Abstract
Introduction
Nucleotide 1311 polymorphism at exon 11 of the glucose-6-phosphate dehydrogenase (G6PD) gene is fairly common in various populations worldwide, especially among Mediterranean populations. In this study, 1311 polymorphism in G6PD-deficient cases was identified by microarray technique.
Material and methods
Four hundred and fifty clinically healthy subjects were screened and 32 cases were found to have G6PD deficiency (7.11%). Our analysis of genomic DNA samples from 32 G6PD-deficient individuals revealed that the number and percentage of subjects who had a C-to-T alteration at nucleotide 1311 were 21 and 4.7% respectively. Given that the frequency of 1311 polymorphism has been reported in previous studies to be fairly high among G6PD-deficient people with the Mediterranean mutation, our data seem to be inconsistent with what we would expect for this particular region.
Results
The highly diverse ethnic background of the Adana population which probably results from the high level of immigration into this part of Turkey may be one of the most sensible explanations for this unexpected finding. Nevertheless, it seems that our results need to be confirmed in larger studies.
Conclusions
The polymorphism studies in the G6PD gene may help us to illuminate the genetic basis of the G6PD deficiency in different regions and in various ethnic groups, and also to discover the influence of a specific polymorphism on the clinical course of the deficiency.
Keywords: 1311 polymorphism, G6PD deficiency, microarray technique
Introduction
Glucose-6-phosphate dehydrogenase (G6PD) enzyme is encoded by the G6PD gene, which is localized on the X chromosome (Xq28), spans over 18 kb, and is composed of 13 exons and 12 introns [1]. The G6PD is expressed in all cells and is responsible for the first reaction of the pentose phosphate pathway in which glucose-6-phosphate is oxidized to 6-phosphogluconolactone with concomitant production of NADPH [2].
Approximately 150 mutations in the G6PD gene have been documented so far, most of them being single-base substitutions leading to amino acid replacements [3]. The G6PD deficiency, inherited as an X-linked disorder, is the most common enzymopathy that produces disease affecting nearly 400 million people worldwide [4]. With more than 300 reported variants, the disease is highly polymorphic. Furthermore, it has been shown to provide protection against malaria, which probably explains its high frequency in some geographical regions [5]. Indeed, G6PD deficiency occurs with an increased rate throughout Africa, Asia, the Mediterranean, and the Middle East [6]. In Turkey, the frequency of G6PD deficiency varies greatly between 0.5% and 20% with the genetic and ethnic background of the population studied [7]. However, the overall incidence was stated to be 0.5% and 8.2% in Turkey and in Adana, the biggest city of the Cukurova Plain located at the southeast coast of the Mediterranean Sea, respectively [7, 8].
Although most patients are asymptomatic, G6PD deficiency can cause a spectrum of diseases including neonatal hyperbilirubinaemia, acute and chronic haemolysis, and so on. The disease is typically more severe in subjects who are homozygous for the deficiency, and these patients generally present with or report a history of neonatal jaundice, often requiring exchange transfusion. A history of infection, food or drug-induced haemolysis is also common [9, 10]. The G6PD-deficient variants are grouped into different classes corresponding with disease severity that varies significantly among racial groups, and severe deficient variants primarily occur in the Mediterranean population [11, 12]. Previous reports indicated that Mediterranean mutation is the main variant throughout Turkey [13], and the silent polymorphism at nucleotide position 1311 (C-T) in exon 11, which would not produce any substitution at the amino acid level, has been shown with a high frequency in various populations, particularly in subjects with the Mediterranean mutation [14].
The G6PD enzyme activity is usually measured via quantitative spectrophotometric enzymatic assay which is based on the detection of the rate of NADPH generation from NADP. A number of other tests are also available but the fluorescent spot test has been considered to be one of the simplest, most reliable and the most sensitive screening tests for G6PD [15]. Furthermore, DNA microarray assays, an alternative method for rapidly genotyping large numbers of mutations, have also been utilized to detect G6PD-deficient individuals in specific populations [16]. In the present study, we report our results of 1311 (C-T) polymorphisms in 32 G6PD-deficient individuals in Adana, Turkey using the microarray technique.
Material and methods
Blood samples were obtained from 450 clinically healthy volunteers living in the Cukurova region of Turkey and collected into EDTA-containing tubes. To diagnose G6PD deficiency, the erythrocyte enzyme activity was measured using Beutler’s test [17], and 32 out of all cases (7.11%) were shown to have G6PD deficiency with an enzyme activity in the range 0-4.9 U/gHb.
We created a microarray-based assay for the detection of 1311 polymorphism in Adana, utilizing electronic microarray technology on the NanoChip™ Molecular Biology Workstation (Nanogen, Inc., San Diego, CA, USA). The fully automated system allows the active deposition and concentration of negatively charged biotinylated molecules on selected test sites using a proprietary semiconductor microchip. The DNA at each pad is then hybridized with specific Cy5- and Cy3-labelled oligonucleotide reporters, complementary to wild-type or mutant sequences. Through well-developed, digital image-processing operations, the array is imaged; fluorescence signals are detected, displayed, and finally evaluated. After thermal stringency was applied, the remaining reporter signals were quantified to determine the genotype of the hybridized genomic DNA sample.
In the first step of the microarray-based assay, DNAs were isolated from white blood cells as described by Poncz et al. to perform molecular diagnostic tests [18]. The PCR mixture contained 75 mM Tris-HCl (pH 8.8), 20 mM (NH4)2SO4, 1.5 mM MgCl2, 0.2 mM of each dNTP, 10 pmol primer mix, 0.75 units of Taq polymerase and 100 ng of genomic DNA in a total volume of 25 µl. Thermocycling was performed on a Perkin Elmer 9600 system (Norwalk, Connecticut, USA) using the following conditions: Subsequent to denaturation at 95°C for 5 min, 35 cycles consisted of 1 min at 95°C, 1 min at 60°C, 1 min at 72°C, and a final extension at 72°C for 10 min were performed.
After purification from excess unincorporated primers and desalting, the PCR products were mixed with histidine, and then electronically addressed. Afterwards, the amplified products were denatured using 0.1 M sodium hydroxide for 3 min and then the cartridge was incubated with stabilizers in a high salt buffer for another 3 min. Following hybridization of probes, a thermal stringency step specific for 1311 polymorphism was carried out in the Reader and hybridization was detected through fluorescence using automated scanning and dedicated software. Restriction fragment length polymorphism (RFLP) was used to confirm the C1311T mutation.
Approval from the institutional Ethics Committee of Cukurova University Medical Faculty was provided and informed consent was obtained from all subjects prior to enrolment in the study.
Results
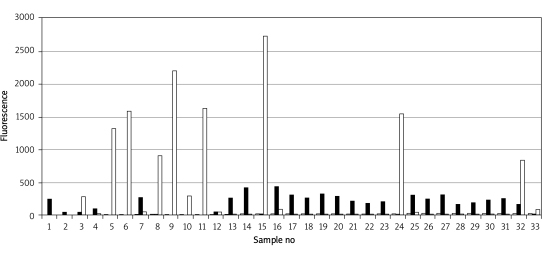
A screening protocol was performed and of 450 subjects, 32 were found to have G6PD deficiency. Genomic DNA samples from these 32 G6PD-deficient individuals were analyzed, and 15 subjects were found to be hemizygous, five cases were homozygous, one case was heterozygous, whereas eleven subjects were shown to be the wild-type (Figure 1). That is, the percentage of the G6PD-deficient individuals with 1311 polymorphism was 4.7% in our study group. G6PD activity, fluorescence signal rate and molecular features of the cases are detailed in Table I and Table II.
Figure 1.
The fluorescence signal rates of the mutant and wild-type individuals for 1311 polymorphism in the microarray analysis
Table I.
The fluorescence signal rates from microarray analysis and the genotype of 1311 polymorphism in the G6PD gene
| Case no. and sex | Red | Green | Rate (R/G) | Prob | Genotype |
|---|---|---|---|---|---|
| 1.align="center">Gd 1311 Control | 237 | 3.33 | 21.72 | Red/red | Control |
| 2. M | 39 | 4.43 | 8.79 : 1 | Red/red | Hemizygous |
| 3. F | 42 | 280.5 | 1 : 6.68 | Green/green | Normal |
| 4. M | 99 | 17.74 | 5.58 : 1 | Red/red | Hemizygous |
| 5. F | 2 | 1331.54 | 1 : 665.77 | Green/green | Normal |
| 6. F | 0 | 1579.89 | Endless | Green/green | Normal |
| 7. M | 272 | 48.31 | 5.63 : 1 | Red/red | Hemizygous |
| 8. M | 14 | 900.26 | 1 : 64.3 | Green/green | Normal |
| 9. M | 0 | 2197.43 | Endless | Green/green | Normal |
| align="center">10. M | 0 | 282.72 | Endless | Green/green | Normal |
| align="center">11. F | 0 | 1637.54 | Endless | Green/green | Normal |
| align="center">12. F | 51 | 51 | 1 : 1 | Red/green | Heterozygous |
| align="center">13. F | 259 | 11.09 | 23.36 : 1 | Red/red | Homozygous |
| align="center">14. F | 418 | 14.41 | 29 : 1 | Red/red | Homozygous |
| align="center">15. F | 15 | 2734.04 | 1 : 182.27 | Green/green | Normal |
| align="center">16. M | 436 | 85.37 | 5.11 : 1 | Red/red | Hemizygous |
| align="center">17. M | 311 | 16.63 | 18.7 : 1 | Red/red | Hemizygous |
| align="center">18. M | 261 | 9.98 | 26.16 : 1 | Red/red | Hemizygous |
| align="center">19. F | 317 | 14.41 | 23.83 : 1 | Red/red | Homozygous |
| 20. M | 282 | 9.98 | 28.26 : 1 | Red/red | Hemizygous |
| align="center">21. F | 207 | 14.41 | 14.36 : 1 | Red/red | Homozygous |
| 22. M | 174 | 9.98 | 17.44 : 1 | Red/red | Hemizygous |
| 23. M | 197 | 6.65 | 29.61 : 1 | Red/red | Hemizygous |
| 24. M | 7 | 1544.41 | 1 : 220.63 | Green/green | Normal |
| 25. M | 296 | 35.48 | 8.34 : 1 | Red/red | Hemizygous |
| 26. M | 235 | 9.98 | 23.55 : 1 | Red/red | Hemizygous |
| 27. F | 304 | 6.65 | 45.7 : 1 | Red/red | Homozygous |
| 28. M | 162 | 6.65 | 24.35 : 1 | Red/red | Hemizygous |
| 29. M | 188 | 6.65 | 28.26 : 1 | Red/red | Hemizygous |
| 30. M | 232 | 7.76 | 29.89 : 1 | Red/red | Hemizygous |
| align="center">31.align="center">M | 246 | 6.65 | 3698 : 1 | Red/red | Hemizygous |
| 32. F | 155 | 838.55 | 1 : 5.41 | Green/green | Normal |
| 33. M | 15 | 88.7 | 1 : 5.91 | Green/green | Normal |
Table II.
G6PD activity, the type of Gd-Med mutation and the genotype of 1311 polymorphisms in the study group
| Case no.andsex | G6PD activity [U/gHb] | Gd-Med mutation | Genotype 1311 |
|---|---|---|---|
| align="center">1.align="center">Control | – | – | Control |
| 2. M | 0.0 | Hemizygous | Hemizygous |
| 3. F | 3.7 | Heterozygous | Normal |
| 4. M | 0.0 | Hemizygous | Hemizygous |
| 5. F | 2.3 | Heterozygous | Normal |
| 6. F | 4.6 | Hemizygous | Normal |
| 7. M | 0.0 | Hemizygous | Hemizygous |
| 8. M | 0.0 | Hemizygous | Normal |
| 9. M | 0.0 | Hemizygous | Normal |
| align="center">10. M | 0.0 | Hemizygous | Normal |
| align="center">11. F | 0.0 | Homozygous | Normal |
| align="center">12. F | 4.1 | Heterozygous | Heterozygous |
| align="center">13. F | 1.5 | Heterozygous | Homozygous |
| align="center">14. F | 3.6 | Heterozygous | Homozygous |
| align="center">15. F | 2.8 | Heterozygous | Normal |
| align="center">16. M | 0.0 | Hemizygous | Hemizygous |
| align="center">17. M | 0.0 | Hemizygous | Hemizygous |
| align="center">18. M | 0.0 | Hemizygous | Hemizygous |
| align="center">19. F | 3.9 | Heterozygous | Homozygous |
| 20. M | 0.0 | Hemizygous | Hemizygous |
| align="center">21. F | 4.9 | Heterozygous | Homozygous |
| 22. M | 0.0 | Hemizygous | Hemizygous |
| 23. M | 0.0 | Hemizygous | Hemizygous |
| 24. M | 0.0 | Hemizygous | Normal |
| 25. M | 0.0 | Hemizygous | Hemizygous |
| 26. M | 0.0 | Hemizygous | Hemizygous |
| align="center">27. F | 2.3 | Heterozygous | Homozygous |
| 28. M | 0.0 | Hemizygous | Hemizygous |
| 29. M | 0.0 | Hemizygous | Hemizygous |
| 30. M | 0.0 | Hemizygous | Hemizygous |
| align="center">31.align="center">M | 0.0 | Hemizygous | Hemizygous |
| 32. F | 4.2 | Heterozygous | Normal |
| 33. M | 0.0 | Hemizygous | Normal |
Discussion
The presence of a mutation, a silent C-to-T change, at nucleotide 1311 in exon 11 of the G6PD gene was first noticed when DNA samples from subjects with G6PD Mediterranean (Gd-Med) mutation were sequenced by De Vita et al. [19]. Since then, 1311 polymorphism has been shown to be quite prevalent among G6PD-deficient people, particularly in those with Gd-Med mutation, and has been reported in many countries all over the world including Portugal, Pakistan, Sardinia, and Tunisia [20-23].
1311 mutation has been considered as a polymorphic marker of the G6PD gene, independent of G6PD deficiency. Nevertheless, studying the 1311 polymorphism in subjects with G6PD Mediterranean mutation could help us to recognize the spread of this mutation [24]. Using sequence analysis, Beutler and Kuhl studied the distribution of the nucleotide polymorphism C1311T in cases with and without G6PD deficiency in diverse populations in 1990. They found that out of 21 Gd-Med individuals from Mediterranean countries, 20 had a T at nucleotide 1311, whereas neither of two Gd-Med cases from the Indian subcontinent had 1311 polymorphism. The investigator concluded that the Gd-Med mutation may have arisen independently in Europe and in Asia, since the vast majority of European subjects with Gd-Med had 1311 polymorphism [14].
Moiz et al. reported that the 1311 polymorphism was seen frequently both in subjects with Gd-Med mutation and in healthy individuals in Pakistan [21]. Similarly, Sukumar et al. reported a high frequency of 1311 polymorphism in Gd-Med and G6PD Kerala-Kalyan Indians [24]. Using the PCR-RFLP technique, Nuchprayoon et al. identified the G6PD mutations in two different ethnic groups in Thailand, and reported that the 1311 polymorphism was common both in G6PD-deficient and in normal cases [25]. According to the data of Ruzzo et al. from Italy, in the normal population of southern Italy, which is at risk for Gd-Med mutation, the 1311 polymorphism was 16.7-20%, whereas in the Marche region, in central Italy, the 1311T frequency was found to be about one half of that found in southern Italy [26]. In Great Britain, the incidence of this silent mutation among British was reported to be 24%, while among Iranian subjects it was 44%. It seems that the incidence of the silent mutation is higher in the population of the Mediterranean basin than among populations of continental Europe [27].
Similar studies were performed by Kurdi-Haidar et al. in 20 unrelated individuals with Gd-Med mutation from Saudi Arabia, Iraq, Iran, Jordan, Lebanon, and Israel. Except for one subject, all of the cases were shown to have the C-to-T change at nucleotide 1311. Kurdi-Haidar et al. concluded that the silent mutation is an independent polymorphism in the Middle East, and that the mutation leading to Gd-Med deficiency probably arose on a chromosome that already carried the silent mutation [11].
Inconsistent with the above-mentioned reports, the rate of 1311 polymorphism among our study group was found to be low, with a percentage of 4.7%. The most reasonable explanation for this may be the extremely diverse ethnic background of the Adana population, which may be due in part to the high level of immigration into this region. The G6PD deficiency is mainly found in malaria-endemic areas, and accordingly, it was hypothesized that G6PD deficiency had arisen as a protective factor against lethal malaria [28]. Consistent with this data, the number of G6PD-deficient cases was reported to be significantly elevated in Adana, which has been reported as one of the malaria-endemic regions in Turkey [8]. By using the microarray technique, Menziletoglu Yildiz et al. revealed in a recent study that the frequency of the Gd-Med mutation was found to be substantially high in subjects with G6PD deficiency in Adana [29]. However, the reason why the number of subjects with 1311 polymorphism was observed to be significantly lower than expected and also below that reported by the other Mediterranean countries may be the very heterogeneous population residing in this area. It is also most likely that many variants of G6PD deficiency may exist in this area due to this highly divergent ethnicity.
The PCR-based assays have been widely utilized to detect the G6PD mutations and 1311 polymorphism. To our knowledge, this study represents the first report of 1311 polymorphism in the Cukurova Region with the microarray technique. The Nanogen® microarray system enabled us to design a new probe to detect the 1311 polymorphism in G6PD-deficient cases, and the results obtained from the newly designed microarray probe were confirmed by the RFLP. We suggest that the microarray technique can be safely utilized not only for analysis of mutations and gene expression, but also for detecting the single nucleotide polymorphisms. Furthermore, it seems that it can also be used to develop new applications as it can be easily modified. After the appropriate infrastructure has been established, the cost of the chemical substance required for the procedure may greatly decrease, particularly when large numbers of samples need to be processed in a short period of time and with a minimum of handling.
In conclusion, studying the polymorphisms of the G6PD gene may facilitate the identification of the genetic basis of G6PD deficiency in different regions and in various ethnic groups. Furthermore, DNA polymorphisms have been shown to be very supportive in the study of the human genome by providing markers for gene mapping as well as linkage analysis [30]. Although the G6PD gene holds a substantial number of polymorphic sites, whether these genetic variants including the nucleotide C1311T polymorphism could help us to understand the differences in disease severity in G6PD-deficient individuals from distinct populations needs to be further clarified by additional investigations.
Acknowledgments
This work was supported by grants from DPT-2005K120320-E and Çukurova University, Grant No. TF2007BAP21.
References
- 1.Beutler E, Westwood B, Kuhl W. Definition of the mutations of G6PD Wayne, G6PD Viangchan, G6PD Jammu, and G6PD 'LeJeune'. Acta Haematol. 1991;86:179–82. doi: 10.1159/000204830. [DOI] [PubMed] [Google Scholar]
- 2.Lehninger AL, Nelson DL, Cox MM. Lehninger principles of biochemistry. 5nd ed. New York: W.H. Freeman Press; 2005. pp. 350–85. [Google Scholar]
- 3.Beutler E, Vulliamy TJ. Hematologically important mutations: glucose-6-phosphate dehydrogenase. Blood Cells Mol Dis. 2002;28:93–103. doi: 10.1006/bcmd.2002.0490. [DOI] [PubMed] [Google Scholar]
- 4.Luzzatto L, Karadimitris A, editors. The molecular basis of anemia. 3nd ed. John Wiley & Sons Press; 2010. Provan Drew. Molecular hematology; pp. 140–64. [Google Scholar]
- 5.Chalvam R, Colah RB, Mohanty D, Ghosh K, Mukherjee MB. Molecular heterogeneity of glucose-6-phosphate dehydrogenase deficiency among the tribals in Western India. Blood Cells Mol Dis. 2009;43:156–7. doi: 10.1016/j.bcmd.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 6.Beutler E. G6PD: population genetics and clinical manifestations. Blood Rev. 1996;10:45–52. doi: 10.1016/s0268-960x(96)90019-3. [DOI] [PubMed] [Google Scholar]
- 7.Altay C, Gümrük F. Red cell glucose-6-phosphate dehydrogenase deficiency in Turkey. Turk J Hematol. 2008;25:1–7. [PubMed] [Google Scholar]
- 8.Yüregir GT, Aksoy K, Arpaci A, Unlükurt I, Tuli A. Studies on red cell glucose-6-phosphate dehydrogenase: evaluation of reference values. Ann Clin Biochem. 1994;31:50–5. doi: 10.1177/000456329403100109. [DOI] [PubMed] [Google Scholar]
- 9.Abdel Fattah M, Abdel Ghany E, Adel A, Mosallam D, Kamal S. Glucose-6-phosphate dehydrogenase and red cell pyruvate kinase deficiency in neonatal jaundice cases in egypt. Pediatr Hematol Oncol. 2010;7:262–71. doi: 10.3109/08880011003639986. [DOI] [PubMed] [Google Scholar]
- 10.Beutler E, Duparc S. Glucose-6-phosphate dehydrogenase deficiency and antimalarial drug development. Am J Trop Med Hyg. 2007;77:779–89. [PubMed] [Google Scholar]
- 11.Kurdi-Haidar B, Mason PJ, Berrebi A, et al. Origin and spread of the glucose-6-phosphate dehydrogenase variant G6PD-Mediterranean in the Middle East. Am Hum Genet. 1990;47:1013–9. [PMC free article] [PubMed] [Google Scholar]
- 12.Frank JE. Diagnosis and management of G6PD deficiency. Am Fam Physician. 2005;72:1277–82. [PubMed] [Google Scholar]
- 13.Oner R, Gumruk F, Acar C, Oner C, Gurgey A, Altay C. Molecular characterization of glucose-6-phosphate dehydrogenase deficiency in Turkey. Haematologica. 2000;85:320–1. [PubMed] [Google Scholar]
- 14.Beutler E, Kuhl W. The NT 1311 polymorphism of G6PD: G6PD Mediterranean mutation may have originated independently in Europe and Asia. Am Hum Genet. 1990;47:1008–12. [PMC free article] [PubMed] [Google Scholar]
- 15.Wang FL, Boo NY, Ainoon O, Wong MK. Comparison of detection of glucose-6-phosphate dehydrogenase deficiency using fluorescent spot test, enzyme assay and molecular method for prediction of severe neonatal hyperbilirubinaemia. Singapore Med J. 2009;50:62–7. [PubMed] [Google Scholar]
- 16.Bang-Ce Y, Hongqiong L, Zhensong L. Rapid detection of common Chinese glucose-6-phosphate dehydrogenase G6PD) mutations by microarray-based assay. Am J Hematol. 2004;76:405–12. doi: 10.1002/ajh.20126. [DOI] [PubMed] [Google Scholar]
- 17.Beutler E. Red cell metabolism: a manuel of biochemical methods. 3rd ed. Orlanda: Grune & Stratton Inc; 1984. pp. 68–71. [Google Scholar]
- 18.Poncz M, Solowiejzk D, Harpel B, Mory Y, Schwartz E, Surrey S. Construction of human gene library from small amounts of peripheral blood. Analysis of beta like globin genes. Hemoglobin. 1982;6:27–36. doi: 10.3109/03630268208996930. [DOI] [PubMed] [Google Scholar]
- 19.De Vita G, Alcalay M, Sampietro M, Cappellini MD, Fiorelli G, Toniolo D. Two point mutations are responsible for G6PD polymorphism in Sardinia. Am J Hum Genet. 1989;44:233–40. [PMC free article] [PubMed] [Google Scholar]
- 20.Rodrigues MO, Freire AP, Martins G, Pereira J, Martins MD, Monteiro C. Glucose-6-phosphate dehydrogenase deficiency in Portugal: biochemical and mutational profiles, heterogeneity, and haplotype association. Blood Cells Mol Dis. 2002;28:249–59. doi: 10.1006/bcmd.2002.0505. [DOI] [PubMed] [Google Scholar]
- 21.Moiz B, Nasir A, Moatter T, Naqvi ZA, Khurshid M. Population study of 1311 C/T polymorphism of glucose 6 phosphate dehydrogenase gene in Pakistan - an analysis of 715 X-chromosomes. BMC Genet. 2009;30:41–8. doi: 10.1186/1471-2156-10-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frigerio R, Sole G, Lovicu M, Passiu G. Molecular and biochemical data on some glucose-6-phosphate dehydrogenase variants from southern Sardinia. Haematologica. 1994;79:319–21. [PubMed] [Google Scholar]
- 23.Daoud BB, Mosbehi I, Préhu C, Chaouachi D, Hafsia R, Abbes S. Molecular characterization of erythrocyte glucose-6-phosphate dehydrogenase deficiency in Tunisia. Pathol Biol. 2008;56:260–7. doi: 10.1016/j.patbio.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 24.Sukumar S, Mukherjee MB, Colah RB, Mohanty D. Molecular basis of G6PD deficiency in India. Blood Cells Mol Dis. 2004;33:141–5. doi: 10.1016/j.bcmd.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 25.Nuchprayoon I, Louicharoen C, Charoenvej W. Glucose-6-phosphate dehydrogenase mutations in Mon and Burmese of southern Myanmar. J Hum Genet. 2008;53:48–54. doi: 10.1007/s10038-007-0217-3. [DOI] [PubMed] [Google Scholar]
- 26.Ruzzo A, Ninfali P, Magnani M. Glucose-6-phosphate dehydrogenase nucleotide 1311 polymorphism in central Italy Marche Region. Enzyme Protein. 1993;47:22–6. doi: 10.1159/000468652. [DOI] [PubMed] [Google Scholar]
- 27.Mortazavi Y, Chopra R, Gordon-Smith EC, Rutherford TR. Frequency of the G6PD nt 1311 C/T polymorphism in English and Iranian populations: relevance to studies of X chromosome inactivation. J Med Genet. 1997;34:1028–9. doi: 10.1136/jmg.34.12.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Motulsky A. Metabolic polymorphisms and the role of infectious diseases in human evolution. Hum Biol. 1960;32:28–62. [PubMed] [Google Scholar]
- 29.Menziletoglu Yildiz S, Yüzbasioglu Ariyurek S, Aksoy K. Detection of Mediterranean mutation in the glucose-6-phosphate dehydrogenase gene with microarray technique. Turk J Biochem. 2010;35:63–6. doi: 10.5114/aoms.2011.24126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Platzer M. The human genome and its upcoming dynamics. Genome Dyn. 2006;2:1–16. doi: 10.1159/000095083. [DOI] [PubMed] [Google Scholar]