Abstract
Introduction
Glutathione S-transferase (GST) is a xenobiotic metabolising enzyme (XME), which may modify susceptibility in certain ethnic groups, showing ethnic dependent polymorphism. The aim of this study was to determine GSTM1, GSTM3 and GSTT1 gene polymorphisms in a Malaysian population in Kuala Lumpur.
Material and methods
Blood or buccal swab samples were collected from 137 Form II students from three schools in Wilayah Persekutuan Kuala Lumpur. Genotyping was done by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP).
Results
Glutathione-S-transferase GSTM3 gene frequencies were 89% for AA, 10% for AB and 1% for BB. The gene frequencies for deleted GSTM1 and GSTT1 were 66% and 18% respectively.
Conclusions
This study suggested that the Malay population is at risk for environmental diseases and provides the basis for gene-environment association studies to be carried out.
Keywords: gene, glutathione S-transferase, polymorphism, Malaysians
Introduction
The glutathione S-transferase (GST) enzyme system constitutes a family of multifunctional enzymes which play an important role in biotransformation and detoxification of many different xenobiotic and endogenous substances [1]. Human cytosolic GSTs are polymorphic, and have ethnic-dependent polymorphism frequencies [2,3]. Of all the GSTs, the Mu and Theta classes have been extensively investigated worldwide.
The glutathione S-transferase Mu-1 (GSTM1) gene polymorphism includes a single base alteration giving rise to the functionally identical GSTM1*A and GSTM1*B alleles; a duplication of the GSTM1 gene; and deletion of the GSTM1 gene resulting in the GSTM1 null allele (GSTM1*0) [4]. The percentage of GSTM1*0 is different among populations as it tends to be higher in Caucasians and Asians than Africans, ranging from 21.7% in Nigerians, 43% in French to 58.3% in Chinese [5].
Glutathione S-transferase Mu-3 (GSTM3) is specifically expressed in the ciliated airway epithelium and is the principal GST-mu class enzyme in bronchoalveolar macrophages. The polymorphism in the GSTM3 gene consisting of two alleles, GSTM3*A and GSTM3*B, is distinguished by a three base pair deletion in intron 6. Frequencies of GSTM3 polymorphism in most available studies showed that homozygous GSTM3*A (wild type) was the most common allele, while homozygous GSTM3*B was less frequent [6,7].
The most common polymorphism in the glutathione S-transferase Theta-1 (GSTT1) gene consists of a deletion of the whole gene (GSTT1*0), resulting in the lack of active enzyme. GSTT1*0 frequencies were 13-26% and 35-52% in Caucasians and Asians respectively [8].
To date, most studies of xenobiotic metabolising enzymes (XME) gene-disease associations have used the case-control design, which had certain disadvantages, such as the following: 1) the selection of cases has not been well-described and controls were not selected from the same source population as the cases; 2) there is a possibility of population stratification effects on the results of population-based case control studies. Population stratification includes differences between groups in ethnic origin, and it can also arise because of differences between groups of similar ethnic origin but between which there has been limited admixture; for example, variations in the frequency of certain genotypes in African Americans appear to be much wider than those observed in subjects of European origin, and therefore the possibility of stratification may be higher. And 3) a small study size is a limitation of many studies that test a priori hypotheses about gene-disease associations, and the number of cases or exposed subjects and the number of controls should be planned prior to the study [9,10].
To overcome these problems in the study of gene-disease association in our population, we did a pilot study to determine the actual genotypic frequencies of these genes in the population.
Material and methods
Subject selection
Two schools were selected by cluster random sampling among Wilayah Persekutuan Kuala Lumpur schools, which were Wangsa Maju Secondary School Section 5 (n = 487), and Taman Melati Setapak Secondary school (n = 408). The total number of Form II students in the two schools was 895.
Inclusion criteria:
male/female 14 years old;
Muslim Malay mother and father;
healthy (no history of hospital admission).
The information sheet about the study, which included the consent form and methodology to extract DNA, was written in Bahasa Malaysia, Chinese and English languages and was distributed to all students by Form II teachers. Two weeks later we collected the sheets and forms to identify the subjects who agreed to take part in the study. A week later the collection of samples commenced.
The total number of sheets returned was 405 and the total number of those who agreed to take part in our study was 138. One sample out of 138 samples was discarded because of error during DNA extraction from blood.
Ethical consideration
The Universiti Kebangsaan Malaysia Research and Ethics committee approved this project. Participants were asked to complete a “Consent Form” stating their willingness to participate in the study and a separate consent form for permission to store participants’ samples for future investigations.
Collection of the sample
Blood
Five to 10 ml of venous blood samples were collected in EDTA tubes and transported to the Medical Molecular Biology Institute of the National University of Malaysia (UMBI-HUKM) within 3 h, and then manual DNA extraction from the blood was immediately carried out as described by Miller et al., 1988 [14].
Buccal swab
This method was done directly by the researchers by using an alcohol-free cytobrush. Two brushes from each subject were collected and transferred to HUKM, UMBI and stored at –20°C until DNA extraction. DNA extraction was done with “3 min” protocol MasterAmp Buccal Swab DNA Extraction Kits (Epicenter, USA) as instructed by the manufacturer.
Quality control
Quality control was carried out to validate specific DNA amplification for the DNA samples extracted from whole blood and buccal swab. Twelve samples were examined separately: seven samples which were randomly selected (blood and buccal swab) from the population in this study (who gave informed consent for blood test), and five samples which were obtained from the staff of our institute who agreed to be volunteers. We amplified the three genes GSTM1, GSTT1, and GSTM3 using the same protocols as mentioned below.
To overcome the problems of high failure rates and misclassification, we used Whole Genome Amplification (GenomePlex®) for buccal swab. This method yielded a qualitative and quantitative DNA compared to DNA extracted from blood. But unfortunately GSTT1 was also difficult to amplify even after WGA, so the size products for these genes were changed by changing the primers as advised by the technical support of WGA from Sigma Company (USA).
Genotyping
The PCR approach for GSTM1 polymorphism is adopted from Fryer et al. [15], GSTM3 as described by Flamant et al. [12]. For GSTT1 blood samples genotyping was done as described by Abdul-Rahman et al. [16], while buccal swab samples GSTT1 amplification from buccal swab was done after changing size products to 112 bp by adapting Suryanarayana et al. [17] with modifications. The PCRs were carried out in a final volume of 25 ml mixture containing 0.5 µmol/l each of the primers, 2.5 mmol/l MgCl2, 200 µmol/l each dNTP, 1X buffer, 2 U of Taq DNA polymerase and 5 µl of DNA template from buccal swab extraction. After initial denaturation at 94°C for 5 min, amplification was obtained with 35 cycles of 94°C for 30 s, 5°C for 30 s and 72°C for 30 s followed by a final extension at 72°C for 10 min.
Results
Quality control results
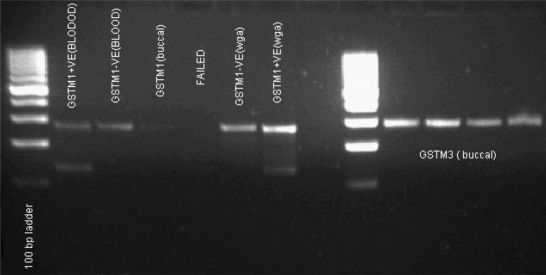
For GSTM3 the results of the reaction were the same for both DNA extracted from blood and buccal swab (Figure 1), while GSTM1 from buccal swab showed high failure rates and some results were difficult to be interpreted as positive GSTM1 (Figure 1). GSTT1 analysis showed a result of a deleted GSTT1 gene (GSTT1*0 genotype) in all the buccal swab samples, while the same samples appeared GSTT1 positive genotype on DNA extracted from blood (Figure 2).
Figure 1.
Comparing GSTM1 amplification from blood, buccal swab and WGA, and GSTM3 amplification from buccal swab without WGA
Figure 2.
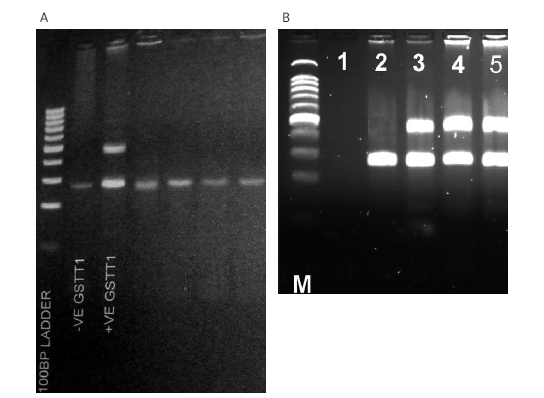
A – GSTT1 amplification from buccal swab (–VE and +VE GSTT1 is from blood used as control) compared to B – same samples from blood (lane 1 is water control)
Response rate
The total number of information sheets distributed to students was 895. Results of participation from each school are shown in Table I. The total response rate of sheets returned out of the total number of sheets distributed was 34.1%, while the proportion of those who disagreed was 65.9%.
Table I.
Number of respondents in two schools in Kuala Lumpur
| Schools visited | Number of sheets distributed | Number of sheets returned [%] | Agreed to participate [%] | Select blood sample | Select buccal swab sample | Did not agree [%] | Gave consent | Did not give consent |
|---|---|---|---|---|---|---|---|---|
| Sekolah Menengah Seksyen 5 (Wangsa Maju) | 487 | 267 (29.8) | 72 (29.8) | 30 | 42 | 195 (48.1) | 93 | 103 |
| Sekolah Menengah Taman Melati, Setapak | 408 | 138 (15.4) | 72 (15.4) | 10 | 56 | 66 (16.2) | 55 | 11 |
| Total | 895 | 405 (45.3) | 138 (34.1) | 40 | 98 | 267 (65.9) | 147 | 120 |
GSTs frequencies
Results of allelic frequencies are shown in Tables II-IV. Hardy-Weinberg Equilibrium (HWE) was examined for GSTM3. Results showed no deviation. As each gene was represented by two alleles, so genotyping of the GSTM1 gene would be GSTM1*A/A, GSTM1*B/B or GSTM1*A/B genotypes. But due to presence of a deletion in this gene, the possible genotypes would be GSTM1*A/A, GSTM1*A/0, GSTM1*B/B, GSTM1*B/0, GSTM1*A/B or GSTM1*0/0. PCR readings impossible to differentiate between GSTM1*A/A, GSTM1*A/0 genotype, as both appear as positive band at the specific locus, are considered to be GSTM1*A wild genotype, while GSTM1*B/B and GSTM1*B/0 are considered as GSTM1*B mutant genotype. Same analysis for GSTT1 gene. Therefore, the HWE was not calculated for these genes.
Table II.
The distribution and frequencies of GSTM3 polymorphism in Malay students
| Genotype | Total no. of students | Genotype frequency | Total no. of alleles | Allelic frequency |
|---|---|---|---|---|
| Wild type A/A | 122 | 0.89 | 244 | 0.94 |
| Heterozygous A/B | 14 | 0.10 | 28 | |
| Homozygous mutant B/B | 1 | 0.01 | 2 | 0.06 |
| Total | 137 | 1.00 | 274 | 1.00 |
Table IV.
The distribution and frequencies of GSTT1 polymorphism in Malay students
| Genotype | Total no. of students | Genotype frequency |
|---|---|---|
| Positive | 113 | 0.82 |
| Null | 24 | 0.18 |
| Total | 137 | 1.00 |
Discussion
Generally, participation in genetic studies has been reported to be lower than in non-genetic studies [18]. A study found that a physician (who had just completed a child’s evaluation) introducing the genetic study to the parents had greater rates of enrolment than introduction done by an unfamiliar individual [19].
Reasons associated with refusal are usually described as loss of confidentiality, feeling that these studies were not important, and fear of venipuncture [19]. In this study, poor participation could be due to the unfamiliarity because the study was introduced to the teachers rather than the parents, loss of confidentiality, and fear of the procedure of sample collection.
High rates of participation in the population were associated with their intention to participate in government studies, high educational level, high socioeconomic status and presence of family disease [20]. However, willingness to participate in genetic studies was higher in Singapore (70.3%) [20] compared to our study (12.5%).
DNA from buccal swab samples was associated with low yield of DNA in comparison with blood. Disappointing results have been reported from a study of GSTs in a paediatric population because many buccal swab specimens contained less DNA, failed repeated attempts at PCR amplification and produced unreliable results [21]. A study of N-acetyl transferase (NAT2) polymorphism reported failure to amplify DNA from buccal swab [22]. In the present study we also encountered the same finding, particularly for GSTT1, where homozygous wild type and heterozygous subjects who showed positive amplification of the gene from blood samples became homozygous null when analysed by buccal swab samples (Figure 2). A possible explanation for this problem may be degradation of DNA during buccal swab extraction that affects the integrity of these loci.
The frequency of homozygous wild type (GSTM3*A) was 71%, heterozygous (GSTM3*AB) 25%, and 4% for homozygous mutant (GSTM3*B) in Caucasian U.K. [6]. These frequencies were similar to other Caucasians in Italy, Romania, Scotland, and Sweden [7]. In another study, performed in India, the frequencies were 82%, 16% and 2% for homozygous wild type (GSTM3*A), heterozygous (GSTM3*AB) and homozygous mutant (GSTM3*B) respectively [23]. Our results in this study were similar to Indians: 89%, 10% and 1%, for GSTM3*A, GSTM3*AB and GSTM3*B, respectively. Our allelic frequencies were in agreement with the Hardy-Weinberg equilibrium.
Frequencies of GSTM1 homozygous null genotype (GSTM1*0) were 0.42-0.60 in Caucasians and 0.16-0.36 in Africans [24]. In Asians, the highest frequency is in Chinese (0.63), while in Japanese and Koreans it is 0.56 [25]. In Malay, our results showed GSTM1*0 frequency in Malay children of 0.66, which was consistent with another study in Malaysia (0.72) [26] and a study of Malays in Singapore [27] that demonstrated 0.62 for the GSTM1*0 genotype.
The GSTM1*A genotype (GSTM1*A/A or GSTM1*A/0) was 0.01 in our study, while heterozygous GSTM1*AB was 0.06. GSTM1*B (GSTM1*B/B or GSTM1*B/0) was higher in our study, i.e. 0.27, than in Malay Singapore, with a value of 0.8 [28]. The predominance of GSTM1*B was also reported in Japanese and Chinese [5].
In our study, GSTT1*0 genotype frequency was 0.18 and this was consistent with that in Malay controls (0.21) [26], while Singaporean Malay showed a higher frequency of GSTT1*0, with a value of 0.38 [27].
Combined deletion of both genes GSTM1 and GSTT1 in our subjects was 0.08, and according to a study in North Americans was 0.06 and in Egyptians 0.08 [29].
Our results for the above discussed genes showed some inconsistency with those reported [26-28] regarding the Malay population, and we attributed these differences to the fact that these previous studies were case-control designed and involved adult controls. We hypothesized that case-control studies for detection of genetic association with particular disease should be based on careful selection of cases and controls [30] depending on the allele variation in the population, not merely matching the age, sex, and exposure. Furthermore, adults do not reflect the true frequency in the population since these genes can affect the paediatric population as mentioned previously. On the other hand, the effect that can be modulated by XME genes cannot be expected making selection of sample from for example; university students, blood donors, or those attending hospitals for periodic check-up (to reflect population frequencies) not appropriate, because the absence of the affected genetic trait may lead them doing higher studies, donating blood, and check-up. Our study objective is to determine the frequencies rather than to look for an association with disease, and selecting school children as a sample will avoid the genetic effect of modulation which could occur later on as in the case if adults are used as a sample.
In conclusion, the Malaysian population has a high rate of deleted GSTM1 gene, and we emphasize quality control in a genetic study for detecting method inconsistency regarding amplification of certain genes from blood and buccal swab samples. This study provides the basis for gene-environment association studies and screening for those at high risk of exposure to xenobiotics.
Table III.
The distribution and frequencies of GSTM1 polymorphism in Malay students
| Genotype | Total no. of students | Genotype frequency |
|---|---|---|
| Wild type A/A or A/0 | 2 | 0.01 |
| Heterozygous A/B | 8 | 0.06 |
| Homozygous mutant B/B or B/0 | 36 | 0.27 |
| Null 0/0 | 91 | 0.66 |
| Total | 137 | 1.00 |
Acknowledgments
We are thankful for the National University of Malaysia (UKM) for supporting this study (FF-213-2005 Dana Fundamental HUKM). Thanks to Madam Norhasyimah Noordin for her lab assistance.
References
- 1.Abel EL, Bammler TK, Eaton DL. Biotransformation of methyl parathion by glutathione-S-transferases. Toxicol Sci. 2004;79:224–32. doi: 10.1093/toxsci/kfh118. [DOI] [PubMed] [Google Scholar]
- 2.Gattas G, Kato J, Soares-Vieria JA, et al. Ethnicity and glutathione-S-transferase (GSTM1/GSTT1) polymorphism in Brazilian population. Braz J Med Bio Res. 2004;37:451–8. doi: 10.1590/s0100-879x2004000400002. [DOI] [PubMed] [Google Scholar]
- 3.Tetlow N, Board P. Functional polymorphism of human glutathione transferase A2. Pharmacogenetics. 2004;14:111–6. doi: 10.1097/00008571-200402000-00005. [DOI] [PubMed] [Google Scholar]
- 4.Tetlow N, Robinson A, Mantle T, Board P. Polymorphism of mu class glutathione transferase. Pharmacogenetics. 2004;14:359–68. doi: 10.1097/00008571-200406000-00005. [DOI] [PubMed] [Google Scholar]
- 5.Hatagima A, Klautau-Guimara~es MN, Penalva SF, Cabello PH. Glutathione S-transferase M1 (GSTM1) polymorphism in two Brazilian populations. Gen Mol Biol. 2000;23:709–13. [Google Scholar]
- 6.Inskip A, Elexperu-Camiruaga J, Buxton N, Dias PS, Macintosh J, Campbell D. Identification of polymorphism at the glutathione S-transferase, GSTM3 locus: evidence for linkage with GSTM1*A. Biochem J. 1995;312:713–6. doi: 10.1042/bj3120713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mozzoni P, De Palma G, Scotti E, Capelletti M, Muttib A. Characterization of GSTM3 polymorphism by real-time polymerase chain reaction with LightCycler. Anal Biochem. 2004;330:175–7. doi: 10.1016/j.ab.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 8.Raimondi S, Paracchini V, Autrup H, et al. Meta- and pooled analysis of GSTT1 and lung cancer: a huge-GSEC review. Am J Epidemiol. 2001;164:1027–42. doi: 10.1093/aje/kwj321. [DOI] [PubMed] [Google Scholar]
- 9.Brockmoller J, Cascorbi I, Kerb R, Sachse C, Roots I. Polymorphisms in xenobiotic conjugation and disease predisposition. Toxicol Lett. 1998;102:173–83. doi: 10.1016/s0378-4274(98)00304-x. [DOI] [PubMed] [Google Scholar]
- 10.Little J, Bradley L, Bray MS, et al. Reporting, appraising, and integrating data on genotype prevalence and gene-disease associations. Am J Epidemiol. 2002;156:300–10. doi: 10.1093/oxfordjournals.aje.a000179. [DOI] [PubMed] [Google Scholar]
- 11.Dieckvoss B, Stanulla M, Schrappe M, et al. Polymorphisms within glutathione S-transferase genes in pediatric non-Hodgkin’s lymphoma. Haematologica. 2002;87:709–13. [PubMed] [Google Scholar]
- 12.Flamant C, Henrion CA, Boelle PV, et al. Glutathione-S-transferase M1, M3, P1, T1 polymorphism and severity of lung disease in children with cystic fibrosis. Pharmacogenetics. 2004;14:295–301. doi: 10.1097/00008571-200405000-00004. [DOI] [PubMed] [Google Scholar]
- 13.Saadat I, Saadat M. The glutathione S-transferase mu polymorphism and susceptibility to acute lymphocytic leukemia. Cancer Lett. 2000;158:43–5. doi: 10.1016/s0304-3835(00)00504-8. [DOI] [PubMed] [Google Scholar]
- 14.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucl Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fryer AA, Zhao L, Alldersea J, Pearson WR, Strange RC. Use of site-directed mutagenesis of allele-specific PCR primers to identify the GSTM1 A, GSTM1 B, GSTM1 A,B and GSTM1 null polymorphisms at the glutathione S-transferase, GSTM1 locus. Biochem J. 1993;295:313–5. doi: 10.1042/bj2950313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abdel-Rahman SZ, Soliman AS, Bondy ML, et al. Polymorphism of glutathione-S-transferase loci GSTM1 and GSTT1 and susceptibility to colorectal cancer in Egypt. Cancer Lett. 1999;142:97–104. doi: 10.1016/s0304-3835(99)00159-7. [DOI] [PubMed] [Google Scholar]
- 17.Suryanarayana V, Deenadayal M, Singh L. Association of CYP1A1 gene polymorphism with recurrent pregnancy loss in the South Indian population. Hum Reproduct. 2004;19:2648–52. doi: 10.1093/humrep/deh463. [DOI] [PubMed] [Google Scholar]
- 18.Matsui K, Kita Y, Ueshima H. Informed consent, participation in, and withdrawal from a population based cohort study involving genetic analysis. J Med Ethics. 2005;31:385–92. doi: 10.1136/jme.2004.009530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mazzacco MM, Sonna NL, Shapiro BK, Pinit A, Resis AL. Effective procedures for conducting genetic prevalence studies with children. J Appl Soc Psychol. 1998;28:23–40. [Google Scholar]
- 20.Wong ML, Chia KS, Wee S, et al. Concerns over participation in genetic research among Malay-Muslims, Chinese and Indians in Singapore: a Focus Group Study. Comm Genet. 2004;7:44–54. doi: 10.1159/000080303. [DOI] [PubMed] [Google Scholar]
- 21.Zheng S, Ma X, Buffler PA, Smith MT, Wiencke JK. Whole genome amplification increases the efficiency and validity of buccal cell genotyping in pediatric populations. Cancer Epidemiol Biomarkers Preven. 2001;10:697–700. [PubMed] [Google Scholar]
- 22.Morton LM, Schenk M, Hein DW, et al. Genetic variation in N-acetyltransferase 1 (NAT1) and 2 (NAT2) and risk of non-Hodgkin lymphoma. Pharmacogenet Genom. 2006;16:537–45. doi: 10.1097/01.fpc.0000215071.59836.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buch SC, Notani PN, Bhisey RA. Polymorphism at GSTM1, GSTM3 and GSTT1 gene loci and susceptibility to oral cancerin an Indian population. Carcinogenesis. 2002;23:803–7. doi: 10.1093/carcin/23.5.803. [DOI] [PubMed] [Google Scholar]
- 24.Garte S, Gaspari L, Alexandrie AK, et al. Metabolic gene polymorphism frequencies in control populations. Cancer Epidemiol Biomarkers Preven. 2001;10:1239–48. [PubMed] [Google Scholar]
- 25.Hamajima N, Takezaki T, Tajima K. Allele frequencies of 25 polymorphisms pertaining to cancer risk for Japanese, Koreans and Chinese. Asian Pacific J Cancer Prevent. 2002;3:197–206. [PubMed] [Google Scholar]
- 26.Makpol S, Ahmad Z, Ahmad H, Ahmad A, Merican I, Wan Ngah Z. Glutathione S-Transferase M1 (GSTM1) and T1 (GSTT1) and cytochrome P450 (CYP2E1) polymorphism and susceptibility to hepatocellular carcinoma in a Malaysian study population. Eur J Scien Res. 2005;7:44–63. [Google Scholar]
- 27.Zhao B, Lee EJ, Wong JY, Yeoh PN, Gong NH. Frequencies of mutant CYP1A1, NAT2 and GSTM1 alleles in normal Indians and Malays. Pharmacogenetics. 1995;5:275–80. doi: 10.1097/00008571-199510000-00002. [DOI] [PubMed] [Google Scholar]
- 28.Lee EJ, Wong JY, Yeoh PN, Gong NH. Glutathione S transferase-theta (GSTT1) genetic polymorphism among Chinese, Malays and Indians in Singapore. Pharmacogenetics. 1995;5:332–4. doi: 10.1097/00008571-199510000-00010. [DOI] [PubMed] [Google Scholar]
- 29.Abdel-Rahman SZ, El-Zein RA, Anwar WA, Au WW. A multiplex PCR procedure for polymorphic analysis of GSTM1 and GSTT1 genes in population studies. Cancer Lett. 1996;107:229–33. doi: 10.1016/0304-3835(96)04832-x. [DOI] [PubMed] [Google Scholar]
- 30.Strange RC, Jones PW, Fryer AA. Glutathione S-transferase: genetics and role in toxicology. Toxicol Lett. 2000;112:357–63. doi: 10.1016/s0378-4274(99)00230-1. [DOI] [PubMed] [Google Scholar]