease, progressive supranuclear palsy and glutathione metabolism in the substantia nigra of patients. Neurosci. Lett. 1986;67:269–274. doi: 10.1016/0304-3940(86)90320-4. [DOI] [PubMed] [Google Scholar]
- 213.Saggu H., Cooksey J., Dexter D., Wells F. R., Lees A., Jenner P., Marsden C. D. A selective increase in particulate superoxide dismutase activity in parkinsonian substantia nigra. J. Neurochem. 1989;53:692–697. doi: 10.1111/j.1471-4159.1989.tb11759.x. [DOI] [PubMed] [Google Scholar]
- 214.de Vries H. E., Witte M., Hondius D., Rozemuller A. J., Drukarch B., Hoozemans J., van Horssen J. Nrf2-induced antioxidant protection: a promising target to counteract ROS-mediated damage in neurodegenerative disease? Free Radical Biol. Med. 2008;45:1375–1383. doi: 10.1016/j.freeradbiomed.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 215.Radunovic A., Porto W. G., Zeman S., Leigh P. N. Increased mitochondrial superoxide dismutase activity in Parkinson's disease but not amyotrophic lateral sclerosis motor cortex. Neurosci. Lett. 1997;239:105–108. doi: 10.1016/s0304-3940(97)00905-1. [DOI] [PubMed] [Google Scholar]
- 216.Yoritaka A., Hattori N., Mori H., Kato K., Mizuno Y. An immunohistochemical study on manganese superoxide dismutase in Parkinson's disease. J. Neurol. Sci. 1997;148:181–186. doi: 10.1016/s0022-510x(96)05339-7. [DOI] [PubMed] [Google Scholar]
- 217.Yoritaka A., Hattori N., Uchida K., Tanaka M., Stadtman E. R., Mizuno Y. Immunohistochemical detection of 4-hydroxynonenal protein adducts in Parkinson disease. Proc. Natl. Acad. Sci. U.S.A. 1996;93:2696–2701. doi: 10.1073/pnas.93.7.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Alam Z. I., Jenner A., Daniel S. E., Lees A. J., Cairns N., Marsden C. D., Jenner P., Halliwell B. Oxidative DNA damage in the parkinsonian brain: an apparent selective increase in 8-hydroxyguanine levels in substantia nigra. J. Neurochem. 1997;69:1196–1203. doi: 10.1046/j.1471-4159.1997.69031196.x. [DOI] [PubMed] [Google Scholar]
- 219.Schapira A. H., Mann V. M., Cooper J. M., Dexter D., Daniel S. E., Jenner P., Clark J. B., Marsden C. D. Anatomic and disease specificity of NADH CoQ1 reductase (complex I) deficiency in Parkinson's disease. J. Neurochem. 1990;55:2142–2145. doi: 10.1111/j.1471-4159.1990.tb05809.x. [DOI] [PubMed] [Google Scholar]
- 220.Schapira A. H., Cooper J. M., Dexter D., Clark J. B., Jenner P., Marsden C. D. Mitochondrial complex I deficiency in Parkinson's disease. J. Neurochem. 1990;54:823–827. doi: 10.1111/j.1471-4159.1990.tb02325.x. [DOI] [PubMed] [Google Scholar]
- 221.Parker W. D., Jr, Parks J. K., Swerdlow R. H. Complex I deficiency in Parkinson's disease frontal cortex. Brain Res. 2008;1189:215–218. doi: 10.1016/j.brainres.2007.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Smigrodzki R., Parks J., Parker W. D. High frequency of mitochondrial complex I mutations in Parkinson's disease and aging. Neurobiol. Aging. 2004;25:1273–1281. doi: 10.1016/j.neurobiolaging.2004.02.020. [DOI] [PubMed] [Google Scholar]
- 223.Simon D. K., Lin M. T., Zheng L., Liu G. J., Ahn C. H., Kim L. M., Mauck W. M., Twu F., Beal M. F., Johns D. R. Somatic mitochondrial DNA mutations in cortex and substantia nigra in aging and Parkinson's disease. Neurobiol. Aging. 2004;25:71–81. doi: 10.1016/s0197-4580(03)00037-x. [DOI] [PubMed] [Google Scholar]
- 224.Trimmer P. A., Borland M. K., Keeney P. M., Bennett J. P., Jr, Parker W. D., Jr Parkinson's disease transgenic mitochondrial cybrids generate Lewy inclusion bodies. J. Neurochem. 2004;88:800–812. doi: 10.1046/j.1471-4159.2003.02168.x. [DOI] [PubMed] [Google Scholar]
- 225.Bove J., Prou D., Perier C., Przedborski S. Toxin-induced models of Parkinson's disease. NeuroRx. 2005;2:484–494. doi: 10.1602/neurorx.2.3.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Bender A., Krishnan K. J., Morris C. M., Taylor G. A., Reeve A. K., Perry R. H., Jaros E., Hersheson J. S., Betts J., Klopstock T., et al. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat. Genet. 2006;38:515–517. doi: 10.1038/ng1769. [DOI] [PubMed] [Google Scholar]
- 227.Ekstrand M. I., Terzioglu M., Galter D., Zhu S., Hofstetter C., Lindqvist E., Thams S., Bergstrand A., Hansson F. S., Trifunovic A., et al. Progressive parkinsonism in mice with respiratory-chain-deficient dopamine neurons. Proc. Natl. Acad. Sci. U.S.A. 2007;104:1325–1330. doi: 10.1073/pnas.0605208103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Arthur C. R., Morton S. L., Dunham L. D., Keeney P. M., Bennett J. P., Jr Parkinson's disease brain mitochondria have impaired respirasome assembly, age-related increases in distribution of oxidative damage to mtDNA and no differences in heteroplasmic mtDNA mutation abundance. Mol. Neurodegener. 2009;4:37. doi: 10.1186/1750-1326-4-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Anglade P., Vyas S., Javoy-Agid F., Herrero M. T., Michel P. P., Marquez J., Mouatt-Prigent A., Ruberg M., Hirsch E. C., Agid Y. Apoptosis and autophagy in nigral neurons of patients with Parkinson's disease. Histol. Histopathol. 1997;12:25–31. [PubMed] [Google Scholar]
- 230.Alvarez-Erviti L., Rodriguez-Oroz M. C., Cooper J. M., Caballero C., Ferrer I., Obeso J. A., Schapira A. H. Chaperone-mediated autophagy markers in Parkinson disease brains. Arch. Neurol. 2010;67:1464–1472. doi: 10.1001/archneurol.2010.198. [DOI] [PubMed] [Google Scholar]
- 231.Stefanis L. Caspase-dependent and -independent neuronal death: two distinct pathways to neuronal injury. Neuroscientist. 2005;11:50–62. doi: 10.1177/1073858404271087. [DOI] [PubMed] [Google Scholar]
- 232.Abou-Sleiman P. M., Healy D. G., Quinn N., Lees A. J., Wood N. W. The role of pathogenic DJ-1 mutations in Parkinson's disease. Ann. Neurol. 2003;54:283–286. doi: 10.1002/ana.10675. [DOI] [PubMed] [Google Scholar]
- 233.Valente E. M., Abou-Sleiman P. M., Caputo V., Muqit M. M., Harvey K., Gispert S., Ali Z., Del Turco D., Bentivoglio A. R., Healy D. G., et al. Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science. 2004;304:1158–1160. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- 234.Geisler S., Holmstrom K. M., Skujat D., Fiesel F. C., Rothfuss O. C., Kahle P. J., Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 235.Poole A. C., Thomas R. E., Andrews L. A., McBride H. M., Whitworth A. J., Pallanck L. J. The PINK1/Parkin pathway regulates mitochondrial morphology. Proc. Natl. Acad. Sci. U.S.A. 2008;105:1638–1643. doi: 10.1073/pnas.0709336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Poole A. C., Thomas R. E., Yu S., Vincow E. S., Pallanck L. The mitochondrial fusion-promoting factor mitofusin is a substrate of the PINK1/parkin pathway. PloS ONE. 2010;5:e10054. doi: 10.1371/journal.pone.0010054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Gegg M. E., Cooper J. M., Chau K. Y., Rojo M., Schapira A. H., Taanman J. W. Mitofusin 1 and mitofusin 2 are ubiquitinated in a PINK1/parkin-dependent manner upon induction of mitophagy. Hum. Mol. Genet. 2010;19:4861–4870. doi: 10.1093/hmg/ddq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Yao D., Gu Z., Nakamura T., Shi Z. Q., Ma Y., Gaston B., Palmer L. A., Rockenstein E. M., Zhang Z., Masliah E., et al. Nitrosative stress linked to sporadic Parkinson's disease: S-nitrosylation of parkin regulates its E3 ubiquitin ligase activity. Proc. Natl. Acad. Sci. U.S.A. 2004;101:10810–10814. doi: 10.1073/pnas.0404161101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Meng F., Yao D., Shi Y., Kabakoff J., Wu W., Reicher J., Ma Y., Moosmann B., Masliah E., Lipton S. A., Gu Z. Oxidation of the cysteine-rich regions of parkin perturbs its E3 ligase activity and contributes to protein aggregation. Mol. Neurodegener. 2011;6:34. doi: 10.1186/1750-1326-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Mortiboys H., Thomas K. J., Koopman W. J., Klaffke S., Abou-Sleiman P., Olpin S., Wood N. W., Willems P. H., Smeitink J. A., Cookson M. R., Bandmann O. Mitochondrial function and morphology are impaired in parkin-mutant fibroblasts. Ann. Neurol. 2008;64:555–565. doi: 10.1002/ana.21492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Gautier C. A., Kitada T., Shen J. Loss of PINK1 causes mitochondrial functional defects and increased sensitivity to oxidative stress. Proc. Natl. Acad. Sci. U.S.A. 2008;105:11364–11369. doi: 10.1073/pnas.0802076105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Wang H. L., Chou A. H., Wu A. S., Chen S. Y., Weng Y. H., Kao Y. C., Yeh T. H., Chu P. J., Lu C. S. PARK6 PINK1 mutants are defective in maintaining mitochondrial membrane potential and inhibiting ROS formation of substantia nigra dopaminergic neurons. Biochim. Biophys. Acta. 2011;1812:674–684. doi: 10.1016/j.bbadis.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 243.Dagda R. K., Cherra S. J., III, Kulich S. M., Tandon A., Park D., Chu C. T. Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. J. Biol. Chem. 2009;284:13843–13855. doi: 10.1074/jbc.M808515200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Hao L. Y., Giasson B. I., Bonini N. M. DJ-1 is critical for mitochondrial function and rescues PINK1 loss of function. Proc. Natl. Acad. Sci. U.S.A. 2010;107:9747–9752. doi: 10.1073/pnas.0911175107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Kim R. H., Smith P. D., Aleyasin H., Hayley S., Mount M. P., Pownall S., Wakeham A., You-Ten A. J., Kalia S. K., Horne P., et al. Hypersensitivity of DJ-1-deficient mice to 1-methyl-4-phenyl-1,2,3,6-tetrahydropyrindine (MPTP) and oxidative stress. Proc. Natl. Acad. Sci. U.S.A. 2005;102:5215–5220. doi: 10.1073/pnas.0501282102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Andres-Mateos E., Perier C., Zhang L., Blanchard-Fillion B., Greco T. M., Thomas B., Ko H. S., Sasaki M., Ischiropoulos H., Przedborski S., et al. DJ-1 gene deletion reveals that DJ-1 is an atypical peroxiredoxin-like peroxidase. Proc. Natl. Acad. Sci. U.S.A. 2007;104:14807–14812. doi: 10.1073/pnas.0703219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Guzman J. N., Sanchez-Padilla J., Wokosin D., Kondapalli J., Ilijic E., Schumacker P. T., Surmeier D. J. Oxidant stress evoked by pacemaking in dopaminergic neurons is attenuated by DJ-1. Nature. 2010;468:696–700. doi: 10.1038/nature09536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Irrcher I., Aleyasin H., Seifert E. L., Hewitt S. J., Chhabra S., Phillips M., Lutz A. K., Rousseaux M. W., Bevilacqua L., Jahani-Asl A., et al. Loss of the Parkinson's disease-linked gene DJ-1 perturbs mitochondrial dynamics. Hum. Mol. Genet. 2010;19:3734–3746. doi: 10.1093/hmg/ddq288. [DOI] [PubMed] [Google Scholar]
- 249.McCoy M. K., Cookson M. R. DJ-1 regulation of mitochondrial function and autophagy through oxidative stress. Autophagy. 2011;7:531–532. doi: 10.4161/auto.7.5.14684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Thomas K. J., McCoy M. K., Blackinton J., Beilina A., van der Brug M., Sandebring A., Miller D., Maric D., Cedazo-Minguez A., Cookson M. R. DJ-1 acts in parallel to the PINK1/parkin pathway to control mitochondrial function and autophagy. Hum. Mol. Genet. 2011;20:40–50. doi: 10.1093/hmg/ddq430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Khandelwal P. J., Herman A. M., Hoe H. S., Rebeck G. W., Moussa C. E. Parkin mediates beclin-dependent autophagic clearance of defective mitochondria and ubiquitinated Aβ in AD models. Hum. Mol. Genet. 2011;20:2091–2102. doi: 10.1093/hmg/ddr091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Nakamura T., Cieplak P., Cho D. H., Godzik A., Lipton S. A. S-nitrosylation of Drp1 links excessive mitochondrial fission to neuronal injury in neurodegeneration. Mitochondrion. 2010;10:573–578. doi: 10.1016/j.mito.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Wang X., Su B., Lee H. G., Li X., Perry G., Smith M. A., Zhu X. Impaired balance of mitochondrial fission and fusion in Alzheimer's disease. J. Neurosci. 2009;29:9090–9103. doi: 10.1523/JNEUROSCI.1357-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Manczak M., Calkins M. J., Reddy P. H. Impaired mitochondrial dynamics and abnormal interaction of amyloid β with mitochondrial protein Drp1 in neurons from patients with Alzheimer's disease: implications for neuronal damage. Hum. Mol. Genet. 2011;20:2495–2509. doi: 10.1093/hmg/ddr139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Nishino I., Fu J., Tanji K., Yamada T., Shimojo S., Koori T., Mora M., Riggs J. E., Oh S. J., Koga Y., et al. Primary LAMP-2 deficiency causes X-linked vacuolar cardiomyopathy and myopathy (Danon disease) Nature. 2000;406:906–910. doi: 10.1038/35022604. [DOI] [PubMed] [Google Scholar]
- 256.Tanaka Y., Guhde G., Suter A., Eskelinen E. L., Hartmann D., Lullmann-Rauch R., Janssen P. M., Blanz J., von Figura K., Saftig P. Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature. 2000;406:902–906. doi: 10.1038/35022595. [DOI] [PubMed] [Google Scholar]
- 257.Maron B. J., Roberts W. C., Arad M., Haas T. S., Spirito P., Wright G. B., Almquist A. K., Baffa J. M., Saul J. P., Ho C. Y., et al. Clinical outcome and phenotypic expression in LAMP2 cardiomyopathy. JAMA, J. Am. Med. Assoc. 2009;301:1253–1259. doi: 10.1001/jama.2009.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Zhu H., Tannous P., Johnstone J. L., Kong Y., Shelton J. M., Richardson J. A., Le V., Levine B., Rothermel B. A., Hill J. A. Cardiac autophagy is a maladaptive response to hemodynamic stress. J. Clin. Invest. 2007;117:1782–1793. doi: 10.1172/JCI27523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Nakai A., Yamaguchi O., Takeda T., Higuchi Y., Hikoso S., Taniike M., Omiya S., Mizote I., Matsumura Y., Asahi M., et al. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat. Med. 2007;13:619–624. doi: 10.1038/nm1574. [DOI] [PubMed] [Google Scholar]
- 260.Liang X. H., Jackson S., Seaman M., Brown K., Kempkes B., Hibshoosh H., Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 261.Decker R. S., Wildenthal K. Lysosomal alterations in hypoxic and reoxygenated hearts. I. Ultrastructural and cytochemical changes. Am. J. Pathol. 1980;98:425–444. [PMC free article] [PubMed] [Google Scholar]
- 262.Huang C., Andres A. M., Ratliff E. P., Hernandez G., Lee P., Gottlieb R. A. Preconditioning involves selective mitophagy mediated by parkin and p62/SQSTM1. PloS ONE. 2011;6:e20975. doi: 10.1371/journal.pone.0020975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Hoffman D. L., Salter J. D., Brookes P. S. Response of mitochondrial reactive oxygen species generation to steady-state oxygen tension: implications for hypoxic cell signaling. Am. J. Physiol. Heart Circ. Physiol. 2007;292:H101–H108. doi: 10.1152/ajpheart.00699.2006. [DOI] [PubMed] [Google Scholar]
- 264.Gurusamy N., Lekli I., Gorbunov N. V., Gherghiceanu M., Popescu L. M., Das D. K. Cardioprotection by adaptation to ischaemia augments autophagy in association with BAG-1 protein. J. Cell. Mol. Med. 2009;13:373–387. doi: 10.1111/j.1582-4934.2008.00495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Bellot G., Garcia-Medina R., Gounon P., Chiche J., Roux D., Pouyssegur J., Mazure N. M. Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol. Cell. Biol. 2009;29:2570–2581. doi: 10.1128/MCB.00166-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Novak I., Kirkin V., McEwan D. G., Zhang J., Wild P., Rozenknop A., Rogov V., Lohr F., Popovic D., Occhipinti A., et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Kroemer G., Marino G., Levine B. Autophagy and the integrated stress response. Mol. Cell. 2010;40:280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Diwan A., Krenz M., Syed F. M., Wansapura J., Ren X., Koesters A. G., Li H., Kirshenbaum L. A., Hahn H. S., Robbins J., et al. Inhibition of ischemic cardiomyocyte apoptosis through targeted ablation of Bnip3 restrains postinfarction remodeling in mice. J. Clin. Invest. 2007;117:2825–2833. doi: 10.1172/JCI32490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Chen N., Karantza-Wadsworth V. Role and regulation of autophagy in cancer. Biochim. Biophys. Acta. 2009;1793:1516–1523. doi: 10.1016/j.bbamcr.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Mathew R., Karp C. M., Beaudoin B., Vuong N., Chen G., Chen H. Y., Bray K., Reddy A., Bhanot G., Gelinas C., et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–1075. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Mariño G., Salvador-Montoliu N., Fueyo A., Knecht E., Mizushima N., López-Otín C. Tissue-specific autophagy alterations and increased tumorigenesis in mice deficient in ATG4C/autophagin-3. J. Biol. Chem. 2007;282:18573–18583. doi: 10.1074/jbc.M701194200. [DOI] [PubMed] [Google Scholar]
- 272.Qu X., Yu J., Bhagat G., Furuya N., Hibshoosh H., Troxel A., Rosen J., Eskelinen E. L., Mizushima N., Ohsumi Y., et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J. Clin. Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Guo J. Y., Chen H. Y., Mathew R., Fan J., Strohecker A. M., Karsli-Uzunbas G., Kamphorst J. J., Chen G., Lemons J. M., Karantza V., et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25:460–470. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Bellodi C., Lidonnici M. R., Hamilton A., Helgason G. V., Soliera A. R., Ronchetti M., Galavotti S., Young K. W., Selmi T., Yacobi R., et al. Targeting autophagy potentiates tyrosine kinase inhibitor-induced cell death in Philadelphia chromosome-positive cells, including primary CML stem cells. J. Clin. Invest. 2009;119:1109–1123. doi: 10.1172/JCI35660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Vazquez-Martin A., Oliveras-Ferraros C., Menendez J. A. Autophagy facilitates the development of breast cancer resistance to the anti-HER2 monoclonal antibody trastuzumab. PLoS ONE. 2009;4:e6251. doi: 10.1371/journal.pone.0006251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Apel A., Herr I., Schwarz H., Rodemann H. P., Mayer A. Blocked autophagy sensitizes resistant carcinoma cells to radiation therapy. Cancer Res. 2008;68:1485–1494. doi: 10.1158/0008-5472.CAN-07-0562. [DOI] [PubMed] [Google Scholar]
- 277.Young A. R., Narita M., Ferreira M., Kirschner K., Sadaie M., Darot J. F., Tavare S., Arakawa S., Shimizu S., Watt F. M., Narita M. Autophagy mediates the mitotic senescence transition. Genes Dev. 2009;23:798–803. doi: 10.1101/gad.519709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Schieke S. M., Phillips D., McCoy J. P., Jr, Aponte A. M., Shen R. F., Balaban R. S., Finkel T. The mammalian target of rapamycin (mTOR) pathway regulates mitochondrial oxygen consumption and oxidative capacity. J. Biol. Chem. 2006;281:27643–27652. doi: 10.1074/jbc.M603536200. [DOI] [PubMed] [Google Scholar]
- 279.Jia W., He Y. W. Temporal regulation of intracellular organelle homeostasis in T lymphocytes by autophagy. J. Immunol. 2011;186:5313–5322. doi: 10.4049/jimmunol.1002404. [DOI] [PubMed] [Google Scholar]
- 280.Mariño G., Fernández A. F., Cabrera S., Lundberg Y. W., Cabanillas R., Rodríguez F., Salvador-Montoliu N., Vega J. A., Germanà A., Fueyo A., et al. Autophagy is essential for mouse sense of balance. J. Clin. Invest. 2010;120:2331–2344. doi: 10.1172/JCI42601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Betin V. M., Lane J. D. Caspase cleavage of Atg4D stimulates GABARAP-L1 processing and triggers mitochondrial targeting and apoptosis. J. Cell Sci. 2009;122:2554–2566. doi: 10.1242/jcs.046250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Komatsu M., Waguri S., Ueno T., Iwata J., Murata S., Tanida I., Ezaki J., Mizushima N., Ohsumi Y., Uchiyama Y., et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Tal M. C., Sasai M., Lee H. K., Yordy B., Shadel G. S., Iwasaki A. Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proc. Natl. Acad. Sci. U.S.A. 2009;106:2770–2775. doi: 10.1073/pnas.0807694106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Stephenson L. M., Miller B. C., Ng A., Eisenberg J., Zhao Z., Cadwell K., Graham D. B., Mizushima N. N., Xavier R., Virgin H. W., Swat W. Identification of Atg5-dependent transcriptional changes and increases in mitochondrial mass in Atg5-deficient T lymphocytes. Autophagy. 2009;5:625–635. doi: 10.4161/auto.5.5.8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Pua H. H., Guo J., Komatsu M., He Y. W. Autophagy is essential for mitochondrial clearance in mature T lymphocytes. J. Immunol. 2009;182:4046–4055. doi: 10.4049/jimmunol.0801143. [DOI] [PubMed] [Google Scholar]
- 286.Zhang J., Randall M. S., Loyd M. R., Dorsey F. C., Kundu M., Cleveland J. L., Ney P. A. Mitochondrial clearance is regulated by Atg7-dependent and -independent mechanisms during reticulocyte maturation. Blood. 2009;114:157–164. doi: 10.1182/blood-2008-04-151639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Mortensen M., Ferguson D. J., Edelmann M., Kessler B., Morten K. J., Komatsu M., Simon A. K. Loss of autophagy in erythroid cells leads to defective removal of mitochondria and severe anemia in vivo. Proc. Natl. Acad. Sci. U.S.A. 2010;107:832–837. doi: 10.1073/pnas.0913170107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Mortensen M., Soilleux E. J., Djordjevic G., Tripp R., Lutteropp M., Sadighi-Akha E., Stranks A. J., Glanville J., Knight S., Jacobsen S. E., et al. The autophagy protein Atg7 is essential for hematopoietic stem cell maintenance. J. Exp. Med. 2011;208:455–467. doi: 10.1084/jem.20101145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Nakahira K., Haspel J. A., Rathinam V. A., Lee S. J., Dolinay T., Lam H. C., Englert J. A., Rabinovitch M., Cernadas M., Kim H. P., et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat. Immunol. 2011;12:222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Sandoval H., Thiagarajan P., Dasgupta S. K., Schumacher A., Prchal J. T., Chen M., Wang J. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454:232–235. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]