Abstract
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) is caused by the production of low-affinity penicillin-binding protein 2a and β-lactamases, which are encoded by mecA and blaZ, respectively. Expressions of the two key genes are mutually regulated by MecI and BlaI. The aim of this study was to design specific anti-mecR1 and anti-blaR1 deoxyribozymes and identify the restoration of susceptibility in MRSA isolates with mecI or blaI or no deletions by interfering with the mutual regulation of mecA and blaZ.
Material and methods
Specific deoxyribozymes were designed by using the program RNA structure 4.6. RNA substrates were obtained by transcription in vitro and used to assess the target cleavage of DNAzymes. Transcription of mecR1-mecA and blaR1-blaZ was analysed by real time RT-PCR. The susceptibility of MRSA was tested.
Results
Specific deoxyribozymes showed efficient catalytic activity to each own substrate mecR1 or blaR1 in vitro and caused the reduction of mecR1 and blaR1 transcription in vivo. Furthermore, simultaneous administration of two DNAzymes to knockdown mecR1 and blaR1 resulted in increased susceptibility of all MRSA strains tested in this study.
Conclusions
These results demonstrated that combined use of the two specific phosphorothioate deoxyribozymes could be a viable and promising strategy to restore the susceptibility of almost all MRSA clinical isolates.
Keywords: methicillin-resistant Staphylococcus aureus, mecR1, blaR1, phosphorothioate deoxyribozyme
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA), a multidrug resistant gram-positive bacterium, is one of the leading causes of hospital-associated infections [1-3]. Vancomycin is one of the few drugs that have remained effective against MRSA. However, recently the susceptibility of MRSA to vancomycin has decreased and vancomycin is losing potency against MRSA [4-6]. Linezolid (LZD) is typically used for the treatment of infections with Gram-positive bacteria, including MRSA and VRSA in the United States [7, 8]. The emergence of linezolid (LZD)-resistant MRSA infections has been reported from the USA, the UK, Brazil and Japan; it warns that we are currently facing a growing shortage of effective antibiotics [7, 9, 10]. The emergence of MRSA resistant to the last-defence antibiotics (vancomycin and linezolid) has created an urgent need to discover alternative anti-MRSA approaches.
The antibiotic resistance to β-lactam in MRSA is mediated by mecA, which codes for the low-affinity penicillin binding protein 2a (PBP 2a), an enzyme working to allow cell-wall synthesis despite the presence of β-lactam antibiotics [11]. Transcription of mecA is regulated by a repressor, mecI, and a sensor/transducer, mecR1 [12]. More than 90% of staphylococcal isolates also produce β-lactamases, the product of blaZ, and contain blaZ regulatory sequences (blaI and blaR1) that are similar in sequence and function to mecA regulators [13]. In addition to regulating blaZ transcription, BlaI also binds to mecA-mecR1 promoter-operator (P-O) sequences and regulates their transcription. Co-regulation of mecA by both MecI and BlaI has been proved in clinical S. aureus isolates [14, 15].
The restoration of susceptibility in MRSA by respective blocking mecR1 and blaR1 is only applicable to strains that harbour wild mecI or blaI [16, 17]. But, mutations and deletions in mecI and deletions of both mecI and mecR1 appear to be common in clinical S. aureus isolates. It has been shown that 96% of clinical isolates with mutant mecI or with a deletion of mecI contain blaI, while the isolates that do not contain blaI all have wild-type mecI sequences [12, 18, 19]. The co-egulation of mecA by both MecI and BlaI has been proved [14, 15, 20]. Therefore, MRSA must have at least one of the two functional mecA regulators. This mutual regulation of mecA by both MecI and BlaI plays a very important role in antibiotic resistance of MRSA.
Based on current understanding of the mechanism of mecA co-regulation, the complete restoration of β-lactam antibiotic susceptibility in MRSA clinical isolates should be achieved by simultaneous blockade of blaR1 and mecR1. In this study, we explored the use of specific anti-mecR1 and anti-blaR1 phosphorothioated deoxyribozymes (PS-DRz1694 and PS-DRz1366) and identified the restoration of susceptibility in MRSA clinical isolates with mecI deletion or blaI deletion or no deletions by simultaneous blockage of MecR1 and BlaR1 mediated signal pathways.
Material and methods
Bacterial strains
Three clinical isolates, wild-type MRSA080302 with blaI only (ΔmecI-blaI), MRSA080305 with mecI only (mecI-ΔblaI) and MRSA080309 with both mecI and blaI (mecI-blaI) were obtained from cultures of sputum and catheter samples from patients in Xijing Hospital (Xi’an, China). MSSA type strain ATCC 29213 was used as a positive control.
Deoxyribozyme design
The program RNAstructure 4.6 was used to design anti-mecR1 and anti-blaR1 10-23 deoxyribozymes. The deoxyribozymes were synthesized by Shanghai Sangon Biological Engineering Technology and Services (Shanghai, China), and were partly phosphorothioated (Table I).
Table I.
Summary of sequences and cleavage sites of DNAzymes used
| DNAzyme sequence (5'-3')a | RNA substrate sequence (5'-3') | Cleavage site (nt) | |
|---|---|---|---|
| DRz1694 | ATTCGCAggctagctacaacgaTGTCTTCGCCTT | AAGGCGAAGACAAUGCGAAU | 1694 |
| DRz1366 | CTTGAGTTGAGggctagctacaacgaCGCAGTAAT | AUUACUGCGACUCAACUCAAG | 1366 |
| DRz5491 | CATAGGCAcgcatgctaacacgaTGTCCGCTTCT |
The 10-23 catalytic motifs of PS-DRzymes are indicated in lower case. The sequences of PS-DNAzymes for the target binding domains are in uppercase, which are designed to be complementary to mecR1 and blaR1 respectively and were all modified with phosphorothioates to increase nuclease resistance
In vitro transcription
The two gene fragments (mecR1 and blaR1) were constructed into pGEM-T vector by Shanghai Sangon Biological Engineering Technology and Services (Shanghai, China), respectively. The two constructed plasmids were linearized by NcoI (Toyobo, Japan). The RiboMAX large scale production system (Promega, Madison, WI, USA) was used for the transcription under the guide of the manufacturer’s instructions. The resulting RNA was purified and concentrated with sodium acetate and chilled ethanol.
PS-DRzyme cleavage assay
To assess the target cleavage of DNAzymes, RNA substrates and DNAzymes were mixed with 20 µl of reaction buffer (50 mM Tris, pH 8.0, 20 mM MgCl2, 0.01% SDS) at 37°C. The reaction was quenched at various time intervals with 50 mmol/l EDTA and the samples were denatured for 10 min at 70°C, and then the uncleaved substrate and products were resolved by electrophoresis on 3% denaturing agarose gel. The gel was incubated in 1×SYBR Gold nucleic acid gel staining solution (Invitrogen, Carlsbad, CA, USA) for 40 min. The band densities were quantitated by the Alpha Imager system (Alpha Innotech, California, USA). The fraction of substrates cleaved by DNAzymes was calculated and plotted against time. All reported kinetic values are means ± SD of at least three independent experiments. Vmax and K M values were determined from the y intercept and slope, respectively, of the best-fit line to a Lineweaver-Burke plot of 1/V vs. 1/[S].
PS-DRzyme delivery and real time RT-PCR
Staphylococcus aureus was cultured overnight and 1 ml of culture was transferred to 100 ml broth medium. The medium was incubated at 37°C until the OD600 reached 0.55-0.65. Cells were centrifuged at 6000 rpm at 4°C for 10 min. The cell pellet was washed twice by re-suspending in 100 ml of sterilized, ice-cold water and centrifuging at 6000 rpm at 4°C for 10 min. Then the pellet was washed an additional four times with 40, 10, 2 and 1 ml of 10% cold glycerol. Finally, the pellet was resuspended in 1 ml of 10% cold glycerol, distributed into 50 ml aliquots. 10 mg/l anti-mecR1 phosphorothioated deoxyribozyme1694 (PS-DRz1694) or 10 mg/l anti-blaR1 phosphorothioated deoxyribozyme1366 (PS-DRz1366) was introduced into competent MRSA strains by electroporation under the following conditions: 25 µf, 900 V, 200 Ω, time constant 3.6-4.2 ms.
The culture of S. aureus was centrifuged at 5000 rpm for 10 min at 4°C and supernatant was decanted. The cell pellet was suspended in 100 µl of lysis solution (50 mM Tris-HCl pH 7.5, 5 mM EDTA pH 8, 50 mM NaCl, 10 mM Tris) with 300 µg of lysozyme (Sigma) and 5 µg of lysostaphin (Sigma) for 30 min at room temperature. The total RNA was extracted from the bacterial lysis with Trizol reagent (Invitrogen, Carlsbad, CA, U.S.A.) following the manufacturer’s instructions, and the RNA samples were treated with DNase I to remove any genomic DNA contamination. The total RNAs were extracted from the bacterial lysis with Trizol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions. The cDNA was synthesized by reverse transcription from 1 µg of each RNA sample using SuperScript III reverse transcriptase (Invitrogen, Carlsbad, CA, USA). The nucleotide sequences for various primers were listed (Table II). PCR was run in a DNA Engine Opticon (MJ Research, Waltham, USA) using SYBR Green I. PCR reagents consisted of: 12.5 µl of SYBR Premix Ex TaqTM (TaKaRa, DRR041S, Japan), 0.5 µl of 50× ROX Reference Dye (TaKaRa, DRR041S, Japan), 0.75 µl of each primer (10 µM) and 1 µl of sample cDNA, in a final volume of 25 µl. Thermal cycling conditions: initial denaturation step at 95°C for 5 min, 50 cycles at 95°C for 10 s, 56°C for 20 s and 72°C for 20 s.
Table II.
Oligonucleotide primers used for PCR
| Genes | Primers | Primer sequence (5’-3’) | Location [bp] | Size [bp] | Annealing temperature [°C] |
|---|---|---|---|---|---|
| mecR1 | Forward | acacgacttcttcggttag | 218-236 | 336 | 58 |
| Reverse | gtacaatttgggatttcact | 534-553 | |||
| mecA | Forward | gcaatcgctaaagaactaag | 553-572 | 225 | 58 |
| Reverse | aatgggaccaacataaccta | 758-777 | |||
| blaR1 | Forward | acaatgaagtagaagccgatagat | 719-742 | 489 | 55 |
| Reverse | gtcggtcaagtccaaaca | 1207-1190 | |||
| blaZ | Forward | agagatttgcctatgcttca | 311-330 | 461 | 56 |
| Reverse | agtatctccgcttttattattt | 771-750 | |||
| 16SrRNA | Forward | gttattagggaagaacatatgtg | 446-468 | 750 | 55 |
| Reverse | ccaccttcctccggtttgtcacc | 1195-1173 | |||
The cDNA of the control group was fold series diluted and the analysis was performed as follows: for each sample, the difference in Ct values (ΔCt) was calculated for target genes subtracting Ct for the reference RNA. ΔCt = Ct(target gene) – Ct(16SrRNA). Relative expression of target genes mRNA was calculated using the comparative Ct method as previously described [17]. The ΔΔCt values were calculated by the following equation: ΔΔCt = ΔCt(treatment) – ΔCt(control). The mean of these ΔΔCt measurements was then used to calculate expression of the test gene (2–ΔΔCt) relative to the reference gene, 16SrRNA, and normalized to the untreated control as follows: relative expression = 2–ΔΔCt. The evaluation of 2–ΔΔCt indicates the fold change in gene expression relative to the untreated control.
Susceptibility testing
After electroporation, the cells were recovered in preheated broth medium containing 6 mg/l of oxacillin at 37°C with 150 rpm agitation for 1 h. The growth determination was carried out as follows: The cells were diluted 104-108 times. Fifty microlitres of diluted cells were spread onto plates of Mueller-Hinton agar containing 6 mg/l of oxacillin, and the plates were incubated for 48 h at 35°C. The colonies were counted for plates with > 10 and < 500 colonies and the total number of CFU (colony forming units) per sample was determined by correcting the colony count for the dilution factor.
To determine the growth curves for MRSA, 100 µl of cell dilution were added to the wells of a 96-well microtitre plate, and the plate was incubated at 35°C with agitation at 120 rpm. The optical density (OD600) of each well was measured with a microplate reader (Bio-Rad Laboratories, Tokyo, Japan) at different time points.
According to the guidelines of the Clinical and Laboratory Standards Institute (CLSI), the minimum inhibitory concentrations (MICs) of oxacillin for MRSA080302, MRSA080305, MRSA080309 and MSSA ATCC29213 were determined by the standard broth dilution method [17, 21].
Statistical analysis
Values are expressed as mean ± SD. One-way ANOVA analysis followed by Student-Newman-Keuls (SNK) test was performed for CFU and RNA expression variables. A value of p < 0.05 was considered statistically significant.
Results
Design and characterization of anti-mecR1 and anti-blaR1 deoxyribozyme
Based on analysis of target mRNA secondary structures by the program RNAstructure 4.6, two optimized deoxyribozymes, PS-DRz1694 targeting mecR1 and PS-DRz1366 targeting blaR1, were generated. The design parameters of ΔGoligo-oligo, ΔGoligo-self, ΔGduplex, ΔGoverall and Tm for PS-DRz1694 were –2.1, 0, –28.0, –17.3 kcal/mol, 75.3°C, respectively. The same parameters for PS-DRz1366 were –3, 0, –25.2, –20.6 kcal/mol and 73.9°C.
The target binding domain sequences of PS-DRz1694 and PS-DRz1366 are shown in Table I; they were designed to be complementary in sequence to nt 1682~1701 of mecR1 (GenBank accession no. X63598) and nt 1357~1377 of blaR1 (GenBank accession no. M62650) in S. aureus respectively. Both PS-DRz1694 and PS-DRz1366 are 34-mers consisting of a central 10-23 catalytic core domain (15 nts) flanked by target-specific binding arms (19 nts), which were modified with phosphorothioates to increase nuclease resistance. Mismatched sequences PS-DRz5491 had been randomly aligned with the same number of bases, in which binding arms were also phosphorothioated.
In vitro cleavage of mecR1 and blaR1
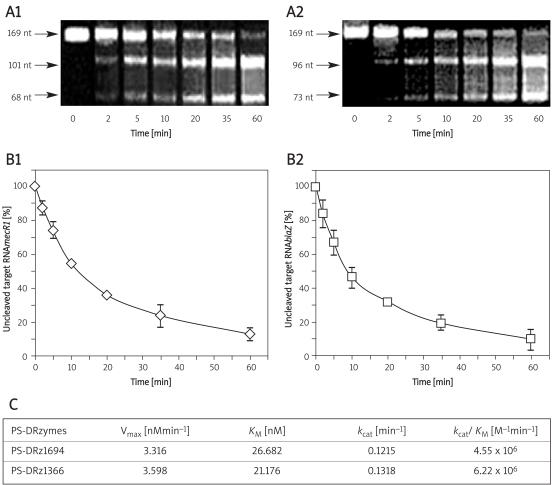
The k cat and K M values for PS-DRz1694 or PS-DRz1366 were determined under a wide range of single turnover conditions with the respective mecR1 or blaR1 RNA substrates. The cleavage products of mecR1 and blaR1 were obtained respectively (Figures 1 A1 , 1 A2). Both DNAzymes exhibited similar catalytic kinetics and the PS-DRz1694 or PS-DRz1366 was able to cleave nearly 90% of mecR1 or blaR1 within 60 min in a cell-free system respectively (Figures 1 B1 , 1 B2). Cleavage of mecR1 RNA by PS-DRz1694 proceeded with the efficiency of k cat/K M of 4.55 × 106 M–1 min–1 and Vmax at 3.316 nM/min. PS-DRz1366 showed k cat/K M value at 6.22 × 106 M–1 min–1 and Vmax at 3.598 nM/min (Figure 1C). These results indicated that anti-mecR1 PS-DRz1694 and anti-blaR1 PS-DRz1366 could effectively cleave RNAmecR1 and RNAblaR1, respectively, and the cleavage efficiency of PS-DRz1366 was higher than that of PS-DRz1694.
Figure 1.
In vitro cleavage activities of PS-DRz1694 and PS-DRz1366. (A1) Cleavage of RNAsmecR1 by PS-DRz1694. (A2) Cleavage of RNAsblaR1 by PS-DRz1366. Time course of the cleavage of in vitro, transcribed mRNAs of mecR1 and blaR1 cleaved by PS-DRz1694 (B1) and PS-DRz1366 (B2), respectively. (C) Kinetic parameters of the PS-DRz1694 and PS-DRz1366 reactions
Real-time quantitation assays for mecA/blaZ and mecR1/blaR1 transcription
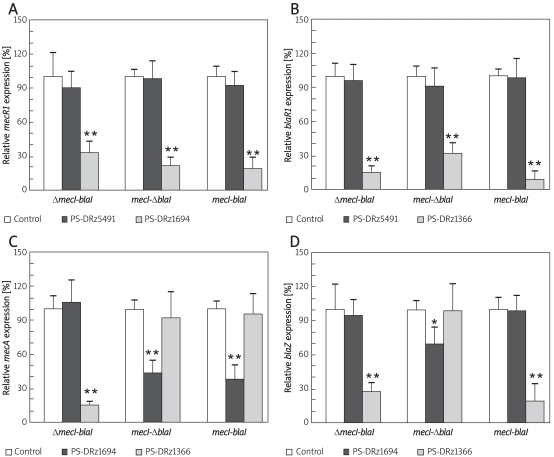
To ascertain whether the blockage of mecR1 or blaR1 also inhibits the expression of downstream gene mecA/blaZ in MRSA isolates, MRSA080302 or MRSA080305 with mecI or blaI deletions, and MRSA080309 with wild types of mecI/blaI, the anti-mecR1 PS-DRz1694 or anti-blaR1 PS-DRz1366 was applied to each clinical isolate of MRSA respectively. The transcriptions of mecR1/blaR1 and mecA/blaZ were detected by real-time PCR.
Compared with the control groups, the relative transcription of mecR1 in three MRSA isolates (MRSA080302, MRSA080305 and MRSA080309) was decreased to 33%, 21%, and 18% of the control values by PS-DRz1694 treatment, respectively (Figure 2A). And the relative transcription of blaR1 of those MRSA isolates was decreased to 14%, 31%, and 9% of the control values by PS-DRz1366 treatment, respectively (Figure 2B). These results demonstrated that mecR1 or blaR1 was blocked specifically by PS-DRz1694 or PS-DRz1366 respectively.
Figure 2.
Comparison of expression of antibiotic resistant genes in three clinical MRSA isolates, MRSA080302 (ΔmecIblaI), MRSA080305 (mecI-ΔblaI) and MRSA080309 (mecI-blaI). Relative mRNA expression of mecR1 (A), blaR1 (B), mecA (C), and blaZ (D)
* p < 0.05,** p < 0.01 vs. control, Δ indicates that the repressor is absent
Compared with the control group, anti-blaR1 PS-DRz1366 treatment caused 85% reduction for mecA expression and 72% of reduction for blaZ expression respectively in the mecI deleted strain MRSA080302 (Figures 2C, 2D). However, the expression of mecA and blaZ was not altered by treatment with anti-mecR1 PS-DRz1694 alone in MRSA080302 (Figures 2C, 2D). A similar inhibition pattern on mecA/blaZ expression was observed in the blaI deleted strain MRSA080305 after anti-mecR1 PS-DRz1694 treatment. The expression of mecA and blaZ in blaI deleted strain MRSA080305 showed 56% (Figure 2C) and 32% (Figure 2D) reduction after PS-DRz1694 treatment. But the expression of mecA and blaZ in MRSA080305 was not altered by PS-DRz1366. Meanwhile, the expression of blaZ or mecA in MRSA080309, a strain harbouring wild type mecI-blaI, was inhibited only by PS-DRz1366 or PS-DRz1694 respectively (Figure 2C, 2D).
Restoration of susceptibility to antibiotic in MRSA clinical isolates
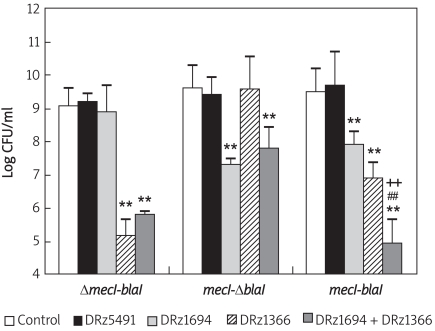
We found that the downregulation of mecR1 and blaR1 by combined administration of anti-mecR1 PS-DRz1694 and anti-blaR1 PS-DRz1366 correlated with the restoration of susceptibility of MRSA clinical isolates to β-lactam antibiotics. The numbers of MRSA080302 colonies in PS-DRz1366 alone or PS-DRz1366 and PS-DRz1694 combination-treated cultures on Mueller-Hinton agar containing oxacillin (6 µg/ml) were reduced by 103- to 104-fold, respectively (Figure 3). The 102- to 103-fold reduction of MRSA080305 colonies was achieved by treatment of either PS-DRz1694 alone or PS-DRz1694 and PS-DRz1366 combination (Figure 3). In MRSA080309, oxacillin gave rise to a 104- or 105-fold striking reduction in CFU after PS-DRz1694 or PS-DRz1366 treatment, respectively (Figure 3). The combination of these two DNAzymes caused stronger effects on CFU reduction and led to a synergistic effect on the reversal of antibiotic resistance of MRSA080309 (Figure 3).
Figure 3.
Effects of PS-DRz1694 or PS-DRz1366 on the growth of three clinical MRSA isolates – MRSA080302 (ΔmecI-blaI), MRSA080305 (mecI-ΔblaI) and MRSA080309 (mecI-blaI). DRz1366 group, DRz1694 group and DRz1694 + DRz1366 group vs. control *p < 0.05, **p < 0.01; DRz1694 + DRz1366 group vs. DRz1694 group, +p < 0.05, ++p < 0.01; DRz1694 + DRz1366 group vs. DRz1366 group, # p < 0.05, ##p < 0.01. The following are different treated groups: control, 10 mg/l PBS; DRz5491, 10 mg/l DRz5491; DRz1694, 10 mg/l DRz1694; DRz1366, 10 mg/l DRz1366; DRz1694 + DRz1366, 10 mg/l DRz1694 + 10 mg/l DRz1366. The data are shown as mean ± SD for 8 samples
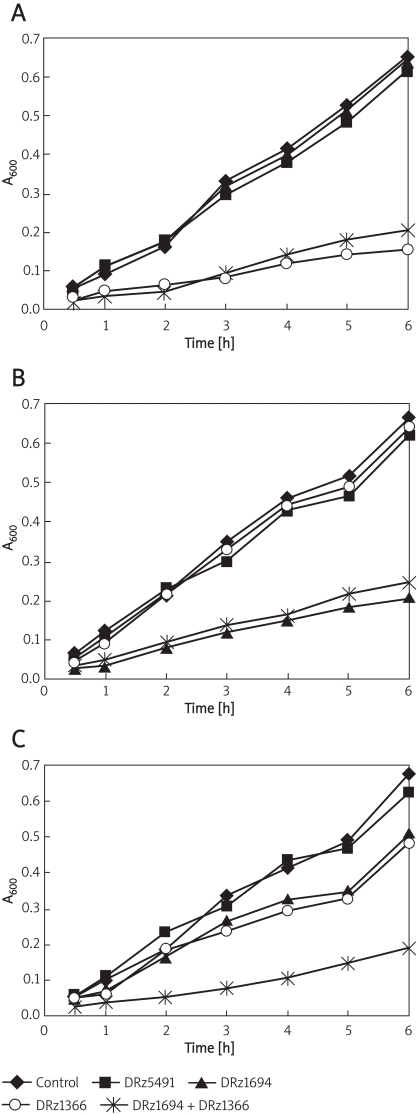
In liquid medium containing oxacillin (6 mg/l), the growth of PS-DRz1366-treated MRSA080302 and PS-DRz1694-treated MRSA080305 cells was inhibited, respectively (Figures 4A, 4B). However, the growth of MRSA080302 was not influenced by treatment with either PS-DRz1694 alone or mismatched PS-DRz5491 (Figures 3, 4A). Similarly, the growth of MRSA080305 was not affected by treatment with either PS-DRz1366 alone or mismatched PS-DRz5491 (Figures 3, 4B). The growth of MRSA080309 was inhibited by treatment with either PS-DRz1694 or PS-DRz1366 alone. Combination treatment of PS-DRz1694 and PS-DRz1366 resulted in the more significant restoration of susceptibility of MRSA080309 to oxacillin (Figures 3, 4C).
Figure 4.
Effects of PS-DRz1694 or PS-DRz1366 on the growth of three clinical MRSA isolates in liquid medium (6 mg/l of oxacillin). The growth of different groups was monitored by using OD600 measurements. (A) The OD600 value of MRSA080302 in different treated groups. (B) The OD600 value of MRSA080305 in different treated groups. (C) The OD600 value of MRSA080309 in different treated groups. The following are different treated groups: control, 10 mg/l PBS; DRz5491, 10 mg/l DRz5491; DRz1694, 10 mg/l DRz1694; DRz1366, 10 mg/l DRz1366; DRz1694 + DRz1366, 10 mg/l DRz1694 + 10 mg/l DRz1366. The data are shown as mean ± SD for 5 samples
Meanwhile, PS-DRz1366 reduced the MIC of oxacillin for MRSA080302 (ΔmecI-blaI) from 1,024 mg/l to 1 mg/l and PS-DRz1694 reduced the MIC of oxacillin on MRSA080305 (mecI-ΔblaI) from 512 mg/l to 2 mg/l (Table III), both of which are within the oxacillin sensitivity range for the MRSA strains on the basis of the interpretive criteria recommended by the CLSI, representing the full restoration of MRSA susceptibility to oxacillin. The PS-DRz1366 or PS-DRz1694 alone did not alter the MIC of oxacillin for MRSA080305 or MRSA080302 respectively (Table III). On MRSA080309 strain harbouring wild type mecI/blaI, PS-DRz1694 or PS-DRz1366 only reduced the MIC of oxacillin from 1024 mg/l to 512 mg/l or from 1024 mg/l to 256 mg/l, respectively, which indicated a partial restoration of susceptibility of MRSA080309 to oxacillin. However, the combined administration of PS-DRz1694 and PS-DRz1366 to MRSA080309 caused a dramatic reduction on MIC of oxacillin from 1024 mg/l to 1 mg/l, which represented a full restoration of MRSA susceptibility to oxacillin. In addition, the MICs of oxacillin on MRSA080302 and MRSA080305 were decreased from 1024 mg/l to 2 mg/l and from 512 mg/l to 4 mg/l respectively under DRz1694 and DRz1366 combination treatment (Table III).
Table III.
MICs of oxacillin in the presence/absense of PS-DRz1694/1366 in MRSA080302, MRSA080305 or MRSA080309 broth culture
| Strain | Competent | Electroporation | PS-DRz1694 [mg/l] | PS-DRz1366 [mg/l] | MIC [mg/l] |
|---|---|---|---|---|---|
| ATCC29213 | − | − | − | − | 0.5 |
| MRSA080302 | − | − | −; | − | 1024 |
| MRSA080302 | + | + | − | − | 1024 |
| MRSA080302 | + | + | 10 | 0 | 1024 |
| MRSA080302 | + | + | 0 | 10 | 1 |
| MRSA080302 | + | + | 10 | 10 | 2 |
| MRSA080305 | − | − | − | − | 512 |
| MRSA080305 | + | + | − | − | 512 |
| MRSA080305 | + | + | 10 | 0 | 2 |
| MRSA080305 | + | + | 0 | 10 | 512 |
| MRSA080305 | + | + | 10 | 10 | 4 |
| MRSA080309 | − | − | − | − | 1024 |
| MRSA080309 | + | + | − | − | 1024 |
| MRSA080309 | + | + | 10 | 0 | 512 |
| MRSA080309 | + | + | 0 | 10 | 256 |
| MRSA080309 | + | + | 10 | 10 | 1 |
Represents competent cells or electroporation processing;
refers to non-competent cells or no electroporation processing. The MIC of oxacillin to Staphylococcus aureus: sensitive (S): MIC ≤ 2 mg/l; resistant (R): MIC ≥ 4 mg/l
Discussion
Methicillin-resistant Staphylococcus aureus is resistant to all commercially available β-lactam antibiotics and also represents a therapeutic challenge, because effective therapeutic options are becoming limited. The expression of antibiotic-resistant genes mecA and blaZ is involved in antibiotic resistance of MRSA. Inhibition of bacterial gene expression by the antisense approach has been proposed as a promising strategy for bacterial infection therapy through targeting specific genes in bacteria [22-25]. Especially, catalytic oligodeoxynucleotides are valuable tools to downregulate the expression of resistant genes in a sequence-specific manner and have been widely applied in vitro and in vivo [26, 27].
In this study, we designed two deoxyribozymes (PS-DRz1694 and PS-DRz1366) specifically targeting mecR1 and blaR1 to restore susceptibility of clinical MRSA isolates. First, the consistency of catalytic activities of deoxyribozyme were demonstrated in vitro and in vivo. With the increase of the reaction time, the substrate mecR1 or blaR1 was cleaved into two cleavage products by the two DNAzymes in vitro, respectively. Then DNAzymes were electroporated into bacteria and the transcription levels of mecR1 and blaR1 were tested. The repression of mecR1 or blaR1 was observed in all MRSA strains treated by anti-mecR1 or anti-blaR1 DNAzyme respectively and showed specific catalytic activity of deoxyribozyme in vivo.
However, the inhibitory efficiency on the mec regulated mecA or bla regulated blaZ expression was different in all MRSA strains. In the blaI deleted strain MRSA080305 (mecI-ΔblaI), expression of both mecA and blaZ was decreased significantly by anti-mecR1 DNAzyme PS-DRz1694, but not by anti-blaR1 DNAzyme PS-DRz1366. Meanwhile in mecI deleted strain MRSA080302 (ΔmecI-blaI), anti-mecR1 DNAzyme PS-DRz1366 caused significant reduction of mecA and blaZ expression. In mecI-blaI non-mutation strain MRSA080309, PS-DRz1366 or PS-DRz1694 only cleaved its target gene, blaR1 or mecR1, and thereafter led to blaZ or mecA repression respectively. The outcomes indicated identically that mecR1 cleaved by PS-DRz1694 could prevent the cleavage of MecI. Similarly, blaR1 cleaved by PS-DRz1366 could merely prohibit the cutting of BlaI. So, we demonstrated that mecR1 and blaR1 are specific for their own homologous repressor and are not interchangeable in repressor cleavage. Furthermore, PS-DRz1366-mediated uncleavage of MecI could effectively inhibit the expression of not only mecA but also blaZ in MRSA080302. It is the same for PS-DRz1694 in MRSA080305. Therefore, we demonstrated that mecA and blaZ can be mutually regulated by either MecI or BlaI, which might be the result of the fact that MecI and BlaI are almost identical and are interchangeable in repression of target gene transcription.
Further investigations on bacteria growth and susceptibility in the presence of oxacillin demonstrated a high correlation between targeted gene repression and MRSA growth suppression. The anti-mecR1 PS-DRz1694 caused repression of mecA and blaZ in MRSA080305. Also, reduction of CFU and inhibition of the growth curve were observed in MRSA080305, but not in mecI deleted strain MRSA080302. In contrast, anti-blaR1 PS-DRz1366 effectively inhibited the expression of mecA and blaZ in MRSA080302, and only inhibited MRSA080302 growth. A high correlation between the down-regulation of blaR1/mecR1 and suppression of MRSA growth was also observed in non-mecI-blaI mutation strain MRSA080309 after treatment with PS-DRz1366 or PS-DRz1694 respectively. As both mecR1-mecI-mecA and blaR1-blaI-blaZ systems play roles in induction of antibiotic resistance in MRSA [13, 15, 28], blockade of each signal pathway only partially restored the susceptibility. The combination treatment of PS-DRz1694 and PS-DRz1366 on MRSA080309 resulted in synergic effects on susceptibility restoration to oxacillin.
It is particularly encouraging that MICs of oxacillin to MRSA080302, MRSA080305 and MRSA080309 were fully restored to values within the sensitivity defining range by PS-DRz1366, PS-DRz1694 and combined administration of these two DNAzymes, respectively (Table III). Although PS-DRz1366 and PS-DRz1694 only worked in certain MRSA strains, more importantly, we demonstrated that simultaneous administration of the two DNAzymes to knockdown mecR1 and blaR1 resulted in increased susceptibility of all MRSA strains tested in this study. This is thought to be accomplished by reducing the level of mecA and blaZ. When the levels of mecA and blaZ are lowered, the MRSA is not protected against oxacillin and the MRSA is killed.
The results of the present study indicate that co-blockade of blaR1-blaZ and mecR1-mecA signal pathways without detecting mecI or blaI mutation is a feasible strategy to restore the susceptibility of MRSA clinical strains to existing β-lactam antibiotics.
Acknowledgments
We thank Mr. Xiuli Xu in Xijing Hospital for help with the clinical MRSA strain. This work was supported by grants from the National Natural Science Foundation of China (grant number 30973666) and the Natural Science Foundation of Shaanxi Province (2002C2-04).
References
- 1.Klevens RM, Morrison MA, Nadle Jl, et al. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. Jama. 2007;298:1763–71. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 2.Bancroft EA. Antimicrobial resistance it's not just for hospitals. JAMA. 2007;298:1803–04. doi: 10.1001/jama.298.15.1803. [DOI] [PubMed] [Google Scholar]
- 3.Lloyd-Smith E, Hull MW, Tyndall MW, et al. Community-associated methicillin-resistant Staphylococcus aureus is prevalent in wounds of community-based injection drug users. Epidemiol Infect. 2010;138:713–20. doi: 10.1017/S0950268810000464. [DOI] [PubMed] [Google Scholar]
- 4.Delaney JA, Schneider-Lindner V, Brassard P, Suissa S. Mortality after infection with methicillin-resistant Staphylococcus aureus (MRSA) diagnosed in the community. BMC Med. 2008;6:2. doi: 10.1186/1741-7015-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neoh HM, Hori S, Komatsu M, et al. Impact of reduced vancomycin susceptibility on the therapeutic outcome of MRSA bloodstream infections. Ann Clin Microbiol Antimicrob. 2007;6:13. doi: 10.1186/1476-0711-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weigel LM, Donlan RM, Shin DH, et al. High-level vancomycin-resistant Staphylococcus aureus isolates associated with a polymicrobial biofilm. Antimicrob Agents Chemother. 2007;51:231–8. doi: 10.1128/AAC.00576-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hill RL, Kearns AM, Nash J, et al. Linezolid-resistant ST36 methicillin-resistant Staphylococcus aureus associated with prolonged linezolid treatment in two paediatric cystic fibrosis patients. J Antimicrob Chemother. 2010;65:442–5. doi: 10.1093/jac/dkp494. [DOI] [PubMed] [Google Scholar]
- 8.Falagas ME, Vardakas KZ. Benefit-risk assessment of linezolid for serious gram-positive bacterial infections. Drug Saf. 2008;31:753–68. doi: 10.2165/00002018-200831090-00004. [DOI] [PubMed] [Google Scholar]
- 9.Morales G, Picazo JJ, Baos E, et al. Resistance to linezolid is mediated by the cfr gene in the first report of an outbreak of linezolid-resistant Staphylococcus aureus. Clin Infect Dis. 2010;50:821–5. doi: 10.1086/650574. [DOI] [PubMed] [Google Scholar]
- 10.Yoshida K, Shoji H, Hanaki H, et al. Linezolid-resistant methicillin-resistant Staphylococcus aureus isolated after long-term repeated use of linezolid. J Infect Chemother. 2009;15:417–9. doi: 10.1007/s10156-009-0727-3. [DOI] [PubMed] [Google Scholar]
- 11.Archer GL, Bosilevac JM. Signaling antibiotic resistance in staphylococci. Science. 2001;291:1915–6. doi: 10.1126/science.1059671. [DOI] [PubMed] [Google Scholar]
- 12.Suzuki E, Kuwahara-Arai K, Richardson JF, Hiramatsu K. Distribution of mec regulator genes in methicillin-resistant Staphylococcus clinical strains. Antimicrob Agents Chemother. 1993;37:1219–26. doi: 10.1128/aac.37.6.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hackbarth CJ, Chambers HF. blaI and blaR1 regulate beta-lactamase and PBP 2a production in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1993;37:1144–9. doi: 10.1128/aac.37.5.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lewis RA, Dyke KG. mecI represses synthesis from the beta-lactamase operon of Staphylococcus aureus. J Antimicrob Chemother. 2000;45:139–44. doi: 10.1093/jac/45.2.139. [DOI] [PubMed] [Google Scholar]
- 15.McKinney TK SV, Craig WA, Archer GL. Transcription of the gene mediating methicillin resistance in Staphylococcus aureus (mecA) is corepressed but not coinduced by cognate mecA and betalactamase regulators. J Bacteriol. 2001;183:6862–8. doi: 10.1128/JB.183.23.6862-6868.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hou Z, Meng JR, Niu C, et al. Restoration of antibiotic susceptibility in methicillin-resistant Staphylococcus aureus by targeting mecR1 with a phosphorothioate deoxyribozyme. Clin Exp Pharmacol Physiol. 2007;34:1160–4. doi: 10.1111/j.1440-1681.2007.04705.x. [DOI] [PubMed] [Google Scholar]
- 17.Hou Z, Meng JR, Zhao JR, et al. Inhibition of beta-lactamase-mediated oxacillin resistance in Staphylococcus aureus by a deoxyribozyme. Acta Pharmacol Sin. 2007;28:1775–82. doi: 10.1111/j.1745-7254.2007.00646.x. [DOI] [PubMed] [Google Scholar]
- 18.Weller TM. The distribution of mecA, mecR1 and mecI and sequence analysis of mecI and the mec promoter region in staphylococci expressing resistance to methicillin. J Antimicrob Chemother. 1999;43:15–22. doi: 10.1093/jac/43.1.15. [DOI] [PubMed] [Google Scholar]
- 19.Kobayashi N, Taniguchi K, Urasawa S. Analysis of diversity of mutations in the mecI gene and mecA promoter/operator region of methicillin-resistant Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob Agents Chemother. 1998;42:717–20. doi: 10.1128/aac.42.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosato AE, Kreiswirth BN, Craig WA, Eisner W, Climo MW, Archer GL. mecA-blaZ corepressors in clinical Staphylococcus aureus isolates. Antimicrob Agents Chemother. 2003;47:1460–3. doi: 10.1128/AAC.47.4.1460-1463.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Serefko A, Malm A. Sensitivity of Candida albicans isolates to caspofungin – comparison of microdilution method and E-test procedure. Arch Med Sci. 2009;5:23–7. [Google Scholar]
- 22.Matveeva OV, Mathews DH, Tsodikov AD, et al. Thermodynamic criteria for high hit rate antisense oligonucleotide design. Nucleic Acids Res. 2003;31:4989–94. doi: 10.1093/nar/gkg710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cairns MJ, Hopkins TM, Witherington C, et al. Target site selection for an RNA-cleaving catalytic DNA. Nat Biotechnol. 1999;17:480–6. doi: 10.1038/8658. [DOI] [PubMed] [Google Scholar]
- 24.Rasmussen LC, Sperling-Petersen HU, Mortensen KK. Hitting bacteria at the heart of the central dogma sequence-specific inhibition. Microb Cell Fact. 2007;6:24. doi: 10.1186/1475-2859-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baum DA, Silverman SK. Deoxyribozymes useful DNA catalysts in vitro and in vivo. Cell Mol Life Sci. 2008;65:2156–74. doi: 10.1007/s00018-008-8029-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fahmy RG, Waldman A, Zhang G, et al. Suppression of vascular permeability and inflammation by targeting of the transcription factor c-Jun. Nat Biotechnol. 2006;24:856–63. doi: 10.1038/nbt1225. [DOI] [PubMed] [Google Scholar]
- 27.Zhang L, Gasper WJ, Stass SA, Ioffe OB, Davis MA, Mixson AJ. Angiogenic inhibition mediated by a DNAzyme that targets vascular endothelial growth factor receptor 2. Cancer Res. 2002;62:5463–9. [PubMed] [Google Scholar]
- 28.Safo MK, Zhao Q, Ko TP, et al. Crystal structures of the BlaI repressor from Staphylococcus aureus and its complex with DNA insights into transcriptional regulation of the bla and mec operons. J Bacteriol. 2005;187:1833–44. doi: 10.1128/JB.187.5.1833-1844.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]