Abstract
Human ABCG2 is a plasma membrane glycoprotein working as a homodimer or homo-oligomer. The protein plays an important role in the protection/detoxification of various tissues and may also be responsible for the multidrug-resistant phenotype of cancer cells. In our previous study we found that the 5D3 monoclonal antibody shows a function-dependent reactivity to an extracellular epitope of the ABCG2 transporter. In the current experiments we have further characterized the 5D3-ABCG2 interaction. The effect of chemical cross-linking and the modulation of extracellular S–S bridges on the transporter function and 5D3 reactivity of ABCG2 were investigated in depth. We found that several protein cross-linkers greatly increased 5D3 labeling in ABCG2 expressing HEK cells; however, there was no correlation between covalent dimer formation, the inhibition of transport activity, and the increase in 5D3 binding. Dithiothreitol treatment, which reduced the extracellular S–S bridge-forming cysteines of ABCG2, had no effect on transport function but caused a significant decrease in 5D3 binding. When analyzing ABCG2 mutants carrying Cys-to-Ala changes in the extracellular loop, we found that the mutant C603A (lacking the intermolecular S–S bond) showed comparable transport activity and 5D3 reactivity to the wild-type ABCG2. However, disruption of the intramolecular S–S bridge (in C592A, C608A, or C592A/C608A mutants) in this loop abolished 5D3 binding, whereas the function of the protein was preserved. Based on these results and ab initio folding simulations, we propose a model for the large extracellular loop of the ABCG2 protein.
Human ABCG2 (also called as MXR/BCRP/ABCP) is a plasma membrane glycoprotein that belongs to the large family of ATP-binding cassette (ABC)3 proteins. ABCG2 mediates the energy-dependent transport of various compounds out of the cell. The protein is abundantly expressed in the intestine, the blood-brain barrier, and the placenta, influencing the absorption and fetal penetration of many toxic agents and food constituents (1). ABCG2 is also present in the liver where it is supposed to have an important role in the excretion of toxic metabolites into the bile (2, 3). ABCG2 is a marker protein of stem cells (4), where its physiological role is not yet clearly understood. It has been documented that ABCG2 expression is up-regulated under hypoxic conditions and that the protein can bind and/or transport porphyrins (5, 6); therefore it may play an important role in the protection of stem cells under hypoxic conditions. Overexpression of ABCG2 has been demonstrated in various tumor cells as well (1), where the transporter may be responsible for the emergence of a multidrug-resistant tumor phenotype that often leads to the failure of chemotherapy treatment in cancer patients.
Because ABCG2 is a half-transporter, bearing only one of each of the characteristic ABC family domains (the ATP-binding domain and transmembrane domain), ABCG2 has to form a homodimer or homo-oligomer to become functionally active (7, 8). The ABCG2 homodimer is covalently linked via a disulfide bond formed by cysteines at position 603, localized in the large ∼55-amino acid-long third extracellular loop (ECL3) of the protein (9, 10). Interestingly, mutation of Cys-603 to Ala, Gly, or Ser does not remarkably influence the expression and functionality of the transporter (9–11). In ECL3, ABCG2 has two other cysteines at positions 592 and 608. These two residues are indicated as forming an intramolecular disulfide bridge that influences plasma membrane targeting and substrate specificity of the transporter (10–13).
Being a stem cell marker protein and one of the most important ABC multidrug transporters, a sensitive method for the detection of ABCG2 expression is of great interest. There are several methods for detecting ABCG2 expression in various cell types (14); however, only a limited number of these use intact cells, which is essential when enrichment and further culturing of ABCG2-expressing cells (e.g. stem cells) is required. One such example is the flow cytometric application of the 5D3 antibody, which allows the easy detection and sorting of ABCG2-expressing intact cells.
The 5D3 monoclonal antibody was generated by immunizing mice with murine cells expressing human ABCG2 (4). This antibody recognizes a yet undefined, extracellular epitope of ABCG2. Previously, we have shown that 5D3 binding strongly depends on the conformation of ABCG2 (15). Namely, inhibition of protein function by the specific inhibitor Ko143 or by using an ABCG2 substrate flavopiridol at a high, inhibitory concentration, as well as ATP depletion of the cells, greatly increases 5D3 binding, called a “5D3 shift” (15). On the other hand, mimicking the ATP-bound state by using a non-hydrolyzable ATP analog, AMP-PNP, or by arresting ABCG2 by sodium orthovanadate significantly reduces 5D3 binding (15). We and others have also demonstrated that 5D3 can inhibit the function of ABCG2 (15, 16). Not only is the 5D3 antibody a good candidate for the detection of ABCG2 in flow cytometry-based assays, but this antibody-protein interaction may also facilitate structural studies at a molecular level, such as in the crystallization of ABCG2. However, because 5D3 reactivity is sensitive to conformational changes of ABCG2, proper assay conditions must be determined and accurately controlled.
The aim of the present study was to further characterize the conditions influencing 5D3 binding to ABCG2. We have analyzed in detail how covalent cross-linkings of two ABCG2 proteins influence 5D3 binding and attempted to unravel the role of the intra- and intermolecular disulfide bonds in 5D3 epitope formation. We found several protein cross-linkers that significantly increased 5D3 binding to ABCG2, resulting in a covalent ABCG2 dimer formation and/or inhibition of transport function. However, we also found a cross-linker that caused a 5D3 shift without covalent cross-linking of the two ABCG2 proteins or without the inhibition of ABCG2 function. When we administered dithiothreitol (DTT) to intact cells to reduce extracellular cysteines, we found that this treatment abolished 5D3 binding without any effect on the transport function. Characterization of 5D3 binding to ABCG2 proteins bearing mutations in extracellular cysteines revealed that the intermolecular S–S bond has only a minor effect on 5D3 binding, but disruption of the intramolecular S–S bridge has a dramatic effect on antibody recognition. Based on these data, we suggest that the epitope of 5D3 is located in the third, large extracellular loop of ABCG2. Additionally, we have generated a model showing the conformation of the third extracellular loop and revealing how conformational changes mediated by the disruption of the extracellular S–S bonds may influence 5D3 epitope formation.
EXPERIMENTAL PROCEDURES
Materials
Protein cross-linkers BM[PEO]3, BMPH, EDC, PMPI, sulfo-EGS, and sulfo-MBS were purchased from Pierce. BXP-21 monoclonal antibody (3) and Ko143 (17) were kind gifts from Drs. George Scheffer and Rik Scheper and from Dr. G. J. Koomen, respectively.
Expression Vectors, Cell Lines, and Cell Culturing
pCIN4 bicistronic mammalian expression vectors containing the cDNAs of ABCG2-R482G, or additional Cys to Ala mutations, were generated as described previously (10). HEK293 cell lines expressing various ABCG2 mutants were generated by transfection of the cells using the FuGENE® 6 (Roche Applied Science). Stable cell lines were obtained by maintaining the cells in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 50 units/ml penicillin, 50 units/ml streptomycin, 5 mmol/liter glutamine, and 0.5 mg/ml G418 (Invitrogen) at 37 °C in 5% CO2. To obtain a cell line showing higher ABCG2-C592A/C608A expression, HEK-C592A/C608A cells were sorted based on rhodamine123 extrusion capacity in a FACSAria flow cytometer. The sorted HEK-C592A/C608A cell line was used throughout this study. Generation of HEK293, A431, or PLB985 cells expressing wild-type ABCG2 was described previously (15, 18, 19).
Generation of 5D3-Alexa647, 5D3-Fab and 5D3-Fab-Alexa647
The 5D3 monoclonal antibody was purified from the supernatant of a hybridoma using affinity chromatography. Fab fragments of the antibody were prepared by papain digestion and separated from Fc fragments and the whole antibody on protein A-Sepharose column as described previously (20). Fab fragments and the monoclonal antibody preparations were more than 97% pure as determined by SDS-PAGE. The antibody and the Fab fragments were labeled with Alexa647 succinimidyl ester (Molecular Probes, Invitrogen) and separated from the unconjugated dye by gel filtration on a Sephadex G-50 column (21). The dye-to-protein labeling ratio was 3.28 and 0.98 for the antibody and Fab preparations, respectively.
Immunodetection of ABCG2
Western Blotting—HEK cells were suspended in a Laemmli buffer containing 2% of the reducing agent β-mercaptoethanol or without it, as indicated in Fig. 1A. Western blot analysis was performed as described previously (22) by using the BXP-21 monoclonal antibody in a 500× dilution and a goat anti-mouse horseradish peroxidase-conjugated secondary antibody (5000× dilution, Jackson ImmunoResearch).
FIGURE 1.
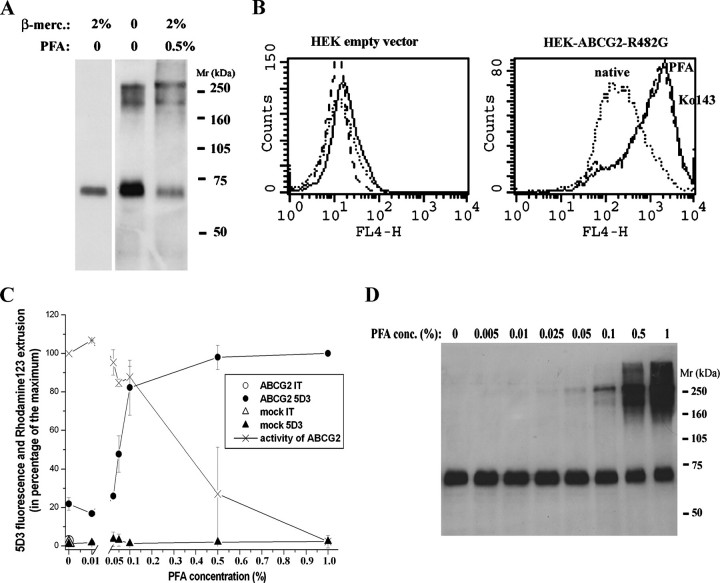
A, effect of PFA fixation on covalent ABCG2 dimer formation. HEK-ABCG2-R482G cells were lysed, dissolved in disaggregation buffer containing the reducing agent β-mercaptoethanol or without it (as indicated on the figure), and subjected to 7.5% SDS-PAGE. ABCG2 in cells fixed with 0.5% PFA prior to cell lysis is also shown. 15 μg of protein was loaded into each lane. ABCG2 was detected by the BXP-21 antibody. B, effect of PFA fixation on 5D3 labeling of ABCG2. HEK293 cells transfected with empty pCIN4 or pCIN4-ABCG2(R482G) were labeled with Alexa647-conjugated 5D3. Fluorescence was detected using a FACSCalibur cytometer. The solid line represents 5D3 binding of 0.5% PFA-treated and the dotted line for nontreated (native) cells. The dashed line represents labeling of cells in the presence of the specific ABCG2 inhibitor, Ko143. C, 5D3 labeling and activity of ABCG2 in cells treated with increasing concentrations of PFA. Mock or pCIN4-ABCG2(R482G)-transfected HEK293 cells were fixed with increasing concentrations of PFA, washed, and then labeled with 5D3 antibody or mouse IgG2b (isotype control (IT)). Fluorescence of goat anti-mouse phycoerythrin-conjugated secondary antibody was detected in a FACSCalibur cytometer. Fluorescence values are shown as the percentage of maximum labeling obtained in ABCG2-expressing cells fixed with 1% PFA and labeled with 5D3. Rhodamine123 uptake is shown in the presence of increasing concentrations of PFA. HEK-ABCG2 cells were fixed with increasing concentrations of PFA, washed, and then incubated with 2 μm rhodamine123 or 2 μm rhodamine123 + 1 μm Ko143. Rhodamine123 fluorescence in living cells was detected in a FACSCalibur cytometer. Rhodamine123 extrusion activity of ABCG2 is shown as the percentage of maximum activity. Activities were calculated as follows: (Mi - M0)/Mi where Mi is the mean rhodamine123 fluorescence in the presence of Ko143 and M0 is the mean fluorescence without Ko143. Shown is the average of two independent experiments. D, Western blot analysis of ABCG2 in HEK cells treated with increasing concentrations of PFA. HEK-ABCG2 cells were fixed with increasing concentrations of PFA or without it (first lane), washed, and then suspended in disaggregation buffer. Samples (each containing 15 μg of protein) were then subjected to 7.5% Laemmli gel electrophoresis and electroblotting. ABCG2 was visualized by the BXP-21 antibody.
Flow Cytometry—5D3 binding in intact cells was examined by suspending HEK cells in HPMI buffer (120 mm NaCl, 5 mm KCl, 400 μm MgCl2, 40 μm CaCl2, 10 mm HEPES, 10 mm NaHCO3, 10 mm glucose, and 5 mm Na2HPO4) containing 0.05% bovine serum albumin (Sigma). Aliquots of the cell suspension (5 × 105 cells in 100 μl) were incubated with Alexa647-conjugated 5D3 antibody (2 μg/ml final concentration) for 45 min at 37 °C. 5D3-Alexa647 binding was determined in a FACSCalibur cytometer at 635 nm excitation and 661/16 nm emission (FL4) wavelengths. When indirect labeling was performed, cells were incubated with unlabeled 5D3 and mouse IgG2b as an isotype control (both used in 1 μg/ml final concentration) for 30 min at 37 °C. After washing, the phycoerythrin-conjugated goat anti-mouse secondary antibody (GAM-PE, Beckman Coulter) was used, and its fluorescence was determined at 488-nm excitation and 585/42-nm emission (FL2) wavelengths. Labeling with 5D3-Fab was carried out the same way as with the whole 5D3 antibody. When labeling was carried out in the presence of an ABCG2 inhibitor (1 μm Ko143 or 5 μm FTC), the cells were preincubated with these agents for 10 min at 37 °C before labeling, and the inhibitors were present throughout the antibody labeling procedure.
Confocal Microscopy
HEK cells were seeded onto 8-well Nunc Lab-Tek II chambered coverglass (Nalge Nunc International) at 3 × 104/well cell density, and grown for 48 h in Dulbecco's modified Eagle's medium containing 10% fetal calf serum. For cell surface labeling, the cells were gently washed with Dulbecco's modified phosphate-buffered saline (DPBS), fixed with 1% paraformaldehyde in DPBS for 15 min at room temperature, and then blocked for 1 h at room temperature in DPBS containing 0.5% bovine serum albumin. The samples were then incubated for 1 h at room temperature with the 5D3 antibody conjugated with allophycocyanin (R&D Systems), diluted 5× in DPBS containing 0.5% bovine serum albumin, and finally washed with DPBS.
For immunostaining of permeabilized cells, samples were gently washed and then fixed with 4% paraformaldehyde in DPBS for 15 min at room temperature. After a few washes with DPBS, the cells were further fixed and permeabilized in prechilled methanol for 5 min at -20 °C. Following further washing steps, the cells were blocked for 1 h at room temperature in DPBS containing 2% bovine serum albumin, 1% fish gelatin, 0.1% Triton-X 100, and 5% goat serum (blocking buffer). The samples were then incubated for 1 h at room temperature with BXP-21 antibody diluted 100× in blocking buffer. After washing with DPBS, the cells were incubated for 1 h at room temperature with Alexa Fluor 488-conjugated goat anti-mouse IgG (H+L) (Molecular Probes) diluted 250× in blocking buffer. As isotype controls, allophycocyanin-conjugated mouse IgG2b (eBioscience) (5 μg/ml) and mouse IgG2a (Dako) (2.5 μg/ml) plus Alexa Fluor 488-conjugated goat anti-mouse IgG (1:250) were used. The stained samples were studied with an Olympus FV500-IX confocal laser scanning microscope using an Olympus PLAPO 60× (1.4) oil immersion objective (Olympus Europa GmbH) at room temperature. Green and deep red fluorescence was acquired above 505 and 650 nm, using excitation at 488 and 633 nm, respectively.
Cellular Dye Uptake and Calculation of ABCG2 Transport Activity
For measurement of ABCG2 activity, 5 × 105 HEK cells were suspended in 100 μl of HPMI buffer containing 2 μm rhodamine123, 5 μm mitoxantrone (MX) or 1 μm pheophorbide A in the presence or absence of 5 μm FTC for 30 min at 37 °C. Cells were then washed and resuspended in ice-cold phosphate-buffered saline, and fluorescence was determined in a FACSCalibur cytometer. Dead cells in the rhodamine123 uptake experiments were excluded based on TOPRO-3 staining (Molecular Probes) and in MX and pheophorbide A transport experiments by propidium iodide (Sigma) staining. Mean fluorescence values measured in the absence (M0) and the presence of inhibitor (Mi) were determined and activity was calculated as follows: (Mi - M0)/Mi.
Measurement of Hoechst 33342 Transport Activity
Hoechst 33342 transport was determined as described previously (23). The effect of 5D3 (12 μg in 100 μl for 3 × 105 cells) on Hoechst 33342 transport was measured as described previously (15). The effect of 5D3-Fab on Hoechst 33342 uptake was determined in the same way, except that 24 or 48 μg of 5D3-Fab was used in 100 μl for 3 × 105 cells.
Treatment of ABCG2 with Cross-linkers or Dithiothreitol—5 × 105 HEK cells were suspended in 100 μl of phosphate-buffered saline containing paraformaldehyde (PFA) (0.001–1%), BM[PEO]3 (0.5 mm), BMPH (1 mm), PMPI (1 mm), EDC (2 mm), sulfo-EGS (2 mm), sulfo-MBS (2 mm), or dithiothreitol (1–50 mm), and incubated at 37 °C for 10 min. After washing with 1 ml of HPMI containing 0.05% bovine serum albumin, 5D3 labeling or functional analysis of ABCG2 was performed as described above. Alternatively, cells were suspended in SDS-sample loading buffer and analyzed by Western blotting (see above). In some experiments PFA fixation and DTT treatment were used in combination. In these cases, cells were fixed first with PFA (or treated with DTT) as described above, washed with 1 ml of HPMI, and then treated with DTT (or fixed with PFA), washed again with 1 ml of HPMI-0.05% bovine serum albumin, and finally labeled with 5D3.
Ab Initio Folding Simulations and Three-dimensional Characterization of the ECL3 of ABCG2
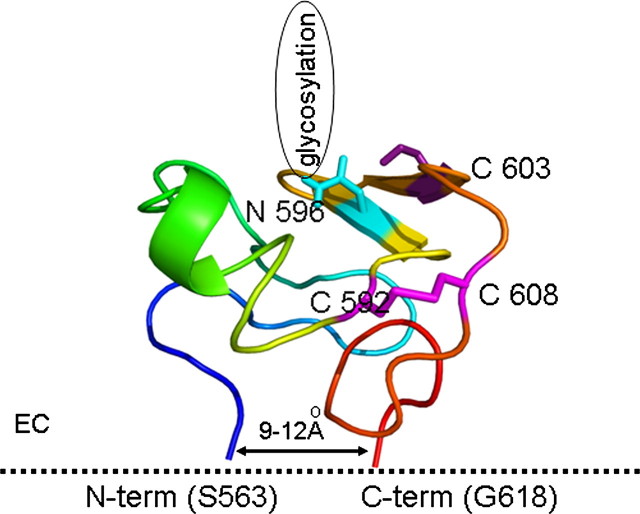
The folding simulations of the third extracellular loop of ABCG2 were performed as described previously (24). Briefly, a linear ECL3 peptide (residues 563–618) was used to generate a large, diverse pool of structures using a dynamic sampling algorithm, discrete molecular dynamics (DMD) (25–28). The search for low energy conformers was performed by replica exchange (28, 29) DMD simulations (26–29) in two steps. In the first round, accessible conformers of ECL3 were sampled within a higher temperature range (0.5–0.78 ε/kb), and a preliminary decoy set composed of structures with low potential energy values (lower than -261 ε) was constructed. Then each decoy in the preliminary set was subjected to a second round of replica exchange DMD, with exchange temperatures in a lower range (0.3–0.58 ε/kb). The final decoy set consisted of 101 low energy structures (lower than -338 ε), which were relaxed in a final step of equilibrium simulation at low temperature (0.2 ε/kb). The distance between the two ends of the ECL3 was set between 9 and 12 Å, based on existing ABC protein crystal structures (30). The S–S distance between Cys-592 and Cys-608 were set to maintain the disulfide bond.
To characterize the major conformations accessed by ECL3, decoys were grouped into clusters using the k-means algorithm88 (MatLab, MathWorks, Inc.) applying Cα root-mean-square deviation as the similarity metric between two structures. We selected a single, representative ECL3 structure for presentation (Fig. 8), which was obtained from the most populated cluster having the glycosylation site and cysteine 603 (responsible for intermolecular dimer interaction) on the surface of the structure.
FIGURE 8.
Model of the third extracellular loop of ABCG2 (amino acids 563–618) obtained by ab initio folding employing discrete molecular dynamics. The structure is colored blue to red from the N to the C terminus. Cysteines involved in intramolecular and intermolecular interactions are represented with sticks and are colored magenta. The glycosylation site (Asn-596) is represented by a blue stick.
RESULTS
Effect of Paraformaldehyde Fixation on ABCG2 Protein 5D3 Antibody Interaction—5D3 is a conformation-sensitive monoclonal antibody that recognizes a yet undefined extracellular epitope of the human ABC half-transporter, ABCG2. PFA generally used cross-linking fixative. Previously, we had found that in a PLB985 cell line expressing ABCG2, PFA fixation (1% final concentration) as well as inhibition of the function of wild-type ABCG2, e.g. by Ko143 or FTC, results in a significant increase in 5D3 labeling (5D3 shift (15)). We found that PFA also increased 5D3 binding in other ABCG2-expressing human cell lines (A431 and HEK). When we analyzed PFA-fixed samples by Western blotting, using the anti-ABCG2 antibody BXP-21 generated against an intracellular epitope of ABCG2, we found that in PFA-fixed cells two higher molecular mass forms of ABCG2 appeared (Fig. 1A, lane 3). These bands have ∼200–250-kDa relative molecular weight that, despite an unusually slow mobility, has been suggested to correspond to the dimeric form of the protein. When the ABCG2 homodimer is linked by a disulfide bridge (8–12, 31), the covalent ABCG2 dimer can be also detected under nonreducing conditions by Western blotting (see Fig. 1A, lane 2).
When we analyzed HEK-ABCG2 cells not treated with any cross-linkers but suspended in a nonreducing buffer (without β-mercaptoethanol), we found the same slower mobility bands as in PFA-fixed samples (Fig. 1A, lane 2). These data suggest that the higher apparent molecular mass forms observed in PFA-treated samples are most probably covalently cross-linked ABCG2 dimers (see more on cross-linkers below).
To analyze whether there is a correlation between covalent dimer formation upon PFA fixation and increased 5D3 binding, and whether PFA cross-linking inhibits ABCG2 function, we treated intact HEK cells expressing ABCG2 (R482G) with increasing concentrations of PFA and analyzed 5D3 binding, ABCG2 function, and covalent dimer formation. Throughout this study we used both the wild-type and the R482G mutant variant of ABCG2 in HEK cells, because the background of the cysteine mutations was this latter variant (10). The R482G mutant also allowed the measurement of transport activity followed by rhodamine123 extrusion, characteristic of the mutant protein. In all functional experiments the R482G protein variant showed the same behavior regarding 5D3 binding as the wild-type ABCG2, that is PFA or Ko143 caused a significant 5D3 shift (Fig. 1B).
When analyzing the effect of increasing amounts of PFA, we found that 5D3 labeling showed a saturating curve. We could already detect a slight increase in 5D3 binding at 0.05% PFA concentration as compared with the untreated cells, and 5D3 labeling reached its maximum when cells were fixed with 0.5–1% PFA (Fig. 1C). Increased antibody binding caused by PFA fixation was specific, as there was no 5D3 labeling in control cells transfected with an empty vector (and having no ABCG2 expression), and the fluorescence in cells incubated with an isotype control mAb did not increase upon PFA fixation (Fig. 1C).
To examine whether fixation-mediated increase in 5D3 labeling of ABCG2 positively correlates with the inhibition of the protein transport function, we also analyzed the effect of PFA on the transport function of ABCG2. Fig. 1C also shows rhodamine123 transport activity (activity factor) of the HEK-ABCG2-R482G cells treated with increasing concentrations of PFA. We found that ABCG2 gradually lost its activity upon PFA fixation, but functional inactivation was observed only in cells fixed with 0.5% or more PFA. Notably, there was already an about 80% increase in 5D3 labeling in the 0.1% PFA-fixed samples, whereas rhodamine123 extrusion was hardly inhibited (less than 20%) at this PFA concentration. When we analyzed PFA-fixed cells by Western blotting (Fig. 1D), we found that the relative amount of covalently linked ABCG2 dimer greatly increased in cells fixed with 0.5% or more PFA; thus, covalent cross-linking seemed to correlate with the inhibited transport function but not with the 5D3 shift.
Effect of Various Protein Cross-linkers on ABCG2 Protein 5D3 Antibody Interaction—To further investigate whether there is a correlation between covalent cross-linking of the ABCG2 monomers and an increased 5D3 binding, we treated ABCG2 expressing cells with different specific protein cross-linkers and analyzed them for 5D3 binding, functionality and dimer formation. We chose several non-cell-permeable protein cross-linkers, reacting with different amino acid side chains on the cell surface (Table 1). As Fig. 2 shows, we found several protein cross-linkers (BM[PEO]3, BMPH, sulfo-MBS, and PMPI) that increased 5D3 binding up to the level observed in FTC or PFA treated cells, whereas they did not affect the labeling of control cells or the fluorescence measured with the isotype antibody control (not shown).
TABLE 1.
Effects of protein cross-linkers on 5D3 binding, transport activity, and covalent dimer formation of ABCG2
| Cross-linker | Side chains cross-linked | Spacer arm length | Increased 5D3 binding | Inhibition of transport function | Cross-linked ABCG2 on Western blot |
|---|---|---|---|---|---|
| Å | |||||
| BM[PEO]3 | SH2–H2 | 14.7 | Yes | Yes | Yes |
| BMPH | CH3–SH2 | 8.1 | Yes | Yes | No |
| EDC | COOH–NH2 | 0 | Yes | No | No |
| Sulfo-EGS | NH2–NH2 | 16.1 | No | No | Yes |
| Sulfo-MBS | NH2–SH2 | 9.9 | Yes | No | Yes |
| PMPI | SH2–OH | 8.7 | Yes | No | Yes |
FIGURE 2.
A, effect of protein cross-linkers on 5D3 labeling and function of ABCG2. HEK-ABCG2-R482G cells were incubated with different protein cross-linkers, washed, and then labeled with 5D3-Alexa647 (left panel) or incubated with 2 μm rhodamine123 (right panel) in the presence or absence of the inhibitor Ko143. Fluorescence was determined by flow cytometry. Left panel, dotted lines represent labeling of mock-transfected native cells, and dashed lines stand for mock-transfected cells treated with cross-linker (or FTC). The solid lines represent 5D3 labeling of native, and heavy solid lines show cross-linker (or FTC)-treated ABCG2-expressing cells. Right panel, rhodamine123 uptake of cross-linker (or FTC) nontreated HEK-ABCG2 cells (solid line) or cross-linker (or FTC)-treated HEK-ABCG2 cells (heavy solid line). Only living cells are shown in the right panel. B, Western blot analysis of the effect of protein cross-linkers on ABCG2. HEK-ABCG2 cells were incubated with different protein cross-linkers, washed, and then suspended in Laemmli buffer. Proteins were separated by 7.5% SDS-PAGE. After electroblotting, ABCG2 was detected by using the BXP-21 antibody. Each lane represents 15 μg of protein (+/0, in the case of PFA, its inhibitory effect depends on the concentration used; see Fig. 1C). sEGS, sulfo-EGS; sMBS, sulfo-MBS.
To examine whether the observed increase in 5D3 binding in these protein cross-linker treated samples was due to the inhibition of the function of ABCG2, we analyzed rhodamine123 uptake in these cells (Fig. 2A, right panel). We found that EDC, sulfo-EGS, sulfo-MBS, and PMPI did not inhibit the transport function of ABCG2, whereas the others abolished rhodamine123 extrusion. We also analyzed protein cross-linker treated cells for covalent ABCG2 dimer formation by Western blotting and found that BMPH and EDC, two compounds that caused a 5D3 shift, did not result in a covalently linked ABCG2 (Fig. 2B). The experiments shown in Fig. 2 were performed in HEK-ABCG2-R482G cells, but experiments repeated in both PLB985 and A431 cells, expressing the wild-type ABCG2 protein, provided the same results (data not shown).
In summary, we found that the effect of cross-linkers was highly variable as to function, 5D3 labeling, and covalent dimer formation (see Table 1). These results suggest that covalent cross-linking of two (or more) ABCG2s is neither sufficient nor necessary for an increased 5D3 binding and/or for transport inhibition. From these data it seems likely that fixation of the epitope on a monomeric ABCG2 protein, even without the inhibition of transport function, may be responsible for an increased 5D3 binding.
Effect of DTT Treatment on 5D3 Binding—ABCG2 has three cysteines localized on the cell surface in the large third extracellular loop of the protein. It has been suggested that Cys-603 is responsible for intermolecular S–S bond formation, whereas the other two cysteines, Cys-592 and Cys-608, form an intramolecular S–S bridge (9–12). To find out whether the extracellular S–S bonds formed by cysteines are important for 5D3 recognition, we treated HEK-ABCG2 cells with the reducing agent DTT, which has a slow or insignificant cellular permeation. We found that treatment with 10 mm DTT caused a significant decrease in 5D3 binding in native (nonfixed) cells; this low 5D3 binding persisted even if the cells were fixed by PFA after the DTT treatment (Fig. 3B). DTT treatment had no effect on the negligible 5D3 labeling of HEK cells transfected with the empty vector (Fig. 3A), nor did it affect the fluorescence of HEK-ABCG2 cells incubated with an isotype control antibody. In parallel experiments we analyzed the effect of DTT on the function of ABCG2 and found that even in cells treated with 10–50 mm DTT, ABCG2 was fully active (data not shown). This latter finding is in harmony with the results of Mitomo et al. (31), who demonstrated that β-mercaptoethanol, another reducing agent, does not influence the functionality of ABCG2.
FIGURE 3.
Effect of DTT treatment on 5D3 binding. HEK293 cells transfected with empty pCIN4 (A) or pCIN4-ABCG2(R482G) (B) were incubated with or without 10 mm DTT, washed, and then labeled with 5D3 or mouse IgG2b and goat anti-mouse phycoerythrin-conjugated secondary antibody. The upper panels represent labeling of PFA nontreated cells (native), and the lower panels show labeling of cells that were fixed with 1% PFA after DTT treatment. Dotted lines, isotype control; dot-dash-dotted lines, isotype control of DTT-treated cells; solid lines, 5D3; dashed lines, 5D3 labeling of DTT-treated cells.
We also examined the expression level and dimerization of ABCG2 in DTT-treated samples by Western blotting. We found that 10 mm DTT treatment resulted in the complete loss of ABCG2 dimer formation observed in samples dissolved in loading buffer without the reducing agent β-mercaptoethanol. However, DTT had no effect on the covalent cross-linking of ABCG2s by PFA (not shown). These results suggested that intact S–S bridges in the ABCG2 protein on the cell surface may play an important role in the 5D3 epitope formation.
Expression, Function, and 5D3 Binding of ABCG2 Variants Carrying Cys-to-Ala Mutations in the Third Extracellular Loop—To investigate whether DTT treatment causes a decrease in 5D3 binding due to the reduction of the intramolecular or intermolecular S–S bonds, or which of these bonds are important for 5D3 epitope formation, we expressed ABCG2 mutants having single Cys-to-Ala changes or the combination of these mutations in HEK cells.
Fig. 4A shows that all Cys-to-Ala mutants (except for C603A/C608A that was expressed in very low amount and exclusively in an underglycosylated form) could be detected by Western blotting, using the ABCG2-specific BXP-21 antibody. The mutants C592A and C603A showed expression levels comparable to that of the wild-type ABCG2, whereas the amount of double mutant C592A/C608A or the triple Ala mutant proteins was about 50% of that seen for the wild-type ABCG2.
FIGURE 4.
A, expression of various ABCG2 mutants carrying Cys-to-Ala changes. HEK cells were transfected with pCIN4 vectors encoding different Cys-to-Ala ABCG2 mutants or R482G (indicated as ABCG2). Cells were lysed, dissolved in disaggregation buffer, and subjected to 7.5% SDS-PAGE. ABCG2 was detectedby the BXP-21 antibody. To demonstrate differences in the expression level of the mutants, various amounts of samples were loaded onto the gel as indicated in the figure. B, mitoxantrone, pheophorbide A, rhodamine123, and Hoechst 33342 transport activity of Cys-to-Ala mutants. HEK cells expressing different ABCG2 mutants or R482G (indicated as ABCG2) were incubated with 5 μm mitoxantrone, 1 μm pheophorbide A, 2 μm rhodamine123, or 1 μm Hoechst 33342 in the absence or presence of 1 μm Ko143. Fluorescence of mitoxantrone, pheophorbide A, and rhodamine123 was detected in a FACSCalibur cytometer, and activity factors were calculated from mean fluorescence values as described under “Experimental Procedures.” Fluorescence due to Hoechst 33342 accumulation was determined in a spectrofluorimeter. The transport rate was determined as described under “Experimental Procedures.” Shown are the average transport activities obtained from two independent experiments.
To analyze whether the ABCG2 cysteine mutants were functional, we performed fluorescent dye uptake experiments by flow cytometry and fluorometry (Fig. 4B). We analyzed the transport of four different fluorescent ABCG2 substrate compounds, MX, pheophorbide A, Hoechst 33342, and rhodamine123 (rhodamine123 is transported only by the R482G ABCG2 mutant). We found that all of the expressed cysteine mutants were functional, bearing dye extrusion capacity, but there was a significant variation in their transport activities relative to the different transported compounds.
Hoechst 33342 and pheophorbide A were transported by all of the mutants (except for C603A/C608A); however, the mitoxantrone transport capacity of the mutants, lacking the intramolecular or both disulfide bonds, was significantly weaker than that of the R482G or C603A variants. There was also a difference in rhodamine123 uptake, with the C592A and C592A/C603A mutants showing practically no transport activity (Fig. 4B). These data are in harmony with the findings of Henriksen et al. (10), who report that some of the Cys-to-Ala mutants have altered substrate specificity.
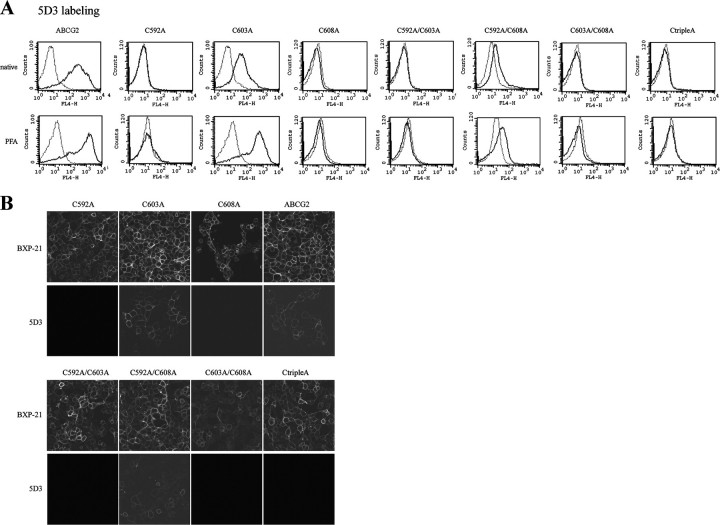
When cells expressing the different Cys-to-Ala mutants were labeled with the 5D3 antibody, we found that only the C603A variant had a clearly detectable 5D3 labeling and the C592A/C608A mutant showed some weak 5D3 binding capacity (Fig. 5A, upper panel). Similar to that seen in the case of the wild-type ABCG2, PFA fixation (Fig. 5A, lower panel) or Ko143 treatment (not shown) of the cells expressing the C603A mutant and the double mutant C592A/C608A resulted in an increased 5D3 binding. However, the other mutant variants did not show any labeling, even in PFA-fixed or Ko143-inhibited samples.
FIGURE 5.
A, 5D3 labeling of HEK cells expressing different Cys-to-Ala mutants of ABCG2. Native (upper panel) or 1% PFA-treated (lower panel) cells were labeled with 5D3-Alexa647. 5D3 fluorescence was detected by flow cytometry. Then solid lines represent 5D3-Alexa647 labeling of cells expressing different ABCG2 mutants, as indicated above the panels, and the dotted lines represent 5D3-Alexa647 labeling of cells transfected with empty vector. B, localization of the ABCG2 Cys-to-Ala mutants. HEK cells expressing different ABCG2 mutants (as indicated above the panels) were fixed in 4% paraformaldehyde, permeabilized in prechilled methanol, and then incubated with the BXP-21 antibody and Alexa Fluor 488-conjugated goat anti-mouse IgG. To label the cell surface epitope of ABCG2, a direct immunofluorescent reaction was performed. The cells were fixed with 1% paraformaldehyde and then incubated with allophycocyanin-conjugated 5D3. The stained slides were analyzed using an Olympus FV500-IX confocal laser scanning microscope. Fluorescence was acquired using the same equipment settings, except the slide representing 5D3 labeling of C592A/C608A was taken at an increased detector voltage.
To rule out the possibility that low protein expression level or protein dislocalization was responsible for the absence of labeling of most of the mutants, and to test whether they could reach the plasma membrane (necessary for recognition by 5D3 antibody), we analyzed ABCG2 immunofluorescence by confocal microscopy.
Fig. 5B shows that all mutants could be detected by the BXP-21 antibody, recognizing an intracellular epitope of ABCG2, and all of them were present in the plasma membrane (except for the hardly expressed C603A/C608A double mutant, which was found, for the most part, intracellularly). However, 5D3 labeling analyzed by confocal microscopy gave the same result as the flow cytometry measurements, that is, only the cells expressing the wild-type ABCG2, C603A, and the C592A/C608A variants (the latter one seen only at increased detector voltage) could bind the 5D3 antibody.
It has been shown that the 5D3 antibody inhibits the transport function of ABCG2 (as judged in a sensitive Hoechst 33342 assay), although this inhibition is only partial and requires high concentrations of the antibody (15). To analyze a potential correlation between 5D3 binding and transport inhibition, we added high concentrations of 5D3 to the HEK cells and then analyzed ABCG2-specific Hoechst 33342 transport activity. We found that, in contrast to a 30–40% inhibition found in the case of the wild-type ABCG2, 5D3 did not influence the Hoechst 33342 transport activity of the C592A/C608A and C592A/C603A/C608A mutants (data not shown). All of these data strongly suggest that the ABCG2 mutant proteins lacking the cysteines required for intramolecular S–S bridge formation are expressed in comparable amounts, reach the cell surface, and work as active transporters in a manner similar to the wild-type ABCG2, but these variants (except for the C592A/C608A mutant showing weak 5D3 binding) are unable to bind the 5D3 antibody.
Effect of DTT on 5D3 Labeling of the Cys-to-Ala Mutants—To test whether decreased 5D3 binding in ABCG2-expressing cells treated with DTT was due to the reduction of the extracellular cysteines, we also examined the effect of DTT on 5D3 labeling of the mutants C603A and C592A/C608A in native or PFA-fixed cells. Fig. 6, A and B, shows that DTT is still effective in the reduction of 5D3 binding in the case of the C603A mutant but has practically no effect on 5D3 labeling of the C592A/C608A mutant. When we analyzed 5D3 fluorescence in ABCG2-expressing HEK cells treated with increasing concentrations of DTT, we found that DTT treatment caused a gradual decrease in 5D3 binding that reached its minimum (almost the fluorescence of the background) at 10–50 mm DTT in ABCG2 and C603A-expressing cells, whereas DTT had no effect on labeling of the C592A/C608A double mutant (Fig. 6C). These experiments suggest that the intramolecular and not the intermolecular S–S bonds are important in 5D3 epitope formation.
FIGURE 6.

A and B, effect of DTT on labeling of the Cys-to-Ala mutants. HEK293 cells expressing different ABCG2 mutants were treated with 10 mm DTT, washed, and then labeled with 5D3-Alexa647 (A). B shows the labeling of samples fixed with 0.5% PFA prior to antibody labeling. Fluorescence was determined in a FACSCalibur cytometer. Dotted lines, mock-transfected HEK; dot-dash-dotted lines, DTT-treated, mock-transfected HEK cells; solid lines, HEK cells expressing different ABCG2 mutants; dashed lines, HEK cells expressing different ABCG2 mutants treated with DTT. C, HEK cells expressing different ABCG2 mutants were treated with increasing concentrations of DTT, fixed with 0. 5% PFA, and labeled with 5D3-Alexa647. 5D3 fluorescence was determined by flow cytometry. Mean values obtained from at least in two independent experiments are shown.
Experiments with 5D3-Fab—To analyze whether the 5D3 shift is caused by the stabilization of the epitopes in a dimer form of ABCG2, and thus an intact, bivalent 5D3 is required for labeling, we incubated HEK-ABCG2 cells with the Alexa647-conjugated Fab fragment of 5D3. We found a lower affinity but specific binding of 5D3 Fab to ABCG2 that was greatly increased by PFA fixation or by Ko143. DTT treatment also reduced Fab binding (Fig. 7). These experiments also suggest that 5D3 binding to ABCG2 depends on the conformation of an epitope found on a monomeric ABCG2.
FIGURE 7.
Direct labeling of HEK cells by Alexa647-conjugated 5D3-Fab. HEK293 cells transfected with the pCIN4 vector or pCIN4 containing the cDNA of ABCG2(R482G) were incubated with 2 μg/ml 5D3-Fab-Alexa647. Fluorescence was determined by flow cytometry. The dotted lines represent labeling of 10 mm DTT-treated cells; solid lines stand for native cells incubated only with 5D3-Fab, heavy solid lines show labeling of 0.5% PFA-fixed cells, and dashed lines indicate fluorescence in the presence of the inhibitor Ko143.
DISCUSSION
ABCG2 is a marker protein of the side population of stem cells and is also important in tumor cells where it can mediate the emergence of a multidrug-resistant phenotype. On one hand, a sensitive method for the detection of low amounts of ABCG2 may allow the enrichment and selection of ABCG2-expressing cells, such as stem cells. On the other hand, monitoring the presence of ABCG2 in tumor samples from patients can help to find the most effective chemotherapy treatment using non-ABCG2 substrate drugs. The conformation-sensitive antibody 5D3 is a good candidate for the detection of ABCG2 for the above mentioned purposes. However, to establish a reliable method it is essential to find the optimum conditions to be able to detect even low amounts of endogenous ABCG2. In our previous paper we showed that 5D3 binding depends on the actual conformation of ABCG2 (15). We could define two ABCG2 conformations based on the 5D3 binding capacity. The high affinity form was observed in ABCG2 inhibited by the specific inhibitor Ko143 in ATP-depleted cells or after fixation of the cells with PFA, whereas stabilization of ABCG2 in a vanadate-trapped form resulted in decreased 5D3 binding, representing a low affinity form.
In this study we have further analyzed how alterations in ABCG2 structure, covalent cross-linking, or changes in the S–S bonds on the external regions of ABCG2 influence 5D3 binding. When analyzing the effects of PFA fixation in HEK-ABCG2 cells, we found that although at higher concentrations PFA abolishes ABCG2 function and results in the formation of an ABCG2 dimer, at lower concentrations, which already cause a 5D3 shift, PFA is neither inhibitory on function nor does it cross-link ABCG2 dimers (Fig. 1, C and D). When we analyzed the effects of specific protein cross-linkers on 5D3 binding to ABCG2, we found that some of these agents increased 5D3 binding while causing a variable loss in transport function or covalent dimer formation.
In fact, EDC resulted in a 5D3 shift without the inhibition of ABCG2 function or covalent formation of an ABCG2 dimer (see Fig. 2, A and B). Thus, there was no general correlation between the 5D3 shift and the inhibition of the function or cross-linking of ABCG2 dimers. Experiments performed with the Fab fragment of 5D3 (see Fig. 7), having only one binding region, also suggested that the epitope is most probably located on an ABCG2 monomer.
The ABCG2 homodimer is linked via a disulfide bond mediated by Cys-603, found in the third extracellular loop of the protein. Additionally, there are two other cysteines (Cys-592 and Cys-608) in the third extracellular loop of ABCG2, suggested to form an intramolecular S–S bridge. When we treated intact HEK-ABCG2 cells with the nonpermeable reducing agent DTT, we found a significant decrease in 5D3 binding with no effect on the ABCG2 transport function (see Figs. 3 and 6), indicating a dramatic effect of the disruption of the extracellular S–S bonds on the 5D3 epitope formation. To find out which disulfide bridge is important for epitope formation, we analyzed different Cys-to-Ala mutants lacking the intermolecular (C603A), the intramolecular (C592A, C608A, C592A/C608A), or both kinds of (C592A/C603A, C603A/C608A, or C592A/C603A/C608A) S–S bonds.
We found that the C603A mutant behaves similarly to ABCG2 having intact S–S bridges. The C603A mutant, which can be expressed in an amount comparable to ABCG2, is found in the plasma membrane (Figs. 4A and 5B). This mutant is fully active and has the same substrate specificity as ABCG2 (Fig. 4B). Cys-603 has a clearly detectable 5D3 binding that can be increased by inhibition of ABCG2 function by Ko143 or treatment with protein cross-linkers. DTT treatment also decreased 5D3 binding of this mutant (Fig. 6), suggesting that this cysteine is not important for the observed effect of S–S bridge reduction.
The single mutants, lacking the intramolecular S–S bond, i.e. C592A, C608A, as well as the C592A/C603A/C608A variant, had clearly detectable expression levels, were present in the plasma membrane, and were functional for active transport with somewhat altered substrate specificities (Figs. 4 and 5). However, these mutants did not show any specific 5D3 binding either in a PFA-fixed or in a Ko143-inhibited form, and moreover, 5D3 did not inhibit their function. These experiments suggest that the intramolecular S–S bridge in the third extracellular loop of ABCG2 has a crucial, either direct or indirect (e.g. stabilizing the proper conformation), role in the formation of the 5D3 epitope as well as in the substrate specificity of the transporter.
The role of the intramolecular S–S bond in 5D3 epitope formation has already been suggested (9, 10). Kage et al. (9) analyzed 5D3 labeling in intact cells expressing various Cys-to-Ser mutants by flow cytometry. They found that C592S and C608S had impaired 5D3 binding; however, these two mutants showed very low expression levels in this study (9). In our experiments we were able to express C592A and C608A mutants in comparable levels to the wild-type ABCG2. Moreover, we demonstrated that even though these mutants were functional and properly localized to the plasma membrane, they could not be labeled with 5D3 even in the presence of an inhibitor or PFA.
Henriksen et al. (10) also suggested the role of the intramolecular S–S bridge of ABCG2 in 5D3 binding; they detected no 5D3 labeling by confocal microscopy for C592A, C608A, and the C592A/C608A double mutant. In harmony with this study, we could not detect any labeling for the single Cys-to-Ala mutants. However, in our hands the C592A/C608A double mutant showed a weak 5D3 binding both in flow cytometry and confocal microscopy, and the 5D3 shift upon PFA or Ko143 treatment could also be observed (Fig. 5). DTT had no effect on the 5D3 labeling of the C592A/C608A variant (Fig. 6), and the excess amount of 5D3 did not inhibit the function of this mutant.
The single Cys-592 or Cys-608 mutants showed an increased cytoplasmic accumulation (9, 10, 13), whereas the simultaneous removal of cysteines 592 and 608 promoted protein stability and proper targeting (10). Thus such a double mutation may allow the development of a favorable conformation within the ABCG2 protein, allowing some 5D3 labeling.
Based on these data we suggest that protein cross-linking most probably stabilizes the epitope of 5D3 present within a single ABCG2 protein. The effect of DTT treatment on 5D3 labeling together with experiments on the extracellular Cys-to-Ala mutants revealed that the third extracellular loop, and especially the intramolecular S–S bond within this region, has a crucial role in 5D3 epitope formation and probably in the substrate interactions of ABCG2.
To visualize the possible molecular arrangement of the third extracellular loop (ECL3) of the ABCG2 protein, we performed a molecular dynamic simulation of the folding of this relatively large protein sequence. Currently, there are no atomic structures available for eukaryotic ABC transporters, and molecular modeling is usually guided by the recently solved structures of bacterial transporters. Although a recent communication offers a homology model for ABCG2 (30), it does not include ECL3, as this loop shows no homology to any ABC protein sequences with a known structure. Data for other ABC transporters, however, suggest that the adjacent transmembrane regions of these proteins are found near each other, within a distance of 9–12 Å. Therefore in our simulation algorithm, we fixed the N- and C-terminal regions of the ECL3 at this distance at the membrane surface. We have also included the information that the cysteines forming intramolecular S–S bonds (Cys-592 and Cys-608), as well as a cysteine involved in the intermolecular S–S bridge formation, are most probably located on the surface of this loop. It is interesting to note, that many of the ABCG-type proteins have this large, conserved, extracellular region, with similar arrangements of cysteines and potential glycosylation sites (11).
The conformation obtained, as seen in Fig. 8, corresponds to these requirements and suggests the presence of a stabilized antiparallel loop with β-sheets in the region of the three cysteines, surrounding the glycosylation site. When either Cys-592 or Cys-608 was replaced by alanines in the in silico model, the structure of the antiparallel loop collapsed and the folding lost its conserved characteristics (data not shown). The combination of the experimental and the simulation results suggest a well defined structure in this area of ECL3, which may be important in mediating ABCG2 interaction with the plasma membrane or other proteins. Moreover, the conformation of this loop seems to modulate the substrate and antibody binding to this membrane transporter.
Acknowledgments
We greatly appreciate the technical help ofÉva Krizsán, Zsuzsanna Andrási, and Judit Kis. We appreciate the kind gifts of Ko143 from Drs. J. D. Allen and G. J. Koomen and anti-ABCG2 BXP-21 antibody obtained from Drs. George Scheffer and Rik Scheper.
This work was supported by grants from the Hungarian Scientific Research Fund (OTKA) (AT 048986 and NK72057) National Research and Development Programmes (NKFP), FP6-INTHER, FP6-MEMTRANS, NEDO, and National Health Council (ETT). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: ABC, ATP-binding cassette; AMP-PNP, adenosine 5′-(β,γ-imidotriphosphate); BM[PEO]3, 1,8-bis-maleimidotriethyleneglycol; BMPH, N-[β-maleimidopropionic acid]hydrazide, trifluoroacetic acid salt; DMD, discrete molecular dynamics; DPBS, Dulbecco's modified phosphate-buffered saline; DTT, dithiothreitol; ECL3, third extracellular loop of ABCG2; EDC, [1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride]; FTC, fumitremorgin C; HEK, human embryonic kidney; MX, mitoxantrone; MXR, mitoxantrone resistance protein; PFA, paraformaldehyde; PMPI, N-(p-maleimidophenyl)isocyanate; sulfo-EGS, ethylene glycol bis(sulfosuccinimidyl succinate); sulfo-MBS, m-maleimidobenzoyl-N-hydoxysuccinimide ester.
References
- 1.Sarkadi, B., Homolya, L., Szakacs, G., and Varadi, A. (2006) Physiol. Rev. 86 1179-1236 [DOI] [PubMed] [Google Scholar]
- 2.Chandra, P., and Brouwer, K. L. (2004) Pharm. Res. (N. Y.) 21 719-735 [DOI] [PubMed] [Google Scholar]
- 3.Maliepaard, M., Scheffer, G. L., Faneyte, I. F., van Gastelen, M. A., Pijnenborg, A. C., Schinkel, A. H., van De Vijver, M. J., Scheper, R. J., and Schellens, J. H. (2001) Cancer Res. 61 3458-3464 [PubMed] [Google Scholar]
- 4.Zhou, S., Schuetz, J. D., Bunting, K. D., Colapietro, A. M., Sampath, J., Morris, J. J., Lagutina, I., Grosveld, G. C., Osawa, M., Nakauchi, H., and Sorrentino, B. P. (2001) Nat. Med. 7 1028-1034 [DOI] [PubMed] [Google Scholar]
- 5.Krishnamurthy, P., Ross, D. D., Nakanishi, T., Bailey-Dell, K., Zhou, S., Mercer, K. E., Sarkadi, B., Sorrentino, B. P., and Schuetz, J. D. (2004) J. Biol. Chem. 279 24218-24225 [DOI] [PubMed] [Google Scholar]
- 6.Krishnamurthy, P., and Schuetz, J. D. (2005) Biometals 18 349-358 [DOI] [PubMed] [Google Scholar]
- 7.Ozvegy, C., Litman, T., Szakacs, G., Nagy, Z., Bates, S., Varadi, A., and Sarkadi, B. (2001) Biochem. Biophys. Res. Commun. 285 111-117 [DOI] [PubMed] [Google Scholar]
- 8.Kage, K., Tsukahara, S., Sugiyama, T., Asada, S., Ishikawa, E., Tsuruo, T., and Sugimoto, Y. (2002) Int. J. Cancer 97 626-630 [DOI] [PubMed] [Google Scholar]
- 9.Kage, K., Fujita, T., and Sugimoto, Y. (2005) Cancer Sci. 96 866-872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henriksen, U., Fog, J. U., Litman, T., and Gether, U. (2005) J. Biol. Chem. 280 36926-36934 [DOI] [PubMed] [Google Scholar]
- 11.Wakabayashi, K., Nakagawa, H., Adachi, T., Kii, I., Kobatake, E., Kudo, A., and Ishikawa, T. (2006) J. Exp. Ther. Oncol. 5 205-222 [PubMed] [Google Scholar]
- 12.Takada, T., Suzuki, H., and Sugiyama, Y. (2005) Pharm. Res. (N. Y.) 22 458-464 [DOI] [PubMed] [Google Scholar]
- 13.Wakabayashi, K., Nakagawa, H., Tamura, A., Koshiba, S., Hoshijima, K., Komada, M., and Ishikawa, T. (2007) J. Biol. Chem. 282 27841-27846 [DOI] [PubMed] [Google Scholar]
- 14.Robey, R. W., Polgar, O., Deeken, J., To, K. W., and Bates, S. E. (2007) Cancer Metastasis Rev. 26 39-57 [DOI] [PubMed] [Google Scholar]
- 15.Ozvegy-Laczka, C., Varady, G., Koblos, G., Ujhelly, O., Cervenak, J., Schuetz, J. D., Sorrentino, B. P., Koomen, G. J., Varadi, A., Nemet, K., and Sarkadi, B. (2005) J. Biol. Chem. 280 4219-4227 [DOI] [PubMed] [Google Scholar]
- 16.Abbott, B. L., Colapietro, A. M., Barnes, Y., Marini, F., Andreeff, M., and Sorrentino, B. P. (2002) Blood 100 4594-4601 [DOI] [PubMed] [Google Scholar]
- 17.Allen, J. D., van Loevezijn, A., Lakhai, J. M., van der Valk, M., van Tellingen, O., Reid, G., Schellens, J. H., Koomen, G. J., and Schinkel, A. H. (2002) Mol. Cancer Ther. 1 417-425 [PubMed] [Google Scholar]
- 18.Robey, R. W., Honjo, Y., Morisaki, K., Nadjem, T. A., Runge, S., Risbood, M., Poruchynsky, M. S., and Bates, S. E. (2003) Br. J. Cancer 89 1971-1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elkind, N. B., Szentpetery, Z., Apati, A., Ozvegy-Laczka, C., Varady, G., Ujhelly, O., Szabo, K., Homolya, L., Varadi, A., Buday, L., Keri, G., Nemet, K., and Sarkadi, B. (2005) Cancer Res. 65 1770-1777 [DOI] [PubMed] [Google Scholar]
- 20.Szollosi, J., Horejsi, V., Bene, L., Angelisova, P., and Damjanovich, S. (1996) J. Immunol. 157 2939-2946 [PubMed] [Google Scholar]
- 21.Spack, E. G., Jr., Packard, B., Wier, M. L., and Edidin, M. (1986) Anal. Biochem. 158 233-237 [DOI] [PubMed] [Google Scholar]
- 22.Ozvegy, C., Varadi, A., and Sarkadi, B. (2002) J. Biol. Chem. 277 47980-47990 [DOI] [PubMed] [Google Scholar]
- 23.Ozvegy-Laczka, C., Hegedus, T., Varady, G., Ujhelly, O., Schuetz, J. D., Varadi, A., Keri, G., Orfi, L., Nemet, K., and Sarkadi, B. (2004) Mol. Pharmacol. 65 1485-1495 [DOI] [PubMed] [Google Scholar]
- 24.Hegedus, T., Serohijos, A. W., Dokholyan, N. V., He, L., and Riordan, J. R. (2008) J. Mol. Biol. 378 1052-1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dokholyan, N. V., Buldyrev, S. V., Stanley, H. E., and Shakhnovich, E. I. (1998) Folding Des. 3 577-587 [DOI] [PubMed] [Google Scholar]
- 26.Ding, F., and Dokholyan, N. V. (2005) Trends Biotechnol. 23 450-455 [DOI] [PubMed] [Google Scholar]
- 27.Ding, F., and Dokholyan, N. V. (2006) PLoS Comput. Biol. 2 e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sugita, Y., and Okamoto, Y. (1999) Chem. Phys. Lett. 314 141-151 [Google Scholar]
- 29.Ding, F., Tsao, D., Nie, H., and Dokholyan, N. V. (2008) Structure (Lond.) 16 1010-1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hazai, E., and Bikadi, Z. (2007) J. Struct. Biol. 162 63-74 [DOI] [PubMed] [Google Scholar]
- 31.Mitomo, H., Kato, R., Ito, A., Kasamatsu, S., Ikegami, Y., Kii, I., Kudo, A., Kobatake, E., Sumino, Y., and Ishikawa, T. (2003) Biochem. J. 373 767-774 [DOI] [PMC free article] [PubMed] [Google Scholar]