Abstract
The Haemophilus influenzae HMW1 adhesin mediates adherence to respiratory epithelial cells, a critical early step in the pathogenesis of H. influenzae disease. In recent work, we demonstrated that HMW1 undergoes glycosylation. In addition, we observed that glycosylation of HMW1 is essential for HMW1 tethering to the bacterial surface, a prerequisite for HMW1-mediated adherence to host epithelium. In this study, we examined HMW1 proteolytic fragments by mass spectrometry, achieved 89% amino acid sequence coverage, and identified 31 novel modification sites. All of the modified sites were asparagine residues, in all but one case in the conventional consensus sequence of N-linked glycans, viz. NX(S/T). Liquid chromatography-tandem mass spectrometry analysis using a hybrid linear quadrupole ion trap Fourier transform ion cyclotron mass spectrometer, accurate mass measurements, and deuterium exchange studies established that the modifying glycan structures were mono- or dihexoses rather than the N-acetylated chitobiosyl core that is characteristic of N-glycosylation. This unusual carbohydrate modification suggests that HMW1 glycosylation requires a glycosyltransferase with a novel activity.
Until recently, glycosylation of proteins was believed to be limited to eukaryotes. However, glycoproteins are now being recognized increasingly in prokaryotes as well, including a variety of pathogenic bacteria. Examples of bacterial glycoproteins include the flagellins of Pseudomonas aeruginosa, Campylobacter jejuni, and Campylobacter coli; the type IV pili of P. aeruginosa, Neisseria gonorrhoeae, and Neisseria meningitidis; and non-pilus adhesins in Chlamydia trachomatis, enterotoxigenic Escherichia coli, and diffusely adhering E. coli (1–9).
Nonencapsulated (nontypeable) strains of Haemophilus influenzae are a common cause of human respiratory tract disease and initiate infection by colonizing the upper respiratory tract. Approximately 75–80% of isolates express two related high molecular weight proteins called HMW1 and HMW2 that promote efficient adherence to respiratory epithelial cells and facilitate the process of colonization (10, 11). The HMW1 and HMW2 adhesins are encoded by highly homologous chromosomal loci that appear to represent a gene duplication event and contain three genes, designated hmw1A, hmw1B, and hmw1C and hmw2A, hmw2B, and hmw2C, respectively (12, 13). HMW1 and HMW2 are synthesized as preproproteins and are secreted by the two-partner secretion system (14–16). Amino acids 1–68 direct the preproproteins to the Sec apparatus, where they are cleaved by signal peptidase I (15). Subsequently, amino acids 69–441 target the proproteins to the outer membrane and interact with the HMW1B or HMW2B outer membrane translocator protein, undergoing removal by an unknown process (15, 16–18). Following translocation across the outer membrane, the mature HMW1 and HMW2 proteins remain noncovalently associated with the bacterial surface, with small amounts released into the culture supernatant (15, 16).
In recent work, we established that HMW1 is a glycoprotein and undergoes glycosylation in the cytoplasm in a process that requires HMW1C and phosphoglucomutase (19). Further analysis revealed that glycosylation of HMW1 appears to protect against premature degradation, analogous to some eukaryotic proteins. In addition, glycosylation appears to influence HMW1 tethering to the bacterial surface, a prerequisite for HMW1-mediated adherence. Based on composition analysis, HMW1 carbohydrate modification includes glucose, galactose, and possibly mannose and corresponds to ∼7–8 kDa of the molecular mass.
In this study, we analyzed HMW1 glycosylation using MALDI-MS/MS2 and nano-LC-linear quadrupole ion trap Fourier transform ion cyclotron MS of endoprotease digests. We obtained 89% sequence coverage of HMW1 and found 31 asparagine residues that were modified with either one or two hexose molecules, including 30 modification sites within sequons that are required for protein N-glycosylation in plant and mammalian cells. The unusual linkage of hexose molecules at asparagine residues suggests that HMW1 glycosylation requires a unique glycosyltransferase with a novel activity.
EXPERIMENTAL PROCEDURES
Protein Purification—The H. influenzae HMW1 protein was purified from a derivative of strain 12 that lacks expression of HMW2. HMW1 was extracted from the bacterial surface as described previously (20) and was then injected onto a Source 15S column in 20 mm MES (pH 6.0). Following elution with an NaCl gradient, fractions containing HMW1 were pooled and then injected onto a butyl 14FF-Sepharose column in 700 mm NH4SO4 and 20 mm MES (pH 6.0). HMW1 was eluted with 400–500 mm NH4SO4 and consolidated for further analysis.
Peptide N-Glycosidase F Treatment of HMW1—A 25-μg aliquot of purified HMW1 was incubated for 4 h at 37 °C with or without 2 units of peptide N-glycosidase F (Glyko, San Leandro, CA) according to the manufacturer's instructions, and samples were resolved on an SDS-polyacrylamide gel and then stained with Coomassie Blue. Fetuin (Sigma) was used as a positive control.
Protease Digestion of HMW1—A 1-μg aliquot of purified HMW1 was precipitated using a 2D Clean Up kit (GE Healthcare).
Tryptic Digest—The precipitated sample was solubilized in 40 μl of 2 m urea and 100 mm Tris (pH 8.5) and subsequently incubated overnight at 37 °C in the presence of 1 μg of trypsin (Sigma).
Glu-C Digest—The precipitated sample was solubilized in 1 m urea, 100 mm Tris 8.5, and 5% acetonitrile and incubated overnight at 30 °C in the presence of 1 μg of Glu-C (Roche Applied Science).
Lys-C Digest—The precipitated sample was solubilized in 8 m urea and 100 mm Tris (pH 8.5) and incubated overnight at 37 °C in the presence of 1 μg of Lys-C (Roche Applied Science).
Trypsin/Glu-C Double Digest—20 μl of the tryptic digest was transferred to a new tube containing 2 μl of acetonitrile, 18 μl of deionized water, and 1 μg of Glu-C and then incubated overnight at 30 °C.
All digestions were acidified to 5% formic acid, and desalted peptides were prepared using a NuTip carbon tip (Glygen) according to the manufacturer's instructions. Peptides were eluted from the tip using 10 μl of 60% acetonitrile and 1% formic acid, and samples were then dried and redissolved in 1% acetonitrile and 1% formic acid for MS analysis.
Mass Spectrometry—The nano-LC-linear quadrupole trap Fourier transform instrument used for these experiments was described in detail elsewhere (21, 22). MS was performed using a linear ion trap Fourier transform mass spectrometer (LTQ-FTMS, Thermo Finnigan, San Jose, CA). The instrument was equipped with a PicoView nanocapillary source (New Objective, Woburn, MA) and was hooked up to a one-dimensional Proteomics Plus NanoLC system (Eksigent, Livermore, CA). The source voltage was set to 2200 V with a capillary voltage of 43 V and a capillary temperature of 200 °C, and the tube lens was 105 V.
Fourier Transform Parameters—The MS mass range was 250–2000 m/z. We employed one full microscan and a full maximum ion time of 1000 ms.
Ion Trap—We used seven MSn microscans in parallel with the full Fourier transform microscan with an MSn maximum ion time of 100 ms.
Autosampler and Liquid Chromatography—A spark autosampler (Endurance, Spark, Plainsboro, NJ) with a 10-μl loop was used for all sample injections. The solvent system consisted of 0.3% formic acid in water (solvent A) and 0.3% formic acid in acetonitrile (solvent B). The gradient employed showed an increase of 2% solvent B/min at a flow rate of 200 nl/min, a starting point of 5% solvent B, and a run time of 60 min. The injected sample amount was 5 μl. A C18 PicoFrit column (peptide II from New Objective) with a column length of 10 cm and an inner diameter of 75 μm was employed for all experiments.
MALDI-MS/MS—All MALDI experiments were performed on the 4700 MALDI TOF/TOF Proteomics Analyzer (Applied Biosystems). MS spectra were acquired in the positive ion mode using the reflectron. The mass range was set to m/z 800–4000 with a focus mass of 1650 Da. 1000 spectra were summed up for each MS spectrum with a set laser energy of 4250 (arbitrary unit). The detection multiplier was set to 1.0 kV, and the final detector voltage was 2.0 kV.
The MS/MS spectra were acquired in the positive ion mode using the metastable suppressor. The parent ion selector was set to ±11 Da. 12,500 individual laser shots were summed up for each MS/MS spectrum with the laser energy set to 5450 (arbitrary unit). The detector multiplier was set to 1.0 kV, and the final detector voltage was 2.225 kV.
Deuterium Replacement Experiment—The deuterium replacement experiments were performed as described previously using α-cyano-4-hydroxycinnamic acid as the MALDI matrix (23, 24).
Analysis of Carbohydrate Composition—To determine the identity of the modifying sugar, a representative tryptic glycopeptide was purified by HPLC using a C18 separation column and was then analyzed for monosaccharide composition by preparing the trimethylsilyl derivative of the methyl glycoside and performing combined gas chromatography-MS analysis as described previously (12). This analysis was performed at the University of Georgia Complex Carbohydrate Research Center (Athens, GA).
RESULTS
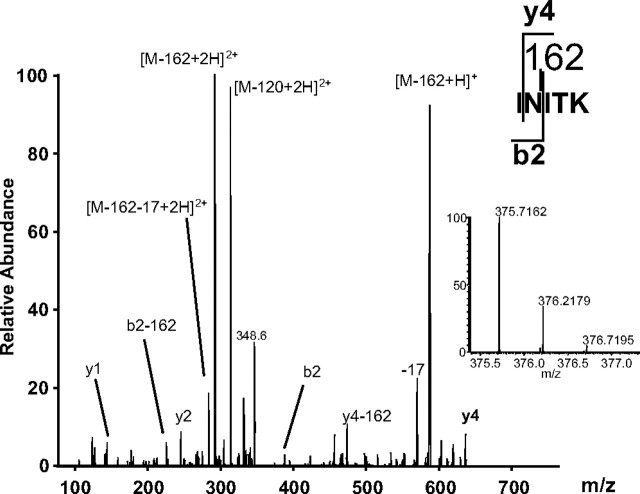
HMW1 Peptides Contain a 162-Da Modification—As a first step toward defining the nature of HMW1 glycosylation, we incubated purified HMW1 with peptide N-glycosidase F, an enzyme that releases asparagine-linked oligosaccharides from glycoproteins. As shown in supplemental Fig. 1, treatment with peptide N-glycosidase F resulted in a significant decrease in the apparent molecular mass of fetuin as assessed by SDS-PAGE, consistent with deglycosylation. In contrast, there was no effect on the apparent molecular mass of HMW1, suggesting O-linkage or an unusual N-linkage of the HMW1 carbohydrate. As a second step, we digested purified HMW1 with trypsin and then analyzed the resulting peptide fragments by LC-MS/MS. Initially, we used the NCBI nr data base and MASCOT software (Matrix Science, San Jose) and allowed for variable modifications of Met residues (oxidation) and all Ser and Thr residues with HexNAc. Using this approach, we found no carbohydrate modifications. However, when we examined the LC-MS/MS results manually, we identified a number of peptides that shared the HMW1 sequence but had a mass 162 Da greater than the calculated mass for the unmodified peptide. As an example, the peptide INITK has a measured mass of the doubly charged peptide of 375.7162 Da, whereas the expected mass of the doubly charged unmodified peptide is 294.6897 Da, consistent with a modification of 162.0532 Da (supplemental Fig. 2). To calculate the chemical formula for modification of 162.0532 Da, we used Xcalibur software (Thermo Fisher Scientific), setting the mass tolerance at 3 ppm and considering the elements carbon, hydrogen, nitrogen, and oxygen. The molecular formulas that agreed with the observed mass ±3 ppm included C6H10O5 (2 ppm) and C5H4N7 (2 ppm). However, C5H4N7 is not a likely chemical formula, leaving C6H10O5, a hexose. Consistent with this conclusion, simulation of the isotopic distribution of peptide INITK modified by a single hexose using IsoPro 3.0 software revealed a good match with the experimentally determined isotopic distribution (supplemental Fig. 2).
To determine the identity of the modifying hexose, we purified the NVTNNNITSHK glycopeptide by HPLC and performed carbohydrate composition analysis by combined gas chromatography-MS. Consistent with our previous examination of mature HMW1 (19), this analysis revealed the presence of glucose (supplemental Fig. 3). At this point, it is difficult to know whether the glucose peak represents the modifying hexose or contaminating sugar.
The 162-Da Modifications of HMW1 are N-Linked at Consensus Sites—To identify the modification sites in HMW1, we examined the tryptic digests by MS/MS. As shown in Fig. 1, the MS/MS spectrum of peptide INITK was dominated by the neutral loss of modification of both the precursor ion and the fragment ions. [M - 162 + 2H]2+ and [M - 162 + H]+ were particularly prominent. Neutral loss of the modification site could also be observed for the fragment ions b2 (b2 - 162) and y4 (y4 - 162). However, the presence of ions b2 and y4 carrying the modification allowed identification of the modification site as the asparagine residue. Interestingly, this residue is part of the consensus sequence that is required for protein N-linked glycosylation, viz. NX(S/T), with X being any amino acid except proline and with the amino acid at position 3 being either serine or threonine.
FIGURE 1.
MS/MS spectrum of peptide INITK allowing the identification of the modification site. The inset in boldface shows the isotopic distribution and accurate mass of the precursor ion.
As summarized in Table 1, analysis of the MS/MS spectrum of each of the other modified tryptic peptides also revealed glycosylation of an asparagine residue at a consensus sequence. The only exception was peptide DTTFNVER, which contains a hexose linked to the asparagine at position 5 (NVE).
TABLE 1.
Analysis of the MS/MS spectrum of each of the modified tryptic peptides
| Sequence | Position | No. of hexoses | Fragment ions | Calculated m/z |
|---|---|---|---|---|
| PDNVSINAETAGR | Asn444 | 0, 1 | b3, y11 | 753.3575 |
| TTLTNTTLESILK | Asn484 | 1, 2 | y9, y9 | 798.9326 + 879.9590 |
| GTFVNITANQR | Asn497 | 1, 2 | y7, y7 | 691.8492 + 772.8756 |
| GANLTIYSGGWVDVHK | Asn546 | 1 | b3, y14 | 939.9652 |
| NISLGAQGNINITAK | Asn560, Asn570 | 1, 1 | b2, y5 | 919.4731 |
| FNNVSLNGTGSGLQFTTK | Asn605, Asn609 | 1, 1 | b3, y12 | 1105.029 |
| FNNVSLNGTGSGLQFTTK | Asn605, Asn609 | 1, 2 | (b4), y12 | 1186.0554 |
| FEGTLNISGK | Asn636 | 1, 2 | (b6), y5, y5 | 614.3080 + 695.3352 |
| VNISMVLPK | Asn642 | 1, 2 | b2, y8, y8 | 581.8230 + 662.8494 |
| YFNVSTGSSLR | Asn773 | 1, 2 | b3, y9, (b3), y9 | 696.8357 + 777.8621 |
| DTTFNVER | Asn709 | 1 | b5, y4 | 572.2619 |
| DLTLNATGGNITLLQVEGTDGMIGK | Asn801, Asn806 | 1, 1 | y16, y21 | 1428.2075 |
| DLTLNATGGNITLLQVEGTDGMIGK | Asn801, Asn806 | 1, 2 | y16 | 1590.2604 |
| KNITFEGGNITFGSR | Asn828, Asn835 | 1, 1 | b9, y14 | 982.976 |
| KNITFEGGNITFGSR | Asn828, Asn835 | 1, 2 | b9, y7, y14 | 1064.0024 |
| AITNFTFNVGGLFDNK | Asn912 | 1, 2 | (y14, b5), (b5, more b ions) | 960.4729 + 1041.4993 |
| GNSNISIAK | Asn928 | 1 | y6 | 533.2748 |
| NLSITTNSSSTYR | Asn952 | 1, 2 | b7, y7, b7, y7 | 803.3838 + 884.4102 |
| NLSITTNSSSTYR | Asn946, Asn952 | 1, 2 | b2, y7 | 965.44 |
| TIISGNITNK | Asn964 | 1, 2 | b6, y5, b6, y5 | 611.8299 + 692.8563 |
| NGDLNITNEGSDTEMQIGGDVSQK | Asn973 | 1 | b6 | 1342.5956 |
| EGNLTISSDK | Asn995 | 0, 1 | b3, y8 | 613.2933 |
| INITK | Asn1004 | 1 | b2, y4 | 375.7158 |
| AGVDGENSDSDATNNANLTIK | Asn1029 | 1, 2 | b17, y5, y5 | 1134.5091 + 1215.5355 |
| LTQDLNISGFNK | Asn1044 | 1, 2 | b6, y7, y7 | 756.3830 + 837.4094 |
| NVTVNNNITSHK | Asn1131 | 0, 1, 2 | y6, y6 | 751.8759 + 832.9023 |
| ITTKTGTTINATTGNVE | Asn1156 | 1, 2 | y8, b10, y8 | 942.4758 + 1023.5023 |
| AANVTLNTTGTLTTVK | Asn1348, Asn1352 | 1, 1 | (b3), b4, y142+, y152+, y10 | 964.9992 |
| GSNINATSGTLVINAK | Asn1366 | 2 | y12 | 942.4758 |
This table lists all peptides with identified modification sites. The modification sites are numbered according to the position of the modified amino acid in the intact protein sequence, beginning with the methionine in the signal peptide. The peptide marked in boldface lettering has a modification that is not part of the NX(S/T) consensus sequon. The underlined letters indicate modification sites, and the boldface letters indicate double modification sites. The table also lists the observed fragments ions that allow the localization of the modification
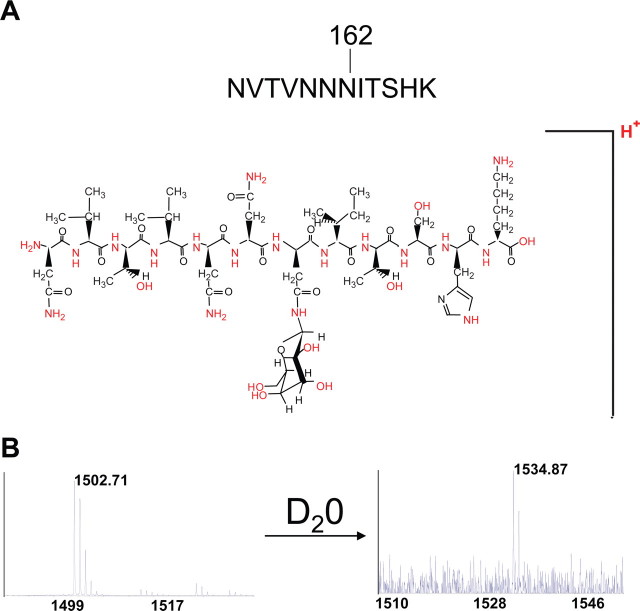
The N-Linked 162-Da Modifications of HMW1 Are Hexose Units—Typically, carbohydrates linked to asparagine residues are N-acetylated. Accordingly, to confirm that the 162-Da modification in HMW1 is truly a hexose unit, we focused on peptide NVTVNNNITSHK and attempted to exchange all active protons for deuterons, performing MALDI-MS and MS/MS analysis before and after deuteration. All together, there are 32 exchangeable protons available in peptide NVTVNNNITSHK: four in the N-terminal asparagine, one in each of the valines, two in each of the threonines, three in each of the other asparagines, one in the isoleucine, two in the serine, two in the histidine, four in the C-terminal lysine, and four in the putative hexose modification (highlighted in Fig. 2A). One proton needs to be added for the positive charge, and one proton needs to be subtracted for the linkage of the hexose to asparagine residues.
FIGURE 2.
Deuterium replacement of peptide NVTVNNNITSHK. A shows the structure of the modified peptide with all active protons that can be exchanged for deuterons (labeled in red). B shows the MALDI mass spectrum of the modified peptide before and after deuteration.
As shown in Fig. 2B, 4700 MALDI TOF/TOF analysis of the deuterated peptide compared with the native peptide demonstrated a mass shift of 32 Da, exactly as predicted. The decrease in the signal-to-noise ratio of the mass spectrum after deuteration can be explained by sample loss due to the repetitive redissolvation of the MALDI matrix/analyte mixture on the MALDI target with deuterium oxide to ensure a deuteration yield in excess of 98%.
To verify that the signal with m/z 1534.87 was indeed the modified peptide of interest, we performed MS/MS analysis of both the native sample and the deuterated sample. As shown in Fig. 3, this analysis confirmed that the deuterated peptide was NVTVNNNITSHK. Of note, the MS/MS spectrum of the deuterated species again had a lower signal-to-noise ratio caused by the repeated redissolvation of the MALDI preparation in deuterium oxide. The modification site in this peptide was the asparagine residue at position 7, in an N-linked consensus sequence, as is evident from the sequence ions, y6 and b7.
FIGURE 3.
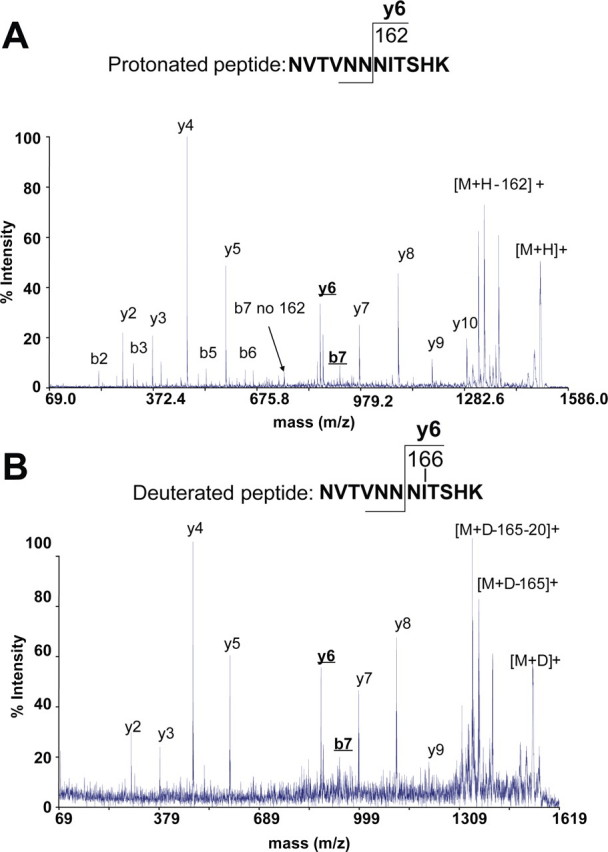
MALDI-MS/MS spectra of the native (A) and deuterated (B) peptide NVTVNNNITSHK allowing the identification of the modification site.
Furthermore, examination of the neutral loss of the modification from the parent ion revealed a loss of 165 Da and a further loss of 20 Da. The further loss of 20 Da is due to water loss (D2O) from the C terminus. Thus, the number of protons that were exchanged with deuterons is in agreement with the proposed presence of a hexose in the modified peptide NVTVNNNITSHK.
Some Glycan Units in HMW1 Are Dihexoses—In an effort to assess the entire HMW1 sequence, we digested purified HMW1 with Lys-C, Glu-C, or both trypsin and Glu-C and then analyzed the resulting peptide fragments by LC-MS/MS. Examination of these digests in combination with the tryptic digest identified peptides accounting for ∼89% of the HMW1 sequence and revealed 31 modification sites.
In considering the total carbohydrate modification of HMW1, we wondered whether there might be peptides with multiple sites of modification or with multiple hexose units at a single site. To address this possibility, we examined peptide fragments for 324-, 486-, and 648-Da modifications and found several additional peptides. As an example, we found that peptide TTLTNTTLESILK contains two hexose moieties at the Asn, within an N-linked consensus sequence (Fig. 4). In an effort to confirm Asn modification in this dihexosylated peptide, we compared the MS/MS spectrum of the modified peptide (Fig. 4A) with the MS3 spectrum on a fragment ion that had undergone neutral loss of 162 Da (Fig. 4B) and the MS3 spectrum on a fragment that had undergone neutral loss of 2 × 162 Da (Fig. 4C). Fragment ions unique to one MSn spectrum were ions with the modification. All together, the 31 modification sites carry 47 hexose units.
FIGURE 4.
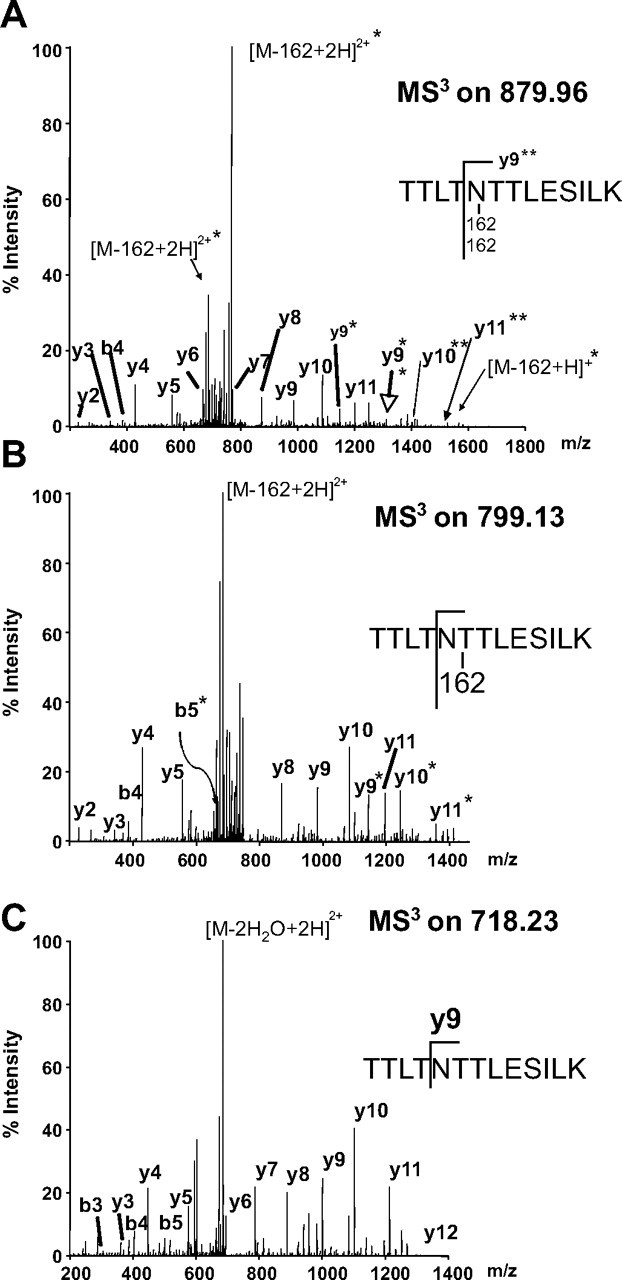
MS2 and MS3 spectra of the doubly modified peptide TTLTNTTLESILK. All fragment ions carrying both modifications are indicated by double asterisks. Fragment ions carrying only one fragmentation site are indicated by single asterisks. A shows the MS2 spectrum of the doubly modified peptide. B shows the MS3 spectrum of [M - 162 + H]+. C shows the MS3 spectrum of [M - 2 × 162 + H]+.
DISCUSSION
In this study, we established that the H. influenzae HMW1 adhesin is glycosylated at multiple asparagines, in all but one case within the well recognized consensus sequence for N-linked glycans, viz. NX(S/T). Interestingly, the modifying carbohydrates at these sites are hexose or dihexose sugars rather than N-acetylated sugars, revealing an unusual carbohydrate modification and suggesting a glycosyltransferase with a novel enzymatic activity capable of transferring hexose moieties to asparagine residues.
Upon initiating MS analysis of HMW1 peptide fragments with the intent to identify glycosylation sites, we encountered two unexpected results. First, we were unable to detect oxonium ions (m/z 163 and m/z 204), the traditional approach used to identify glycopeptides. Second, we found that the hexose units in the glycosylated peptides were very labile under MS conditions, especially in peptides with multiple glycosylation sites. To circumvent these challenges, we performed MS3 experiments driven by the neutral loss of 81 Da (one hexose, doubly charged ion) and 162 Da (two hexoses, doubly charged ion, or one hexose, singly charged ion). Comparing the MSn spectra of peptides carrying a decreasing number of hexoses, we were able to detect fragment ions that were unique and contained the modification site. Combining this technique with the creation of the custom modification of hexose/hexoses bound to asparagine for MASCOT Database searches, we found 47 hexose units.
In considering glycoproteins in general, two types of glycosylation exist, defined on the basis of the glycan linkage. In particular, carbohydrate modifications are either O- or N-linked. O-Linkage occurs at the hydroxyl oxygen on the side chain of a serine or threonine residue (or rarely a tyrosine residue), whereas N-linkage occurs at the amide nitrogen on the side chain of an asparagine residue. O-Linked carbohydrates are either hexose or N-acetylated amino sugars. In contrast, in all cases in eukaryotes and with rare exceptions in prokaryotes, N-linked carbohydrates contain N-acetylated sugars (25, 26). Accordingly, the glycosylation of HMW1 is distinctly unusual, consisting of N-linked hexose units at asparagine residues. It will be especially intriguing to identify the glycosyltransferase responsible for this novel modification.
It is interesting that all but one of the glycosylated asparagine residues in HMW1 reside in a consensus sequon, flanked by any amino acid except proline and then either threonine or serine (NX(S/T)). In the case of the exception, the glycosylated asparagine is flanked by a valine and then a glutamic acid, representing a new glycosylation sequon. In future studies, we will examine whether the same glycosylation machinery is involved in modifying this site.
In a recent study, Kowarik et al. (27) reported that the sequon for N-glycosylation of the C. jejuni AcrA glycoprotein by the PglB oligosaccharyltransferase is (D/E)YNX(S/T), corresponding to the consensus sequon with an N-terminal extension. Review of the modification sites in HMW1 reveals no example of modification at the (D/E)YNX(S/T) sequon. Furthermore, there is significant variability in the residues N- and C-terminal to the NX(S/T) sequon at modification sites in HMW1, arguing against a clear consensus extension.
In earlier work (19), we found that insertional inactivation of either the hmw1C or pgmB gene resulted in elimination of HMW1 glycosylation and a decrease in quantity of HMW1 and tethering of HMW1 to the bacterial surface, dramatically reducing HMW1-mediated adherence. In this context, it is interesting that HMW1 glycosylation involves modification with 47 hexose units at 31 different sites in the protein, roughly accounting for the change in molecular mass associated with HMW1C function. At this point, it remains unclear whether HMW1 stability and tethering to the bacterial surface require modification at all of these sites or at only a subset. As a corollary, it is unclear whether modification at some sites is required for HMW1 stability and modification at other sites is required for HMW1 tethering to the bacterial surface.
The HMW1 and HMW2 adhesins were first identified based on their role as major targets of the serum antibody response during acute infection. With this information in mind, Barenkamp (20) examined the utility of these proteins as potential vaccine antigens. In studies using the chinchilla acute otitis media model, he observed that immunization with purified HMW1 resulted in partial protection against middle ear infection with H. influenzae. In ongoing work, we are comparing the immunogenicity and protective effect of glycosylated and nonglycosylated HMW1.
In summary, we have established that glycosylation of the H. influenzae HMW1 adhesin involves an unusual carbohydrate modification and presumably requires a glycosyltransferase with a novel enzymatic activity. We anticipate that further characterization of HMW1 glycosylation will provide important insights into glycoproteins in both prokaryotes and eukaryotes.
Supplementary Material
Acknowledgments
We thank Dr. Don Rempel, Dr. Henry Rohrs, and Adam Brustkern for support with the maintenance of the linear trap quadrupole Fourier transform instrument.
This work was supported by United States Public Health Service Grants RO1 DC-02873 (to J. W. S.), 2-P41-RR05351 (to the University of Georgia Complex Carbohydrate Research Center), and P41-RR00954 (to the Washington University Mass Spectrometry Resource) and by the William M. Keck Foundation. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–5.
Footnotes
The abbreviations used are: MALDI-MS/MS, matrix-assisted laser desorption ionization tandem mass spectrometry; LC, liquid chromatography; MES, 4-morpholineethanesulfonic acid; HPLC, high pressure liquid chromatography.
References
- 1.Brimer, C., and Montie, T. (1998) J. Bacteriol. 180 3209-3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doig, P., Kinsella, N., Guerry, P., and Trust, T. J. (1996) Mol. Microbiol. 19 379-387 [DOI] [PubMed] [Google Scholar]
- 3.Castric, P. (1995) Microbiology (Read.) 141 1247-1254 [DOI] [PubMed] [Google Scholar]
- 4.Virji, M. (1997) Gene (Amst.) 192 141-147 [DOI] [PubMed] [Google Scholar]
- 5.Kuo, C., Takahashi, N., Swanson, A. F., Ozeki, Y., and Hakomori, S. (1996) J. Clin. Investig. 98 2813-2818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lindenthal, C., and Elsinghorst, E. A. (1999) Infect. Immun. 67 4084-4091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benz, I., and Schmidt, M. A. (2001) Mol. Microbiol. 40 1403-1413 [DOI] [PubMed] [Google Scholar]
- 8.Schmidt, M. A., Riley, L. W., and Benz, I. (2003) Trends Microbiol. 11 554-561 [DOI] [PubMed] [Google Scholar]
- 9.Szymanski, C. M., and Wren, B. W. (2005) Nat. Rev. Micobiol. 3 225-237 [DOI] [PubMed] [Google Scholar]
- 10.St. Geme, J. W., III, Falkow, S., and Barenkamp, S. J. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 2875-2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.St. Geme, J. W., III, Kumar, V. V., Cutter, D., and Barenkamp, S. J. (1998) Infect. Immun. 66 364-368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barenkamp, S. J., and St. Geme, J. W., III (1994) Infect. Immun. 62 3320-3328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buscher, A. Z., Burmeister, K., Barenkamp, S. J., and St. Geme, J. W., III (2004) J. Bacteriol. 186 4209-4217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacob-Dubuisson, F., Locht, C., and Antoine, R. (2001) Mol. Microbiol. 40 306-313 [DOI] [PubMed] [Google Scholar]
- 15.Grass, S., and St. Geme, J. W., III (2000) Mol. Microbiol. 36 55-67 [DOI] [PubMed] [Google Scholar]
- 16.St. Geme, J. W., III, and Grass, S. (1998) Mol. Microbiol. 27 617-630 [DOI] [PubMed] [Google Scholar]
- 17.Barenkamp, S. J., and Leininger, E. (1992) Infect. Immun. 60 1302-1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Surana, N. K., Grass, S., Hardy, G. G., Li, H., Thanassi, D. G., and St. Geme, J. W., III (2004) Proc. Natl. Acad. Sci. U. S. A. 101 14497-14502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grass, S., Buscher, A. Z., Swords, W. E., Apicella, M. A., Barenkamp, S. J., Ozchlewski, N., and St. Geme, J. W., III (2003) Mol. Microbiol. 48 737-751 [DOI] [PubMed] [Google Scholar]
- 20.Barenkamp, S. J. (1996) Infect. Immun. 64 1246-1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.King, J. B., Gross, J., Lovly, C. M., Rohrs, H., Piwnica-Worms, H., and Townsend, R. R. (2006) Anal. Chem. 78 2171-2181 [DOI] [PubMed] [Google Scholar]
- 22.King, J. B., Gross, J., Lovly, C. M., Piwnica-Worms, H., and Townsend, R. R. (2007) Rapid Commun. Mass Spectrom. 21 3443-3451 [DOI] [PubMed] [Google Scholar]
- 23.Gross, J., Leisner, A., Hillenkamp, F., Hahner S., Karas, M., Schäfer, J., Lützenkirchen, F., and Nordhoff, E. (1998) J. Am. Soc. Mass Spectrom. 9 866-878 [DOI] [PubMed] [Google Scholar]
- 24.Gross, J., Hillenkamp, F., Wan, K. X., and Gross, M. L. (2001) J. Am. Soc. Mass Spectrom. 12 180-192 [DOI] [PubMed] [Google Scholar]
- 25.Kupcu, Z., Marz, L., Messner, P., and Sletyr, U. B. (1984) FEBS Lett. 173 185-190 [DOI] [PubMed] [Google Scholar]
- 26.Messner, P., and Sletyr, U. B. (1988) FEBS Lett. 228 317-320 [DOI] [PubMed] [Google Scholar]
- 27.Kowarik, M., Young, N. M., Numao, S., Schulz, B. L., Hug, I., Callewaert, N., Mills, D. C., Watson, D. C., Hernandez, M., Kelly, J. F., Wacker, M., and Aebi, M. (2006) EMBO J. 25 1957-1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.