Abstract
Single-chain variable fragment (scFvs) antibodies are small polypeptides (∼26 kD) containing the heavy (VH) and light (VL) immunoglobulin domains of a parent antibody connected by a flexible linker. In addition to being frequently used in diagnostics and therapy for an increasing number of human diseases, scFvs are important tools for structural biology as crystallization chaperones. Although scFvs can be expressed in many different organisms, the expression level of an scFv strongly depends on its particular amino acid sequence. We report here a system allowing for easy and efficient cloning of (i) scFvs selected by phage display and (ii) individual heavy and light chain sequences from hybridoma cDNA into expression plasmids engineered for secretion of the recombinant fragment produced in Drosophila S2 cells. We validated the method by producing five scFvs derived from human and murine parent antibodies directed against various antigens. The production yields varied between 5 and 12 mg monomeric scFv per liter of supernatant, indicating a relative independence on the individual sequences. The recombinant scFvs bound their cognate antigen with high affinity, comparable with the parent antibodies. The suitability of the produced recombinant fragments for structural studies was demonstrated by crystallization and structure determination of one of the produced scFvs, derived from a broadly neutralizing antibody against the major glycoprotein E2 of the hepatitis C virus. Structural comparison with the Protein Data Bank revealed the typical spatial organization of VH and VL domains, further validating the here-reported expression system.
Keywords: crystallization, Drosophila S2, expression system, monomeric, scFv
Introduction
A single-chain variable fragment (scFv) is a portion of a monoclonal antibody (MW ∼26 kD), in which the variable immunoglobulin domains of the heavy (VH) and light (VL) chains are connected with a flexible linker into a single polypeptide chain (Bird et al., 1988; Huston et al., 1988). Thus, an scFv contains the entire antigen-binding region, and hence the specificity, of the parent antibody (Bird et al., 1988; Huston et al., 1988; Sandhu, 1992; Holliger and Hudson, 2005). The two variable domains are tethered together in either order (VH–VL or VL–VH), with a linker that is usually 10–25 amino acids in length, spanning the 35–40 Å distance from the C-terminus of one V domain to the N-terminus of the other (Filpula et al., 1996; Weisser and Hall, 2009). The length of this linker plays a crucial role for the oligomeric state of the soluble purified scFv (Arndt et al., 1998; Filpula et al., 1996), the most common linker being a 15mer (Gly4Ser)3 (Huston et al., 1988; Weisser and Hall, 2009).
Importantly, scFvs usually bind their cognate antigens with affinity similar to that of the parent antibody (Bird et al., 1988; Huston et al., 1988, 1996; Skerra and Plückthun, 1988; Weisser and Hall, 2009)–if the avidity effect of the bivalency of the latter is taken into account. This is due to the identical 3D arrangement of the variable domains. This feature, combined with the small size of scFvs, has made them attractive candidates for a wide range of applications, including therapeutics, medical imaging and diagnostics (Begent et al., 1996). scFvs and scFv-based antibody fragments are currently in pre-clinical and clinical trials to treat human diseases ranging from heart disease to melanoma, and also for use in medical imaging (reviewed in Holliger and Hudson, 2005). In addition, scFvs have shown promise in drug delivery systems and for the targeting of gene therapy vectors (Glasgow et al., 2009; Eisenstein, 2011).
The compact fold of scFvs, composed of two immunoglobulin domains each consisting of nine strands forming two tightly packed β-sheets stabilized by an intrachain disulfide bond, provides additional protein surfaces that can help to form a crystal lattice and thus promote crystallization of macromolecules (Kovari et al., 1995; Griffin and Lawson, 2011). Such an approach is particularly useful when dealing with proteins that are difficult to crystallize, such as heavily glycosylated proteins, or multidomain proteins with relatively flexible interdomain connections. Moreover, the compactness of scFvs makes them ideal candidates for structural studies on antigen–antibody binding, such as the characterization of the neutralization mechanism of certain monoclonal antibodies in viral infection (Hwang et al., 2006; Sui et al., 2009). These studies would be extremely difficult to perform using full-length bivalent antibody molecules, and are sometimes hindered by the flexibility of the elbow angle between variable and constant domains in Fab (fragment antigen-binding) fragments.
Various expression systems for scFvs have been reported, including Escherichia coli, yeast, fungi, plants, insect and mammalian cells (Bird et al., 1988; Wu et al., 1993; Jost et al., 1994; Ridder et al., 1995; Brocks et al., 1997); reviewed in Verma et al. (1998) and Weisser and Hall (2009). Numerous studies have been performed to evaluate quantity and quality of scFvs produced in different expression systems. The advantages and drawbacks of the most frequently used expression systems are summarized in Table I. Importantly, in spite of numerous expression systems that have been explored for scFv expression, the expected effort required to produce large quantities of an scFv derived from a particular parent antibody strongly depends on the individual amino acid sequence (Verma et al., 1998). Given that it is important to consistently obtain large quantities of pure scFv for structural analyses, as well as for diagnostic or therapeutic applications, we decided to further explore the use of the Drosophila Schneider 2 system for expression of scFvs. This system has been previously used to produce scFvs, but the reported expression levels varied by almost two orders of magnitude (0.2–20 mg/l) (Mahiouz et al., 1998; Reavy et al., 2000). High-yield expression of full-length monoclonal antibodies and Fab-fragments in S2 cells has also been reported (Johansson et al., 2007a; Backovic et al., 2010). The Drosophila S2 expression system is relatively easy to handle and is based upon logarithmically growing healthy cell lines that have been transfected to achieve stable expression of recombinant protein. This is in contrast to the production in insect cells using a recombinant baculovirus, which makes a lytic infection and induces considerable stress to the infected cell. Baculoviral-aided expression can therefore lead to saturation of the endoplasmic reticulum quality control system of the cell, especially when overexpressing proteins with complex, disulfide-stabilized folds. In such cases, baculovirus-induced cell lysis results in dumping of protein that has not gone through the ER quality control into the medium, resulting in a mixture of folded and partially folded protein in the supernatant.
Table I.
Advantages and disadvantages of different expression systems for scFvs
| Expression system | Advantages | Disadvantages |
|---|---|---|
| Bacterial expression (E.coli) |
|
|
| Yeast expression (Saccharomyces cerevisiae, Kluyveromyces lactis, Pichia pastoris) |
|
|
| Plants |
|
|
| Insect cells |
|
|
| Mammalian cells |
|
|
We undertook to establish an expression system for scFvs that would allow easy cloning of the variable domains of heavy and light chain and production of large amounts of correctly processed and secreted scFvs using Drosophila S2 cells. We created a panel of scFvs from both human and murine parent antibodies directed against various antigens. In this paper we discuss in particular the production of five of the recombinant proteins-scFv 3H5, derived from a murine antibody to Npro of bovine viral diarrhea virus (BVDV); scFv 8B9 and scFv 6A5, both derived from murine antibodies to glycoprotein E2 of BVDV; and scFvs 1:7 and A8, both derived from human antibodies to hepatitis C virus glycoprotein E2 (Allander et al., 2000).
Materials and methods
Construction of pMT-based scFv expression vector
The pMT-scFv-Strep vector was constructed based on the pT350 vector reported previously (Krey et al., 2010), containing a Drosophila metallothionein (MT) promoter, a BiP signal sequence and an enterokinase (EK) cleavage site followed by a double Strep tag (NH2-DDDDKAGWSHPQFEKGGGSGGGSGGGSWSHPQFEK-COOH). First we introduced NcoI and NotI sites by polymerase chain reaction (PCR) using oligonucleotides 5′-TTTTTTTTCCATGGCCCCGAGCGAGAGGCCAAC AAAGG-3′ and 5′-AAAAAAAAAGCGGCCGCAGACGATGACGATAAGGCCGGTTG-3′ replacing BglII and BstBI sites used for cloning. In a subsequent PCR step KpnI and NheI sites were inserted using oligonucleotides 5′-GGAGGAGCTAGCAAAAAAGCGGCCGCAGACGATGACG-3′ and 5′-TCCCGAGCCGCCGGTACCTTTTTTCCATGGCCCCGAGCGAG-3′. Finally, the complete linker sequence GGS(GGGGS)2GGG was inserted by PCR using oligonucleotides 5′-CCACCCGATCCTCCTCCTCCCGAGCCGCCGGTACCTTTTTTCCA-3′ and 5′-TGGTGGTAG CGGAGGAGGAGCTAGCAAAAAAGCGGCCGCAGACGATG-3′.
Cloning of pMT-scFv-#-Strep constructs
The respective DNA sequences of the individual parent antibodies have either been reported previously (1:7, A8; Allander et al., 2000) or were obtained from sequencing of the respective hybridoma cell line (8B9, 6A5, 3H5). The VH and VL of the individual scFvs were inserted into the pMT-scFv-Strep vector in two successive rounds of cloning. A VL gene was amplified by PCR using either cDNA of hybridoma cells (8B9, 6A5) or synthetic genes, codon optimized for Drosophila melanogaster (3H5, 1:7, A8), as template. The amplified segment was cloned into pMT-scFv-Strep vector using NheI and NotI. Insertion of the VL gene into the resulting pMT-scFv-#-VL-Strep constructs was confirmed by sequencing. The respective VH gene was subsequently amplified by PCR using either cDNA of hybridoma cells (8B9, 6A5) or synthetic genes, codon optimized for D.melanogaster (3H5, 1:7, A8), as template. The amplified VH sequence was cloned into the newly created VL-containing pMT-scFv-#-VL-Strep vector using NcoI and KpnI. The resulting pMT-scFv-#-Strep constructs were sequenced to confirm insertion of both the VL and the VH. A full list of oligonucleotides used for cloning of VH and VL genes is included as Supplementary material (Table S1).
Transfection of the cells and expression of the scFv
Transfection of Drosophila S2 cells was done as described before (Johansson et al., in press). Briefly, we used Effectene (Qiagen, Hilden, Germany) according to the manufacturer's recommendations, to transfect parental S2 cells with 2 µg of the respective pMT-scFv-#-Strep plasmids. A second plasmid, encoding puromycin acetyltransferase, was cotransfected as dominant selectable marker. Stable scFv expressing cell lines were selected by addition of 8 µg/ml Puromycin (Invivogen, San Diego, USA) to the culture medium 72 h after transfection. Adaptation of the cell lines to serum free Insect Xpress media was performed stepwise as recommended by Invitrogen.
Expression and purification of scFvs
For large-scale production of scFvs the cells were cultured in spinner flasks and induced with 4 μM CdCl2 at a density of at least 7 × 106 cells/ml. After 7–10 days at 28°C cells were pelleted and the scFv was purified by affinity chromatography from the supernatant using a StrepTactin Superflow column (IBA, Goettingen, Germany) followed by size exclusion chromatography (SEC) using a Superdex200 column (GE Healthcare, Uppsala, Sweden). Pure protein was quantified using adsorption at UV280 nm and concentrated to ∼10 mg/ml.
Refolding of dimeric scFvs
Dimeric scFvs were diluted to ≤100 μg/ml and dissociated by overnight dialysis in dialysis buffer (0.3 M NaCl, 20 mM Tris pH 8.0) containing 8 M urea. Subsequently, the dimeric fragments were refolded by a stepwise dialysis overnight at 4°C against 1 l dialysis buffer containing decreasing concentrations of urea (8, 6, 4, 2, 1, 0.5 and 0 M). The buffers containing 1 and 0.5 M urea were supplemented with 0.4 M l-arginine. The oligomerization state of the refolded scFvs was analyzed by SEC using a Superdex™ 200 column.
SEC analysis of antigen-scFv complexes
Twenty micrograms of scFv (final concentration 0.2 mg/ml) and 20–35 μg (final concentration 0.2–0.35 mg/ml) of antigen (depending on its molecular weight), were incubated as isolated proteins as well as in complex for 24 h at 4°C followed by analysis on a Superdex™ 200 5/150GL column (column volume 3 ml, GE Healthcare).
Crystallization of scFv 1:7, data collection, structure determination and refinement
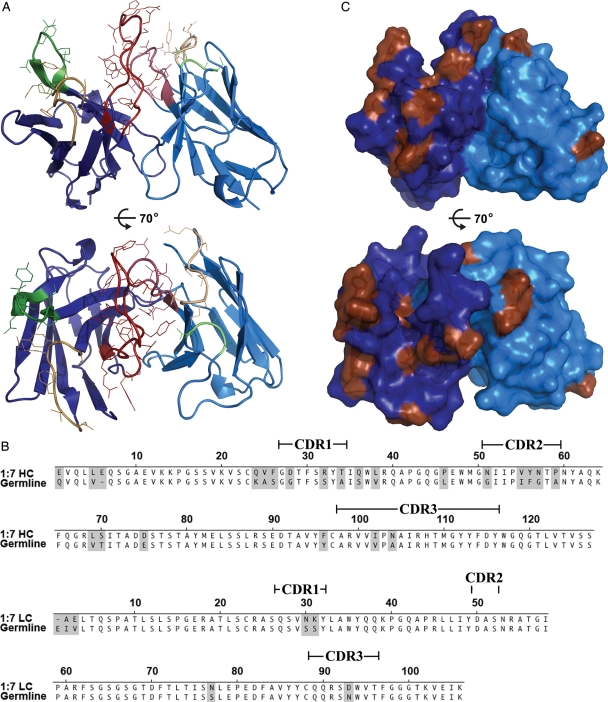
Crystals of scFv 1:7 were grown at 293 K using the sitting-drop vapor-diffusion method. Crystallization was performed using a Mosquito robot (TTP LabTech Ltd, Royston, UK) in a drop containing 800 nl protein (7 mg/ml in 10 mM TRIS pH 8.0, 150 mM NaCl) mixed with 800 nl reservoir solution containing 100 mM TRIS pH 8.5, 1550 mM (NH4)2SO4 and 200 mM Li2SO4. Diffraction quality crystals appeared after 1 week and were flash frozen in mother liquor containing 25% (v/v) glycerol. Diffraction data were collected at the Proxima1 beam line of Synchrotron Soleil and processed with XDS (Kabsch, 2010). Scaling and reduction of the data were performed using Pointless (Evans, 2005) and programs from the CCP4 suite (Collaborative Computational Project, 1994). The structure was determined by molecular replacement using Phaser (McCoy et al., 2007), an scFv derived from a human antibody served as model (PDB 3FKU; Sui et al., 2009). Model building was performed using Coot (Emsley et al., 2010) and refinement was done using AutoBuster (Bricogne et al., 2010). Figure 4 was prepared with Pymol (http://www.pymol.org). The atomic coordinates of the structural model and the corresponding structure factors were deposited in the Protein Data Bank with ID 3U6R.
Fig. 4.
Crystal structure of scFv 1:7. The crystal structure of scFv 1:7 (A) viewed from the side (upper panel) and from the top (lower panel). Framework regions are colored in dark blue (HC) or light blue (LC) and the CDRs according to the immunogenetics (IMGT) nomenclature are colored in yellow (CDR1), green (CDR2) and red (CDR3). The side chains of the residues in the CDRs are shown as lines. (B) The 1:7 amino acid sequence was aligned with the closest homologous germline sequence suggested by IMGT V-QUEST and junction analysis (Lefranc et al., 2009). Positions of somatic mutations during antibody maturation are shaded, bars indicate the positions of the CDRs according to the IMGT nomenclature. (C) Mapping of the somatic mutations (brown) on the molecular surface of scFv 1:7 from the side (upper panel) and from the top (lower panel).
Results and discussion
Design of the expression plasmid
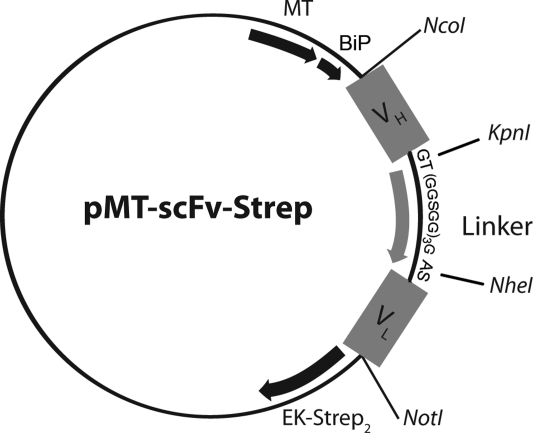
The pMT-scFv-Strep vector was designed based upon an expression plasmid previously described (Krey et al., 2010) that contained an inducible Drosophila MT promoter, followed by a Drosophila BiP signal sequence, which leads to efficient translocation of the protein into the ER. At the C-terminus, this plasmid contains a double Strep-tag for efficient affinity purification and an EK cleavage site upstream of the double Strep-tag to allow for its specific removal for therapeutic applications or structural studies.
The pMT-scFv-Strep was designed to contain the VH and VL of a parent antibody in this order joined by a linker sequence (Fig. 1) that is meant to span the 35–40 Å between the C-terminus of VH and the N-terminus of VL. The major requirement for this linker is sufficient flexibility, a rationale that is followed in the most commonly used linker sequences (Huston et al., 1991). Linker sequences are typically 10–25 amino acids, shorter linker sequences have been shown to favor the formation of dimers or higher multimers (Huston et al., 1988; Arndt et al., 1998; Filpula et al., 1996; Weisser and Hall, 2009). To reduce the probability of oligomer formation, a 16-residue GGGGS-linker was designed flanked by two restriction sites coding for residues that provide a high degree of flexibility, adding up to a final linker length of 20 residues. We designed the vector to be compatible (5′ and 3′ restriction sites NcoI and NotI) to the pHEN2 plasmid frequently used for phage display selection of high-affinity antibodies (Griffiths et al., 1994) allowing for efficient transfer of selected antibodies into the pMT-scFv-Strep vector. In parallel, the linker sequence is framed by cloning sites for directional cloning of the VH and VL facilitating easy cloning of scFv fragments from hybridoma cDNA. The design of this vector permitted efficient and rapid two-step cloning of all VH and VL scFv genes as described in the Materials and methods section, generating the expression plasmids pMT-scFv-3H5-Strep, pMT-scFv-6A5-Strep, pMT-scFv-8B9-Strep, pMT-scFv-1:7-Strep and pMT-scFv-A8-Strep, respectively.
Fig. 1.
Schematic representation of the pMT-scFv-Strep expression vector. Cloning sites for the variable region of the heavy chain (VH) and the variable region of the light chain (VL) are marked as well as the linker sequence GT(GGSGG)3GAS including two restriction sites allowing for the directional cloning of (VH) and (VL), coding for the bold, underlined residues. The MT promoter and the BiP signal sequence are also marked. In addition, the vector contains an EK cleavage site allowing removal of the double Strep-tag in frame directly downstream of the VL-coding sequence.
Expression of scFvs in Drosophila S2 cells
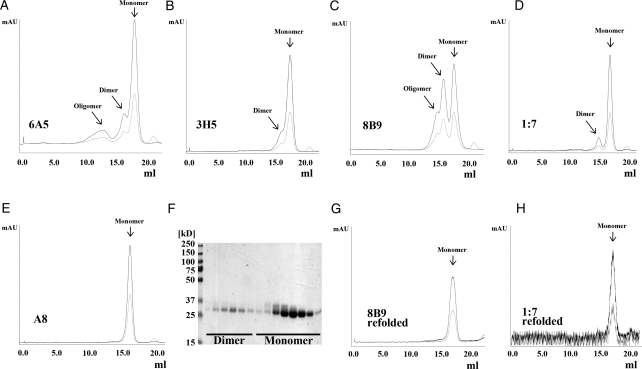
In order to generate stable S2 cell lines expressing the recombinant scFvs, S2 cells were transfected with the respective pMT-scFv-#-Strep plasmid together with a dominant-selectable marker (an expression plasmid encoding Puromycin-Acetyltransferase; Iwaki et al., 2003). After selection, cells were adapted to serum-free Insect Xpress medium (Lonza, Basel, Switzerland), amplified, and protein production was induced by the addition of 4 μM CdCl2 as described in the Materials and methods section. We purified the secreted scFvs to homogeneity from the supernatant by affinity chromatography, with yields varying between 5 and 12 mg/l supernatant. Subsequently, we subjected the eluate to SEC separating monomeric and dimeric scFv species (Fig. 2A–E). All five scFvs from human and mouse origin expressed a majority of monomeric scFv, as judged by SEC followed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS–PAGE) analysis under non-reducing conditions and Coomassie blue staining (Fig. 2F). In a typical SEC profile, the majority of the protein elutes at a volume corresponding to a monomer. However, depending on the individual sequence, minor peaks corresponding to dimeric and higher oligomeric scFvs could be observed (Fig. 2A–D).
Fig. 2.
Purification of human and murine scFvs. ScFvs 6A5 (A), 3H5 (B), 8B9 (C), 1:7 (D) and A8 (E) were affinity purified and subsequently SEC was performed using a Superdex 200 column. Chromatograms show absorption at 280 nm (black) and 254 nm (light grey) and reveal for the majority of the scFvs a major peak corresponding to a monomeric scFv (∼26 kD). In addition, particularly for scFv 8B9 (C), but less pronounced also for scFvs 6A5, 3H5 and 1:7 (A, B, D) a second peak was observed, which corresponds to a dimeric scFv (or diabody). For scFv 8B9 and scFv 6A5 (A, C) a minor peak was detected corresponding to oligomeric scFv. (F) SDS–PAGE analysis of SEC elution fractions of scFv 1:7 under non-reducing conditions confirms the diabodies to be indeed non-covalent dimers. Fractions corresponding to diabodies of scFvs 8B9 and 1:7 were pooled separately, refolded as described in the Materials and methods section and analyzed again by SEC. The SEC profiles of refolded scFvs 8B9 (G) and 1:7 (H) clearly demonstrate a shift of the elution peak of the diabodies toward a monomeric scFv indicating an efficient and successful refolding.
Refolding of dimeric scFvs into monomers
The linker between the two variable domains allows the two variable regions to form the authentic heterodimer; however, it also opens up the possibility that the VH from one scFv molecule may associate with the VL from another molecule, resulting in a linear, antiparallel dimeric molecule or even forming higher oligomers (Holliger et al., 1993). Dimeric scFvs, also known as diabodies, have therapeutic applications of their own. The extreme flexibility in the Fv angle of these oligomeric forms (Holliger and Hudson, 2005) is, however, undesirable for structural studies.
Several studies have investigated the use of different refolding procedures to scFvs (reviewed in Sinacola and Robinson, 2002). To show that the dimeric protein produced in S2 cells represents diabodies rather than aggregated or misfolded protein, we pooled the dimer-containing fractions of those scFvs exhibiting an elution pattern suggestive of dimeric protein and applied a stepwise dialysis refolding approach adapted from (Sinacola and Robinson, 2002). Dimeric scFv at a concentration of 20–50 μg/ml was dialyzed against 8 M urea in the absence of reducing agents to dissociate oligomers. The urea concentration was then slowly lowered to 0 to favor the formation of monomers over dimers or multimers; l-arginine was added at the end of the process, at the lowest urea concentrations, to promote refolding and discourage the formation of aggregates (Chen et al., 2009).
SEC analysis of the refolded scFv revealed that dimeric scFvs 8B9 and 1:7 were entirely refolded into monomers (Fig. 2G and H) in the absence of any reducing agents. This indicates that the original dimeric species already contained the correct disulfide connectivity in both heavy and light chains, likely representing diabodies.
ScFvs interact with their cognate antigen
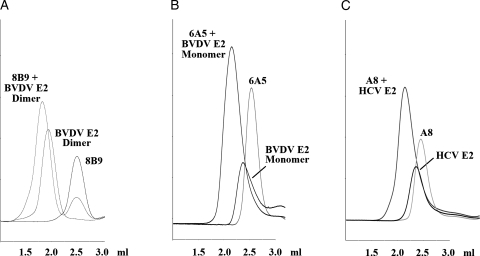
The parent antibodies of scFvs 8B9, 6A5 and A8 each bind their cognate antigen BVDV E2 and HCV E2, respectively, with high affinity as suggested by the stability of all three antibody–antigen complexes in SEC (data not shown). To demonstrate that the affinity of the monomeric scFvs to their cognate antigen was similar to that of the full-length antibody we used SEC to assess the ability of the scFvs to interact with its cognate antigen. Each scFv and its respective cognate antigen were mixed and subsequently analyzed by SEC. All three complexes of scFvs with their cognate antigen were eluted considerably earlier than the respective individual proteins (Fig. 3), indicating a stable complex formation, as observed for the parent antibodies. This was also observed for the scFv 8B9 after being subjected to the stepwise dialysis refolding procedure (data not shown), indicating that the monomeric scFv resulting from the refolding procedure also displays an antigen-binding capacity similar to the parent antibody.
Fig. 3.
Functional characterization of recombinant scFvs. Complex formation between scFvs 8B9 (A), 6A5 (B) and A8 (C) and their respective cognate antigen was analyzed by SEC analysis using a Superdex 200 5/150 column (CV 3 ml). The scFv, the respective antigen and a mixture of the two were loaded to the column (in three different runs) (scFv ∼26 kD, HCV E2 ∼50 kD, BVDV E2 monomer ∼50 kD, BVDV E2 dimer ∼100 kD, complexes with antigen monomer ∼80 kD, complex with antigen dimer ∼160 kD). The peak corresponding to the free scFv in the chromatogram of the scFv-antigen complex for scFv 8B9 (A) is likely due to a molar excess of scFv used in this experiment.
Crystal structure of scFv 1:7
One of the potential applications of a recombinant scFv is the use in structural studies to facilitate crystallization of interesting macromolecules that would not crystallize otherwise. One example of such a molecule is the major glycoprotein E2 of the Hepatitis C virus (HCV), the crystal structure of which has still not been reported despite being the focus of intense research efforts worldwide. One possible strategy to promote crystallization of this multidomain, heavily glycosylated envelope protein is cocrystallization in complex with scFvs. To demonstrate the suitability of scFvs produced in the reported S2 cell expression system for structural studies, we determined the crystal structure of the scFv 1:7, which is derived from a broadly neutralizing anti-HCV E2 antibody previously described (Allander et al., 2000; Johansson et al., 2007b). Diffraction quality crystals of the unliganded scFv, belonging to space group C2221, could be grown within 1 week. These crystals diffracted to 2.7 Å on a synchrotron source. We determined the crystal structure of the scFv using the molecular replacement method; details and statistics of the data collection, processing and refinement are given in Table II.
Table II.
Details and statistics of data collection, processing and refinement for scFv 1:7 crystals
| 1:7 scFv | |
| Data collection | |
| Space group | C2221 |
| Cell dimensions | |
| a, b, c (Å) | 105.49, 105.74, 105.60 |
| α, β, γ (°) | 90.0, 90.0, 90.0 |
| Resolution (Å) | 43.11–2.67 (2.81–2.67) |
| Rmerge | 0.084 (0.459) |
| I/σ I | 11.5 (1.9) |
| Completeness (%) | 92.5 (54.1) |
| Redundancy | 3.5 (2.4) |
| Refinement | |
| No. reflections | 15793 |
| Rwork/Rfree | 0.193/0.232 |
| No. atoms | |
| Protein | 3604 |
| B-factors | |
| Protein | 50.58 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.01 |
| Bond angles (°) | 1.19 |
The scFv 1:7 displays the standard compact fold of two heterodimerizing immunoglobulin domains, VH and VL, each containing nine β-strands tightly packed in two β-sheets (Fig. 4A). The dimer interface between VH and VL buries ∼558 Å2 of solvent accessible surface, which is similar to the buried surface area computed for the variable region interface of other scFvs (e.g. PDB 1qok, 2ghw) or Fab molecules (e.g. PDB 2xqy, 3n9g) of human and murine origin. A structural comparison to the database using the DALI-server (Holm and Rosenström, 2010) followed by superposition with the four closest structural neighbors (data not shown) revealed no distortion between VH and VL. Taken together, this set of evidence strongly suggests a 3D arrangement of the scFv 1:7 that is identical to the variable region of the parent antibody and thus further validates the expression system presented in this study.
Antibody diversity is generated by the combinatorial association of V, D and J segments, which in addition becomes further diversified at the actual junctions (VL–JL, VH–D and D–JH) due to imprecise joining and addition of ‘N region’ nucleotides. Somatic mutation, possibly driven by antigenic selection, contributes further to antibody diversity and leads to increased affinity and specificity as antibody maturation occurs (Tonegawa, 1983; French et al., 1989). The closest homologous germline genes for the 1:7 antibody suggested by immunogenetics (IMGT) V-QUEST and junction analysis (Lefranc et al., 2009) are IGHV1-69*01 F, IGHJ4*02 F and IGHD2-2*01 F in the heavy chain and IGKV3-11*01 F and IGKJ4*01 F in the light chain. The same heavy chain V gene (IGHV1-69*01 F) was found in an exceptional high frequency in anti-HCV-E2 antibodies (Allander et al., 2000; Chan et al., 2001), suggesting a preferential usage of the same V gene in the specific immune response to the HCV E2 glycoprotein. Antibodies derived from IGHV1-69*01 F have been reported to undergo somatic mutations during antigenic selection, with high frequency located in or close to the complementarity determining region (CDR) regions, indicating a positive selection for antigen binding (Chan et al., 2001). We aligned the 1:7 VH and VL amino acid sequence to the respective germline sequences (Fig. 4B) and identified somatic mutations acquired during antibody maturation. Subsequently, we mapped those mutations on the surface of the scFv structure (Fig. 4C). Mutations at the very N-terminus were disregarded, since they are very often due to the primer sequences that have been used to isolate the antibody cDNA. We found 3 mutations in the light chain and 20 mutations in the heavy chain, the majority of which clusters in or around the H1 and H2 loop, while only 2 somatic mutations are found in the long H3 loop. In the light chain two mutations are located on the L1 loop and one in the L3 loop. These data suggest a binding mode of this broadly neutralizing antibody to its cognate antigen that is predominantly mediated by H1, H2 and H3 loops and supported by the L1 loop.
The fact that the crystal structure of the scFv 1:7 could be obtained relatively easily, demonstrates that the scFvs produced in our expression system are applicable for the use in structural studies. We expressed five different scFvs derived from human and murine origin, including codon optimized as well as hybridoma-derived DNA sequences and for all five we obtained expression levels between 5 and 12 mg/l supernatant. This suggests that the strong sequence dependence that has been described for other expression systems like E.coli and mammalian cells (Jost et al., 1994; Verma et al., 1998; Weisser and Hall, 2009) is not observed in the S2 cell expression system described here. We conclude that our system is a very powerful tool, particularly for those antibodies that remain–for yet unknown reasons–difficult to obtain in large-enough yields with other expression systems.
Supplementary data
Conflict of interest
The human antibodies A8 and 1:7 are protected in patents owned by Molecules of Man AB, a spinoff company based on discoveries made at Karolinska Institutet, Sweden. M.A.A.P. has a financial interest in the company as shareholder.
Funding
A.A.G. benefited from a Fulbright-Hays fellowship. This work was supported by the ANRS and the ANR grant ANR-2010-BLAN-1211 01 to F.A.R., in addition to the recurrent Institut Pasteur and CNRS support to F.A.R.; the Swedish Foundation for Strategic Research (Cell Factory and Infection &Vaccines programs), the Swedish Cancer Society and the Swedish Research Council to M.A.A.P. Funding to pay the Open Access publication charges for this article was provided by the Institut Pasteur.
Supplementary Material
Acknowledgements
We thank Patrick Weber of the PF6 for help with the crystallogenesis and Andrew Thompson from the beamline PROXIMA 1 at the synchrotron Soleil for help with data collection. We also thank Scott A.Jeffers for excellent advice during protein purification and Joseph J.Cockburn and Scott A.Jeffers for helpful discussions.
References
- Allander T., Drakenberg K., Beyene A., Rosa D., Abrignani S., Houghton M., Widell A., Grillner L., Persson M.A. J Gen Virol. 2000;81:2451–2459. doi: 10.1099/0022-1317-81-10-2451. [DOI] [PubMed] [Google Scholar]
- Arndt K.M., Müller K.M., Plückthun A. Biochemistry. 1998;37:12918–12926. doi: 10.1021/bi9810407. [DOI] [PubMed] [Google Scholar]
- Backovic M., Johansson D.X., Klupp B.G., Mettenleiter T.C., Persson M.A.A., Rey F.A. Protein Eng Des Sel. 2010;23:169–174. doi: 10.1093/protein/gzp088. [DOI] [PubMed] [Google Scholar]
- Begent R.H., Verhaar M.J., Chester K.A., et al. Nat Med. 1996;2:979–984. doi: 10.1038/nm0996-979. [DOI] [PubMed] [Google Scholar]
- Bird R.E., Hardman K.D., Jacobson J.W., et al. Science. 1988;242:423–426. doi: 10.1126/science.3140379. [DOI] [PubMed] [Google Scholar]
- Bricogne G., Blanc E., Brandl M., et al. Cambridge, UK: Global Phasing Ltd; 2010. [Google Scholar]
- Brocks B., Rode H.J., Klein M., Gerlach E., Dübel S., Little M., Pfizenmaier K., Moosmayer D. Immunotechnology. 1997;3:173–184. doi: 10.1016/s1380-2933(97)00011-0. [DOI] [PubMed] [Google Scholar]
- Chan C.H., Hadlock K.G., Foung S.K., Levy S. Blood. 2001;97:1023–1026. doi: 10.1182/blood.v97.4.1023. [DOI] [PubMed] [Google Scholar]
- Chen J., Liu Y., Li X., Wang Y., Ding H., Ma G., Su Z. Protein expression and purification. 2009;66:82–90. doi: 10.1016/j.pep.2009.02.004. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. [Google Scholar]
- Eisenstein M. Nat Biotechnol. 2011;29:107–109. doi: 10.1038/nbt.1768. [DOI] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P. Acta Crystallogr D Biol Crystallogr. 2005;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- Filpula D., McGuire J., Whitlow M. In: Antibody engineering: a practical approach. McCafferty J., Hoogenboom H., Chiswell D., editors. Oxford: Oxford University Press; 1996. pp. 253–268. [Google Scholar]
- French D.L., Laskov R., Scharff M.D. Science. 1989;244:1152–1157. doi: 10.1126/science.2658060. [DOI] [PubMed] [Google Scholar]
- Glasgow J.N., Mikheeva G., Krasnykh V., Curiel D.T. PLoS ONE. 2009;4:e8355. doi: 10.1371/journal.pone.0008355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin L., Lawson A. Clin Exp Immunol. 2011;165:285–291. doi: 10.1111/j.1365-2249.2011.04427.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths A.D., Williams S.C., Hartley O., et al. EMBO J. 1994;13:3245–3260. doi: 10.1002/j.1460-2075.1994.tb06626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliger P., Hudson P.J. Nat Biotechnol. 2005;23:1126–1136. doi: 10.1038/nbt1142. [DOI] [PubMed] [Google Scholar]
- Holliger P., Prospero T., Winter G. Proc Natl Acad Sci USA. 1993;90:6444–6448. doi: 10.1073/pnas.90.14.6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L., Rosenström P. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huston J.S., Levinson D., Mudgett-Hunter M., et al. Proc Natl Acad Sci USA. 1988;85:5879–5883. doi: 10.1073/pnas.85.16.5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huston J.S., Margolies M.N., Haber E. Adv Protein Chem. 1996;49:329–450. doi: 10.1016/s0065-3233(08)60493-3. [DOI] [PubMed] [Google Scholar]
- Huston J.S., Mudgett-Hunter M., Tai M.S., McCartney J., Warren F., Haber E., Oppermann H. Methods Enzymol. 1991;203:46–88. doi: 10.1016/0076-6879(91)03005-2. [DOI] [PubMed] [Google Scholar]
- Hwang W.C., Lin Y., Santelli E., Sui J., Jaroszewski L., Stec B., Farzan M., Marasco W.A., Liddington R.C. J Biol Chem. 2006;281:34610–34616. doi: 10.1074/jbc.M603275200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwaki T., Figuera M., Ploplis V.A., Castellino F.J. Biotechniques. 2003;35:482–484. doi: 10.2144/03353bm08. 486. [DOI] [PubMed] [Google Scholar]
- Johansson D.X., Drakenberg K., Hopmann K.H., Schmidt A., Yari F., Hinkula J., Persson M.A.A. J Immunol Methods. 2007a;318:37–46. doi: 10.1016/j.jim.2006.08.017. [DOI] [PubMed] [Google Scholar]
- Johansson D.X., Krey T., Andersson O. In: Antibody engineering: methods and protocols. Chames P., editor. Totowa, N.J.: Humana Press; (in press) [Google Scholar]
- Johansson D.X., Voisset C., Tarr A.W., Aung M., Ball J.K., Dubuisson J., Persson M.A.A. Proc Natl Acad Sci USA. 2007b;104:16269–16274. doi: 10.1073/pnas.0705522104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost C.R., Kurucz I., Jacobus C.M., Titus J.A., George A.J., Segal D.M. J Biol Chem. 1994;269:26267–26273. [PubMed] [Google Scholar]
- Kabsch W. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovari L.C., Momany C., Rossmann M.G. Structure. 1995;3:1291–1293. doi: 10.1016/s0969-2126(01)00266-0. [DOI] [PubMed] [Google Scholar]
- Krey T., d'Alayer J., Kikuti C.M., et al. PLoS Pathog. 2010;6:e1000762. doi: 10.1371/journal.ppat.1000762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefranc M.-P., Giudicelli V., Ginestoux C., et al. Nucleic Acids Res. 2009;37:D1006–D1012. doi: 10.1093/nar/gkn838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahiouz D.L., Aichinger G., Haskard D.O., George A.J. J Immunol Methods. 1998;212:149–160. doi: 10.1016/s0022-1759(98)00007-6. [DOI] [PubMed] [Google Scholar]
- McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reavy B., Ziegler A., Diplexcito J., Macintosh S.M., Torrance L., Mayo M. Protein Expr Purif. 2000;18:221–228. doi: 10.1006/prep.1999.1191. [DOI] [PubMed] [Google Scholar]
- Ridder R., Schmitz R., Legay F., Gram H. Biotechnology (NY) 1995;13:255–260. doi: 10.1038/nbt0395-255. [DOI] [PubMed] [Google Scholar]
- Sandhu J.S. Crit Rev Biotechnol. 1992;12:437–462. doi: 10.3109/07388559209114235. [DOI] [PubMed] [Google Scholar]
- Sinacola J.R., Robinson A.S. Protein Expr Purif. 2002;26:301–308. doi: 10.1016/s1046-5928(02)00538-7. [DOI] [PubMed] [Google Scholar]
- Skerra A., Plückthun A. Science. 1988;240:1038–1041. doi: 10.1126/science.3285470. [DOI] [PubMed] [Google Scholar]
- Sui J., Hwang W.C., Perez S., et al. Nature Struct Mol Biol. 2009;16:265–273. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonegawa S. Nature. 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- Verma R., Boleti E., George A.J. J Immunol Methods. 1998;216:165–181. doi: 10.1016/s0022-1759(98)00077-5. [DOI] [PubMed] [Google Scholar]
- Weisser N.E., Hall J.C. Biotechnol Adv. 2009;27:502–520. doi: 10.1016/j.biotechadv.2009.04.004. [DOI] [PubMed] [Google Scholar]
- Wu X.C., Ng S.C., Near R.I., Wong S.L. Biotechnology (NY) 1993;11:71–76. doi: 10.1038/nbt0193-71. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.