Abstract
Gun4 is a porphyrin-binding protein that activates magnesium chelatase, a multimeric enzyme catalyzing the first committed step in chlorophyll biosynthesis. In plants, GUN4 has been implicated in plastid-to-nucleus retrograde signaling processes that coordinate both photosystem II and photosystem I nuclear gene expression with chloroplast function. In this work we present the functional analysis of Gun4 from the cyanobacterium Synechocystis sp. PCC 6803. Affinity co-purification of the FLAG-tagged Gun4 with the ChlH subunit of the magnesium chelatase confirmed the association of Gun4 with the enzyme in cyanobacteria. Inactivation of the gun4 gene abolished photoautotrophic growth of the resulting gun4 mutant strain that exhibited a decreased activity of magnesium chelatase. Consequently, the cellular content of chlorophyll-binding proteins was highly inadequate, especially that of proteins of photosystem II. Immunoblot analyses, blue native polyacrylamide gel electrophoresis, and radiolabeling of the membrane protein complexes suggested that the availability of the photosystem II antenna protein CP47 is a limiting factor for the photosystem II assembly in the gun4 mutant.
The three major tetrapyrrole end products chlorophyll (Chl),3 heme, and phycobilins are synthesized in a branched metabolic pathway with protoporphyrin IX (PIX) as the last common precursor (1–4). The iron branch insertion of Fe2+ into PIX by ferrochelatase leads to the formation of protoheme, whereas in the magnesium branch magnesium chelatase catalyzes the chelation of Mg2+ into PIX, thereby directing tetrapyrroles into Chl synthesis.
Cyanobacteria and plants accumulate various tetrapyrrole species in different quantities in the cell. Unlike plants, which use Chl-containing antenna complexes for light harvesting, the major light-harvesting antennae of cyanobacteria consist of phycobiliproteins, which contain covalently bound phycobilines originating from heme. Thus, in cyanobacteria PIX might be directed into the iron branch to a much larger extent than in plants. Regulation of the PIX distribution at the branch point between the magnesium and iron pathways as well as coordination of tetrapyrrole and cognate apoprotein synthesis are substantial in order to avoid accumulation of photoreactive intermediates during tetrapyrrole synthesis. For instance, in plants the genes coding for the light-harvesting Chl-a/b-binding proteins (LHCB) and for Chl biosynthesis proteins are simultaneously up-regulated in the light (5–7).
In a search for Arabidopsis thaliana mutants that are deregulated in the communication between plastids and the nucleus several gun (genome uncoupled) mutants have been identified (8). The GUN2, GUN3, GUN4, and GUN5 genes encode proteins that are involved in tetrapyrrole biosynthesis (9, 10). Two of the gene products, GUN2 (heme oxygenase) and GUN3 (biliverdin reductase), function in heme degradation. GUN5 encodes the ChlH subunit of the magnesium chelatase complex (9), whereas GUN4 encodes a protein that interacts with magnesium chelatase (10). GUN4-related genes were found only in photosynthetic organisms. In plants GUN4 either is encoded in the nucleus or retained in the chloroplast genome as is the case with several algae, where it was previously designated as ycf53.
All cyanobacterial genomes sequenced so far contain one or more gun4-related genes, with the exception of the ancient cyanobacterium Gloeobacter violaceus PCC7421. An A. thaliana GUN4 mutant forms white-yellow tissue under standard conditions or pale green tissue in dim light (10). The A. thaliana Gun4 protein was co-purified with the ChlH subunit of magnesium chelatase and binds PIX as well as magnesium protoporphyrin IX (MgPIX) (10). In vitro enzyme assays revealed a dramatically increased efficiency of the enzymatic Mg2+ insertion into PIX in the presence of Gun4, especially at low Mg2+ concentrations (10, 11). Accordingly, the crystal structures of Gun4 proteins from two cyanobacterial species bare a specific porphyrin binding site supporting a function of Gun4 in tetrapyrrole trafficking (11, 12).
In a previous report we described an incompletely segregated gun4 mutant of Synechocystis sp. PCC 6803 (hereafter Synechocystis 6803), arguing for substantial roles of Gun4 in cell metabolism (13). Here, we succeeded in generating a fully segregated gun4 mutant strain in the glucose-tolerant (GT) Synechocystis 6803 wild-type background. The analysis of this strain revealed that the absence of Gun4 strongly decreased Chl biosynthesis and completely abolished photoautotrophic growth. These mutant properties correlated with a drastically impaired assembly of the two photosystems (PS). Among all Chl-binding proteins the accumulation of the PSII subunit CP47 was most strongly affected in the gun4 mutant. The regulatory interaction between Chl synthesis and PS accumulation is discussed.
EXPERIMENTAL PROCEDURES
Culture Conditions—Synechocystis 6803 wild-type (GT) and mutant strains were grown at 30 °C in BG-11 medium (14) containing 5 mm glucose under continuous irradiance of 5 μmol of photons m–2 s–1 (low light intensity) or 50 μmol of photons m–2s–1 (medium light intensity) in rotating Erlenmeyer flasks. For microaerobic growth, the strains were grown under medium light in a 1% CO2, 99% N2 atmosphere. The GT strain originates from the laboratory of W. Vermaas (Arizona State University). Transformants of Synechocystis 6803 were selected on media with increasing concentrations of kanamycin (5 μg up to 40 μgml–1). The gun4 mutant strain was prepared as described in Wilde et al. (13). Complete segregation of the mutants was confirmed by PCR using primers 5′-CTTCCGCTGGATCACCTTTA-3′ and 5′-GGTAATGACTTCCCCGAAGA-3′.
Construction of a Synechocystis 6803 Strain Expressing the 3×FLAG-tagged Gun4 Protein—The coding sequence of the Synechocystis 6803 gun4 gene was amplified using the primers 5′-CATATGTCTGATAATTTGACCGAACTCTCC-3′ and 5′-AGATCTTTACCAACCGTATTGGGACC-3′. An NdeI site that overlaps with the ATG start codon of the gun4 gene was introduced by the forward primer. The PCR product was subcloned into the pDrive vector (Qiagen, Hilden, Germany). Two complementary oligonucleotides encoding the 3×FLAG peptide (Sigma-Aldrich) were hybridized, thus creating NdeI-compatible overhangs, and then ligated into the NdeI site of the pDrive-gun4 construct. The resultant construct was verified by sequencing. Using NdeI and BglII restriction enzymes, a DNA fragment encoding the FLAG-tagged Gun4 was excised and then ligated into pSK9 vector for chromosomal integration under the control of the copper dependent petJ promoter (15), which supports low level expression in BG-11 medium. gun4 mutant cells were transformed with the resultant vector pSKgun4FLAG. Transformants were selected on BG-11 agar plates containing 7 μg ml–1 chloramphenicol. For strong expression of the fusion protein under the control of the petJ promoter, cells were grown for at least 7 days in BG-11 medium lacking copper in the trace metal mix.
Determination of Enzyme Activity—Cell cultures (800 ml) were pelleted and disrupted with glass beads in PBS, pH 7.5, and cell debris was removed by centrifugation. Samples were sub-divided for assays of magnesium chelatase and ferrochelatase activities, which were performed according to Papenbrock et al. (16). A typical assay mix contained 1 mm dithiothreitol (DTT), 0.1% bovine serum albumin, 25 mm ATP, 20 mm MgCl2, and 100 μm PIX for magnesium chelatase or 1 mm DTT, 1% Tween 80, 2.5 mm palmitic acid, 10 mm ZnSO4, and 50 μm PIX for ferrochelatase. The reactions were started by the addition of PIX dissolved in DMSO, and the assay mixtures were further incubated at 28 °C. Reactions were stopped after 0, 20, 40, and 60 min by the addition of an equal volume of methanol to the reaction mixture followed by freezing in liquid nitrogen. After the initial methanol extraction all samples were extracted twice with acetone, methanol, 0.1 n NH4OH (10:9:1; v/v/v). Reaction products were quantified by HPLC using authentic porphyrin standards.
Determination of Porphyrins—Steady state levels of magnesium porphyrins were analyzed in the exponential growth phase of the cell culture (OD750 nm ∼ 0.5–0.9). Pelleted cells were resuspended in methanol and incubated on ice for 15 min. After a short centrifugation, supernatants were collected, and the pellets were resuspended in methanol, acetone, 0.1 n NH4OH (10:9:1; v/v/v). Supernatants were combined after centrifugation at 13,000 × g for 5 min and prepared for HPLC analysis. Extracted porphyrinogens were oxidized by the addition of 5 μl of 1 m acetic acid and 5 μl of 2-butanone peroxide per 200 μl of extract and separated by HPLC (Agilent-1100, Agilent, Waldborn, Germany) on a RP 18 column (Novapak C18, 4-μm particle size, 3.9 × 150 mm; Millipore, Eschborn, Germany) at a flow rate of 1 ml min–1. Porphyrins were eluted with a linear gradient of solvent B (90% methanol, 0.1 m ammonium acetate, pH 5.2) in solvent A (10% methanol, 0.1 m ammonium acetate, pH 5.2) as follows: 0 to 100% in 7 min followed by 100% solvent B for 17 min. The eluate was monitored by fluorescence detection. The excitation and emission wavelengths were, respectively: for Proto, 405 and 625 nm; for ZnProto, 416 and 589 nm; for MgProto and MgProtoME, 420 and 595 nm, respectively. Porphyrins were identified and quantified using authentic standards (Fluka, Germany; Porphyrin Products, UT). For quantification of PIX precipitated in growth medium, 100 ml of culture at OD750 ∼ 0.4 were filtered through 4-μm cellulose filters to collect precipitated pigments in growth media. The filter was frozen in liquid nitrogen, powdered in a 2-ml Eppendorf tube, and dissolved in 0.5 ml of alkaline acetone. Acetone was then extracted with 250 μl of hexane to remove Chl. PIX was diluted 50× in DMSO and quantified spectrofluorometrically using an authentic standard (Sigma).
Radioactive Labeling of Cells and Preparation of Membranes—Radiolabeling of cells using a mixture of [35S]Met and [35S]Cys (Trans-label, MP Biochemicals, Irvine, CA; final activity, 400 μCi ml–1) was performed at 30 °C for 20 min under illumination with 125 μmol of photons m–2 s–1 as described in a previous study (17). For preparation of membrane and cytosolic protein fractions, cells (OD750 ∼ 0.5) were washed and resuspended in thylakoid buffer containing 25 mm Hepes, pH 7.4, 10 mm MgCl2, 5 mm CaCl2, and 20% glycerol. The cell suspension was mixed with glass beads and broken in a Mini Bead Beater, and the resulted homogenate was divided into two halves. To prepare a whole cell extract, the first part was solubilized by 1% β-dodecyl maltoside (DM) and centrifuged at 32,000 × g for 10 min at 4 °C to discard unbroken cells. The second part was centrifuged at 100,000 × g for 30 min at 4 °C, and the supernatant containing soluble proteins was transferred to a new tube. Pelleted membranes were washed 3 times in thylakoid buffer and resuspended in the same buffer containing 1% DM, and unbroken cells were discarded by centrifugation. To prove whether proteins are tightly bound to the membrane, an additional washing step with 2 m NaCl was applied.
Protein Analyses—Analysis of membrane proteins under native conditions was performed by Blue Native (BN)-PAGE using the buffer system of Schägger and von Jagow (18). The isolated membranes were resuspended in 25 mm MES/NaOH, pH 6.5, containing 10 mm CaCl2, 10 mm MgCl2, and 25% glycerol. The membranes were then solubilized with DM (DM: Chl = 40 for WT and 160 for gun4–, respectively, w/w) and analyzed at 4 °C in a 4–16% polyacrylamide gel. Protein composition of complexes was then assessed by electrophoresis in a denaturing 12–20% linear gradient polyacrylamide gel containing 7 m urea (19). The whole lanes from the BN gel were excised, incubated for 30 min in 25 mm Tris/HCl, pH 7.5, containing 1% SDS (w/v), and placed on the top of the denaturing gel; two lanes were analyzed in a single denaturing gel. Proteins separated in the gel were either stained by Coomassie Blue or transferred onto a membrane for immunodetection. One-dimensional SDS-PAGE was carried out as described by Jänsch et al. (20). For immunodetection proteins were transferred from gels onto nitrocellulose or polyvinylidene difluoride membranes and immunolabeled with specific antibodies. The primary antibodies used in the study were raised in rabbits against (i) residues 58–86 of the spinach D1 polypeptide, (ii) residues 380–394 of barley CP47, (iii) whole recombinant Gun4 and ferrochelatase proteins from Synechocystis 6803 overexpressed in Escherichia coli using the pQE80 expression vector (Qiagen) and purified with nickel-nitrilotriacetic acid-agarose. The antibodies against Synechocystis 6803 ChlH and ChlD subunits of the magnesium chelatase were kindly provided by Prof. Neil C. Hunter (University of Sheffield). Protein signals were visualized using the Immobilon Western (Millipore, Bedford, MA) or Supersignal West Pico (Pierce) chemiluminescence detection systems. Purification of 3×FLAG-tagged Gun4 from Synechocystis 6803 was performed from solubilized whole cell extracts as previously described (21).
Electron Microscopy—Cells were washed twice in 50 mm phosphate buffer, pH 7.2, and fixed in a solution containing 2% glutaraldehyde in 50 mm phosphate buffer for 2 h. This was followed by 3 washes in phosphate buffer and post-fixated in 1% OsO4 in 50 mm phosphate buffer for 1 h. Finally, the cells were washed 3 times and embedded in 2% agarose. The segments of agarose with cells were dehydrated in isopropyl alcohol and embedded in Spurr's resin. Thin sections were cut with a diamond knife and post-stained for 30 min with 5% aqueous uranyl acetate followed by lead citrate for 80 s. The samples were examined with a Jeol 1010 electron microscope equipped with CCD camera MegaView III and image processing software Analysis (3.1.110).
Absorption Spectra and Pigment Measurements—Absorption spectra of whole cells were measured with a UV-2401 PC spectrophotometer equipped with an integrating sphere (Shimadzu). Chl was extracted from cell pellets (10 ml, OD750 ∼ 0.5) with 100% methanol, and its content was measured spectrophotometrically (22).
77 K Fluorescence Emission Spectra—77 K fluorescence emission spectra were measured in liquid nitrogen with an Aminco spectrofluorimeter (Spectronic Unicam) using membranes washed twice with excess of thylakoid buffer. 100 μl of the sample at Chl concentration of 14 μgml–1 were used for the measurement with 2 μm rhodamine as an internal standard. The spectra were normalized to rhodamine peak (573 nm) and multiplied by relative Chl:absorbance ratio for individual strains to indicate their Chl fluorescence per cell. 77 K fluorescence emission spectra were collected using excitation wavelength 435 nm to excite Chla.
RESULTS
Mutant Characterization—We had previously analyzed the role of Synechocystis 6803 Gun4 in tetrapyrrole biosynthesis using a gun4 mutant strain that was not fully segregated under different growth conditions. These gun4 mutant cells had exhibited lower magnesium and ferrochelatase activities accompanied by the accumulation of PIX, the substrate of both enzymes. In the present study we used a GT Synechocystis 6803 strain for mutational analysis. This different lineage of the original Synechocystis 6803 strain is widely used for the dissection and analysis of photosynthetic mutants. Fully segregated PSI, PSII, and Chl biosynthesis mutants could be generated in this genetic background (23, 24).
PCR analysis (Fig. 1) revealed that gun4 mutant in the GT substrain of Synechocystis 6803 (hereafter designated gun4–) was completely segregated. In contrast to previous results obtained with the PCC strain of Synechocystis 6803 (13), no chromosomal wild-type copies of the gun4 gene were detected. Gun4 inactivation was confirmed by immunoblot analysis, demonstrating the complete lack of Gun4 in protein extracts from the mutant (compare Fig. 3A). Because Gun4-deficient cells lost the ability to grow photoautotrophically, all media were supplemented with 5 mm glucose to support the growth of the gun4 mutant cells. At a medium light intensity (50 μmol of photons m–2 s–1) gun4 mutant cells grew slowly, lost more than 90% of Chl, and accumulated large amounts of PIX (Table 1). In addition, PIX also strongly accumulated in the growth medium leading to the formation of brown porphyrin aggregates. When these aggregates were separated from cells by filtration, more than eight times more PIX was found in gun4– cells compared with the wild type (data not shown). In addition, mutant cells lost substantial amounts of thylakoid membranes (Fig. 2B) at medium light intensity. In contrast, at low light intensity (5 μmol of photons m–2 s–1) growth of the gun4 mutant on glucose (Table 1) was improved, its thylakoid structure (Fig. 2C) rather resembled that of wild type, and endogenous accumulation of PIX was decreased (Table 1). However, even under low light conditions, the Chl content in the mutant was only about 25% that of the wild-type level (Table 1), and the content of the Chl precursor MgPIX monomethyl ester was strongly reduced (Table 1). Similar effects were observed when the gun4 mutant was grown under microaerobic conditions at medium light intensity. Chl content in the mutant increased from 8 to 15%, and the cells contained more thylakoid membranes than under aerobic conditions (Fig. 2D). The negative influence of both light and aerobic conditions on the formation of thylakoid membranes and Chl contents in the gun4 mutant suggests increased light sensitivity and photooxidative damage in the absence of Gun4 at medium light intensity. To prove whether oxidative stress contributes to the gun4– phenotype even at low light intensities, the mutant was grown microaerobically under these light conditions. No further increase in Chl accumulation was observed compared with aerobic growth at low light, and the Chl content in the mutant remained at 25% that of the wild-type level. In addition, the amount of Chl also was not enhanced in gun4 mutant cells, grown aerobically in the dark (data not shown). It was concluded that low light intensity did not photosensitize the mutant cells and did not cause light-dependent oxidative stress. Thus, strains were grown at low light intensities for further experiments.
FIGURE 1.
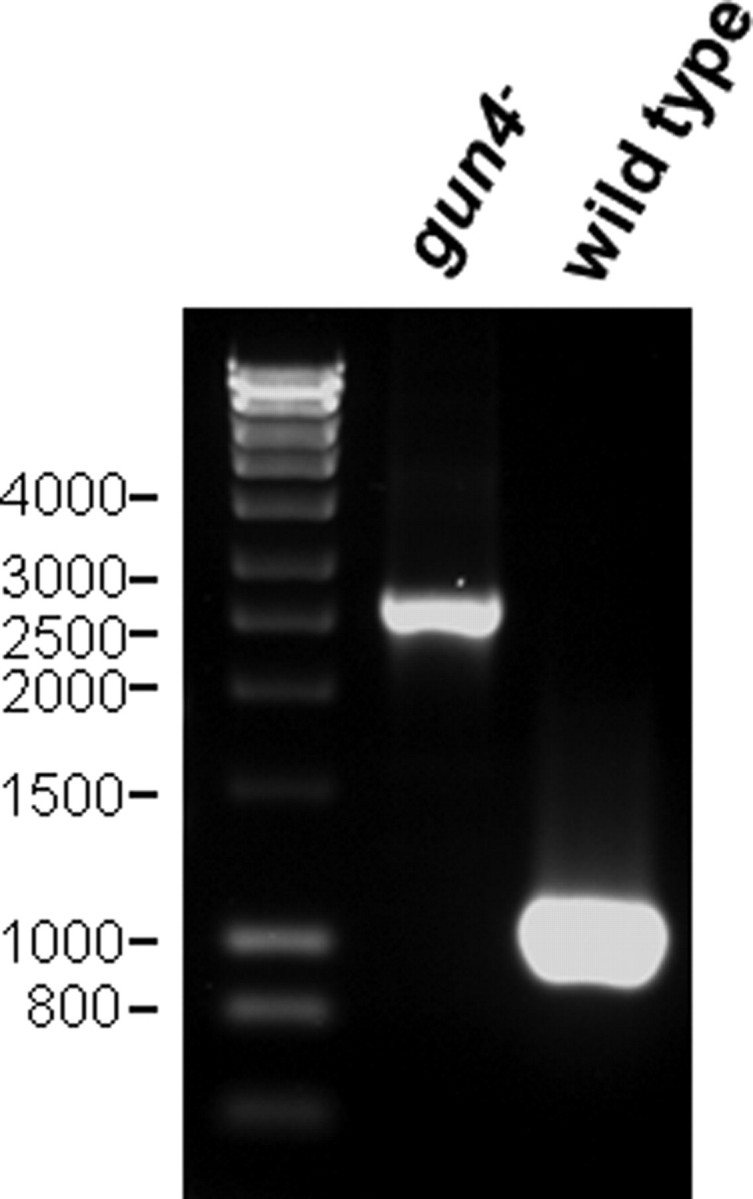
Inactivation of the gun4 gene in Synechocystis 6803 (GT strain). Wild-type and mutant gene copies were amplified using total chromosomal DNA from wild-type and gun4– strains as templates. PCR amplification of the gun4 gene yielded a correct 1.1-kilobase pair product in the wild type. The mutant gun4 copy gave rise to a larger PCR fragment of 2.6 kilobase pairs due to the insertion of the kanamycin resistance cassette (1.5 kilobase pairs). The sizes of the DNA marker fragments are indicated on the left.
FIGURE 3.
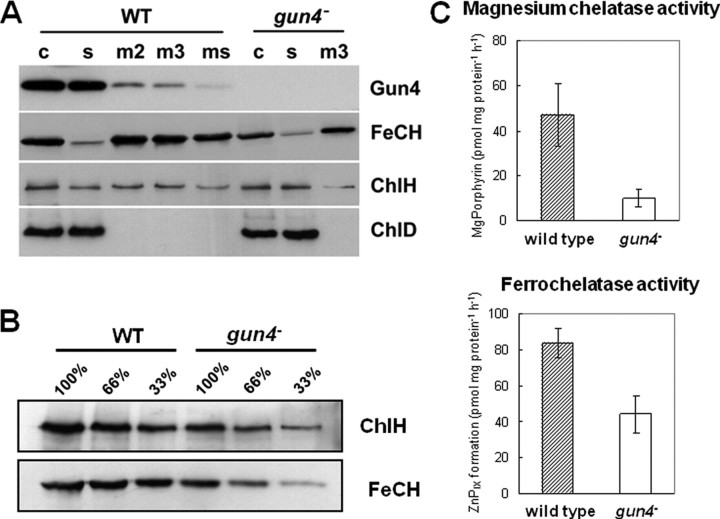
Localization, quantification, and activities of both chelatases. A, subcellular localization of Gun4, ferrochelatase (FeCH), and the magnesium chelatase ChlH and ChlD subunits. Membranes have been washed 2 (m2) or 3 (m3) times in thylakoid buffer followed by an additional wash with 2 m NaCl in thylakoid buffer (ms). Membrane fractions (m), crude (c), and soluble extracts (s) of wild-type and gun4 mutant cells were separated by SDS-PAGE and transferred to nitrocellulose membranes. The amount of Chl corresponding to 150 μl of cells at A730 = 1 was loaded per each lane. Polyclonal antibodies used for the immunoblots are shown on the right. B, accumulation of the ChlH subunit of magnesium chelatase and ferrochelatase in the crude cell extract were determined by immunodetection. Numbers indicate relative protein loading based on the amount of cells. C, activities of magnesium and ferrochelatase measured in wild-type and gun4 mutant cells.
TABLE 1.
Phenotypical analysis of gun4 mutant in comparison to the wild type
Values represent the mean ± S.D. of at least three independent experiments using different cultures harvested in logarithmic growth phase.
|
Strain |
Low light
intensitya |
Medium light
intensityb |
||
|---|---|---|---|---|
| Wild type | gun4- | Wild type | gun4- | |
| Doubling time (h) | 19 ± 0.5 | 21 ± 0.5 | 10 ± 0.2 | 25 ± 0.5 |
| Chlorophyll (μg·mg protein-1) | 11.3 ± 1.1 | 2.7 ± 0.2 | 11.8 ± 1.0 | 0.9 ± 0.4 |
| Phycocyanin (μg·mg protein-1) | 153 ± 36 | 154 ± 32 | 163 ± 45 | 131 ± 43 |
| Protoporphyrin IX (pmol·mg protein-1) | 17.2 ± 10.9 | 61.6 ± 51.2 | 4.7 ± 2.0 | 205.0 ± 100 |
| Mg-protoporphyrin IX (pmol·mg protein-1) | 1.17 ± 0.46 | 0.15 ± 0.06 | 1.00 ± 0.22 | 0.16 ± 0.06 |
| Mg-protoporphyrin IX monomethyl ester (pmol·mg protein-1) | 4.51 ± 0.87 | 0.98 ± 0.51 | 3.88 ± 1.12 | 0.72 ± 0.33 |
5 μmol of photons m-2 s-1 in medium containing 5 mm glucose.
50 μmol of photons m-2 s-1 in medium containing 5 mm glucose.
FIGURE 2.
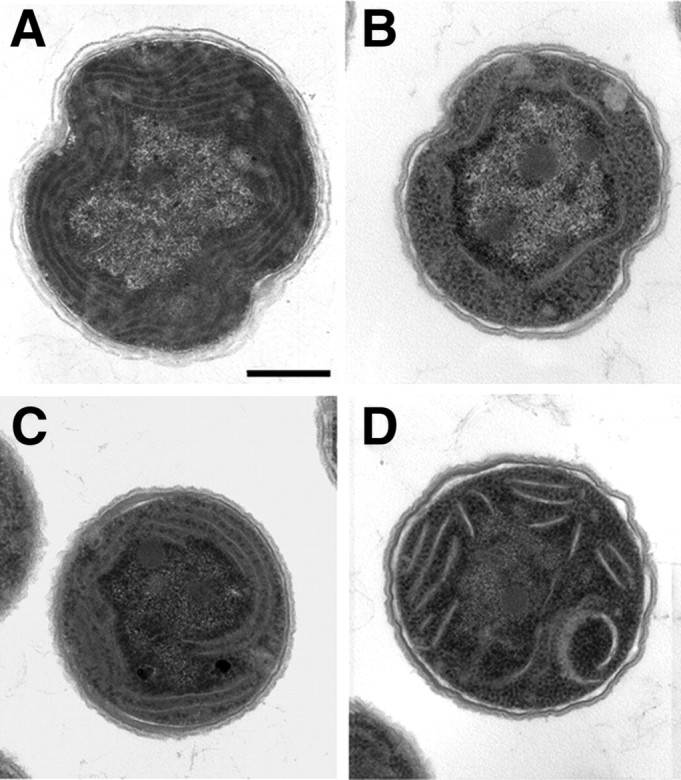
Transmission electron microscopy of Synechocystis 6803 cells. A, wild-type cells grown aerobically under medium light conditions. B, gun4 mutant cells grown aerobically under medium light intensity. C, gun4 mutant cells grown aerobically under low light intensity. D, gun4 mutant cells grown microaerobically under medium light conditions. Scale bar = 500 nm.
Localization of Gun4, Ferrochelatase, and Magnesium Chelatase in the Cyanobacterial Cell—Gun4 is proposed to function at the branch point of Chl and heme biosynthesis. To localize Gun4 and the two metal chelatases in cyanobacterial cells, whole cell extracts of wild-type and gun4 mutant cells were fractionated. Proteins of the cytosolic and membrane fractions were separated by SDS-PAGE, blotted, and probed with antibodies. Fig. 3A shows that the ferrochelatase is tightly bound to the membrane, as there is no significant decrease in signal intensity after additional washing of the membranes or treatment with 2 m NaCl. In contrast, the ChlH subunit of magnesium chelatase was detected both in the soluble and in the membrane fractions, whereas the ChlD subunit of the magnesium chelatase appears to be exclusively located in the soluble fraction. Gun4 was mainly detected as a soluble protein and appeared in the membrane fraction only to a minor degree. The substantially lower signal of Gun4 in the membrane fraction after treatment with 2 m NaCl suggests peripheral association of this protein with membranes (Fig. 3A).
In the fully segregated gun4 mutant no immune-reacting protein band was detectable using the anti-Gun4 antiserum (Fig. 3A). Interestingly, in contrast to an equal distribution of wild type ChlH subunit of the magnesium chelatase between soluble and membrane fractions, in the gun4 mutant this protein seems be localized mainly in the soluble fraction (Fig. 3A). Intriguingly, gun4– cells contained unequivocally lower amounts of ChlH and ferrochelatase (Fig. 3B) than the wild type. This finding corroborates the decreased activities of both enzymes in vivo (Fig. 3C). Whereas the loss of ferrochelatase activity was proportional to the loss of protein, loss of magnesium chelatase activity could not entirely be attributed to the ChlH deficiency, supporting a direct role of Gun4 for retaining the enzyme activity (10).
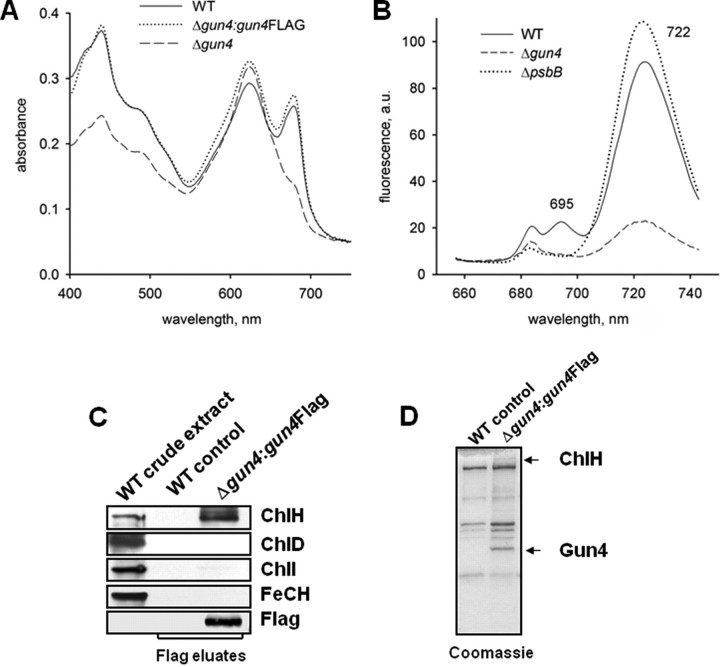
Affinity Purification of 3×FLAG-tagged Gun4—A possible explanation for the decreased activity of both chelatases in the gun4 mutant might be their physical interaction with Gun4. To identify and specify interacting protein partners of Gun4, gun4 mutant cells were engineered to express a 3×FLAG-tagged Gun4 protein. For this purpose we used a vector harboring the petJ promoter, which is induced by copper deficiency and a platform for stable insertion of the construct into the Synechocystis 6803 chromosome. Absorption spectra of whole cells confirmed that expression of 3×FLAG-Gun4 complemented the gun4 mutation (Fig. 4A), indicating the generation and activity of a functional fusion protein. The 3×FLAG-Gun4 fusion protein was expressed during growth in copper-deficient medium and immunoprecipitated with anti-FLAG M2 affinity gel (Sigma-Aldrich) as described (21). The 3×FLAG-Gun4 protein was eluted by the addition of a 3×FLAG peptide and separated by SDS-PAGE (Fig. 4, C and D). Cells expressing the non-tagged Gun4 were used as the negative control. Among the proteins co-purified with Gun4, only ChlH was identified by immunoblot analysis (Fig. 4C). The other magnesium chelatase subunits ChlI and ChlD as well as ferrochelatase did apparently not associate with Gun4. Immunoblot analysis demonstrates that the magnesium chelatase subunits ChlH and ChlD as well as the ferrochelatase were present in similar amounts in cells expressing 3×FLAG Gun4 and control cells (data not shown). In addition, in vitro pulldown assays did not indicate an interaction between recombinant Gun4 and ferrochelatase (data not shown). No other putative interacting proteins were detected in the eluate fraction from the gun4–:gun4FLAG strain after affinity purification of the 3×FLAG-Gun4 protein (Fig. 4D).
FIGURE 4.
Phenotype of gun4 mutant cells and affinity purification of Gun4. A, absorption spectra of Synechocystis 6803 wild-type (WT) and mutant cells lacking Gun4 (gun4–) or expressing a FLAG-tagged version of Gun4 (Δgun4:gun4FLAG). Cells were grown with 5 mm glucose in the medium. Peaks at 620 and 682 nm represent phycocyanin and Chl absorption, respectively. Spectra of whole cells were measured with a UV-2401 PC spectrophotometer (Shimadzu, Kyoto, Japan). B, semiquantitative 77 K fluorescence emission spectra of thylakoid membranes from Synechocystis 6803 wild-type and mutant cells lacking Gun4 (Δgun4) or CP47 (ΔpsbB). Rhodamine was used as an internal standard, and the spectra were normalized to indicate Chl fluorescence per cell. Peaks at 685/695 and 722 nm represent PSII and PSI complexes, respectively. a.u., absorbance units. C, affinity purification of 3×FLAG-Gun4 and identification of magnesium and ferrochelatase (FeCH) subunits by immunodetection. Solubilized extracts of gun4 mutant cells engineered to express 3×FLAG-Gun4 were immunoprecipitated with anti-FLAG M2-agarose (Sigma-Aldrich). FLAG-Gun4 complexes were eluted with 3×FLAG peptide, resolved by SDS-PAGE, stained with Coomassie (D) or blotted onto a nitrocellulose membrane and immunodetected (C). Wild-type eluate was used as a control.
Absence of Gun4 Affects Accumulation of Both PSII and PSI—The drastically reduced Chl content in the gun4 mutant (Table 1) was necessarily associated with lower amounts of the Chl-binding protein complexes PSII and PSI. Indeed, semiquantitative measurements of 77 K fluorescence spectra of membranes using rhodamine as the internal standard revealed an almost complete loss of PSII complexes and a large decrease in the amount of PSI complexes in gun4– cells compared with the wild type (Fig. 4B). The pronounced loss of PSII complexes containing the CP47 protein (PsbB) was indicated by the absence of the 695-nm emission peak in gun4–, similar to the ΔpsbB deletion mutant (25), which served as reference in this study (Fig. 4B).
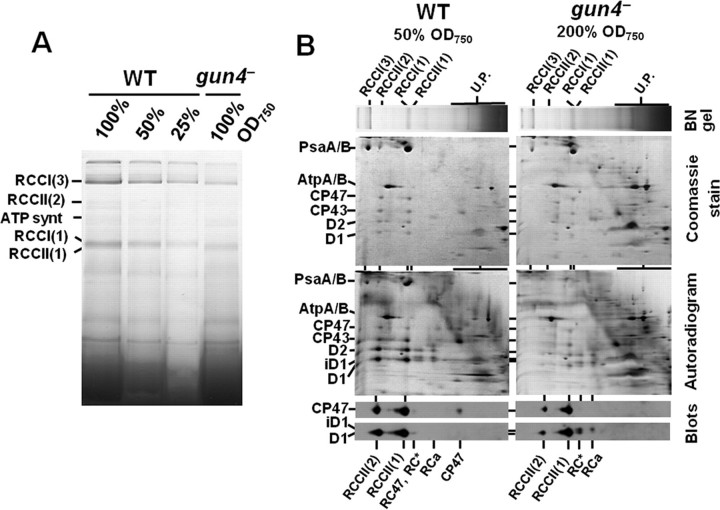
The Chl deficiency in gun4– gave us a unique opportunity to investigate consequences of very limited Chl supply for assembly and stability of Chl-binding proteins under standard light conditions. Therefore, we performed a detailed comparison of protein complexes of membranes isolated from wild-type and gun4 mutant cells applying BN-PAGE (Fig. 5A). Apparently, on the per cell basis the contents of both PSI (RCCI) and PSII (RCCII) complexes were dramatically decreased in gun4 mutant cells compared with wild-type cells (about 25% PSI and maximally 10% PSII of the wild-type level). The PSI trimer was the prevailing form in both strains, but the PSI trimer/PSI monomer ratio was apparently decreased in the mutant. In addition, the relative content of the dimeric form of PSII core complex (RCCII(2)) was lower in the mutant than in the wild type. The two-dimensional gel separation of proteins from wild-type membranes and the corresponding immunoblots revealed the presence of the CP47 protein in the core complex lacking CP43 (RC47) and in the region of unassembled proteins. Both free CP47 and CP47 integrated in RC47 were missing in thylakoids of gun4–. Moreover, the mutant accumulated two distinct complexes lacking CP47 previously designated as RC* and RCa (26). These complexes contain mature D1 or its partially processed form iD1, D2, cytochrome b 559, and PsbI and are typical for strains impaired in synthesis and/or stability of CP47 (17, 26). Thus, the results suggest that the insufficient accumulation of CP47 limits the assembly and accumulation of complete PSII core complexes. In contrast to the Chl-containing PSs, the ATP synthase was present in both strains in equal amounts showing that accumulation of membrane protein complexes lacking Chl was not changed after inactivation of the gun4 gene.
FIGURE 5.
Analysis of membrane protein complexes and their assembly in wild-type and gun4 mutant cells. A, membrane protein complexes from wild-type and gun4 mutant labeled in vivo with 35[S]Met/Cys were separated in 4–16% BN gel as described under “Experimental Procedures.” Amounts of Chl corresponding to designated amounts of cells were loaded per each lane. B, two-dimensional analysis of membrane protein complexes separated in A. Equal amounts of Chl corresponding to the half (50% A750) and double (200% A750) amounts of wild-type and gun4– cells were loaded, respectively. Lanes from the BN-gel were excised and placed on top of the 12–20% denaturating gel. The two-dimensional gels were either stained by Coomassie Blue (Coomassie gels), dried, and exposed to phosphorimaging plates (Autoradiograms) or electroblotted onto a polyvinylidene difluoride membrane that was immunodecorated using antibodies raised against D1 and CP47 proteins (blots). Designation of complexes: RCCI(3) and RCCI(1), trimeric and monomeric PSI complexes, respectively; RCCII(2) and RCCII(1), dimeric and monomeric PSII core complexes, respectively; RC47, the monomeric PSII core complex lacking CP43; RC* and RCa, reaction center complexes accumulating in PSII mutants lacking CP47 (24); U.P., unassembled proteins; iD1 designates D1 processing intermediate.
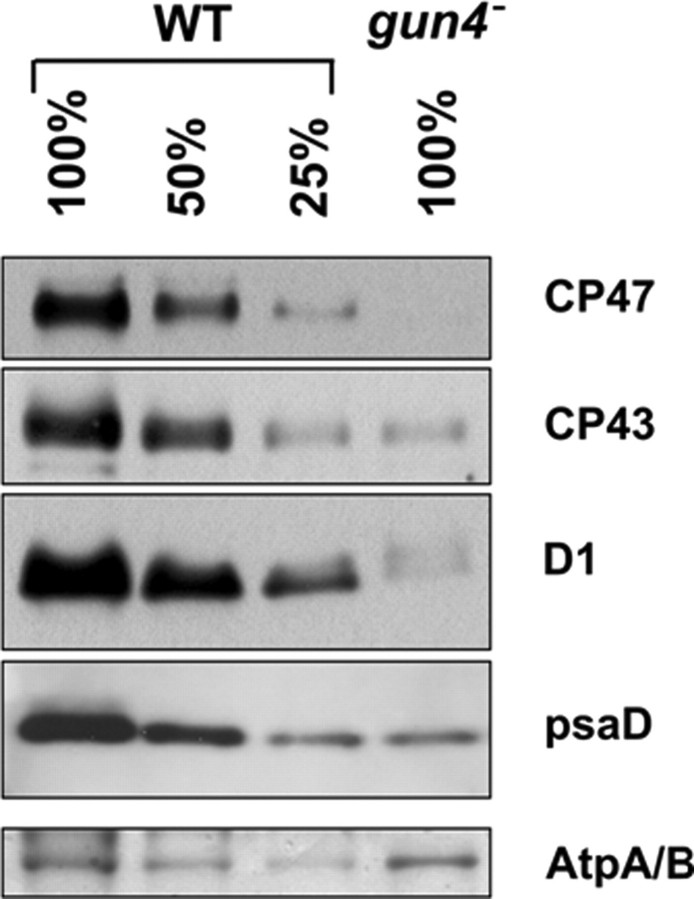
The differential effect of the gun4 deletion on accumulation of Chl-binding proteins was further supported by semiquantitative immunoblot analysis (Fig. 6). Consistent with the BN-PAGE data, the ATP synthase was present in similar amounts in wild type and gun4–, whereas the amount of the PSI protein PsaD corresponded to about 25% that of the wild-type level. Among the PSII proteins, CP43 accumulated to 25% that of the wild-type level, whereas the D1 protein and especially CP47 were hardly detectable. Negligible cellular contents of CP47 corresponded with a non-detectable oxygen-evolving capacity of PSII, and no variable fluorescence was observed in the gun4 mutant (not shown).
FIGURE 6.
Semiquantitative immunoblot analysis of PSII and PSI subunits in wild type and gun4 mutant. Wild-type and gun4 mutant membrane fractions were separated by SDS-PAGE and transferred to a polyvinylidene difluoride membrane. Different amounts of Chl corresponding to designated amounts of cells were loaded per each lane. Immunodetection was performed using polyclonal antibodies raised against the PSII subunits CP47, CP43, and D1, and the PSI subunit PsaD. ATPase band stained on the membrane with Ponceau red indicates protein loading.
Because two-dimensional BN/SDS-PAGE was performed with thylakoids isolated from radioactively labeled cells, the analysis also provided information on the synthesis of PSII subunits and their assembly into complexes in both strains (Fig. 5B, autoradiograms). In wild-type cells the labeled D1 and D2 subunits were mainly found in RCCII(1) and RCCII(2), and their distribution between both complexes was approximately equal. Small amounts of labeled D1 and D2 (about 10% of the overall levels) were detected in the above-mentioned complexes RC* and RCa. However, these RC complexes accumulated only transiently, quickly disappeared during the chase (data not shown), and were not detected by immunoblot analysis. The inner PSII antennae CP43 and CP47 were labeled less strongly and were present in the core complexes as well as in the fraction of unassembled proteins. In this fraction the labeling of CP43 was particularly strong. The labeling pattern in gun4– thylakoids was different. The autoradiogram revealed that all four large PSII proteins in the assembled core complex RCCII(1) were labeled weakly and nearly to the same extent. The labeling of these proteins in RCCII(2) was hardly detectable. About 70% of the labeled D2, D1, and its partially processed form, iD1, were found in the RC* and RCa complexes. The remaining 30% were found in the core complexes (Fig. 5B, see Ref. 26). Moreover, the labeled proteins in RC* and RCa were detectable even after the 30 min of the radioactive chase (data not shown), showing that the complexes are not easily transformed into the core complexes. In the fraction of unassembled proteins, a strong labeling of CP43, but no labeling of CP47, was observed. The results confirmed that the insufficient synthesis of CP47 most probably limits the assembly and accumulation of the complete PSII core complexes. Similar transcript levels of the CP47-encoding gene psbB in wild type and gun4– (not shown) showed that the limitation in the accumulation of CP47 occurred at the posttranscriptional level.
DISCUSSION
In a previous work we did not succeed in preparing a fully segregated cyanobacterial gun4 mutant in Synechocystis 6803. However, generation of such a mutant was feasible in a GT substrain of Synechocystis 6803. The gun4 mutant was further examined for the function of Gun4 in tetrapyrrole biosynthesis and consecutive processes of protein assembly of the photosynthetic apparatus. Although A. thaliana GUN4 was reported to be nonessential for plant growth (10), here we show that the complete loss of gun4 expression in a cyanobacterium prevents photoautotrophic growth. The Chl content in the gun4 mutant grown at medium light intensities was dramatically reduced to about 8% that of the wild-type amount. At low light intensities the gun4 mutant accumulated about 25% that of the wild-type Chl content. The better viability of the gun4– cells under low light or microaerobic conditions may be explained by the increased accumulation of PIX in this mutant (Table 1) as PIX is known to cause photooxidative damage (27). Recently, a very similar phenotype has been observed in a Synechocystis 6803 mutant that also accumulated large quantities of PIX, resulting from a mutation in the ferrochelatase gene (28). However, at low light intensities under microaerobic conditions no further improvement in Chl accumulation was observed, with the gun4 mutant accumulating about 25% of the wild-type Chl content. This suggests that the low light gun4– phenotype can be mostly attributed to the lack of Chl precursors and not to enhanced photooxidative stress.
In agreement with the predicted function of Gun4 (10), the absence of this protein adversely affected the magnesium chelatase activity resulting in a limited supply of magnesium porphyrins for Chl synthesis (Table 1). In Synechocystis 6803, the Gun4 protein is present mainly as a soluble protein with a small fraction bound to the membrane (Fig. 3A). This is consistent with the situation described in A. thaliana, where the majority of GUN4 was localized in the stroma and clearly smaller amounts in the envelope and thylakoid membranes (10). Moreover, the same authors reported that unlike the soluble GUN4, which probably is present as a monomer, the thylakoid-bound GUN4 was identified as part of a >500 kDa complex also containing the ChlH subunit (10). We have confirmed that cyanobacterial Gun4 is associated with ChlH as well (Fig. 4C) and showed that the accumulation of ChlH is influenced by absence of Gun4 (Fig. 3B). Interestingly, the proportion of the membrane-bound ChlH seemed to be decreased in the gun4 mutant as well (Fig. 3A). Therefore, Gun4 is possibly important for association of ChlH with the membrane, maybe as a part of a membrane-bound magnesium chelatase complex.
The absence of the Gun4 protein also resulted in a decreased accumulation of ferrochelatase. Because no direct physical interaction between Gun4 and ferrochelatase was detected, the lack of Chl in gun4– may influence the cellular content of ferrochelatase. The plant ferrochelatase as well as its cyanobacterial counterpart contains a highly conserved Chl binding motif within the C-terminal domain (2). Thus, we cannot exclude that a putative Chl binding to this domain is important for the synthesis or stability of ferrochelatase.
The gun4 mutant provided a new opportunity to analyze the effect of Chl deficiency on photosynthetic membrane proteins in cyanobacterial cells grown under standard light conditions. Previously, a comparably detailed study in the cyanobacterium Plectonema boryanum revealed that a mutation in chlL, which encodes a subunit of the light-independent protochlorophyllide oxidoreductase (29), leads to Chl deficiency during growth in nearly complete darkness. However, the consequences of Chl deficiency in the dark might significantly differ from those in the light, which is supported by the data of Kada et al. (29). The depletion of PSI Chl proteins was much faster than the depletion of PSII in the chlL– strain. We assume that this difference is associated with the different stabilities of PSI and PSII Chl proteins in the dark and under illumination, respectively. Recently, it has been shown that the half-life of PSII-bound Chl in illuminated cells is remarkably shorter than that of the PSI-bound Chl (30). Within PSII, the turnover rate of unassembled CP43 is slower than that of other PSII Chl-binding proteins. In the absence of several PSII proteins, CP43 still accumulates to a larger extent (17). In the gun4– mutant the cellular level of CP43 corresponded to the level of PSI proteins, both of which were reduced to about 25% that of the wild-type level. Moreover, the unassembled protein was clearly visible in the Coomassie-stained two-dimensional gel (Fig. 5B). Immunoblot analyses detected the incompletely processed form of D1, which is referred to as iD1, as a component of the RC complexes in the gun4 mutant but not in wild-type cells (Fig. 5B, blot). The presence of the incompletely processed form of the D1 protein was reported to be closely related to Chl deficiency, underlying the hypothesis that maturation of D1 occurs after binding of Chl to the protein (31). Here we show that the incompletely processed form of D1 is present in the RC complexes and that the effect of Chl deficiency on the D1 maturation is indirect and related to the absence of CP47. Not until CP47 binds to the RC complexes can the complete and fast D1 maturation proceed.
Despite the presence of RC complexes, the subsequent PSII assembly intermediate RC47, which usually is formed from RC complexes after binding of CP47 (17), is absent in the gun4 mutant. Because unassembled CP47 is not detectable in the mutant either (Fig. 5B, blot), it is clear that the accumulation of CP47 is strongly compromised in the mutant. A similar effect was detected in a CP47 mutant that harbors a few amino acid changes in the E-loop of the protein (32). This mutation resulted in a lowered cellular amount of functional PSII accompanied by an increased accumulation of RC complexes. Possibly, Chl deficiency limits the translation of CP47 to a greater extent than the translation of D1 due to the requirement of co-translational Chl binding to the CP47 apoprotein.
In conclusion, our results demonstrate that Gun4 plays an essential role in the magnesium branch of tetrapyrrole biosynthesis and dramatically affects the Chl content in the cell. 77 K fluorescence emission spectra as well as protein analyses demonstrated that Chl deficiency arising from the inactivation of gun4 affects accumulation of both photosystems. The more extensive deficiency of PSII most probably was related to the insufficient accumulation of CP47. This Chl-binding protein appears to be the protein most sensitive to changes in the Chl availability and may, therefore, represent an important target for the mutual regulation of tetrapyrrole biosynthesis and formation of new PSII complexes.
Acknowledgments
We thank Prof. C. Neil Hunter (University of Sheffield) for the antibodies against the subunits of magnesium chelatase, Gisa Baumert and Kerstin Kaminski for technical assistance, and Dennis Dienst for critical reading of the manuscript. The vector psk9 was a kind gift of Prof. S. Zinchenko (Moscow State University).
This work was supported by Deutsche Forschungsgemeinschaft Grants SFB429 and TPA8 (to A. W. and B. G.), by Institutional Research Concept AV0Z50200510, by Ministry of Education of the Czech Republic project MSM6007665808 (to J. K.), and by Grant Agency of the Czech Academy of Sciences project IAA500200713 (to R. S. and M. T.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: Chl, chlorophyll; PIX, protoporphyrin IX; PS, photosystem; DM, β-dodecyl maltoside; BN-PAGE, blue native PAGE; GT, glucose-tolerant; HPLC, high performance liquid chromatography; MES, 4-morpholineethanesulfonic acid.
References
- 1.Papenbrock, J., and Grimm, B. (2001) Planta 213 667–681 [DOI] [PubMed] [Google Scholar]
- 2.Vavilin, D. V., and Vermaas, W. F. (2002) Physiol. Plant 115 9–24 [DOI] [PubMed] [Google Scholar]
- 3.Grossman, A. R., Lohr, M., and Im, C. S. (2004) Annu. Rev. Genet. 38 119–173 [DOI] [PubMed] [Google Scholar]
- 4.Beale, S. I. (2005) Trends Plant Sci. 10 309–312 [DOI] [PubMed] [Google Scholar]
- 5.Herrin, D. L., Battey, J. F., Greer, K., and Schmidt, G. W. (1992) J. Biol. Chem. 267 8260–8269 [PubMed] [Google Scholar]
- 6.Plumley, G. F., and Schmidt, G. W. (1995) Plant Cell 7 689–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brusslan, J. A., and Peterson, M. P. (2002) Photosynth. Res. 71 185–194 [DOI] [PubMed] [Google Scholar]
- 8.Susek, R. E., Ausubel, F. M., and Chory, J. (1993) Cell 74 787–799 [DOI] [PubMed] [Google Scholar]
- 9.Mochizuki, N., Brusslan, J. A., Larkin, R., Nagatani, A., and Chory, J. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 2053–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Larkin, R. M., Alonso, J. M., Ecker, J. R., and Chory, J. (2003) Science 299 902–906 [DOI] [PubMed] [Google Scholar]
- 11.Davison, P. A., Schubert, H. L., Reid, J. D., Iorg, C. D., Heroux, A., Hill, C. P., and Hunter, C. N. (2005) Biochemistry 44 7603–7612 [DOI] [PubMed] [Google Scholar]
- 12.Verdecia, M. A., Larkin, R. M., Ferrer, J. L., Riek, R., Chory, J., and Noel, J. P. (2005) PLoS Biol 3 e151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilde, A., Mikolajczyk, S., Alawady, A., Lokstein, H., and Grimm, B. (2004) FEBS Lett. 571 119–123 [DOI] [PubMed] [Google Scholar]
- 14.Rippka, R., Desrulles, J., Waterbury, J. B., Herdman, M., and Stanier, R. Y. (1979) J. Gen. Microbiol. 11 419–436 [Google Scholar]
- 15.Tous, C., Vega-Palas, M. A., and Vioque, A. (2001) J. Biol. Chem. 276 29059–29066 [DOI] [PubMed] [Google Scholar]
- 16.Papenbrock, J., Mock, H. P., Kruse, E., and Grimm, B. (1999) Planta 208 264–273 [Google Scholar]
- 17.Komenda, J., Reisinger, V., Muller, B. C., Dobakova, M., Granvogl, B., and Eichacker, L. A. (2004) J. Biol. Chem. 279 48620–48629 [DOI] [PubMed] [Google Scholar]
- 18.Schägger, H., and von Jagow, G. (1991) Anal. Biochem. 199 223–231 [DOI] [PubMed] [Google Scholar]
- 19.Komenda, J., Lupinkova, L., and Kopecky, J. (2002) Eur. J. Biochem. 269 610–619 [DOI] [PubMed] [Google Scholar]
- 20.Jänsch, L., Kruft, V., Schmitz, U. K., and Braun, H. P. (1996) Plant J. 9 357–368 [DOI] [PubMed] [Google Scholar]
- 21.Dühring, U., Irrgang, K. D., Lünser, K., Kehr, J., and Wilde, A. (2006) Biochim. Biophys. Acta 1757 3–11 [DOI] [PubMed] [Google Scholar]
- 22.Porra, R. J., Thompson, W. A., and Kriedmann, P. E. (1989) Biochim. Biophys. Acta 975 384–394 [Google Scholar]
- 23.Shen, G., and Vermaas, W. F. J. (1994) J. Biol. Chem. 269 13904–13910 [PubMed] [Google Scholar]
- 24.He, Q., Brune, D., Nieman, R., and Vermaas, W. (1998) Eur. J. Biochem. 253 161–172 [DOI] [PubMed] [Google Scholar]
- 25.Eaton-Rye, J. J., and Vermaas, W. F. J. (1991) Plant Mol. Biol. 17 1165–1177 [DOI] [PubMed] [Google Scholar]
- 26.Dobakova, M., Tichy, M., and Komenda, J. (2007) Plant Physiol. 145 1681–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang, H., Inokuchi, H., and Adler, J. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 7332–7336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sobotka, R., McLean, S., Zuberova, M., Hunter, C. N., and Tichy, M. (2008) J. Bacteriol. 190 2086–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kada, S., Koike, H., Satoh, K., Hase, T., and Fujita, Y. (2003) Plant Mol. Biol. 51 225–235 [DOI] [PubMed] [Google Scholar]
- 30.Vavilin, D., and Vermaas, W. (2007) Biochim. Biophys. Acta 1767 920–929 [DOI] [PubMed] [Google Scholar]
- 31.He, Q., and Vermaas, W. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 5830–5835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sobotka, R., Komenda, J., Bumba, L., and Tichy, M. (2005) J. Biol. Chem. 280 31595–31602 [DOI] [PubMed] [Google Scholar]