Abstract
Down syndrome (DS), resulting from trisomy of chromosome 21, is the most common live-born human aneuploidy. The phenotypic expression of trisomy 21 produces variable, though characteristic, facial morphology. Although certain facial features have been documented quantitatively and qualitatively as characteristic of DS (e.g., epicanthic folds, macroglossia, and hypertelorism), all of these traits occur in other craniofacial conditions with an underlying genetic cause. We hypothesize that the typical DS face is integrated differently than the face of non-DS siblings, and that the pattern of morphological integration unique to individuals with DS will yield information about underlying developmental associations between facial regions. We statistically compared morphological integration patterns of immature DS faces (N = 53) with those of non-DS siblings (N = 54), aged 6–12 years using 31 distances estimated from 3D coordinate data representing 17 anthropometric landmarks recorded on 3D digital photographic images. Facial features are affected differentially in DS, as evidenced by statistically significant differences in integration both within and between facial regions. Our results suggest a differential affect of trisomy on facial prominences during craniofacial development.
Keywords: trisomy 21, facial correlation, craniofacial morphology, facial development, 3dMD
Down syndrome (DS), first described in the medical literature more than 100 years ago (Down, 1866), is caused by an extra copy of chromosome 21 (LeJeune et al., 1959). Trisomy 21 causes gene-dosage imbalance for genes located on chromosome 21 (Reeves et al., 2001) and represents an extreme form of copy number variation (Makino and McLysaght, 2010). Chromosome 21 contains about 1.5% of the human genome, spans 47 million nucleotides, and includes between 300–400 protein coding genes depending on the method of estimation used (Hattori et al., 2000; Gardiner et al., 2003; Megarbane et al., 2009). DS occurs in all ethnic backgrounds and socioeconomic classes at a frequency of about 1:700 live births, and a marked maternal age effect has been noted (CDCP, 2006; Kuppermann et al., 2006). Despite being the most common live-born human chromosomal anomaly, the scientific understanding of how developmental processes are altered to produce DS phenotypes is limited (Sommer and Henrique-Silva, 2008; Wiseman et al., 2009).
The DS face always exhibits some degree of dysmorphology (Farkas et al., 1985, 2002; Farkas, 2001a, b) representing a composite of changes in qualitative and quantitative phenotypic characteristics (Table 1). These attributes involve every craniofacial sensory organ and most tissue classes (e.g., skin, muscle, skeletal, and neural); yet the effects are not uniform within a single DS individual, nor are they consistent across DS individuals. Early pioneering analyses of DS craniofacial morphology frequently focused on characterizing how and why DS faces differ from typically developing faces. However, these studies often produced conflicting conclusions, because the technologies available for answering developmental questions were restricted at that time, and studies were limited by the unsuitability of control groups, small sample sizes, and inconsistent methods of analysis.
TABLE 1.
Qualitative and quantitative craniofacial characteristics associated with trisomy 21 (Shapiro et al., 1967; Shapiro, 1969, 1970, 1975, 1983, 1992, 2001; Frostad et al., 1971; Fink et al., 1975; O’Riordan and Walker, 1979; Fischer-Brandies et al., 1986; Allanson et al., 1993; Ferrario et al., 2004a, b)
| Almond-shaped eyes | Smaller head circumference |
| Epicanthic folds | Brachycephaly (short, broad head) |
| Smaller orbital height | Insufficient or absent sinus development |
| Smaller interorbital distances | Smaller and flatter cranial base |
| Reduced orbital width | Increased variance in facial linear distances |
| Midfacial hypoplasia | Tooth eruption pattern differences and dysmorphology |
| Missing or small nasal bones | Increased variance in palate dimensions |
| Mandibular prognathism | Ear dysmorphology |
The purpose of this study is to determine the effects of trisomy 21 upon covariance patterns of facial measures to increase our understanding of how this chromosomal change alters developmental mechanisms underlying the emergence of DS facial phenotypes. We hypothesize that beyond the local anatomical changes produced by trisomy 21, covariance patterns of the typical DS face are different than faces of non-DS siblings, and that analysis of these patterns will yield information about how trisomy 21 affects the underlying developmental associations between facial regions. We address this hypothesis by comparing patterns of morphological integration for measurements of facial features in individuals with DS and unaffected siblings of individuals with DS using three-dimensional (3D) photogrammatic data, morphometric methods of analysis, and known biological relationships.
The question of how and why DS faces develop differently is addressed using morphological integration, a method of analysis that has not previously been used to investigate developmental aspects of the DS facial phenotype. Morphological integration, evaluated by statistical correlation or covariation among phenotypic traits, refers to the relationships among morphological elements (Olson and Miller, 1958; Cheverud, 1996). Analyses of morphological integration are useful for demarcating subsets of phenotypic traits that strongly covary, which can occur due to shared function and/or developmental origin (Olson and Miller, 1958). During facial embryogenesis several facial prominences must become established and fuse in a coordinated fashion to produce the bone, skin, musculature, and craniofacial organs required for a functioning craniofacial complex (Larson, 1997; Helms and Schneider, 2003; Chai and Maxson, 2006). Under the model of morphological integration, the strength of covariation among measures within each region of the face is expected to be stronger than the covariation among measures from different regions of the face. Because trisomy 21 alters craniofacial development in unrelated individuals to produce an immediately recognizable and characteristic facial morphology, patterns of integration can be compared between samples to better define how trisomy 21 alters the covariation structure of the developing face. Since the most profound difference between individuals with DS and their non-DS siblings is the presence or absence of an extra copy of chromosome 21, differences in facial integration between these samples provides clues to the alterations in craniofacial morphogenesis caused by trisomy 21.
MATERIALS AND METHODS
A cross-sectional sample of 3D facial images (N = 107) was acquired for analysis. All images used in this study were collected with protocols approved by the Pennsylvania State University Institutional Review Board (IRB #23283). The study sample consists of individuals with DS (N = 53) and individuals who are siblings of an individual with DS, but who do not have DS themselves (hereafter referred to as non-DS siblings; N = 54). This sample was acquired from 2006 to 2010 at several venues: annual National Down Syndrome Congresses (Atlanta, GA, Boston, MA, Sacramento, CA, and Lake Buena Vista, FL, respectively) and the 2008 Pennsylvania Summer Special Olympics (University Park, PA). The age range of participants in this sample is 6–12 years (Table 2). Ages were calculated from birthdates provided by parents at the time of image acquisition. No attempt was made to control for ethnic/ancestral background; however, the great majority of the sample is Caucasian.
TABLE 2.
Age distribution of the DS individuals (n = 53) and non-DS siblings (n = 54) used in this study. Mean and standard deviation for each sample are provided. This sample includes 19 DS sibling pairs (one DS individual and one non-DS sibling), three DS sibling trios (one DS individual and two non-DS siblings), four non-DS sibling pairs (each has a sibling with DS that is not in this study), 31 isolated DS individuals (no siblings in this study), and 21 isolated non-DS siblings (no sibling in this study, but has a sibling with DS). Within this sample, there is one known individual with mosaic DS, two pairs of dizygotic twins (one twin pair consists of two non-DS siblings and the other consists of one DS individual and one non-DS sibling), and one set of fraternal triplets (one DS individual and two non-DS siblings)
| Age | DS | Non-DS Siblings |
|---|---|---|
| 6 years | 6 | 6 |
| 7 years | 10 | 5 |
| 8 years | 6 | 12 |
| 9 years | 12 | 10 |
| 10 years | 6 | 10 |
| 11 years | 7 | 4 |
| 12 years | 6 | 7 |
| Mean | 7.57 | 7.71 |
| St. Dev. | 2.44 | 2.98 |
Data were collected using the 3dMDface photogrammatic system (http://3dmd.com/). This portable camera system incorporates images acquired simultaneously by six cameras, each at a different viewpoint, and merges images into a single 3D facial image. This is accomplished automatically using random scatterpoint clouds, superimposition of the various images using a registration system, and 3D visualization software developed by 3dMD engineers (3dMDpatient version 4.0). During photo capture, volunteers were asked to adopt a neutral facial expression. A single anterior image was taken, and two lateral images were taken from the right and left side at ~30° deviation from the frontal position. Additional pictures were taken when neutral facial expressions were not maintained throughout the photographic process. 3D images were immediately batch processed for each of these views and checked to make sure that surfaces of interest were visible in the acquired images.
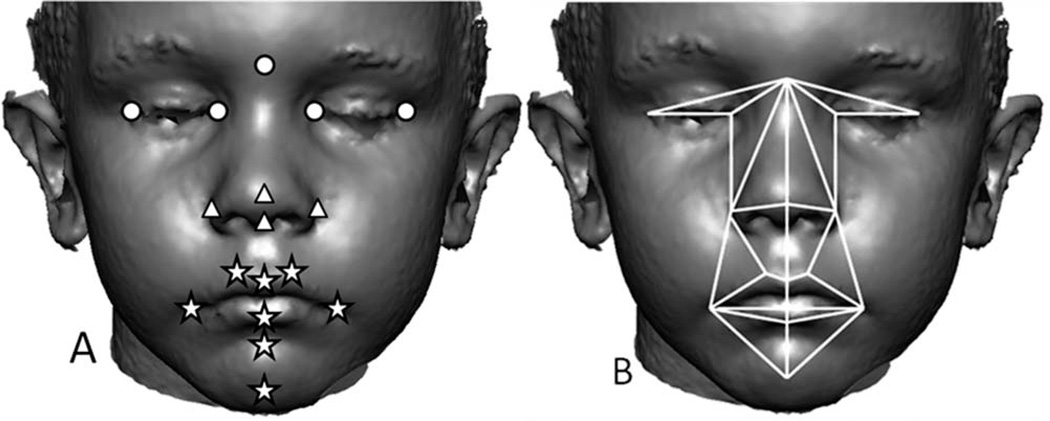
3dMDpatient was used to record the 3D coordinates of 17 standard landmarks from each image twice (Fig. 1A). Several studies have determined that 3dMD facial images are an accurate 3D representation of the face, and that the 3D locations of anatomical landmarks located on these images are precise, reliable, and unbiased representations of facial topography (Aldridge et al., 2005; Weinberg et al., 2006; Wong et al., 2008). Measurement error was estimated by calculating the standard deviation of the x, y, and z coordinates of landmarks that had been collected five times from a single image. This was done for the images of five separate individuals drawn at random from the overall sample with at least 24 h between landmarking sessions to avoid memory bias in landmark placement. Standard deviations of landmark coordinates along the x, y, and z axes were averaged to calculate mean measurement error. Mean measurement error was estimated to be 0.29 mm (0.26 mm along the x-dimension, 0.30 mm along the y-dimension, and 0.31 mm along the z-dimension) and is considered very low. For each individual, we checked for gross landmark coordinate errors (e.g., mislabeling right and left side landmarks) and then averaged landmark coordinates from two separate digitizing episodes to minimize any potential effects of measurement error for each individual. Average coordinate locations of landmarks were used for analyses.
Fig. 1.
A) The image depicts 17 anatomical landmarks that were used in this analysis to represent the three facial regions: circles (upper face), triangles (midface), and stars (lower face) shown on a 3dMD image. 3D coordinates of landmark locations were determined using 3dMDpatient (v4.0). Several medial and bilateral landmarks were used in this study: nasion, pronasale, subnasale, labiale superius, labiale inferius, sublabiale, pogonion, endocanthion, exocanthion, alar curvature, crista philtra landmark, and chelion. Anatomical definitions of landmarks can be found on our laboratory website (http://getahead.psu.edu/.) B) The 31 linear distances used in this study shown on a 3dMD image. The midline represents six nonoverlapping linear distances.
In this study, we are not testing a specific hypothesis of integration or facial modularity. Rather, we are exploring patterns of covariation in facial metrics to determine if differences in integration occur in the DS face relative to the typically developing non-DS sibling face. Consequently, we divided the face into traditional, developmentally based anatomical regions of upper, middle, and lower face (Fig. 1A). Any differences revealed in analysis will provide information about how typical patterns of integration are altered to produce the characteristic DS face and will yield information about underlying developmental associations between facial regions.
We compare patterns of integration in the DS and non-DS sibling samples using a bootstrap-based method (Cole and Lele, 2002; Richtsmeier et al., 2006; Richtsmeier and DeLeon, 2009). Using 3D coordinates of 17 landmark locations recorded from 3dMD photos, 31 linear distances were estimated for analysis (Fig. 1B). These linear distances were chosen to cover the facial regions of interest adequately without redundancy. Because there is size variation within the two samples, linear distances were natural log transformed prior to computing correlation matrices (Lele and Richtsmeier, 2001). Using these scaled distances a correlation matrix was calculated for the DS sample and for the non-DS sibling sample.
Further calculations were carried out using MIBoot, a Windows-based software package (Cole, 2002) that statistically evaluates differences in the elements of two correlation or covariance matrices using a nonparametric bootstrap approach. Using the correlation matrices estimated for each sample, a correlation-difference matrix was estimated by subtracting the elements of the DS correlation matrix from the corresponding elements of the correlation matrix calculated for the non-DS siblings. If the correlations between anatomical structures are the same for the two samples, then the correlation difference matrix consists of zeros. If they are not similar, the bootstrap is used to calculate confidence intervals for each correlation difference (α ≤ 0.10) (Richtsmeier et al., 2006). If a confidence interval does not include zero (the expected value under the null hypothesis of similarity), then we reject the null hypothesis of equal associations for that particular linear distance pair (Richtsmeier et al., 2006). Using this method, we tested whether or not patterns of facial integration are similar between DS individuals and non-DS siblings.
Potential confounding factors
A few confounding factors could affect our results and how these results relate to previous investigations of DS craniofacial morphology. First, individuals with DS sometimes have an open-mouth posture when their faces are relaxed due to lax musculature, and reduced mandibular and maxillary size. Although we have made efforts to collect multiple images from each individual and to use the best image for analysis, it may be the case that subtle differences in mouth or jaw posture will affect our results. Second, we measured soft-tissue in 3D, whereas many previous studies focused on osseous characters using 2D lateral cephalograms. While 2D measurements are useful for relatively flat geometric forms, these same measurements can be deficient surrogates for 3D metrics in studies of craniofacial form (Adams et al., 2004). Moreover, bone acts as an anchor for soft-tissue that can be variable in shape and size especially in children with DS. Consequently, it is unlikely that our 3D results from soft-tissue will precisely track findings from prior investigations of osseous craniofacial morphology in DS. Third, many previous studies used analyses that assume normal distributions for metric characters, which could be a poor assumption given that these measurements are from an aneuploid population. The bootstrap analysis used here does not require an assumption of normality in the distribution of data or adjustments for multiple testing (Richtsmeier et al., 2005). Fourth, rather than picking typical individuals from the population, we attempted to match individuals with DS to their own siblings or to individuals who have a sibling with DS to control for variation that occurs due to shared genetic background.
RESULTS
Magnitudes of correlation
Summary statistics for correlation values for both samples are provided in Table 3. Both the mean and the mean of the absolute value of correlations were calculated. While the DS sample exhibits a smaller overall mean correlation value than the non-DS sibling sample, this trend is reversed for the mean of the absolute value of correlations calculation. This is primarily a function of the larger number of negative correlation values found in the DS sample compared with the non-DS sibling sample. For both the mean and the mean of the absolute value correlation summaries, the standard deviation of correlation values is higher in the DS sample.
TABLE 3.
Summary statistics for correlation values of paired linear distances in each sample
| All correlation values | Absolute value of correlation values | |||||
|---|---|---|---|---|---|---|
| Sample | Mean r2 value | Standard deviation | Range | Mean r2 value | Standard deviation | Range |
| DS | 0.27 | 0.259 | −0.362–0.959 | 0.303 | 0.338 | 0.001–0.959 |
| Non-DS siblings | 0.32 | 0.235 | −0.386–0.963 | 0.219 | 0.207 | 0.001–0.963 |
Mean correlation value, standard deviation, and range of correlation values for all paired linear distances from the DS and non-DS sibling sample. Mean correlation values (x̄ = Σx/N) are provided along with the means of the absolute values of correlation values (x̄abs = Σ|x|/N).
Using the three facial regions (upper, middle, and lower), we statistically compared patterns of integration between DS and non-DS siblings. For both samples, the mean correlation values and the mean of the absolute value of correlation values are organized by facial region and provided in Table 4. Comparisons between the samples show that mean correlation values are always relatively higher for the non-DS sibling sample, when comparing homologous mean values for within or between facial regions. When the absolute values of the correlations are used, the results are more variable due to the high number of negative correlations for DS faces. These observations indicate that the faces of DS individuals are integrated differently than that of non-DS siblings.
TABLE 4.
Mean correlation values by facial region and sample
| Mean correlation values | ||||||
|---|---|---|---|---|---|---|
| DS | Non-DS siblings | |||||
| Upper face | Midface | Lower face | Upper face | Midface | Lower face | |
| Upper face | 0.571 | 0.594 | ||||
| Midface | 0.411 | 0.598 | 0.473 | 0.636 | ||
| Lower face | 0.172 | 0.211 | 0.251 | 0.254 | 0.253 | 0.288 |
| Mean of the absolute value of correlations | ||||||
| Upper face | 0.571 | 0.594 | ||||
| Midface | 0.411 | 0.636 | 0.473 | 0.636 | ||
| Lower face | 0.19 | 0.24 | 0.322 | 0.263 | 0.258 | 0.338 |
Elements of the matrix in the top half of this table represent mean correlation values (x̄ = Σx/N) estimated using all linear distance pairs within (diagonal elements) and between (off-diagonal elements) all facial regions separately for each sample. The bottom half of this table shows the mean of the absolute value of correlation coefficients (x̄abs = Σ |x|/N) estimated within (diagonal elements) and between (off-diagonal elements) all facial regions for each sample.
Furthermore, based upon our model of morphological integration, we expected to find that within region comparisons covary more strongly than between region comparisons within each sample, and this is precisely what we see for both samples in Table 4. Thus, for each between region comparison within each sample, the value is lower than the values for within region comparisons of the regions making up the between region comparison. For example, the mean DS correlation value for between region integration of the upper face and midface (i.e., 0.411) is less than the correlation value for within region integration of the upper face or the midface (i.e., 0.571 and 0.598, respectively). This trend is consistent for all facial regions (see Table 4). This lends support to the premise of the morphological integration model that developmental relationships have been altered by gene-dosage imbalance caused by trisomy 21, which manifests as changes in morphological integration in DS.
Pattern of correlation differences
Statistical comparison of the correlation matrices for DS and non-DS siblings revealed that 47 out of 422 linear distance pairs (11.14%) are significantly different by confidence interval testing (α ≤ 0.10). Statistically significant differences are found for linear distances within the lower face region and for comparisons between the lower, middle, and upper face regions; however, no significant differences between samples were found for correlations of linear distances within the upper face and midface. Significant correlation differences are summarized by the direction of correlation values in each sample as follows: positive correlation associations in both samples (+/+; N = 23), negative associations in both samples (−/−; N = 2), negative association in the DS sample and a positive association in the non-DS sibling sample (−/+; N = 22), and positive association in the DS sample and a negative association in the non-DS sibling sample (+/−; N = 0). Many of the significant differences are found around the lower and midfacial region of the face.
Regional comparisons of correlation patterns
Lower face
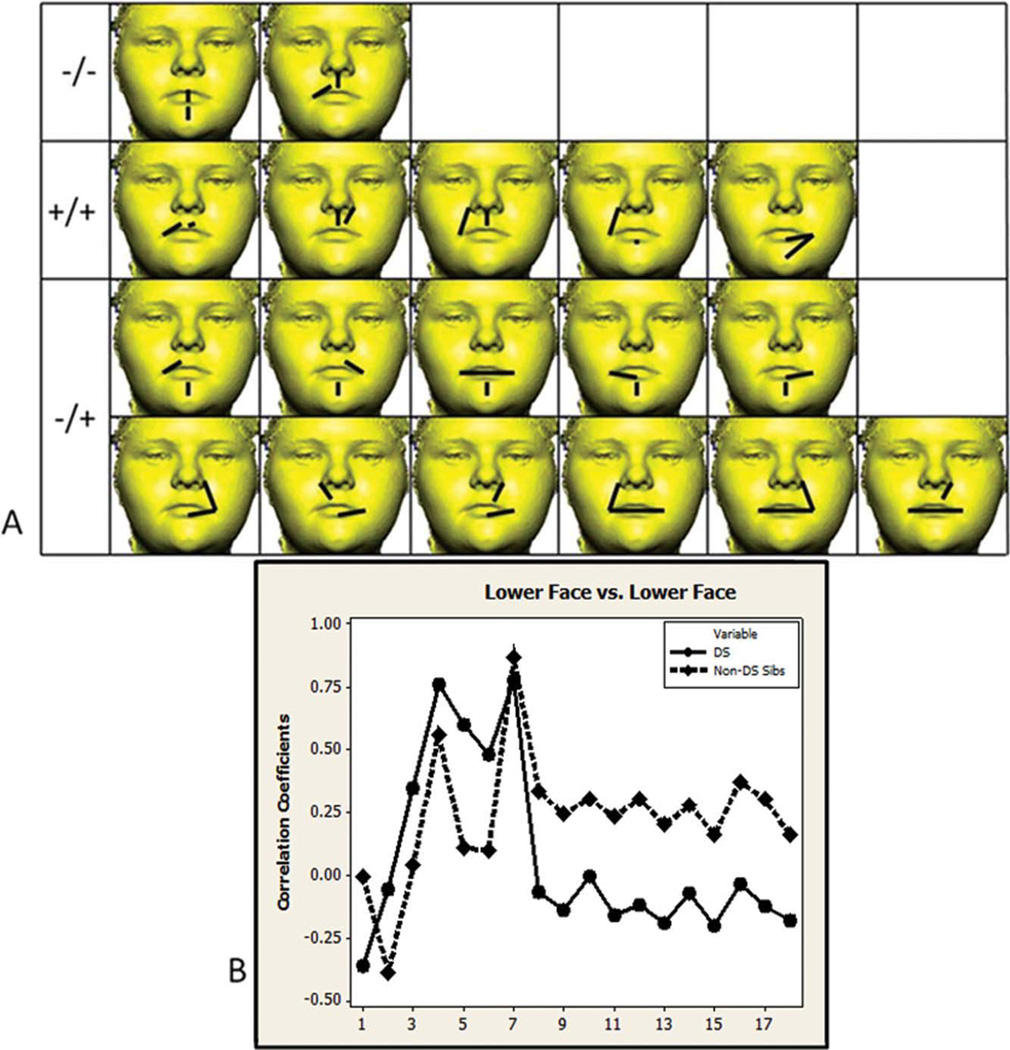
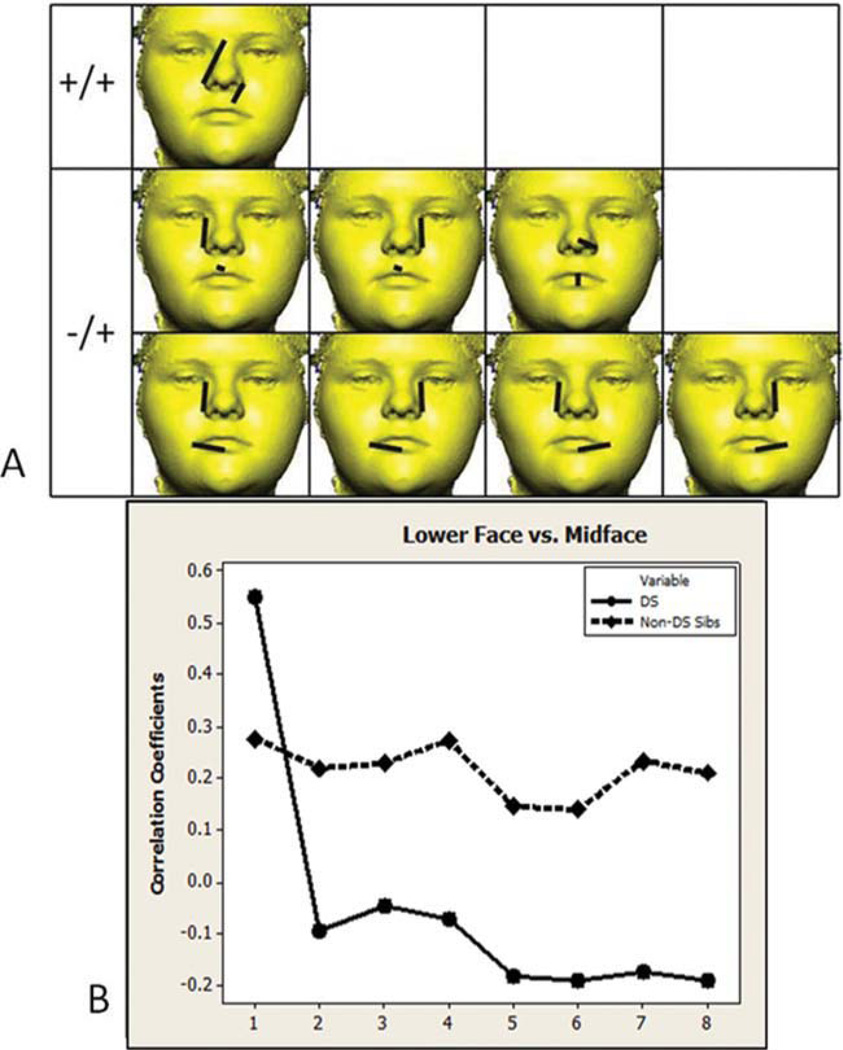
Of the 136 unique linear distance pairs from the lower face, 18 (13.24%) are significantly different in the DS and non-DS sibling sample (see Fig. 2). Two correlations are negative in both samples (−/−) and involve correlations between height measures of the upper lip, mouth, and lower chin, and width measures of the upper lip. Five correlations are positive in both samples (+/+) and involve associations between height measures of the upper lip and chin, and width measures of the lower lip. Eleven correlation differences are negative in the DS sample and positive in the non-DS sibling sample (−/+) and involve correlations between height measures of the upper lip and chin, and width measures of the mouth and lower lip. Comparisons within the lower face region between samples are the most variable in terms of the direction of correlation as evidence by the five +/+, eleven −/+, and two −/− significantly different correlation comparisons. Almost all significantly different correlation values are lower in the DS sample and follow a similar pattern when graphed as corresponding values in the non-DS sibling sample (see Fig. 2B), suggestion a proportional difference in integration for the lower face region of individuals with DS.
Fig. 2.
Lower face region. A) Eighteen linear distance pairs that show significantly different associations in the two samples are shown grouped by the direction of correlation that each sample exhibits (negative in both samples −/−, positive in both samples +/+, negative in the DS sample and positive in the non-DS sibling sample −/+). The paired linear distances shown involve measurements of chin height, lip width and height, and cutaneous upper lip height. B) Significantly different correlation values are graphed in the same order as distances shown in the picture matrix starting with the upper left corner reading from left to right across each row. Circles and solid lines represent the DS sample. Diamonds and dashed lines represent the non-DS sibling sample. Homologous linear distance pairs between samples are located directly above or below each other in this figure and in the following figures. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Middle and upper face
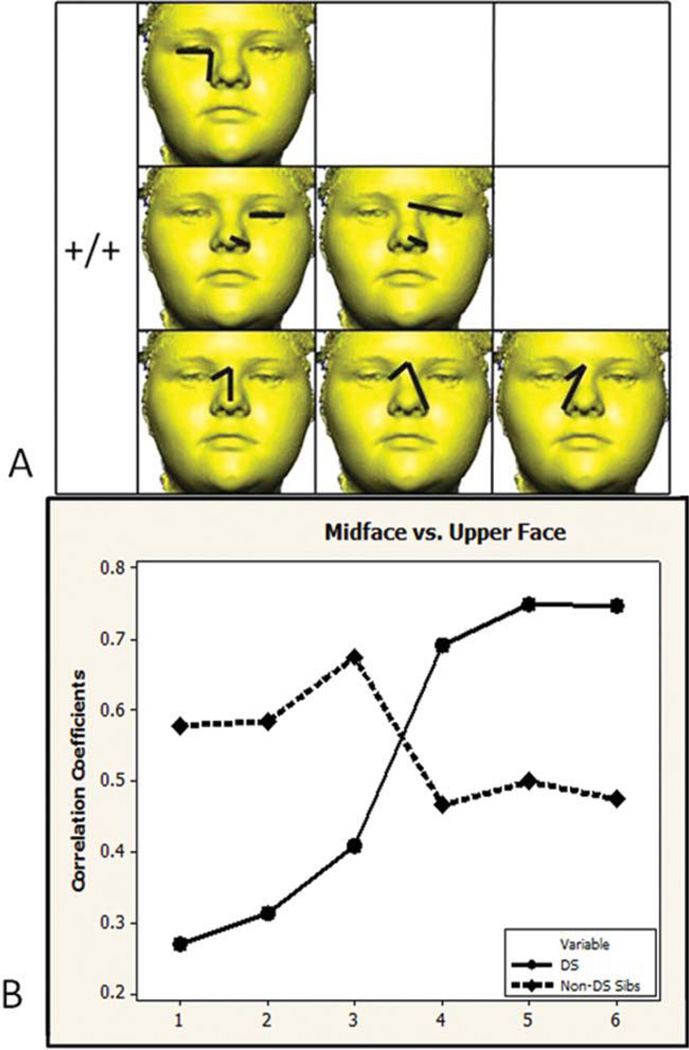
Of the 48 unique linear distance pairs from the midface and upper face, 6 (12.5%) are significantly different between the samples (see Fig. 3). The correlations for all six linear distance pairs are positive in both samples (+/+). Significant differences involve correlations between measures of nose height and width with measurements of eye width and upper facial width. Correlation values are higher in the DS sample for half of the significant correlation differences and lower for the other half (see Fig. 3B).
Fig. 3.
Midface and upper face. A) Six linear distance pairs that show significantly different associations in the two samples are grouped by direction of correlation. The paired linear distances shown involve measures of nose height and width with measurements of eye width and upper facial width. B) Significantly different correlation values are graphed in the same order as distances shown in the picture matrix. Circles and solid lines represent the DS sample. Diamonds and dashed lined represent the non-DS sibling sample. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Lower and upper face
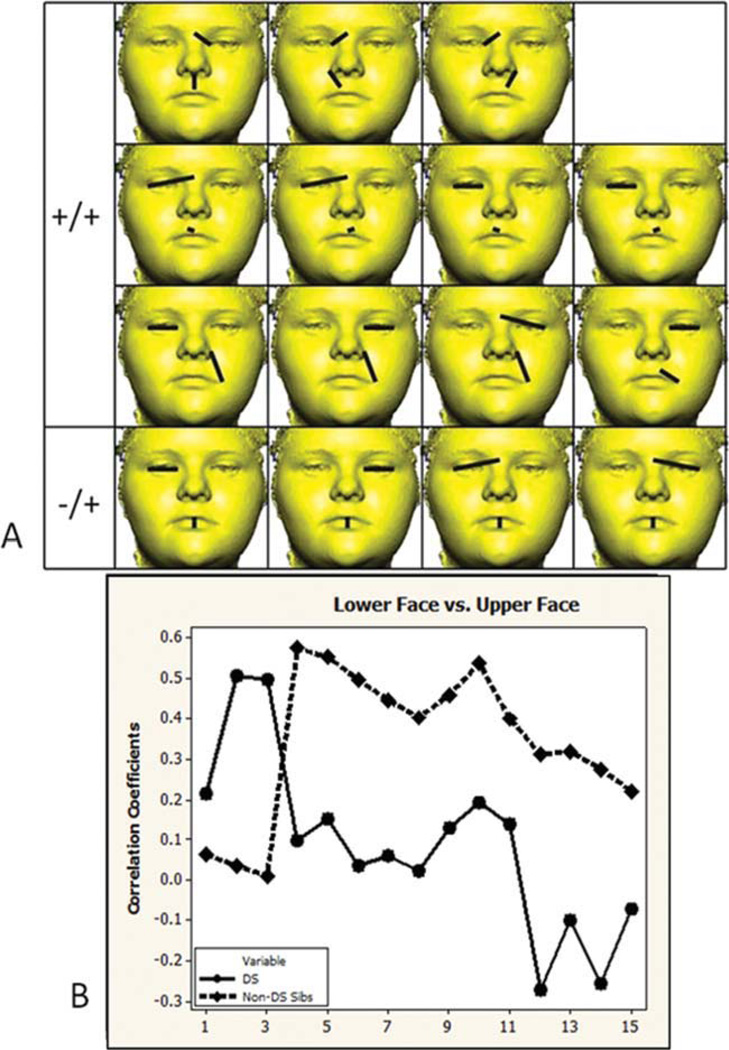
Of the 102 unique linear distance pairs from the lower face and upper face, 15 (14.71%) are significantly different in the DS and non-DS sibling sample (see Fig. 4). Eleven correlation differences are positive in both samples (+/+) and involve correlations between height and width measures of the upper lip, and eye width and upper facial width. Four correlation differences are negative in the DS sample and positive in the non-DS sibling sample (−/+) and involve correlations between measures of mouth height with eye width and upper facial width. Almost all significantly different correlation values are lower in the DS sample and do not follow a similar pattern when graphed as corresponding values in the non-DS sibling sample (see graph in Fig. 4B).
Fig. 4.
Lower face and upper face. A) Fifteen linear distance pairs that show significantly different associations in the two samples are grouped by the direction of correlation that each sample exhibits (positive +/+, negative in the DS sample and positive in the non-DS sibling sample −/+). The paired linear distances shown involve measures lips and cutaneous upper lip width and height, and eye measures of width and upper facial width. B) Significantly different correlation values are graphed in the same order as distances shown in the picture matrix. Circles and solid lines represent the DS sample. Diamonds and dashed lined represent the non-DS sibling sample. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Lower and middle face
Of the 136 unique linear distance pairs from the lower face and midface, 8 (5.88%) are significantly different in the DS and non-DS sibling sample (see Fig. 5). One correlation difference is positive in both samples (+/+) and involves an association of height of the upper lip and nose, which is stronger in the DS sample. Seven correlations are negative in the DS sample and positive in the non-DS sibling sample (−/+) and involve correlations between various measures of lip height and width with measures of midfacial height and nasal ala length. Although a single significantly different correlation value is higher in the DS sample, the other seven significantly different values are consistently lower in the DS sample and follow a similar pattern when graphed as corresponding values in the non-DS sibling sample (see Fig. 5B), suggestion a proportional difference in integration for the lower face and upper face regions of individuals with DS.
Fig. 5.
Lower face and midface region. A) Eight linear distance pairs that show significantly different associations in the two samples are grouped by the direction of correlation that each sample exhibits (positive +/+, negative in the DS sample, and positive in the non-DS sibling sample −/+). The paired linear distances shown involve measures of lip height and width, and measures of nose height, nasal ala width, and midfacial height. B) Significantly different correlation values are graphed in the same order as distances shown in the picture matrix. Circles and solid lines represent the DS sample. Diamonds and dashed lined represent the non-DS sibling sample. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
DISCUSSION
In this study, we compared patterns of morphological integration of facial dimensions in individuals with DS and siblings of individuals with DS to reveal patterns underlying developmental associations among different parts of the face. Patterns of association of linear distances within the upper face region and within the middle face region do not appear to be affected by trisomy 21 as the overall pattern and magnitude of associations of linear distances within these regions is not significantly different between the DS and non-DS sibling samples. However, patterns of association of linear distances within the lower face and between facial regions are significantly different between the two samples. Localized differences in integration of facial features between DS and non-DS siblings are depicted in Figures 3–6, and the overall trend that we see is one of similar patterns of correlation (see graphs from Figs. 3–6) in both samples for about half of the correlation comparisons within the lower face and between the lower, middle, and upper face. In most instances significantly different correlation values between the two samples involve correlation values that have smaller magnitudes in the DS sample compared with the non-DS sibling sample. When a significant difference involves a negative correlation, that value is associated with the DS sample 92% of the time.
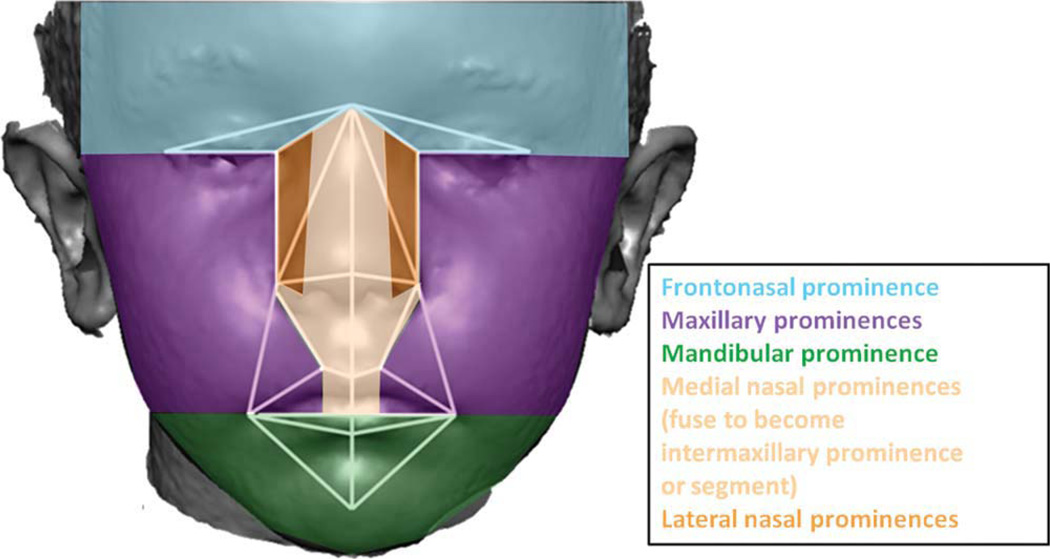
Fig. 6.
Derivatives of the facial prominences that form during craniofacial development. The 31 linear distances used in analysis are shown on a face divided into regions based upon their association with the facial prominences of the embryo.
Although several craniofacial investigations of DS craniofacial morphology have been carried out (Shapiro et al., 1967; Shapiro, 1969, 1970, 1975, 1983, 1992, 2001; Frostad et al., 1971; Fink et al., 1975; O’Riordan and Walker, 1979; Farkas et al., 1985, 2002; Fischer-Brandies et al., 1986; Allanson et al., 1993; Farkas, 2001a,b; Ferrario et al., 2004a,b), this is the first to investigate morphological integration of soft-tissues of the DS face. Previous studies have found midfacial hypoplasia and reductions in mandibular size in DS individuals. The few analyses of soft-tissue morphology indicate that DS faces are smaller overall (Ferrario et al., 2005), while exhibiting a reduced nasal and lower lip surface area, and an increased upper lip area (Ferrario et al., 2004b). Our study used a fundamentally different approach to the analysis of facial morphology, localizing significant differences in facial integration between DS and non-DS faces. The most striking differences are found around the oronasal region, as evidenced by mean correlation value differences, significantly different correlation values within and between regions of the face, and differences in the direction of correlation values (+ or −) in each sample.
One interpretation of the differences in correlations that we have found is that gene-dosage imbalance, caused by trisomy 21, alters the way in which facial prominences are integrated during embryonic craniofacial development, potentially by altering aspects of prominence formation, growth, movement, or fusion during craniofacial development. The linear distances associated with the oronasal region of the face that we measured are derived from several paired facial prominences (see Fig. 6). Many signaling pathways have been shown experimentally to play important roles during craniofacial morphogenesis by contributing to global and local temporally specific patterning of the face during development (Hu et al., 2003; Marcucio et al., 2005; Brugmann et al., 2006, 2007; Donner and Williams, 2006; Levi et al., 2006; Foppiano et al., 2007; Szabo-Rogers et al., 2008; Feng et al., 2009; Hu and Marcucio, 2009; Nakatomi et al., 2010; Roybal et al., 2010). Disruptions or changes in signaling can change relationships between parts of the face during morphogenesis by altering the size, shape, or relative location of facial prominences, or the timing of their appearance, thereby producing a variety of craniofacial dysmorphologies.
Our findings suggest that gene dosage imbalance may alter the timing, sequence, or boundaries of specific morphogenetic events taking place during craniofacial morphogenesis. This view is supported by recent research using Ts65Dn mouse models for DS (Roper et al., 2009) that show reduced mitosis, proliferation, delamination, and migration of cranial neural crest cells into the head and face, which eventually give rise to the facial prominences, thereby establishing trisomy 21 as a neurocristopathy. Our results provide evidence that gene dosage imbalance does not affect the facial prominences equally, supporting the idea that trisomy 21 genes differentially affect developing facial structures rather than causing a generalized disruption to development as previously suggested (Shapiro, 1983). Currently, it is not known if age-specific dosage effects alter the phenotype at specific points in development. Longitudinal investigations of DS facial development may help to elucidate how dosage effects alter developmental trajectories. Future investigations using DS mouse models should seek to elucidate how gene-dosage imbalance affects specific facial prominences and how facial prominences interact during development to produce the characteristic facial morphology associated with DS.
ACKNOWLEDGMENTS
Grant sponsor: NIH; Grant numbers: R01-HD038384, R01-DE018500, R01-DE018500-S1; Grant sponsor: NSF; Grant number: GRF-053135.
This work was supported in part by a grant from the National Institute of Child Health & Human Development (RHR) and the National Institute of Dental and Craniofacial Development (JTR). The content is solely the responsibility of the authors and does not necessarily represent the official views of these Institutes of the National Institutes of Health.
LITERATURE CITED
- [Accessed on 18 June, 2011];3dMDface photogrammatic system. Available at: http://3dmd.com/
- Adams GL, Gansky SA, Miller AJ, Harrell WE, Jr, Hatcher DC. Comparison between traditional 2-dimensional cephalometry and a 3-dimensional approach on human dry skulls. Am J Orthod Dentofac Orthop. 2004;126:397–409. doi: 10.1016/j.ajodo.2004.03.023. [DOI] [PubMed] [Google Scholar]
- Aldridge K, Boyadjiev SA, Capone GT, DeLeon VB, Richtsmeier JT. Precision and error of three-dimensional phenotypic measures acquired from 3dMD photogrammetric images. Am J Med Genet A. 2005;138A:247–253. doi: 10.1002/ajmg.a.30959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allanson JE, O’Hara P, Farkas LG, Nair RC. Anthropometric craniofacial pattern profiles in Down syndrome. Am J Med Genet. 1993;47:748–752. doi: 10.1002/ajmg.1320470530. [DOI] [PubMed] [Google Scholar]
- Brugmann SA, Goodnough LH, Gregorieff A, Leucht P, ten Berge D, Fuerer C, Clevers H, Nusse R, Helms JA. Wnt signaling mediates regional specification in the vertebrate face. Development. 2007;134:3283–3295. doi: 10.1242/dev.005132. [DOI] [PubMed] [Google Scholar]
- Brugmann SA, Kim J, Helms JA. Looking different: understanding diversity in facial form. Am J Med Genet A. 2006;140:2521–2529. doi: 10.1002/ajmg.a.31361. [DOI] [PubMed] [Google Scholar]
- CDCP. Center for disease control and prevention: improved national prevalence estimates for 18 selected major birth defects—United States: 1999–2001. Morbidity Mortality Weekly Rep. 2006;54:1301–1305. [PubMed] [Google Scholar]
- Chai Y, Maxson RE., Jr Recent advances in craniofacial morphogenesis. Dev Dyn. 2006;235:2353–2375. doi: 10.1002/dvdy.20833. [DOI] [PubMed] [Google Scholar]
- Cheverud J. Developmental integration and the evolution of pleiotropy. Am Zool. 1996;36:44–50. [Google Scholar]
- Cole T., III . MIBoot Windows-based software for bootstrap-based comparison of morphological integration patterns. Richtsmeier Lab; 2002. [Accessed on 18 June, 2011]. Available at: http://getahead.psu.edu. [Google Scholar]
- Cole TM, III, Lele S. Bootstrap-based methods for comparing morphological integration patterns. Am J Phys Anthropol. 2002;117 Suppl 34:55. [Google Scholar]
- Donner AL, Williams T. Frontal nasal prominence expression driven by Tcfap2a relies on a conserved binding site for STAT proteins. Dev Dyn. 2006;235:1358–1370. doi: 10.1002/dvdy.20722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Down JL. Observations on an ethic classification of idiots. Clinical Lectures and Reports by the Medical and Surgical Staff. 1866;3:259–262. [Google Scholar]
- Farkas LG. Surface anatomy of the face in Down’s syndrome; anthropometric proportion indices in the craniofacial regions. J Craniofac Surg. 2001a;12:519–526. doi: 10.1097/00001665-200111000-00003. [DOI] [PubMed] [Google Scholar]
- Farkas LG. Surface anatomy of the face in Down’s syndrome; linear and angular measurements in the craniofacial regions. J Craniofac Surg. 2001b;12:373–380. doi: 10.1097/00001665-200107000-00011. [DOI] [PubMed] [Google Scholar]
- Farkas LG, Katic MJ, Forrest CR. Surface anatomy of the face in Down’s syndrome: age-related changes of anthropometric proportion indices in the craniofacial regions. J Craniofac Surg. 2002;13:368–374. doi: 10.1097/00001665-200205000-00002. [DOI] [PubMed] [Google Scholar]
- Farkas LG, Munro IR, Kolar JC. Abnormal measurements and disproportions in the face of Down’s syndrome patients: preliminary report of an anthropometric study. Plast Reconstr Surg. 1985;75:159–169. doi: 10.1097/00006534-198502000-00002. [DOI] [PubMed] [Google Scholar]
- Feng W, Leach SM, Tipney H, Phang T, Geraci M, Spritz RA, Hunter LE, Williams T. Spatial and temporal analysis of gene expression during growth and fusion of the mouse facial prominences. PLoS One. 2009;4:e8066. doi: 10.1371/journal.pone.0008066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrario VF, Dellavia C, Zanotti G, Sforza C. Soft tissue facial anthropometry in Down syndrome subjects. J Craniofac Surg. 2004a;15:528–532. doi: 10.1097/00001665-200405000-00037. [DOI] [PubMed] [Google Scholar]
- Ferrario VF, Dellavia C, Colombo A, Sforza C. Three-dimensional assessment of nose and lip morphology in subjects with Down syndrome. Ann Plast Surg. 2004b;53:577–583. doi: 10.1097/01.sap.0000130702.51499.6b. [DOI] [PubMed] [Google Scholar]
- Ferrario VF, Dellavia C, Serrao G, Sforza C. Soft tissue facial angles in Down’s syndrome subjects: a three-dimensional non-invasive study. Eur J Orthod. 2005;27:355–362. doi: 10.1093/ejo/cji017. [DOI] [PubMed] [Google Scholar]
- Fink GB, Madaus WK, Walker GF. A quantitative study of the face in Down’s syndrome. Am J Orthod. 1975;67:540–553. doi: 10.1016/0002-9416(75)90299-7. [DOI] [PubMed] [Google Scholar]
- Fischer-Brandies H, Schmid RG, Fischer-Brandies E. Craniofacial development in patients with Down’s syndrome from birth to 14 years of age. Eur J Orthod. 1986;8:35–42. doi: 10.1093/ejo/8.1.35. [DOI] [PubMed] [Google Scholar]
- Foppiano S, Hu D, Marcucio RS. Signaling by bone morphogenetic proteins directs formation of an ectodermal signaling center that regulates craniofacial development. Dev Biol. 2007;312:103–114. doi: 10.1016/j.ydbio.2007.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frostad WA, Cleall JF, Melosky LC. Craniofacial complex in the trisomy 21 syndrome (Down’s syndrome) Archs Oral Biol. 1971;16:707–722. doi: 10.1016/0003-9969(71)90116-6. [DOI] [PubMed] [Google Scholar]
- Gardiner K, Fortna A, Bechtel L, Davisson MT. Mouse models of Down syndrome: how useful can they be? Comparison of the gene content of human chromosome 21 with orthologous mouse genomic regions. Gene. 2003;318:137–147. doi: 10.1016/s0378-1119(03)00769-8. [DOI] [PubMed] [Google Scholar]
- Hattori M, Fujiyama A, Taylor TD, Watanabe H, Yada T, Park HS, Toyoda A, Ishii K, Totoki Y, Choi DK, et al. The DNA sequence of human chromosome 21. Nature. 2000;405:311–319. doi: 10.1038/35012518. [DOI] [PubMed] [Google Scholar]
- Helms JA, Schneider RA. Cranial skeletal biology. Nature. 2003;423:326–331. doi: 10.1038/nature01656. [DOI] [PubMed] [Google Scholar]
- Hu D, Marcucio RS. A SHH-responsive signaling center in the forebrain regulates craniofacial morphogenesis via the facial ectoderm. Development. 2009;136:107–116. doi: 10.1242/dev.026583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D, Marcucio RS, Helms JA. A zone of frontonasal ectoderm regulates patterning and growth in the face. Development. 2003;130:1749–1758. doi: 10.1242/dev.00397. [DOI] [PubMed] [Google Scholar]
- Kuppermann M, Learman LA, Gates E, Gregorich SE, Nease RF, Jr, Lewis J, Washington AE. Beyond race or ethnicity and socioeconomic status: predictors of prenatal testing for Down syndrome. Obstet Gynecol. 2006;107:1087–1097. doi: 10.1097/01.AOG.0000214953.90248.db. [DOI] [PubMed] [Google Scholar]
- Larson WJ. Human embryology. New York: Churchill Livingstone; 1997. [Google Scholar]
- LeJeune J, Gauthier M, Turpin R. Etudes des chromosomes somatiques de neuf enfants mongoliens. CR Acad Sci. 1959;248:1721–1722. [PubMed] [Google Scholar]
- Lele SR, Richtsmeier JT. An invariant approach to statistical analyses of shape. New York: Chapman and Hall/CRC; 2001. [Google Scholar]
- Levi G, Mantero S, Barbieri O, Cantatore D, Paleari L, Beverdam A, Genova F, Robert B, Merlo GR. Msx1 and Dlx5 act independently in development of craniofacial skeleton, but converge on the regulation of Bmp signaling in palate formation. Mech Dev. 2006;123:3–16. doi: 10.1016/j.mod.2005.10.007. [DOI] [PubMed] [Google Scholar]
- Makino T, McLysaght A. Ohnologs in the human genome are dosage balanced and frequently associated with disease. Proc Natl Acad Sci U S A. 2010;107:9270–9274. doi: 10.1073/pnas.0914697107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcucio RS, Cordero DR, Hu D, Helms JA. Molecular interactions coordinating the development of the forebrain and face. Dev Biol. 2005;284:48–61. doi: 10.1016/j.ydbio.2005.04.030. [DOI] [PubMed] [Google Scholar]
- Megarbane A, Ravel A, Mircher C, Sturtz F, Grattau Y, Rethore MO, Delabar JM, Mobley WC. The 50th anniversary of the discovery of trisomy 21: the past, present, and future of research and treatment of Down syndrome. Genet Med. 2009;11:611–616. doi: 10.1097/GIM.0b013e3181b2e34c. [DOI] [PubMed] [Google Scholar]
- Nakatomi M, Wang XP, Key D, Lund JJ, Turbe-Doan A, Kist R, Aw A, Chen Y, Maas RL, Peters H. Genetic interactions between Pax9 and Msx1 regulate lip development and several stages of tooth morphogenesis. Dev Biol. 2010;340:438–449. doi: 10.1016/j.ydbio.2010.01.031. [DOI] [PubMed] [Google Scholar]
- O’Riordan MW, Walker GF. Dimensional and proportional characteristics of the face in Down’s syndrome. J Dent Handicapped. 1979;4:6–9. [PubMed] [Google Scholar]
- Olson EC, Miller RL. Morphological integration. Chicago: University of Chicago Press; 1958. [Google Scholar]
- Reeves RH, Baxter LL, Richtsmeier JT. Too much of a good thing: mechanisms of gene action in Down syndrome. Trends Genet. 2001;17:83–88. doi: 10.1016/s0168-9525(00)02172-7. [DOI] [PubMed] [Google Scholar]
- Richtsmeier J, Cole T, III, Lele S. Landmark morphometrics and the analysis of variation. In: Hallgrímsson B, editor. Variation: a central concept in biology. Boston: Elsevier Academic Press; 2005. pp. 49–68. [Google Scholar]
- Richtsmeier JT, Aldridge K, DeLeon VB, Panchal J, Kane AA, Marsh JL, Yan P, Cole TM., III Phenotypic integration of neurocranium and brain. J Exp Zool B Mol Dev Evol. 2006;306B:360–378. doi: 10.1002/jez.b.21092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richtsmeier JT, DeLeon VB. Morphological integration of the skull in craniofacial anomalies. Orthod Craniofac Res. 2009;12:149–158. doi: 10.1111/j.1601-6343.2009.01448.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper RJ, Van Horn JF, Cain CC, Reeves RH. A neural crest deficit in Down syndrome mice is associated with deficient mitotic response to Sonic hedgehog. Mech Dev. 2009;126:212–219. doi: 10.1016/j.mod.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roybal PG, Wu NL, Sun J, Ting MC, Schafer CA, Maxson RE. Inactivation of Msx1 and Msx2 in neural crest reveals an unexpected role in suppressing heterotopic bone formation in the head. Dev Biol. 2010;343:28–39. doi: 10.1016/j.ydbio.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro B. Prenatal dental anomalies in mongolism: comments on the basis and implications of variability. Ann NY Acad Sci. 1970;171:562–577. [Google Scholar]
- Shapiro B. Amplified developmental instability in Down’s syndrome. Ann Hum Genet Lond. 1975;38:429–437. doi: 10.1111/j.1469-1809.1975.tb00632.x. [DOI] [PubMed] [Google Scholar]
- Shapiro B. Down syndrome—a disruption of homeostasis. Am J Med Genet. 1983;14:241–269. doi: 10.1002/ajmg.1320140206. [DOI] [PubMed] [Google Scholar]
- Shapiro BL. A twin study of palatal dimensions partitioning genetic and environmental contributions to variability. Angle Orthod. 1969;39:139–151. doi: 10.1043/0003-3219(1969)039<0139:ATSOPD>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Shapiro BL. Development of human autosomal aneuploid phenotypes (with an emphasis on Down syndrome) Acta Zool Fennica. 1992;191:97–105. [Google Scholar]
- Shapiro BL. Developmental instability of the cerebellum and its relevance to Down syndrome. J Neural Transm Suppl. 2001;(61):11–34. doi: 10.1007/978-3-7091-6262-0_2. [DOI] [PubMed] [Google Scholar]
- Shapiro BL, Gorlin RJ, Redman RS, Bruhl HH. The palate and Down’s syndrome. N Engl J Med. 1967;276:1460–1463. doi: 10.1056/NEJM196706292762603. [DOI] [PubMed] [Google Scholar]
- Sommer CA, Henrique-Silva F. Trisomy 21 and Down syndrome: a short review. Braz J Biol. 2008;68:447–452. doi: 10.1590/s1519-69842008000200031. [DOI] [PubMed] [Google Scholar]
- Szabo-Rogers HL, Geetha-Loganathan P, Nimmagadda S, Fu KK, Richman JM. FGF signals from the nasal pit are necessary for normal facial morphogenesis. Dev Biol. 2008;318:289–302. doi: 10.1016/j.ydbio.2008.03.027. [DOI] [PubMed] [Google Scholar]
- Weinberg SM, Naidoo S, Govier DP, Martin RA, Kane AA, Marazita ML. Anthropometric precision and accuracy of digital three-dimensional photogrammetry: comparing the Genex and 3dMD imaging systems with one another and with direct anthropometry. J Craniofac Surg. 2006;17:477–483. doi: 10.1097/00001665-200605000-00015. [DOI] [PubMed] [Google Scholar]
- Wiseman FK, Alford KA, Tybulewicz VL, Fisher EM. Down syndrome-recent progress and future prospects. Hum Mol Genet. 2009;18:R75–R83. doi: 10.1093/hmg/ddp010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JY, Oh AK, Ohta E, Hunt AT, Rogers GF, Mulliken JB, Deutsch CK. Validity and reliability of craniofacial anthropometric measurement of three-dimensional digital photogrammetric images. Cleft Palate Craniofac J. 2008;45:232–239. doi: 10.1597/06-175. [DOI] [PubMed] [Google Scholar]