Abstract
The large subunit rRNA in eukaryotes contains an unusually dense cluster of 8–10 pseudouridine (Ψ) modifications located in a three-helix structure (H37–H39) implicated in several functions. This region is dominated by a long flexible helix (H38) known as the “A-site finger” (ASF). The ASF protrudes from the large subunit just above the A-site of tRNA binding, interacts with 5 S rRNA and tRNA, and through the terminal loop, forms a bridge (B1a) with the small subunit. In yeast, the three-helix domain contains 10 Ψs and 6 are concentrated in the ASF helix (3 of the ASF Ψs are conserved among eukaryotes). Here, we show by genetic depletion analysis that the Ψs in the ASF helix and adjoining helices are not crucial for cell viability; however, their presence notably enhances ribosome fitness. Depleting different combinations of Ψs suggest that the modification pattern is important and revealed that loss of multiple Ψs negatively influences ribosome performance. The effects observed include slower cell growth (reduced rates up to 23% at 30 °C and 40–50% at 37 °C and 11 °C), reduced level of the large subunit (up to 17%), impaired polysome formation (appearance of half-mers), reduced translation activity (up to 20% at 30 °C and 25% at 11 °C), and increased sensitivity to ribosome-based drugs. The results indicate that the Ψs in the three-helix region improve fitness of a eukaryotic ribosome.
A common feature of ribosomal RNAs is the presence of several types of modified nucleotides that are formed at the precursor level, perhaps before transcription is completed. Three general classes of modification occur: isomerization of uridine to pseudouridine (Ψ),3 methylation of ribose groups (Nm), and methylation of different positions in base moieties (mN). The Ψ nucleotides constitute the largest single subset of modification, and these and the other modifications are heavily concentrated in regions of the ribosome known or predicted to be functionally important (1, 2). The conserved nature of the distribution patterns argues that the modifications have important roles in ribosome structure and function. Consistent with this view, it has been shown that blocking the formation of Ψ or Nm at the global level in yeast rRNA creates a nearly lethal growth phenotype (3, 4).
Most such modifications in eukaryotes are formed by small nucleolar ribonucleoprotein complexes (snoRNPs), and synthesis was disrupted in these latter experiments by point mutations in the snoRNP protein that catalyzes the modification reaction (3, 4). In general, loss of single rRNA modifications from yeast or Escherichia coli has had only minor or no detectable affect on growth, indicating that most individual modifications provide beneficial effects that are not essential (1, 5, 6). The bases of these benefits are all but unknown, because only a few studies have been extended beyond examining effects on growth. In one such case, depleting different combinations of 1–6 Ψs from the peptidyl transferase center region in yeast revealed both negative, synergistic effects on growth rate for multiple depletions and a substantial reduction (20%) in translation rate with loss of a particular modification implicated in tRNA binding at the A-site (7). Another study has shown that depleting modifications from an intersubunit bridge region in the large subunit (helix H69) impairs ribosome accumulation (8).
The present study is of this new generation of modification depletion analyses, in which modifications are blocked in a systematic, combinatorial fashion from a selected rRNA region and effects are examined at the levels of ribosome synthesis and function. Region-specific depletions of this type became possible and practical with the advent of high resolution crystal structures of the ribosome, and the discoveries that most Ψ and Nm modifications in eukaryotes (and archaea) are formed by snoRNPs. The snoRNP complexes contain site-specific guide snoRNAs, and genetic depletion of the snoRNA blocks formation of the corresponding snoRNP (9–13). Here, we characterize the importance of a remarkable cluster of Ψ modifications in the large subunit (LSU) rRNA of Saccharomyces cerevisiae. Indeed, the region of interest is the richest in Ψs in rRNA and is also known to be functionally important.
The target region is located in domain II of yeast 25 S rRNA and consists of consecutive helices H37, H38, and H39 (E. coli numbering). The three helices are organized into a highly folded, compact structure in mature ribosomes and likely have interrelated functions (Fig. 1 and see below). On this basis we reasoned the three helices should be examined as one structural unit. Helices H37 and H39 are relatively short, whereas H38 is a long hinged helix. The hinge in H38 is a kink-turn, and the portion from the hinge to the distal loop (∼100 Å) is best known as the A-site finger (ASF), because it lies above the A-site of tRNA binding in the large subunit (14, 15). The terminal 30% portion of the ASF protrudes from the central protuberance and participates in formation of an intersubunit bridge (B1a) (Fig. 1A) (16). This bridge was proposed to be an essential path for signal transmission between functional centers in the small and large subunits (17, 18). The distal part of the ASF helix also contacts the elbow of tRNA (D loop) in the A-site (19–21). Notably, this portion is often disordered in three-dimensional structure models, reflecting its dynamic nature (Fig. 1D) (22, 23).
FIGURE 1.
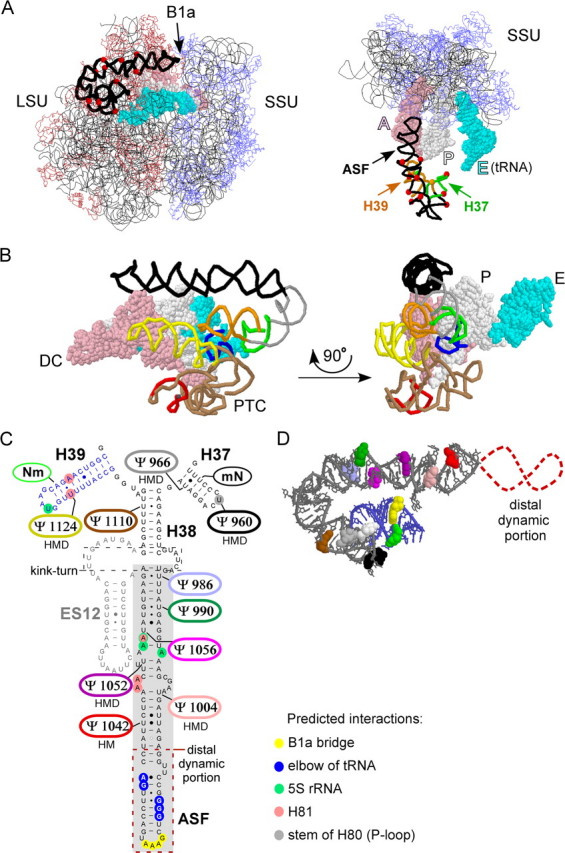
Helix H38 and flanking helices H37 and H39 form a compact structure that contains 10 Ψs. A, the helices and Ψs(red dots) are highlighted in the context of the 70 S ribosome of T. thermophilus. Left, view from the E-site of the ribosome; right, view from the top of the ribosome (most of the LSU is hidden). B1a is an intersubunit bridge involving the ASF; A, P, and E identify tRNAs at the corresponding binding sites. B, alternate perspectives of the H37–H39 region, without the remaining rRNA backbone. Left, relationship to the decoding center (DC) region of the small subunit (SSU) and the peptidyl transferase center (PTC) region of the LSU; right, 90° rotation. The color code: ASF (black); H37 (green); H39 (orange); peptidyl transferase center (brown); A-loop (red); P-loop (navy blue); H89 (yellow); tRNA in the A-site (pink); tRNA in the P-site (white); tRNA in the E-site (cyan). C, distribution of the 10Ψs. Six are highly conserved and occur in the same positions in human, mouse, and D. melanogaster (H, M, and D). The highlighted segment of H38 (gray box) corresponds to the ASF, which is defined by the tip of H38 and an internal kink-turn. An expansion segment, ES12 (sequence in gray) (50), does not exist in H. marismortui and eubacterial ribosomes. Positions of predicted interactions for the three-helix region are indicated according to data from ribosome crystal structures for E. coli and H. marismortui (22, 23). These include contacts with: the SSU (B1a bridge), the elbow of tRNA, 5 S rRNA, H81, and the stem of H80 (P-loop). D, the deduced three-dimensional arrangement of the 10 Ψs in the yeast H37–H39 region as derived from the crystal structure of the H. marismortui LSU. The color scheme is the same as in panel C. The dynamic distal portion of the ASF is not visualized in the crystal structure and is depicted here as a broken red line. Information about the three-dimensional visualization of the ASF and helices H37 and H39 is provided in the supplemental material.
The location of the ASF above the A-site and its contact with tRNA at this site suggests that it may have a role in tRNA movement in the ribosome, in addition to subunit joining. Consistent with this activity, point mutations in the ASF of yeast altered ribosome affinity for aminoacyl-tRNA and decoding fidelity (18). Interestingly, eubacterial ribosomes lacking the terminal portion of the ASF exhibited increases in translation activity and tRNA translocation (from the A- to P-site) in vitro (24).
Support for the prediction that the three-helix cluster H37–H39 might function in some connected fashion comes from crystal structures of the ribosome showing the helices are organized into a compact unit, with apparent couplings between helices. A more centrally located portion of the ASF, as well as the loop of H39, interacts with 5 S rRNA (21–23, 25). Both the ASF and H39 make contact with H81. Additionally, H81 interacts with the P-loop, which interacts directly with the CCA end of P-site bound tRNA. However, the loop of H37 makes contact with the stem of H80 (P-loop) (22, 26). The three-dimensional structures make clear that the ASF and adjoining helices have a close relationship to the peptidyl transferase center and tRNAs in the A- and P-binding sites (Fig. 1B, and see instruction for three-dimensional visualization in the supplemental materials). This information adds strong support for suggestions made earlier that the ASF may have overlapping roles in inter-subunit communication and tRNA accommodation and movement.
In S. cerevisiae, helices H37–H39 contain a total of 10 Ψ modifications, making it the richest region of Ψs in yeast rRNA. High frequencies of Ψ occur in the corresponding regions of other rRNAs as well, in particular other eukaryotes (27, 28). Indeed, 6 of the 10 Ψs in the yeast 3-helix region are conserved in higher eukaryotes (Fig. 1C). In yeast the Ψs are distributed as follows: six occur in the middle and lower portions of the ASF helix (the distal ∼30% is free of modifications). One Ψ occurs at the base of H38 (H39 side). H37 contains two Ψs, and H39 has one Ψ (Fig. 1C). The fact that the Ψs in the ASF occur only in the lower (proximal) two-thirds of the helix suggests that these modifications are most beneficial in this portion. The Ψs in H37, H39, and the segment of H38 below the kink-turn can be expected to affect the structures of these helices too and perhaps alter their interactions. Interestingly, the nucleotides in H37 and H39 that interact with the stem of H80 (P-loop) and H81, correspond to the Ψs conserved in eukaryotes (Fig. 1C) (22).
We tested the functional importance of the yeast modifications by blocking the formation of specific Ψs individually and in combinations and searched for effects on ribosome synthesis and activity. Modification reactions were blocked by genetically disrupting expression (or activity) of the snoRNAs that guide the modifications of interest. The results demonstrate that the Ψs in the ASF region are indeed important for ribosome function. The most substantial effects occurred with loss of multiple Ψs and the specific distribution patterns are important.
EXPERIMENTAL PROCEDURES
Yeast Strains, Plasmids, and Oligonucleotides—The initial set of 13 test strains (supplemental Fig. S1) was constructed by sequential disruption of snoRNA coding sequences for snR5, snR8, snR33, snR44, and snR81, using PCR products (29). Construction of the improved strains (Table 1) involved two types of manipulations: 1) complementing disrupted snoRNA coding units with plasmids that express the same snoRNA, but with Ψ guide elements for the ASF region specifically inactivated, and 2) restoring a protein (Rps22B) lost on disruption of a snoRNA coding unit embedded in an intron of the same mRNA, as well as a Ψ in U2 snRNA targeted by a different snoRNA (supplemental Fig. S2). In this last situation, complementation was achieved with a plasmid-borne variant of this locus in which the snoRNA coding segment (snR44) was replaced with an artificial variant that restores two Ψs that are not part of the study. One Ψ (located in 18 S rRNA) is normally specified by snR44, the other (in U2 snRNA) is specified by a guide motif in snR81 that was moved into snR44, to replace a guide motif that targets a Ψ to the region of interest (supplemental Fig. S2D). Because genetic markers were limiting in our strategy, complementations were achieved with one or two plasmids bearing two or three expression cassettes for snoRNA genes (supplemental Figs. S2, B and D). Descriptions of the plasmid constructions and genotypes of the initial set of strains and improved strains are in the supplemental material.
TABLE 1.
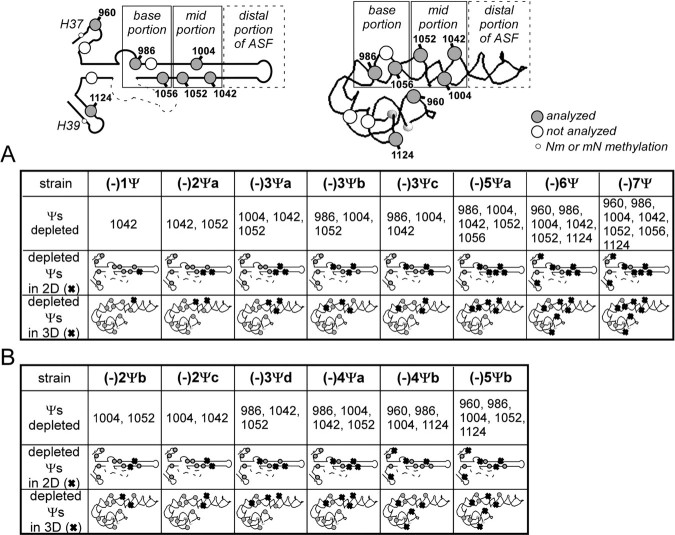
Modification patterns of Ψ-depletion strains examined The structure of the H37–H39 region is shown in two- and three-dimensional formats, with the locations of the Ψ modification highlighted. The ASF has been dissected into three portions, corresponding to base- and mid-portions that contain Ψs and a (dynamic) distal portion that lacks modifications. Modification maps are shown for a preliminary set of eight depletion strains used in these studies (A) and six additional strains used to refine and analysis (see text) (B).
General Procedures—Media and culturing methods have been described (29), as have methods for preparing yeast total RNA (30) and Northern blot analysis of snoRNAs (31). The intensity of ethidium bromide staining of RNA was quantified with Multi Gauge software using the Kodak EDAS 290 imaging system. The presence of Ψ was detected by primer extension screening of total RNA pretreated with 1-cyclohexyl-3-(morpholinoethyl)carbodiimide metho-p-sulfonate (32).
Growth Studies—Growth rate measurements were carried out in triplicate in SD medium without adenine and uracil and supplemented with 60 mg/liter each of tryptophan, histidine, and leucine. Synthetic medium SD is 0.67% yeast nitrogen base (without amino acids) and 2% glucose; supplemented here with adenine, uracil, and a complete mixture of amino acids at concentrations described elsewhere (29).
Growth Competition Assays—Competition growth studies were done with selective liquid medium, i.e. SD without adenine, uracil, and supplemented with 60 mg/liter each of tryptophan, histidine, and leucine was used. All steps were performed as reported elsewhere (7), with cell counting done after plating samples of the co-culture onto SD without adenine, uracil and SD without adenine, uracil, and leucine.
Antibiotic Sensitivity Test—Sensitivity to anisomycin, sparsomycin, and paromomycin was analyzed by a disk diffusion assay (13). Here, 20 μl of the drug (1 mg/ml anisomycin, 0.5 mg/ml sparsomycin, and 250 mg/ml paromomycin) was dispensed onto a sterile 0.25-inch paper filter disk (Whatman) on solid medium that had been seeded with the strain to be analyzed (0.3 ml of cells at A600 of 0.4 per plate, well spread). The plates were incubated at 30 °C.
In Vivo [35S]Methionine Incorporation—For analysis of translation rate, cells were grown in selective medium SD (without adenine, uracil, and methionine) at 30 °C to an A600 of 0.8. Amino acid incorporation assays were done essentially as reported elsewhere (13). For measurement of translation rate at a lower temperature, cells were cultured at 30 °C to A600 of 0.8, and then transferred for 1 h to 11 °C. Triplicate samples were analyzed for each strain at each time point, and average values for each point are shown. The analysis was repeated three times.
Sucrose Gradient Analysis—Polysome and ribosomal subunits patterns were examined as reported previously (13). When analyzing polysome patterns, cell lysates were normally prepared from cells cultured at 30 °C to A600 of 0.8. For lower (and higher) temperatures, cells were grown to A600 of 0.8 at 30 °C and then shifted to 11 °C for 1 h, or to A600 of 0.7, and then shifted to 37 °C for 45 min. Lysates were prepared in 10 mm Tris-HCl (pH 7.4), 100 mm NaCl, 30 mm MgCl2, 100 μg/ml cycloheximide, and 200 μg/ml heparin. Ribosome patterns were examined on linear 7–47% sucrose gradients with centrifugation at 39,000 rpm for 2.5 h in a Beckman SW41-Ti rotor (4 °C). To analyze the relative amounts of subunits, cell lysates were prepared in 50 mm Tris-HCl (pH 7.4), 50 mm NaCl, and 1 mm dithiothreitol, and the subunits were fractioned on linear 7–47% sucrose gradients with centrifugation at 41,000 rpm for 3 h in a Beckman SW41Ti rotor (4 °C). The polysome, monosome, and subunit patterns were determined by monitoring absorbance at 254 nm with a UV-5 detector (ISCO).
Computational Analysis of Ribosome Structure—The crystal structure of the 70 S Thermus thermophilus (PDB: 1GIY and 1GIX (21)) and 50 S subunit of the Haloarcula marismortui ribosome (PDB: 1FFK (22)) were visualized and analyzed with bioinformatics tools available from the Fournier laboratory through the University of Massachusetts-Amherst website (12), in conjunction with Protein Explorer (33). The two-dimensional and three-dimensional modification maps of human 28 S rRNA can be found at a complementary website described elsewhere (28).
RESULTS
Depletion of Ψs in the ASF Region: a Two-stage Strategy—The ten Ψs in helices H37, H38, and H39 are guided by eight different snoRNAs. Among these, four also guide Ψs to other regions of the ribosome, one guides a Ψ to U2 snRNA, and one is encoded in an intron of a gene for a ribosomal protein (Rps22B). This complexity requires more tedious depletion approaches with re-engineering of coding regions for several snoRNAs. Because of these complications our depletion analysis was carried out in two stages.
In the first stage, we sequentially disrupted coding sequences of five snoRNAs, which together target the formation of 7 of the 10 Ψs in the H37–H39 region (supplemental Fig. S1). In a second stage of analysis we created an improved strain collection that only lacked pseudouridines in the region of interest. Construction of new strains included restoring the missing ribosomal protein and the Ψs located outside of the test region, in both cases using expression plasmids. The multiguide snoRNAs were engineered to contain only the desired guide elements (supplemental Fig. S2).
Subsequent analyses focused on strains with different Ψ contents and patterns in the ASF alone or in combination with Ψ depletions from H37 and H39. Rather than attempting to examine all combinations of depletions, we sought to discover modification effects that would be informative within the confines of a practical study. Our goal was to determine if the Ψsinthe greater ASF region are, in fact, important, and if so, to obtain good insights into the importance of the content and pattern of Ψs. Fuller descriptions would be a matter for the future, when results from sensitive functional assays can be correlated with changes in ribosome structure that accompany Ψ modification.
Most of the functional results that follow were derived from eight principal test strains. Where indicated, results are included for six other strains developed in the course of the study for further refinement. The depletion patterns for the eight primary strains are shown schematically in Table 1A, and those used in the refinement analyses are depicted in Table 1B.
Both the Number and Pattern of Ψs Influence Cell Growth and Ribosome Level—We focused initially on H38, because the ASF segment is already known to be functionally important. The six Ψs in the lower two-thirds of the ASF portion are distributed somewhat evenly (Fig. 1, C and D). As noted above no modifications are present in the distal stem-loop portion. Three of the modifications, in the mid-portion of the ASF, are positionally conserved in several other organisms, including humans (Ψ1004, Ψ1042, and Ψ1052, Fig. 1C). Because of their implied importance, we evaluated the Ψs in this segment first. Two test strains were examined, one lacking the three conserved Ψs in the mid-portion of the ASF, and the other lacking these and two additional Ψs in the base-portion; the strains are designated (-)3Ψa and (-)5Ψa, respectively (Table 1A).
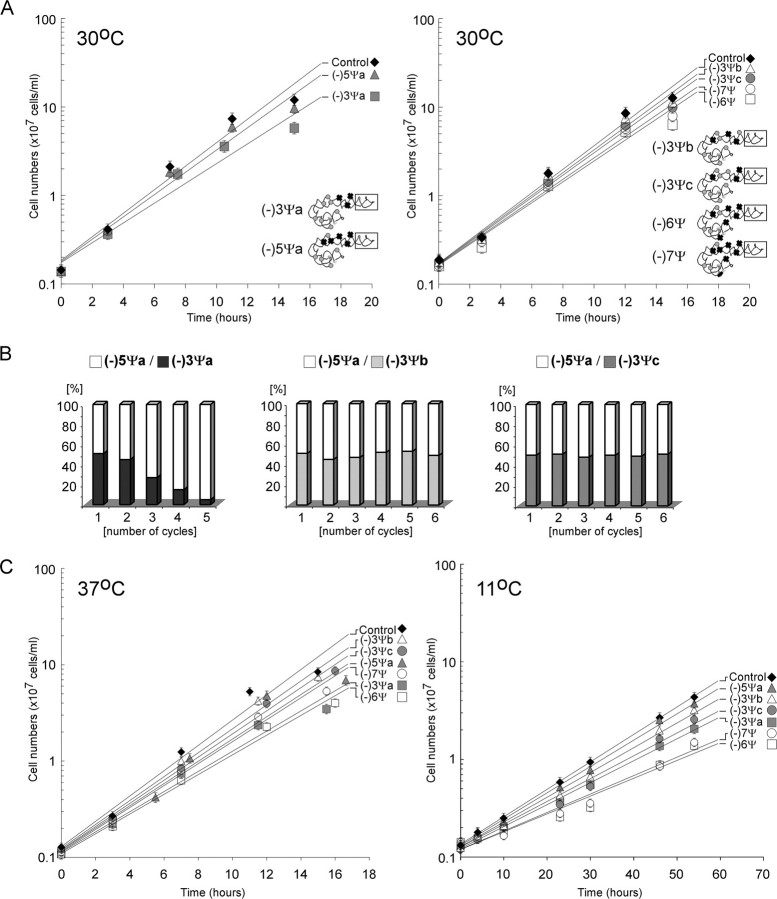
In growth rate analyses in selective liquid medium at 30 °C, which is normally optimal temperature, the strain lacking the three conserved Ψs grew slower than control cells (doubling times of ∼170 min versus ∼130 min in the exponential phase) (Fig. 2A, left panel). When two of the Ψs were restored to this strain (Ψ1004 and Ψ1052, but not Ψ1042), a growth curve similar to the control cells (Ys602 with empty marker plasmids) was obtained, i.e. strain (-)1Ψ (data not shown). Interestingly, the strain lacking 5 Ψs was less impaired for growth (generation time of ∼145 min versus ∼170 min), despite that fact that the three conserved Ψs are also missing from this strain. In a competition growth experiment, the (-)3Ψa strain was out-competed by the (-)5Ψa strain in a mixed culture (Fig. 2B, left panel). Because the growth of the (-)5Ψa cells is less impaired, the results suggest the slower growth of the (-)3Ψa cells is due to a combination of loss of the conserved Ψs and the pattern of remaining Ψs. We note that strains (-)3Ψa and (-)5Ψa were created from two different parental strains, and these unexpected results could occur as an indirect consequence of constructing multiply deleted strains.
FIGURE 2.
Growth is impaired in cells lacking multiple Ψs in the ASF and H37 and H39. A, growth rates of mutants at 30 °C in liquid medium. Left, cells missing three and five Ψs in the ASF; right, cells missing other combinations of Ψs, as follows: two other mutants lacking three Ψs from the ASF, and two mutants lacking either four or five Ψs from the ASF plus one Ψ each from H37 and H39. B, competitive growth analysis of ASF mutant (-)5Ψa mixed with ASF mutants (-)3Ψa, (-)3Ψb, and (-)3Ψc. C, exponential growth rates for different mutants at 37 °C and 11 °C in liquid medium.
To gain additional insight we also analyzed strains with other combinations of depletions. In addition to strain (-)1Ψ, which lacks only Ψ1042, we created strain (-)2Ψa lacking two Ψs (Ψ1042 and Ψ1052) in the mid-portion of the ASF, two strains (-)3Ψb and (-)3Ψc missing three Ψs in the ASF, but in different combinations than strain (-)3Ψa. Additionally, we also created two strains (-)6Ψ and (-)7Ψ lacking 6 and 7 Ψs from the three helices (described in Table 1A and see below). These new strains contain one or two plasmids expressing wild-type or mutant snoRNAs to create particular patterns of Ψ depletions. We note that (-)1Ψ and (-)2Ψa are strains in which Ψs have been restored by complementation with plasmid-encoded guide snoRNAs (see “Experimental Procedures” and supplemental material).
No effect on growth rate was seen for the ASF mutant lacking two conserved Ψs, i.e. (-)2Ψa (data not shown). However, slightly slower growth rates were observed for two strains lacking three Ψs from the ASF, in combinations that differ from the strain that lacks only the conserved Ψs, i.e. the (-)3Ψa strain. These variants lack two conserved Ψs: Ψ1004 and Ψ1052 in (-)3Ψb, and Ψ1004 and Ψ1042 in (-)3Ψc, and one, Ψ986, from the base portion of the ASF. Unlike mutant (-)3Ψa these mutants have growth rate profiles similar to that of the (-)5Ψa strain (Fig. 2A, right panel). In competition growth experiments with the (-)5Ψa strain both strains comprised about half of the mixed culture after six cycles (Fig. 2B, middle and right panels).
Interestingly, increasing the number of depletions in the three-helix region to a total of six and seven Ψs resulted in slower growth (doubling time of ∼160 min), but to a similar extent seen for the strain missing the three conserved Ψs, i.e. mutant (-)3Ψa. The depletions in the (-)6Ψ and (-)7Ψ cells include depletions of four and five Ψs in the ASF, and additional depletions in the flanking helices (Fig. 2A, right panel). Thus, extending the depletion series to the single Ψs in the neighboring helices H37 and H39 did not increase growth impairment significantly nor exceed the effect seen for the slowest growing (-)3Ψa strain. Taken together, the results suggest that the Ψs in the three-helix region are not crucial for yeast cell growth; however, certain combinations of depletions can notably reduce the growth rate. Although not all combinations of depletions have been examined, the Ψs in the ASF are important, including the conserved modifications, and the magnitude of the effects depends on the patterns of the depleted and residual modifications.
Because of the RNA stabilizing effects of Ψ we also examined the growth properties of the 8 second-stage test strains (Table 1A) at more extreme temperatures, specifically 37 °C and 11 °C; normal culturing temperature for laboratory strains of S. cerevisiae is 30 °C. No significant differences in growth rate were seen at either temperature for the (-)1Ψ and (-)2Ψa strains (data not shown). However, strains missing three and more Ψs exhibited slower growth at both temperatures relative to control cells (Fig. 2C). The latter reductions in growth rate ranged from slight to ∼40–50% for both 37 °C and 11 °C, with the greatest effect seen for the (-)6Ψ and (-)7Ψ cells, which have Ψ depletions in all three helices. Interestingly, at the lower temperature the strain missing five of the six modifications from the ASF, i.e. the (-)5Ψa strain, exhibits smaller growth impairment (∼10%). We note that the (-)5Ψa and (-)7Ψ strains were derived from a common parental strain and differ only in the content of plasmid-encoded snoRNA genes needed to achieve the desired modification patterns.
To assess whether the differences in growth rates between strains (-)5Ψa and (-)7Ψ may result from reduced levels of ribosomal subunits, we examined the steady-state content of 18 S and 25 S rRNAs in these strains, as well as for mutant (-)3Ψa, lacking only the conserved Ψs. Comparisons were made by gel electrophoresis using total RNA prepared from approximately equal numbers of cells cultured at 11 °C to A600 =∼0.8. No significant difference from control cells was evident for the (-)3Ψa and (-)5Ψa cells. However, for the (-)7Ψ cells, the steady-state levels of both 18 S and 25 S rRNAs were substantially reduced by ∼30 and ∼50%, respectively, compared with control cells at the same temperature (supplemental Fig. S3A).
Analysis of the rates of rRNA processing by pulse-chase labeling with [3H]methionine did not reveal differences for the (-)3Ψa, (-)5Ψa, and (-)7Ψ strains relative to control cells (supplemental Fig. S3B). Thus, the reduced amount of 18 S rRNA for the (-)7Ψ mutant could reflect a decrease in small subunit production caused by the deficiency in large subunits needed to form active ribosomes. The greater deficiency in 25 S rRNA could stem from defects in large subunit production and increased turnover. Defects in either or both processes could arise from Ψ-loss, because of impaired folding, assembly, or activity of the LSU. In subsequent studies we also investigated the translation activity of mutated ribosomes in lower temperature, and, indeed, this activity was reduced (see below). The reduction may result from a decrease in ribosome level revealed in polyribosome profile analysis (see below).
Taken together, the results of the growth analysis show that the cluster of Ψs in the three-helix ASF region protects the large subunit at lower temperature and presumably at higher temperature as well. Here too, both the content and pattern of Ψs are important. Loss or abnormal distributions of Ψs could influence subunit stability and function. Defects could be minor in optimal temperature conditions, but greater at lower and higher temperatures.
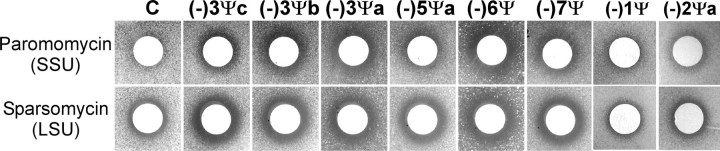
Cell Sensitivity to Ribosome-based Drugs Increases with Ψ Loss—Screening for changes in drug sensitivity was further examined with the refined strain collection (Table 1A), for paromomycin, anisomycin, and sparsomycin. The three antibiotics bind and act in different ways; paromomycin binds to the SSU, and anisomycin and sparsomycin bind to the LSU. None of the drugs is known to interact directly with the ASF region; however, changes in this region could have indirect effects on drug interference. In these tests, a filter paper disk containing drug was placed on medium seeded with a test strain, and interference was reflected as a zone of growth inhibition around the disk. The size of the zone indicates the degree of drug sensitivity.
No significant difference in growth was seen for any strain tested with anisomycin (data not shown). However, small increases in sensitivity were apparent with sparsomycin (LSU) and paromomycin (SSU) for all strains missing three and more Ψs (Fig. 3). Normal sensitivity was observed when all but one or two Ψs were restored with complementing plasmids (Fig. 3, see strains (-)1Ψ and (-)2Ψa). In liquid medium the concentration of sparsomycin, that inhibited growth by ∼50%, was ∼30 μm for all strains missing three and more Ψs, ∼35 μm for (-)2Ψa, and ∼40 μm for (-)1Ψ and control cells. For paromomycin, the inhibitory concentrations were ∼3.2 mm for (-)6Ψ, ∼4 mm for (-)3Ψa–c and (-)7Ψ cells, ∼4.9 mm for (-)5Ψ, ∼5.7 mm for (-)1Ψ and (-)2Ψa, and ∼6.5 mm for control cells.
FIGURE 3.
Growth inhibition by paromomycin and sparsomycin increases with loss of multiple Ψs from the ASF or ASF plus H37 and H39. C, control cells.
Paromomycin interacts with the decoding center in the SSU and induces translational misreading, whereas sparsomycin binds to the peptidyl transferase center, where it induces conformational changes and triggers tRNA translocation (34–36). The increased sensitivities to these drugs provide additional evidence that the mutant ribosomes are altered in both structure and activity and further imply that the function of the ASF region is tightly coupled to other (but not all) events related to the decoding process and translation (and see “Discussion”).
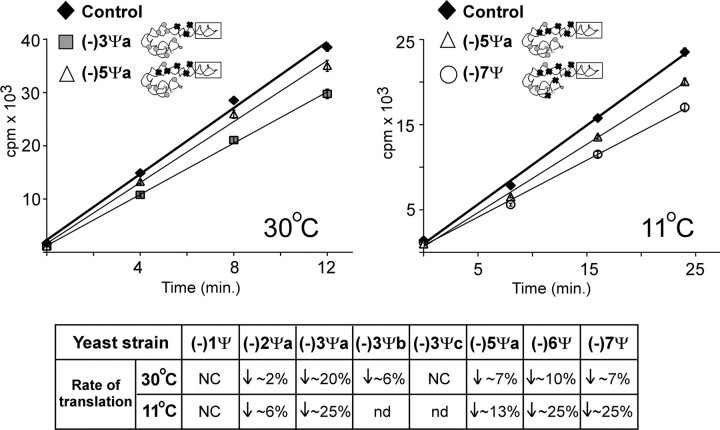
Translation Activity Is Altered in Cells Deficient in Ψs from the ASF Region—Protein synthesis activity of mutant ribosomes was examined by [35S]methionine incorporation into total protein, at 30 °C and 11 °C. Among the eight test strains (Table 1A), the (-)3Ψa mutant showed a significant reduction of ∼20% in the rate of incorporation at 30 °C; several other mutants exhibited smaller decreases of ∼2–10% at this temperature (Fig. 4). When test strains were shifted from 30 °C to 11 °C for 1 h, the rate of translation was found to be reduced relative to control cells by ∼25% for the (-)3Ψa, (-)6Ψ, and (-)7Ψ mutants, and by ∼13% for the (-)5Ψa cells. Because the cells were incubated at 11 °C for only 1 h, it seemed likely that the content of ribosomal subunits did not change significantly, and that the reduced incorporation rates indicate impairment in ribosome activity. Indeed total RNA isolated from cells cultured at 30 °C and cells incubated for 1 h at 11 °C did not reveal differences in the level of 18 S and 25 S (data not shown). Re-introducing all but one Ψ (strain (-)1Ψ) restored full activity at 11 °C; similarly, restoring all but two Ψs (strain (-)2Ψa) yielded ∼94% activity (Fig. 4).
FIGURE 4.
Loss of Ψs in the ASF impairs translation. Activity was measured by incorporation of [35S]methionine into total protein. Results are shown only for ASF mutants (-)3Ψa and (-)5Ψaat30°C(left), and strains (-)5Ψa and (-)7Ψ at 11 °C (right). Results for all mutants examined are summarized in the table below. NC, no change; downward arrow, decrease in translation rate (%); nd, not determined.
Although the effects were not fully dissected in our study, the results indicate that the conserved Ψs and others in the greater ASF region affect ribosome activity, especially at the lower temperature condition. Because the ASF is involved in forming the intersubunit bridge B1a, the decreased incorporation rates could reflect defects in subunit joining during ribosome formation.
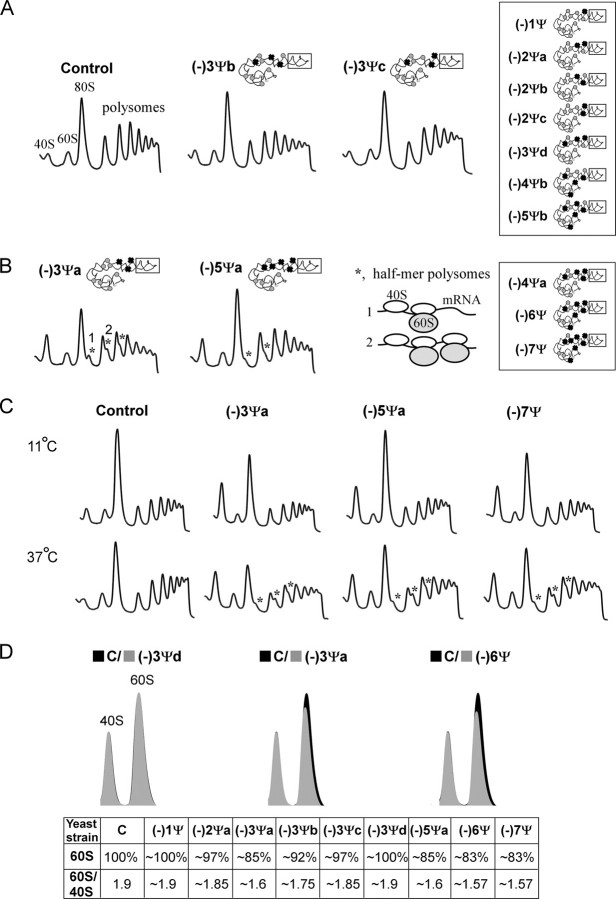
Polysome Patterns and Ribosomal Subunit Ratios Are Altered with Loss of Ψs—Polyribosome profiles were analyzed in sucrose gradients to assess the ability of the Ψ-deficient large subunits to form active ribosomes at 30 °C, 11 °C, and 37 °C (Fig. 5). Small decreases in polysome content and the presence of half-mer signals were observed at the optimal temperature condition (30 °C) for various strains lacking 3–5 Ψs from the ASF alone (Fig. 5, A and B), and strains lacking 6–7 Ψs distributed over the larger three-helix region (data not shown). When cells were incubated at 37 °C for 45 min the half-mer effect was even more pronounced. However, no half-mers were evident when strains were cultured for 1 h at 11 °C, although some strains showed a reduction in the 80 S signal (Fig. 5C).
FIGURE 5.
Loss of Ψs from the ASF region can impair polysome formation and accumulation of large subunits. The effects depend on the depletion pattern, as evident for different ASF mutants lacking 3 Ψs(A and B). Polysome defects are reflected by the appearance of half-mer polysomes and by a decrease in the level of 60 S subunits. A, normal polysome patterns at 30 °C are shown for control cells and two strains missing three Ψs in the ASF, i.e. (-)3Ψb and (-)3Ψc. Similar profiles were obtained for several other test strains (identified in-frame, right side), including: (-)1Ψ and (-)2Ψa (see supplemental Fig. S4), and (-)2Ψb, (-)2Ψc, (-)3Ψd, (-)4Ψb, and (-)5Ψb (not shown). B, half-mer polysome patterns at 30 °C. The half-mer species are identified with stars and correspond to polysomes with 80 S ribosomes plus one small subunit lacking a large subunit. Results are shown only for ASF mutants (-)3Ψa and (-)5Ψa. Other strains exhibiting this effect are (-)4Ψa, (-)6Ψ, and (-)7Ψ (identified in-frame, right side; data not shown). Elevated signals in the small subunit region are presumed to be a mixture of free small subunits (40 S) and initiation complexes (48 S). C, polysome profiles for mutants cultured at 11 °C for 1 h and at 37 °C for 45 min. The half-mer signals disappear when cells are shifted from 30 °C to the lower temperature, but increase when shifted to 37 °C. D, effects of Ψ loss on the relative abundance of subunits. Subunit ratios (60 S/40 S) were determined for cells at 30 °C. Results are shown for three mutants and summarized for six others. The relative content of 60 S subunits is compared with control cells (C, 100%).
A half-mer polyribosome consists of an mRNA bound to a single 40 S small subunit, followed downstream by one or more 80 S ribosomes. Accumulation of this intermediate reflects a defect in subunit joining during translation initiation (37). This behavior can reflect impaired interaction of the subunits, but it can also occur when the level of 60 S subunits is limiting (38–40). Indeed, we observed some deficiencies in the relative abundance of large subunits (reduced 15–17%) on examining cell extract on sucrose gradients in the absence of magnesium. Our analysis focused on the appearance of half-mers and reduction in LSU content, because these effects are more readily apparent than changes in polysome levels; the latter are less obvious and difficult to quantify.
The half-mer effect was observed for several mutant strains and encouraged us to extend our analysis to six additional mutant strains with new combinations of Ψ depletions (summarized in Table 1B). Half-mer polysomes were seen altogether in five strains at 30 °C, as follows: one of four strains lacking three Ψs in different combinations, i.e. strain (-)3Ψa that lacks only the conserved Ψs, two strains lacking 4 and 5 Ψs in the ASF, i.e. (-)4Ψa and (-)5Ψa, and strains lacking 6 and 7 Ψs distributed among all three helices, i.e. (-)6Ψ and (-)7Ψ (Fig. 5, A and B); results are shown for strains (-)3Ψa–c and (-)5Ψa.
Notably, patterns were similar to that of the control strain when Ψs were partially restored to generate complementation strains (-)1Ψ and (-)2Ψa that are lacking one (Ψ1042) and two (Ψ1042 and Ψ1052) of the three conserved Ψs in the ASF, respectively (supplemental Fig. S4). These data are consistent with our results showing that lack of the three conserved Ψsin mutant (-)3Ψa causes impaired growth and translational activity, and an increase in sensitivity to ribosome based drugs. However, lack of one or two of the conserved Ψs caused no such effect for mutants (-)1Ψ and (-)2Ψa, respectively. No half-mer effect was seen for two strains, (-)2Ψb and (-)2Ψc, lacking two of the conserved Ψs in different combinations from strain (-)2Ψa (data not shown). This situation implies that at least one of the conserved Ψs is needed for normal polysome formation. Taken together, polysome profiles without half-mer effects were observed in nine cases, for mutants (-)1Ψ, (-)2Ψa–c, (-)3Ψb–d, (-)4Ψb, and (-)5Ψb. In all these strains one or two Ψs in the mid-portion of the ASF are present, suggesting that at least one conserved Ψ is necessary in this part of the helix for polysome formation to proceed normally.
We also analyzed the polysome profiles at higher and lower temperature for four strains with half-mer signals at 30 °C, i.e. (-)3Ψa, (-)5Ψa, (-)6Ψ, and (-)7Ψ. The cells were grown at 30 °C and exposed to 37 °C and 11 °C for a short period of time, i.e. 45 min and 60 min, respectively. Interestingly, stronger half-mer signals were observed at 37 °C, however, this effect was not evident at 11 °C (Fig. 5C; not shown for mutant (-)6Ψ). Profiles similar to control cells were observed at both 11 °C and 37 °C for partially restored Ψs, i.e. strains (-)1Ψ and (-)2Ψa (supplemental Fig. S4).
The half-mer polysome profiles showed an imbalance between free 40 S and free 60 S subunits for all three temperatures, with ratios of 40 S:60 S ∼2:1 at 30 °C and 37 °C, and ∼4:1 at 11 °C. A high signal in the 40 S region could represent a mixture of free 40 S subunits and 48 S initiation complexes (small subunit-mRNA complexes with no full ribosomes present) (41). The imbalance may also result from a deficiency in large subunits. No decrease in 25 S rRNA was apparent on agarose gel analysis for strains (-)3Ψa, (-)5Ψa, and (-)7Ψ examined at 30 °C (data not shown). However, deficiencies in the relative abundance of large subunits were evident when cell extracts were fractionated on sucrose gradients in the absence of Mg2+, a condition that causes subunits to be dissociated.
Comparing the patterns to those for a control strain and normalizing to an equivalent number of cells, the data show the level of 60 S subunits was reduced by ∼15–17% in several mutants, i.e. (-)3Ψa, (-)5Ψa, (-)6Ψ, and (-)7Ψ (Fig. 5D). The ratio of 60 S/40 S dropped from 1.9 in control cells to ∼1.6 (Fig. 5D). However, when all but one or two Ψs were restored, in strains (-)1Ψ and (-)2Ψa, the level of 60 S subunits increased and normal polysome profiles were observed (supplemental Fig. S4). The profiles that did not reveal half-mer signals showed no change in large subunit content, i.e. mutants (-)1Ψ and (-)3Ψd or only a small decrease of ∼3–8%, i.e. (-)2Ψa, (-)3Ψb, and (-)3Ψc (Fig. 5D). Imbalances in subunit contents were observed at the same level as in 30 °C when strains (-)3Ψa, (-)5Ψa, (-)6Ψ, and (-)7Ψ were exposed for short periods of time to 37 °C (45 min) and 11 °C (60 min) (data not shown).
Taken together, the half-mer effect may result from an imbalance of subunits due to a shortage of 60 S subunits (15–17%). However, this defect may also reflect a delay in subunit joining during formation of full ribosomes. This latter possibility is supported by our observation that half-mer signals increase when cells are exposed to higher temperature but disappear at lower temperature, whereas no change in subunit content was observed. These differences might result from altered rates of subunit association and dissociation in lower and higher temperature. A delay in ribosome formation could be suppressed at lower temperature due to a decrease in translation activity and lower demand for ribosome formation. In addition, reduced levels of 80 S signals (∼30%) seen at 11 °C for mutants (-)3Ψa, (-)6Ψ, and (-)7Ψ, indicate that ribosome formation is impaired, but this effect was not seen for (-)5Ψa cells (Fig. 5C, not shown for (-)6Ψ). This effect is consistent with other results, where mutants (-)3Ψa, (-)6Ψ, and (-)7Ψ grew slower and exhibited a greater decrease in translational activity at 11 °C, whereas the (-)5Ψa strain was less impaired in this temperature condition.
DISCUSSION
The results of the depletions for 7 of the 10 Ψs in helices H37, H38, and H39 show that this unusual cluster substantially improves both stability of the large subunit and normal ribosome performance. The greatest effects were observed with depletions of three modifications located in the mid-portion of the ASF (Ψs 1004, 1042, and 1052). These modifications appear to be highly conserved in evolution and occur in the same positions in human and mouse 28 S rRNA. We found that at least one of these three Ψs is needed, because blocking any two did not have a significant effect. Analysis of different depletion patterns revealed that Ψ distribution is important and that the modifications benefit both stability and function of the LSU. One interesting question that lingers from our study is the possibility that imbalances in the normal or optimal Ψ pattern might be more deleterious than the loss of Ψs in this region. This was not examined as our system of genetic disruption relies on convenient marker genes and the number available is limiting. We also note that, while richness in Ψs is a hallmark of the ASF region, the corresponding structures in eubacteria T. thermophilus do not possess any Ψ modifications, whereas E. coli has only one Ψ, in H39 (28).
The observed decreases in growth rate and translation activity are caused by a reduced level of active ribosomes in the cell. This condition reflects a combination of a small decrease in the level of large subunits (∼15%) and a reduced ability of the mutated LSU to form 80 S ribosomes during initiation of translation. Increased sensitivity of the cells to ribosome-based drugs is also a strong indication of potentially deleterious changes in ribosome structure that accompany Ψ depletion. The negative effects presumably result from rRNA structure changes that deviate from an “improved” or more optimal state created by a subset or all of the modifications. Those changes, which are predicted to be minor, could be amplified at the levels of subunit stability and function, and when cells are exposed to stress such as higher or lower temperatures and the presence of antibiotics.
Where reductions in large subunit content occurred we did not observe any correlation with the number of modifications depleted, indicating that here too the effects are tied to the distribution of depleted and residual Ψs. In addition to loss of beneficial modifications altered distributions could be deleterious, for example, by impeding proper folding and assembly of the mutant LSU and consequently reducing its stability or activity. Results from a pulse-chase analysis of rRNA processing at 30 °C for mutants (-)3Ψa, (-)5Ψa, and (-)7Ψ showed no differences in the rate of rRNA processing (data not shown). Thus, the reduced level of 60 S subunits seems most likely to result from defects in the subunit itself. Faster turnover could stem from quality control processes related to ribosome synthesis or translation activity. Interestingly, it has been shown that point mutations in the mid-portion of the prokaryotic ASF caused a lethal phenotype, and this was presumed to be caused by impaired assembly of the large subunit (42).
The importance and role of the ASF in ribosome function has been examined in several studies. The helix is localized within the central protuberance, a very dynamic region of the large subunit that is associated with tRNA movement through the ribosome. The ASF itself seems to move up and down independently through the flexibility of the kink-turn in the base of this helix (15, 43). Such motion has been proposed to harmonize its dynamic relationship with the neighboring B1b bridge, above the P-site, which is connected with the ASF through 5 S rRNA (15). The clearest function of the ASF is formation of this particular intersubunit bridge with the small subunit, above the A-site for tRNAs entering the ribosome. Cryoelectron microscopy showed that, during the ratchet-like motion associated with tRNA translocation, bridge B1a moves up and down (43, 44).
It has been proposed that the ASF is involved in translocation of tRNA from the A-site to the P-site and may act as an attenuator during this process (15, 23, 24, 45). Consistent with this possibility E. coli ribosomes lacking the dynamic end of the ASF exhibit increases in translation and tRNA translocation rates in vitro, with a slight decrease in translation fidelity (17, 24). Also revealing are results from in vivo studies in yeast with mutations in the distal region of the ASF. The mutations were at sites involved in B1a bridge formation and interaction with the elbow of tRNA bound to the A-site. Effects included increased stop codon read-through and increased ribosomal affinity to aminoacyl-tRNA (18). The intersubunit bridge does not seem to be required for subunit association, but it has been suggested that it could participate in signal transmission that is important for ribosome function (17, 18, 46). In this context, interactions of the ASF with 5 S rRNA and H81 could be important couplings in such a communication system (17, 42).
The richness and density of Ψs in eukaryotic H37, H38, and H39 could be related to the dynamic nature of the central protuberance of the ribosome during its association with tRNA, because the latter moves through the ribosome. In particular, the cluster of Ψs could optimize the interactions of the various components. The Ψs present in helices H37 (Ψ960) and H39 (Ψ1124) could be directly involved in interactions with the stem of H80 (P loop) and loop of H81, as disclosed for the corresponding unmodified uridines in H. marismortui 50 S (22). Studies of modification effects on small model RNAs have shown that Ψ has a stabilizing effect on helical segments (47–49), and Ψ has been proposed to function as “molecular glue” to reduce conformational flexibility (1). We suggest this could be the main effect for Ψs in the ASF, especially those present in the mid-portion. Interestingly, the ASF in human and mouse possess an additional Ψ in this region, which corresponds to unmodified U1039 in yeast, located at the border with the dynamic end-region of the ASF (27, 28).
The location and pattern of the Ψs in the ASF suggest a possible role in modulating the motion of the ASF, where it protrudes from the LSU. Although the independent motion of the tip could be important for ASF function, its intrinsic motion in unmodified RNA might have a deleterious effect on the other parts of H38. Thus, modifications in the mid-portion of the ASF could protect this region from inefficient, possibly disruptive motion. Depleting three Ψs within the ASF had little or no effect provided at least one of the three conserved Ψs was still present. Loss of the conserved Ψs, and perhaps in combination with others might have a negative effect on ASF function, such as interaction with 5 S rRNA or H81. The less deleterious effect of removing five Ψs, in the (-)5Ψa mutant, might allow more productive motion of the ASF.
Although the ASF is involved in forming the intersubunit bridge, loss of the B1a bridge interaction in prokaryotic and yeast cells had only a minor influence on subunit joining, presumably because of the cooperative nature of this process (17, 18, 46). In the present study we interpret the appearance of half-mer polysomes in Ψ-deficient cells to reflect a possible delay in association of mutant large subunits with the SSU-mRNA initiation complex. Such a delay could result from altered dynamics of the central protuberance region and reduced ability of this region to interact with the head of the small subunit.
Finally, the three-helix unit could be part of a communication system between the decoding center in the small subunit and the peptidyl transferase center and P-sites in the large subunit. Signal transmission could involve the B1a bridge and play a key role in controlling tRNA movement through the ribosome, before and after peptide bond formation and translocation. In this context, the Ψs in the three-helix structure could be important for intersubunit signal transmission in eukaryotic ribosomes.
Supplementary Material
This work was supported by United States Public Health Services Grant GM19351 (to M. J. F.) and by Association Française contre les Myopathies Grant 12652 and Association pour la Recherche sur le Cancer Grant 3849 (to J. P. R.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1, Figs. S1–S4, text, and references.
Footnotes
The abbreviations used are: Ψ, pseudouridine; snoRNP, small nucleolar ribonucleoprotein complex; snRNA, small nuclear RNA; snoRNA, small nucleolar RNA; LSU, large subunit; ASF, A-site finger; SSU, small subunit.
References
- 1.Ofengand, J. (2002) FEBS Lett. 514 17-25 [DOI] [PubMed] [Google Scholar]
- 2.Decatur, W. A., and Fournier, M. J. (2002) Trends Biochem. Sci. 27 344-351 [DOI] [PubMed] [Google Scholar]
- 3.Tollervey, D., Lehtonen, H., Jansen, R., Kern, H., and Hurt, E. C. (1993) Cell 72 443-457 [DOI] [PubMed] [Google Scholar]
- 4.Zebarjadian, Y., King, T., Fournier, M. J., Clarke, L., and Carbon, J. (1999) Mol. Cell Biol. 19 7461-7472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conrad, J., Sun, D., Englund, N., and Ofengand, J. (1998) J. Biol. Chem. 273 18562-18566 [DOI] [PubMed] [Google Scholar]
- 6.Parker, R., Simmons, T., Shuster, E. O., Siliciano, P. G., and Guthrie, C. (1988) Mol. Cell Biol. 8 3150-3159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.King, T. H., Liu, B., McCully, R. R., and Fournier, M. J. (2003) Mol. Cell 11 425-435 [DOI] [PubMed] [Google Scholar]
- 8.Liang, X. H., Liu, Q., and Fournier, M. J. (2007) Mol. Cell 28 965-977 [DOI] [PubMed] [Google Scholar]
- 9.Kiss-Laszlo, Z., Henry, Y., Bachellerie, J. P., Caizergues-Ferrer, M., and Kiss, T. (1996) Cell 85 1077-1088 [DOI] [PubMed] [Google Scholar]
- 10.Ni, J., Tien, A. L., and Fournier, M. J. (1997) Cell 89 565-573 [DOI] [PubMed] [Google Scholar]
- 11.Ganot, P., Bortolin, M. L., and Kiss, T. (1997) Cell 89 799-809 [DOI] [PubMed] [Google Scholar]
- 12.Piekna-Przybylska, D., Decatur, W. A., and Fournier, M. J. (2007) RNA 13 305-312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Decatur, W. A., Liang, X. H., Piekna-Przybylska, D., and Fournier, M. J. (2007) Methods Enzymol. 425 283-316 [DOI] [PubMed] [Google Scholar]
- 14.Stark, H., Orlova, E. V., Rinke-Appel, J., Junke, N., Mueller, F., Rodnina, M., Wintermeyer, W., Brimacombe, R., and van Heel, M. (1997) Cell 88 19-28 [DOI] [PubMed] [Google Scholar]
- 15.Razga, F., Koca, J., Sponer, J., and Leontis, N. B. (2005) Biophys. J. 88 3466-3485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Culver, G. M., Cate, J. H., Yusupova, G. Z., Yusupov, M. M., and Noller, H. F. (1999) Science 285 2133-2136 [DOI] [PubMed] [Google Scholar]
- 17.Sergiev, P. V., Kiparisov, S. V., Burakovsky, D. E., Lesnyak, D. V., Leonov, A. A., Bogdanov, A. A., and Dontsova, O. A. (2005) J. Mol. Biol. 353 116-123 [DOI] [PubMed] [Google Scholar]
- 18.Rakauskaite, R., and Dinman, J. D. (2006) Mol. Cell Biol. 26 8992-9002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rinke-Appel, J., Junke, N., Osswald, M., and Brimacombe, R. (1995) RNA (N. Y.) 1 1018-1028 [PMC free article] [PubMed] [Google Scholar]
- 20.Osswald, M., Doring, T., and Brimacombe, R. (1995) Nucleic Acids Res. 23 4635-4641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yusupov, M. M., Yusupova, G. Z., Baucom, A., Lieberman, K., Earnest, T. N., Cate, J. H., and Noller, H. F. (2001) Science 292 883-896 [DOI] [PubMed] [Google Scholar]
- 22.Ban, N., Nissen, P., Hansen, J., Moore, P. B., and Steitz, T. A. (2000) Science 289 905-920 [DOI] [PubMed] [Google Scholar]
- 23.Schuwirth, B. S., Borovinskaya, M. A., Hau, C. W., Zhang, W., Vila-Sanjurjo, A., Holton, J. M., and Cate, J. H. (2005) Science 310 827-834 [DOI] [PubMed] [Google Scholar]
- 24.Komoda, T., Sato, N. S., Phelps, S. S., Namba, N., Joseph, S., and Suzuki, T. (2006) J. Biol. Chem. 281 32303-32309 [DOI] [PubMed] [Google Scholar]
- 25.Dokudovskaya, S., Dontsova, O., Shpanchenko, O., Bogdanov, A., and Brimacombe, R. (1996) RNA (N. Y.) 2 146-152 [PMC free article] [PubMed] [Google Scholar]
- 26.Nissen, P., Ippolito, J. A., Ban, N., Moore, P. B., and Steitz, T. A. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 4899-4903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ofengand, J., and Bakin, A. (1997) J. Mol. Biol. 266 246-268 [DOI] [PubMed] [Google Scholar]
- 28.Piekna-Przybylska, D., Decatur, W. A., and Fournier, M. J. (2008) Nucleic Acids Res. 36 D178-D183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaiser, C., Michaelis, C., and Mitchell, A. (1994) Methods in Yeast Genetics, Cold Spring Harbor Laboratory Press, Plainview, NY
- 30.Balakin, A. G., Schneider, G. S., Corbett, M. S., Ni, J., and Fournier, M. J. (1993) Nucleic Acids Res. 21 5391-5397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chanfreau, G., Rotondo, G., Legrain, P., and Jacquier, A. (1998) EMBO J. 17 3726-3737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bakin, A., and Ofengand, J. (1993) Biochemistry 32 9754-9762 [DOI] [PubMed] [Google Scholar]
- 33.Martz, E. (2002) Trends Biochem. Sci. 27 107-109 [DOI] [PubMed] [Google Scholar]
- 34.Yonath, A. (2005) Annu. Rev. Biochem. 74 649-679 [DOI] [PubMed] [Google Scholar]
- 35.Bashan, A., Agmon, I., Zarivach, R., Schluenzen, F., Harms, J., Berisio, R., Bartels, H., Franceschi, F., Auerbach, T., Hansen, H. A., Kossoy, E., Kessler, M., and Yonath, A. (2003) Mol. Cell 11 91-102 [DOI] [PubMed] [Google Scholar]
- 36.Fredrick, K., and Noller, H. F. (2003) Science 300 1159-1162 [DOI] [PubMed] [Google Scholar]
- 37.Helser, T. L., Baan, R. A., and Dahlberg, A. E. (1981) Mol. Cell Biol. 1 51-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baronas-Lowell, D. M., and Warner, J. R. (1990) Mol. Cell Biol. 10 5235-5243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petitjean, A., Bonneaud, N., and Lacroute, F. (1995) Mol. Cell Biol. 15 5071-5081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eisinger, D. P., Dick, F. A., and Trumpower, B. L. (1997) Mol. Cell Biol. 17 5136-5145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Asano, K., Shalev, A., Phan, L., Nielsen, K., Clayton, J., Valasek, L., Donahue, T. F., and Hinnebusch, A. G. (2001) EMBO J. 20 2326-2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yassin, A., and Mankin, A. S. (2007) J. Biol. Chem. 282 24329-24342 [DOI] [PubMed] [Google Scholar]
- 43.Frank, J. (2003) Biopolymers 68 223-233 [DOI] [PubMed] [Google Scholar]
- 44.Gabashvili, I. S., Agrawal, R. K., Spahn, C. M., Grassucci, R. A., Svergun, D. I., Frank, J., and Penczek, P. (2000) Cell 100 537-549 [DOI] [PubMed] [Google Scholar]
- 45.Moazed, D., and Noller, H. F. (1987) Biochimie (Paris) 69 879-884 [DOI] [PubMed] [Google Scholar]
- 46.Liiv, A., and O'Connor, M. (2006) J. Biol. Chem. 281 29850-29862 [DOI] [PubMed] [Google Scholar]
- 47.Davis, D. R. (1995) Nucleic Acids Res. 23 5020-5026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Durant, P. C., and Davis, D. R. (1999) J. Mol. Biol. 285 115-131 [DOI] [PubMed] [Google Scholar]
- 49.Sumita, M., Desaulniers, J. P., Chang, Y. C., Chui, H. M., Clos, L., 2nd, and Chow, C. S. (2005) RNA (N. Y.) 11 1420-1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gerbi, S. A. (1996) in Ribosomal RNA: Structure, Evolution, Processing, and Function in Protein Biosynthesis (Zimmermann, R. A., and Dahlberg, A. E., eds) pp. 71-87, CRC Press, Boca Raton, FL
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.