Abstract
It has been shown that transforming growth factor β1 (TGF-β1) is critical in the generation of CD4+CD25+Foxp3+-inducible regulatory T cells (iTregs) from naïve CD4+T cells. However, in contrast to natural Tregs, TGF-β1-induced iTregs rapidly lose both Foxp3 expression and suppression activity. We found that TGF-β1-induced Foxp3 levels were maintained by the addition of the anti-interleukin 4 (IL-4) antibody or by STAT6 gene deletion. Thus, IL-4 is an important suppressor of Foxp3 induction, and T helper 2 development is a major cause for the disappearance of iTreg during long culture. Using promoter analysis in EL4 cells and primary T cells, we identified a silencer region containing a STAT6 binding site. STAT6 binding to this site reduced TGF-β1-mediated Foxp3 promoter activation and chromatin modification. Retinoic acid has also been shown to suppress loss of Foxp3 induced by TGF-β1. Retinoic acid in the presence of TGF-β1 reduced STAT6 binding to the Foxp3 promoter and enhanced histone acetylation, thereby reverting the effect of IL-4. We propose that antagonistic agents for neutralizing IL-4 could be a novel strategy to facilitate inducible Treg cell generation and the promotion of tolerance in Th2-dominated diseases such as allergy.
CD4+CD25+Foxp3+ regulatory T cells (Tregs)3 are crucial for the maintenance of immunological tolerance (1, 2). Treg produced in the thymus (natural Treg (nTreg)) constitutes 3–6% of CD4+ T cells (3). More recent studies have shown that Foxp3 may also be induced in CD4+Foxp3– T cells in vivo during some immune responses or in vitro after stimulation of Foxp3– cells in the presence of TGF-β1 (inducible Treg (iTreg)) (4–6). Such a conversion has been demonstrated in vivo in thymectomized mice whose CD25– T cells in the periphery could be converted into Foxp3+CD25+ T cells by continuous low dose antigen stimulation (7). In addition, TGF-β1 has a strong potential of Foxp3 induction in CD4+CD25– naïve T cells in vitro (8). Recently, an enhancer in the Foxp3 gene in which NFAT and Smad3 bind and cooperatively induce Foxp3 expression was identified (9).
Although the differentiation, function, and survival of Treg cells are regulated by Foxp3, the details of the molecular mechanism responsible for inducing Foxp3 gene expression and its modulation are poorly understood. Various factors have been shown to modulate the generation of Foxp3+ iTregs in vitro. IL-2 is an essential factor for iTreg generation (10, 11), whereas inflammatory cytokine IL-6 suppresses iTreg but enhances Th17 generation (12). IL-4 has also been found to suppress iTreg induction (6, 13). Recently, retinoic acid (RA) has been discovered as a potent inducer and preserver of Foxp3 in iTregs (14–18). Unlike that in nTregs, Foxp3 expression in iTregs has been shown to be transient both in vitro and in vivo (8, 19). However, the molecular mechanisms for the suppression of Foxp3 induction by IL-6 and IL-4 as well as those for transient induction of Foxp3 have not been clarified.
There are two other subsets of regulatory helper T cells, Type 1 regulatory (Tr1) and Th3. Tr1 cells specifically produce IL-10 and, to a lesser extent, TGF-β, which are induced in vitro by repeated stimulation with IL-10, by immature dendritic cells, or by a combination of vitamin D3 and dexamethasone (20). Th3 cells were originally thought to be responsible for oral tolerance and to mainly produce large amounts of TGF-β (21). In vitro differentiation of Th3-type cells from Th0 precursors has been shown to be enhanced by culture with TGF-β, IL-4, IL-10, and anti-IL-12 (22, 23). Because Foxp3+T cells have been shown to produce higher levels of TGF-β than Foxp3– T cells (24) and RA has been implicated in the generation of gut-homing Foxp3+ T cells (17), Th3 may be identical to iTreg. However, no precise comparison has been made between these two types of cells.
It has been difficult to study transcriptional regulation using primary T cells because (i) the population of iTreg cells from naïve T cells is dependent on both proliferation and differentiation, (ii) the proportion of iTreg is affected by the generation of other types of helper T cells, such as Th1 and Th2, and (iii) factors from Th1 and Th2 cells modify the Foxp3 levels. We therefore generated a model system for studying the induction and maintenance of Foxp3 gene expression in the lymphoma EL4 T cell line in combination with promoter analysis in primary T cells.
We found that TGF-β1-mediated iTreg induction was transient due to gradual expansion of Th2 cells that overwhelmed iTregs. We identified the TGF-β1-responsive enhancer region and found a particular silencer region of the Foxp3 promoter containing a STAT6 binding site. In contrast, RA enhanced TGF-β1-mediated histone acetylation of this region even in the presence of IL-4. Collectively, these data strongly suggest that STAT6 is an important regulator of Foxp3 induction and Foxp3 levels determine the iTreg/Th2 balance.
EXPERIMENTAL PROCEDURES
Mice—C57B/6 mice were purchased from Clea Japan, Inc. (Tokyo, Japan). STAT6-deficient mice were from D. M. Kubo (RCAI, Yokohama, Japan). T cell-specific STAT3-deficient mice were from Dr. K. Takeda (Osaka University, Osaka, Japan). Six- to 12-week-old mice were used as experimental animals. All experiments were approved by the Animal Ethics Committee of Kyushu University.
Cell Preparation and Culture—CD4+CD25– T cells were isolated from spleens and lymph nodes by negative selection using magnetic beads (Milteny Biotech) (typically >95% purity). For differentiation, 1 × 106 CD4+CD25– T cells were cultured with the plate-bound anti-CD3 antibody (1 μg/ml) and the soluble anti-CD28 antibody (0.5 μg/ml) in the presence of 0.2 ng/ml recombinant murine IL-2 (Peprotech) and the anti-IFN-γ antibody (10 μg/ml). To induce iTreg differentiation, 2 ng/ml recombinant human TGF-β1 was added to the culture. CD4+CD25+ nTregs were purified by a cell sorter using fluorescein isothiocyanate anti-CD4 and phycoerythrin anti-CD25 from pre-isolated CD4+ T cells that were purified from mouse spleens and lympho nodes with negative selection using magnetic beads. Flow cytometric analysis and enzyme-linked immunosorbent assay were performed as described (25).
Suppression Assay—For the suppression assays, freshly isolated 1 × 105 CD4+CD25– T cells (responder) and CD25+ T cells expanded by cytokines (suppressors) were cultured with irradiated whole spleen cells (1 × 105) at indicated suppressor/responder cell ratios with the 1.0 μg/ml anti-CD3 antibody. The number of responder cells was fixed (1 × 105) and that of suppressors was varied. [3H]Thymidine was added for the last 16 h of a 72-h assay.
Construction—PCR was done to generate the Foxp3 promoter plasmid using mouse genomic DNA as a template. A 5.2-kb KpnI-SacII fragment corresponding to nucleotides from –3523 to +1627 relative to the determined transcriptional starting site of the Foxp3 gene and a 5.0-kb SacII-XhoI fragment corresponding to nucleotides from +1628 to +6668 were subcloned into the pGV-basic 2 vector (TOYOINKI) and designated as the pFoxp3 reporter. Reporter plasmids, including a series of deletion mutants of the Foxp3 promoter, were generated by PCR. Mutations in a putative STAT6 binding site (TTCCTCTAAA to TTCATGGGGC) and RA response element (AGGTCA to AAGGCA) were introduced by PCR. The subcloned PCR products were sequenced to confirm that the products were the authentic promoter fragments. pFLAG-CMV-STAT6VT (V547A/T548A) mutants were introduced by PCR. pcDNA3-human RARα and pcDNA3-human RXRα were described previously (26).
Transfection and Luciferase Assay—EL4 (4 × 105) and 293T (2 × 105) cells were seeded on 6-well plates and transfected with various amounts of expression vectors, the Foxp3 promoter plasmid, and the β-galactosidase plasmid by FuGENE 6 HD. Cells were harvested in a 30-μl lysis buffer. The luciferase assay was performed using a luciferase substrate kit (Promega), and luciferase activity was read using a Packard luminometer. The luciferase activity was normalized by the internal control β-galactosidase activity and is shown as the means ± S.D. of three experiments. For the chromatin immunoprecipitation (ChIP) assay, EL4 cells were transfected with FLAG-tagged STAT6VT by Amaxa and then incubated. After 2 h, the cells were stimulated with or without TGF-β1 and RA for 24 h. The cells were then subjected to Western blotting, to confirm the expression of STAT6VT, and to the ChIP assay.
Real-time PCR—Real-time PCR and ChIP assay were performed as described (27, 28). Primer lists are shown in supplemental Table S1.
Nuclear Extract Preparation and DNA Affinity Precipitation Assay—Nuclear extract preparation was performed as described (29). The biotinylated oligonucleotides (putative STAT6 binding site in intron I, 5′-GATCGCTTTTTTCCTCTAAACTGC-3′), was mixed with 200 μg of nuclear extract containing poly(dI·dC) (15 μg) in 500 μl of DNA precipitation buffer (20 mm Hepes-KOH, pH 7.9, 1 mm MgCl2, 80 mm KCl, 0.2 mm EDTA, 10% glycerol, 0.5 mm dithiothreitol, 0.1% Triton X-100, 1 mm phenylmethylsulfonyl fluoride, protease inhibitor mixture), and the mixture was incubated for 30 min at 4 °C. Then, 50 μl of streptavidin Dynabeads (Dynal) were added with mixing by rotation for 30 min at 4 °C. The Dynabeads were collected with magnet and washed twice with DNA precipitation buffer. The trapped proteins were analyzed by SDS-PAGE followed by immunoblotting with the anti-STAT6 Ab (Santa Cruz Biotechnology).
Statistical Analyses—The Student's paired two-tailed t test was used. Values of p < 0.05 were considered significant. All error bars shown in this article are S.E.
RESULTS
Comparison of iTregs and Th3 Generated in Vitro—We compared in vitro generated iTreg Th3. Naive CD4+CD25– T cells were isolated from mouse spleen and activated by plate-bound anti-TCR antibody, anti-IFN-γ antibody in the presence of IL-2 (Th0 condition), TGF-β + IL2 (iTreg condition), or TGF-β + IL-2 + IL-4 + IL-10 (Th3 condition). After 3 days of culture, the Foxp3 levels were compared using intracellular fluorescence-activated cell sorter. As shown in Fig. 1A, the population of Foxp3+ T cells was ∼40–70% under the iTreg condition but <15% under the Th3 condition. Suppression assay was performed to examine the regulatory activity against proliferation of naïve T cells (Fig. 1B). T cells that expanded under the iTreg condition could suppress the proliferation of naïve responder T cells. However, T cells that expanded under the Th3 condition were not anergic and suppressive, probably because of the higher contamination of CD4+CD25+ effector T cells. Because IL-4 and IL-10 were included in Th3 conditions, but not in iTreg conditions, we examined which one was responsible for the suppression of Foxp3 induction. As shown in Fig. 1C, IL-4, but not IL-10, suppressed Foxp3 levels. The effect of IL-4 was much stronger than that of IL-6. IFN-γ showed no effect on TGF-β-mediated Foxp3 induction (data not shown). These data indicate that the so-called in vitro Th3 condition, a combination of TGF-β, IL-4, and IL-10, seems not to be suitable for Foxp3 high T cell induction.
FIGURE 1.
Comparison of iTreg and Th3 in vitro. A, Foxp3 expression. Naïve T cells were cultured under the Th0/iTreg/Th3 conditions for 3 days. T cells were stained with anti-CD25 and anti-Foxp3 antibodies and analyzed by flow cytometry. B, suppression assay; freshly isolated CD4+CD25– T cells (responder) and in vitro differentiated Th0/iTreg/Th3 cells (suppressor) were cultured with irradiated whole spleen cells at the indicated suppressor/responder cell ratios in the presence of 1 μg/ml anti-CD3 Ab for 72 h. Proliferation was assessed by [3H]thymidine uptakes during the final 12 h. One representative experiment of two is shown. C, naïve T cells from WT mice were cultured with anti-TCR, anti-IFN-γ Ab, murine IL-2, and the indicated cytokines for 3 or 5 days. Foxp3 expression was analyzed by flow cytometry. One representative experiment of three is shown.
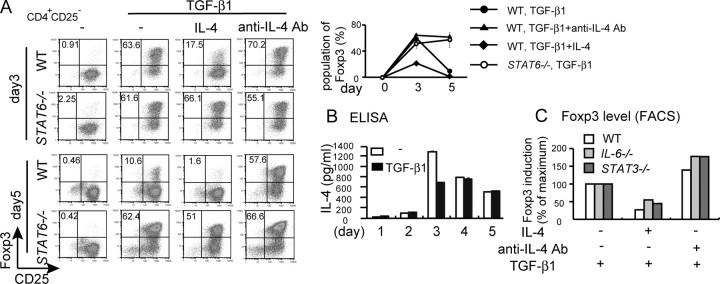
IL-4/STAT6 Suppresses TGF-β1-mediated iTreg Induction in Vitro—Upon stimulation with anti-CD3 antibody and TGF-β1, Foxp3 expression was induced in primary CD4+CD25– T cells. However, the Foxp3+ fraction reached maximum levels on day 3 and then decreased after 2 additional days of culture even in the presence of TGF-β1 and IL-2 (Fig. 2A). To examine whether endogenous IL-4 is responsible for the repression of Foxp3 levels, we investigated the effect of the anti-IL-4 antibody and STAT6 deficiency (Fig. 2A). In the presence of the anti-IL-4 antibody, the population of Foxp3+ was enhanced on day 3 and maintained thereafter (Fig. 2A). Similarly, TGF-β1-induced Foxp3 levels were higher in STAT6–/– T cells than in WT T cells, and Foxp3 was maintained after day 3 in STAT6–/– T cells (Fig. 2A). The Foxp3 levels were maintained even on day 7 by IL-4 depletion or STAT6 deficiency (data not shown). IL-4 levels in the culture supernatants were up-regulated after 3 days of culture and maintained thereafter (Fig. 2B), suggesting that IL-4 from Th2 cells overruled TGF-β1-mediated iTreg differentiation. Exogenous IL-4 reduced Foxp3 mRNA levels in iTregs but not in nTregs (supplemental Fig. S1).
FIGURE 2.
IL-4/STAT6 suppresses TGF-β1-mediated iTreg induction. A, naïve T cells from WT and STAT6–/– mice were cultured with anti-CD3 Ab and TGF-β1 in the presence or absence of IL-4 or anti-IL-4 Ab for 3 or 5 days. Foxp3 expression was analyzed by flow cytometry. The percentage of the Foxp3+ population on days 3 and 5 is shown as a graph on the right. Data indicate mean ± S.D. of triplicate samples from three mice. B, IL-4 levels in the culture of Th0 or iTreg conditions for the indicated periods. Data indicate the mean ± S.D. of triplicate cultures. ELISA, enzyme-linked immunosorbent assay. C, naïve T cells from WT, IL-6–/–, and STAT3 conditional knock-out mice were cultured under the iTreg condition in the presence or absence of IL-4 or anti-IL-4 Ab for 3 days. Foxp3 expression was analyzed by flow cytometry. Foxp3 induction was expressed as the -fold change. FACS, fluorescence-activated cell sorter.
It has been demonstrated that IL-6/STAT3 and IL-2/STAT5 are potent mechanisms for the suppression and enhancement of iTregs, respectively (10, 12). Therefore, we examined the involvement of IL-6 and IL-2 in IL-4-mediated iTreg repression. IL-4 still suppressed Foxp3 induction in IL-6–/– T cells as well as STAT3-deficient T cells from T cell-specific STAT3 conditional knock-out mice (Fig. 2C). The anti-IL-4 antibody also enhanced the Foxp3-positive fraction in IL-6- or STAT3-deficient T cells (Fig. 2C). Furthermore, a high level of exogenous IL-2 (20 ng/ml) did not overcome the suppressive effect of IL-4 (data not shown). These data suggest that the suppressive effect of IL-4 is independent of the IL-6/STAT3 pathway as well as IL-2.
IL-4 Repressed TGF-β1-mediated Chromatin Modification of the Foxp3 Promoter—We then investigated the molecular mechanism for the suppression of Foxp3+ T cell induction by IL-4. Recently, GATA3 was shown to suppress Foxp3 mRNA induction (13). However, we found that GATA3 mRNA was undetectable at 24 h of culture when Foxp3 was repressed by IL-4 (supplemental Fig. S2A). IL-4 did not affect TGF-β1-induced Smad2 phosphorylation either (supplemental Fig. S2B). IL-4 also showed no effect on Smad transcriptional activity in T cells transfected with the Smad binding element reporter (data not shown). Therefore, we hypothesized that IL-4/STAT6 directly inhibits Foxp3 transcription by modulating chromatin structure.
To examine this hypothesis, we performed ChIP assay. Various pairs of primers were designed to amplify 19 individual PCR fragments covering the entire promoter region (Fig. 3A), and the ChIP assay was performed with the antibody specific for acetylated histone H4. Recently, the TGFβ-1-enhanced element of the Foxp3 promoter was identified (9). ChIP assay for histone modification revealed that TGF-β1 induced histone H4 acetylation of this enhancer region (region 10) in primary T cells (Fig. 3A). However, IL-4 did not affect the levels of histone acetylation of this region (Fig. 3A, lower left panel). Therefore, we examined the effect of IL-4 on the chromatin remodeling of the overall promoter region (Fig. 3A, upper panel). TGF-β1 induced histone acetylation in various regions, including the STAT5 binding site (11) and the cyclic AMP response element-binding protein binding site (30) of the Foxp3 promoter. However, IL-4 also did not affect histone acetylation in these regions.
FIGURE 3.

Identification of the IL-4/STAT6-dependent silencer region of the Foxp3 promoter. A, naïve T cells were cultured under the Th0 or iTreg conditions in the absence or presence of IL-4. 48 h later, cells were subjected to the ChIP assay with anti-rabbit IgG or anti-acetylated histone H4 Ab. The histone acetylation levels were determined by real-time PCR. Each plot represents the normalized value to the input. Data are representative of at least two independent experiments with similar results. Data of the ChIP assay for PCR 10 and 11 are shown as a graph in the lower panels. B, DNA affinity precipitation assay. T cells expanded with anti-TCR Ab and IL-2 cells were restimulated with (+) or without (–) IL-4 for 1 h. Extracts of cells were incubated with the biotinylated oligonucleotide corresponding to the putative STAT6 binding site, and then the isolated DNA-protein complexes were analyzed by immunoblotting with anti-STAT6 Ab. C, EL4 cells were transfected with the empty vector or FLAG-tagged STAT6VT (an active form of STAT6). Chromatins from transfected cells were subjected to ChIP assay using anti-FLAG Ab. The binding of STAT6VT to the putative STAT6 binding site located in the Foxp3 promoter region was determined by real-time PCR using the final DNA extractions. PCR product 11 was electrophoresed and visualized in the upper panel. D, induction of Foxp3 mRNA in EL4 cells. EL4 cells cultured with plate-bound anti-CD3 Ab in the presence or absence of TGF-β1 and IL-4 for 12 h. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. E, promoter assay in EL4 cells with the Foxp3 promoter reporter constructs. After transfection, cells were stimulated with plate-bound anti-CD3 Ab and TGF-β1 for 12 h. The luciferase activities normalized by the β-galactosidase activity and expressed as the -fold change to control cultures defined as 1.0 are shown. F, effects of point mutations introduced into the putative STAT6 binding site on the Foxp3 promoter reporter activity. EL4 cells transfected with WT or mutant pFoxp3 luciferase plasmids were stimulated with TGF-β1 with or without IL-4, and luciferase activity was measured after 10 h. The luciferase activities normalized by the β-galactosidase activity are shown as the mean ± S.D. of three independent samples. One representative experiment of three independent experiments is shown.
We noticed that, in the region from +2459 to +2866 (PCR11), histone acetylation was strongly induced by TGF-β1 but blocked by IL-4 treatment (Fig. 3A, lower right panel). This region contains one putative STAT6 binding site (TTCNNNNG/AAA). To demonstrate that STAT6 can bind to this element sequence, we performed an in vitro binding assay. The nuclear extracts from IL-4-treated or -untreated T cells were incubated with a biotinylated oligonucleotide and then precipitated with streptavidin beads. We confirmed STAT6 binding to this sequence only after treatment with IL-4 (Fig. 3B). We have tried to show the binding of endogenous STAT6 to this region by ChIP assay. However, all the antibodies we tested were not suitable for ChIP assay. Then, we transfected FLAG-tagged STAT6 into EL4 T cells and immunoprecipitated with the anti-FLAG antibody. We confirmed the binding of FLAG-tagged STAT6 to this region in EL4 cells (Fig. 3C).
Suppression of Foxp3 Promoter Activation by STAT6—To investigate the effect of IL-4 on TGF-β1-mediated Foxp3 induction more directly, we first characterized the Foxp3 promoter using EL4 T cells. EL4 is a tumor cell line, but it maintains many T cell properties, and it has been used for studying TGF-β1-mediated Foxp3 transcriptional regulation (9). As shown in Fig. 3D, we first confirmed that TGF-β1 induced Foxp3 mRNA and IL-4 repressed this induction in EL4 cells. To investigate Foxp3 transcription, we cloned the 10.2-kb promoter region of the Foxp3 gene containing exons 1 to 3. The 10.2-kb fragment was fused to the luciferase expression vector and introduced into EL4 cells. The Foxp3 promoter activity was strongly enhanced by TGF-β1 and the anti-CD3 antibody, whereas a deletion mutant (+2114 to +2190) lacking two putative Smad binding elements (9) did not respond to TGF-β1 (Fig. 3E). As shown in Fig. 3F, this Foxp3 promoter-reporter was activated by TGF-β1 and repressed by IL-4. Mutations were then introduced to the putative STAT6 binding sequence, and the reporter activity was compared (Fig. 3F, upper panel). Although the TGF-β1 response was unaffected by the disruption of the STAT6 binding element, IL-4-mediated suppression was severely impaired (Fig. 3F). These data supported our notion that this region is the IL-4/STAT6-responsive silencer of the Foxp3 promoter.
Retinoic Acid Inhibited IL-4-mediated Foxp3 Repression—Our data suggest that IL-4 from Th2 cells suppressed Foxp3 induction and therefore overwhelmed iTreg. Recently, RA was shown to remarkably enhance Foxp3 expression and increase iTregs (14). Foxp3+T cells did not decrease even after 5 days of culture in the presence of RA and TGF-β1 (Fig. 4A), which was similar to those in T cells depleted with IL-4 or STAT6 (Fig. 2A). As shown in Fig. 4B, RA suppressed IL-4 production and GATA3 expression in primary T cell under iTreg conditions regardless of the presence or absence of IL-4.
FIGURE 4.
RA suppressed Th2 development in the presence of TGF-β1. A, naïve T cells were cultured under the Th0/iTreg condition with the indicated cytokines, 10 nm RA, with or without IL-4 for 3 days. The Foxp3 levels were determined by fluorescence-activated cell sorting, and the percentage of Foxp3+ cells is shown in the graph. B, effect of RA on Th2 development. Naïve T cells were cultured under the indicated conditions for 3 days, and then the cells were restimulated with plate-bound anti-CD3 Ab for 24 h. IL-4 production was determined by enzyme-linked immunosorbent assay (ELISA). IL-4 and GATA3 mRNA expression was determined by real-time PCR.
Like primary T cells, IL-4 did not reduce Foxp3 mRNA levels in the presence of RA in EL4 cells (Fig. 5A). Furthermore, IL-4-mediated suppression of the Foxp3 promoter activity was canceled by RA (Fig. 5B). To investigate the molecular mechanism, we performed ChIP assay. As shown in Fig. 5C, RA reverted the effect of IL-4 on histone deacetylation. RA enhanced histone acetylation of region 11 even in the presence of IL-4. As a result, RA prevented STAT6 binding to the intron 1 of the Foxp3 gene (Fig. 5D). To identify the RA-responsive element in the Foxp3 promoter region, we performed reporter assay in 293T cells transiently transfected with RARα and RXRα. Reporter assay using a series of deletion mutants revealed that the RA-responsive element was present between +2114 and +2350 (Fig. 5E, upper panel). This region was 300 bp upstream of the STAT6 binding site and contained one RA response element. RA response was severely impaired by the disruption of this RA response element (Fig. 5E, lower panel). These data suggest that RARα/RXRα binds to the Foxp3 promoter, thereby inducing chromatin remodeling in the region, including STAT6 binding sites.
FIGURE 5.
RA inhibited IL-4-mediated Foxp3 repression. A, EL4 cells were cultured with plate-bound anit-CD3 Ab and the indicated cytokines or RA for 12 h. The Foxp3 mRNA level was determined by real-time PCR, and the band intensity was quantified. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. B, EL4 cells transfected with the pFoxp3-luciferase plasmid were stimulated with the indicated cytokines or RA for 10 h. One representative experiment of three independent experiments is shown. C, naïve T cells were cultured under the Th0/iTreg condition and, where indicated, with or without IL-4 or RA. After 48 h, the cells were subjected to ChIP assay with anti-rabbit IgG or anti-acetylated histone H4 Ab. Histone acetylation was determined by real-time PCR. D, effect of RA on STAT6 binding to the Foxp3 promoter region. EL4 cells were transfected with the empty vector or FLAG-tagged STAT6VT. 24 h after transfection, expression of FLAG-tagged STAT6VT was detected by Western blotting. Cells were stimulated with or without TGF-β1 and RA for 12 h. Chromatins from transfected cells were subjected to ChIP assay using anti-FLAG Ab. The binding of STAT6VT to the putative STAT6 binding site located in the Foxp3 promoter region was determined by real-time PCR. E, localization of the RA response element (RARE) in the Foxp3 promoter. 293T cells were transiently transfected with plasmid containing various fragments of the Foxp3 promoter region and β-galactosidase expression vector with or without RA receptor α (RARα) and RXRα. 24 h after transfection, luciferase activity and β-galactosidase activity were measured. Luciferase activity was normalized with β-galactosidase activity. Data indicate mean ± S.D. of triplicate cultures.
DISCUSSION
In this study, we found that Foxp3+iTregs by TGF-β1 were transient and overwhelmed by Th2 after long term culture. This is because IL-4 suppressed Foxp3 expression by recruiting STAT6 to the silence region. In contrast, RA abrogated the suppressive activity of IL-4 on Foxp3 expression. STAT6 inhibits TGF-β1-mediated Foxp3 induction by a direct binding to the Foxp3 promoter, which is reverted by RA.
TGF-β1 has been shown to be an immunoregulatory cytokine and to induce Foxp3 in T cells. We showed here that Foxp3 suppresses Th2 development. Similarly, Th1 development has been shown to be suppressed by TGF-β1 (31). Induction of Foxp3 by TGF-β1 is a mechanism for immune suppression by TGF-β1, while a high level of IL-4 inhibits Foxp3+iTreg development by suppressing Foxp3 expression. This may be necessary for eliciting immune responses against foreign antigens and microbes. Therefore, Foxp3 levels regulated by TGF-β1 and IL-4 may determine either immune response or tolerance.
We identified a TGF-β1-responsive enhancer region and an IL-4-responsive silencer region of the Foxp3 promoter. TGF-β1 plus TCR stimulation induced open conformation of chromatins of the Foxp3 promoter in various regions, whereas IL-4/STAT6 induced closed chromatin conformation of the silencer region. After completion of this study, Tone et al. (9) reported the enhancer element for TGF-β1 where Smad3 binds. This is exactly the same region we identified (Fig. 3, A and E). However, we noticed that IL-4/STAT6 negatively regulates Foxp3 expression without affecting this enhancer region.
We found that IL-4 suppressed TGF-β1-induced Foxp3 expression via binding of STAT6 to the Foxp3 promoter silencer region. On the other hand, IL-4 had no effect on Foxp3 levels in CD4+CD25+ nTregs (supplemental Fig. S1), which is consistent with a recent report (32). Recently, GATA3 as well as T-bet has been implicated in the suppression of iTreg development (13). However, we noticed that IL-4 suppressed Foxp3 mRNA at 24 h of stimulation before the appearance of GATA3. Although we could not rule out the possibility that both STAT6 and GATA3 independently inhibit Foxp3 promoter activity, we propose that STAT6 directly represses Foxp3 promoter activity at the early lineage determination stage.
The mechanism of STAT6-mediated repression of the Foxp3 promoter activity has not been clarified yet. Although coactivator proteins CBP/p300 as well as p100 have been implicated in the regulation of transcription-promoting activity in all STATs (33, 34), IL-4 can concomitantly suppress the expression of a number of genes, including the κ light chain (35), FcγRI (36), IL-8, and E-selectin. Bennett et al. (37) demonstrated that IL-4 induces STAT6 binding to the promoter of the E-selectin gene in human vascular endothelial cells. This IL-4-induced STAT6 binding suppressed tumor necrosis factor α-induced expression of the E-selectin gene. STAT6 was found to compete for binding of NF-κB to a region in the E-selectin gene promoter. Similarly, STAT6 may inhibit Smad binding to the promoter region of Foxp3. Alternatively, STAT6 may directly recruit co-repressors such as histone deacetylases. However, such interactions have not been reported so far. We also found that RA re-opened the chromatin structure of the Foxp3 promoter (data not shown). The molecular mechanism by which RA induces the dissociation of STAT6 from the Foxp3 promoter remains to be investigated.
RA has been shown to suppress Th1 development while promoting Th2 development (38, 39). Thus, suppression of Th2 development by RA occurred specifically in the presence of TGF-β1. We showed that in EL4 cells, RA enhanced IL-4 production in the absence of TGF-β1 but suppressed it in the presence of TGF-β1. Foxp3 has been shown to suppress NFAT and NF-κB transcription factors, which inhibit IL-4 and IFN-γ production (32). We could not obtain any evidence showing that Foxp3 directly suppresses STAT6 activity. Therefore, Foxp3 may inhibit Th2 development by suppressing TCR-mediated signals or GATA3 activity. In addition, RA may inhibit IL-4-mediated Foxp3 repression. We found that RA also canceled the suppressive effect of IL-4 on TGF-β1-induced histone acetylation of the Foxp3 silencer region (data not shown). RA may induce dissociation of STAT6 from the silencer region, resulting in the restored expression of Foxp3. However, the molecular mechanism of such chromatin remodeling is not clear at present.
Our study provides a new understanding of the tolerance development controlled by a type-2 immune response. TGF-β1 with RA agonists could be therapeutic approaches facilitating tolerance induction, particularly in Th2-mediated diseases such as allergy.
Supplementary Material
Acknowledgments
We thank T. Yoshioka, S. Sasaki, N. Kinoshita, and M. Ohstu for technical assistance and Y. Nishi for manuscript preparation.
This work was supported by special grants-in-aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation, the Takeda Science Foundation, the Clinical Research Foundation, the Kato Memorial Foundation, Suzuken Memorial Foundation, the Naito Foundation, the Nakatomi Foundation, the Yakuruto Bioscience Foundation, Japan Intractable Disease Research Foundation, the Mitsubishi Pharama Research Foundation, and the Princess Takamatsu Cancer Research Fund. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
Footnotes
The abbreviations used are: Treg, regulatory T cell; nTreg, natural Treg; iTreg, inducible Treg; TGF, transforming growth factor; IL, interleukin; RA, retinoic acid; IFN, interferon; RAR, RA receptor; RXR, retinoid X receptor; ChIP, chromatin immunoprecipitation; Ab, antibody; WT, wild type; Th, T helper; TCR, T cell receptor.
References
- 1.Schwartz, R. H. (2005) Nat. Immunol. 6 327–330 [DOI] [PubMed] [Google Scholar]
- 2.Hori, S., Nomura, T., and Sakaguchi, S. (2003) Science 299 1057–106112522256 [Google Scholar]
- 3.Nelson, B. H. (2004) J. Immunol. 172 3983–3988 [DOI] [PubMed] [Google Scholar]
- 4.Zheng, S. G., Wang, J. H., Gray, J. D., Soucier, H., and Horwitz, D. A. (2004) J. Immunol. 172 5213–5221 [DOI] [PubMed] [Google Scholar]
- 5.Chen, W., Jin, W., Hardegen, N., Lei, K. J., Li, L., Marinos, N., McGrady, G., and Wahl, S. M. (2003) J. Exp. Med. 198 1875–1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fu, S., Zhang, N., Yopp, A. C., Chen, D., Mao, M., Zhang, H., Ding, Y., and Bromberg, J. S. (2004) Am. J. Transplant. 4 1614–1627 [DOI] [PubMed] [Google Scholar]
- 7.Apostolou, I., and von Boehmer, H. (2004) J. Exp. Med. 199 1401–1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Selvaraj, R. K., and Geiger, T. L. (2007) J. Immunol. 178 7667–7677 [DOI] [PubMed] [Google Scholar]
- 9.Tone, Y., Furuuchi, K., Kojima, Y., Tykocinski, M. L., Greene, M. I., and Tone, M. (2008) Nat. Immunol. 9 194–202 [DOI] [PubMed] [Google Scholar]
- 10.Davidson, T. S., DiPaolo, R. J., Andersson, J., and Shevach, E. M. (2007) J. Immunol. 178 4022–4026 [DOI] [PubMed] [Google Scholar]
- 11.Yao, Z., Kanno, Y., Kerenyi, M., Stephens, G., Durant, L., Watford, W. T., Laurence, A., Robinson, G. W., Shevach, E. M., Moriggl, R., Hennighausen, L., Wu, C., and O'Shea, J. J. (2007) Blood 109 4368–4375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bettelli, E., Carrier, Y., Gao, W., Korn, T., Strom, T. B., Oukka, M., Weiner, H. L., and Kuchroo, V. K. (2006) Nature 441 235–238 [DOI] [PubMed] [Google Scholar]
- 13.Wei, J., Duramad, O., Perng, O. A., Reiner, S. L., Liu, Y.-J., and Qin, F. X.-F. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 18169–18174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mucida, D., Park, Y., Kim, G., Turovskaya, O., Scott, I., Kronenberg, M., and Cheroutre, H. (2007) Science 317 256–260 [DOI] [PubMed] [Google Scholar]
- 15.Coombes, J. L., Siddiqui, K. R., Arancibia-Carcamo, C. V., Hall, J., Sun, C. M., Belkaid, Y., and Powrie, F. (2007) J. Exp. Med. 204 1757–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun, C. M., Hall, J. A., Blank, R. B., Bouladoux, N., Oukka, M., Mora, J. R., and Belkaid, Y. (2007) J. Exp. Med. 204 1775–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benson, M. J., Pino-Lagos, K., Rosemblatt, M., and Noelle, R. J. (2007) J. Exp. Med. 204 1765–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schambach, F., Schupp, M., Lazar, M. A., and Reiner, S. L. (2007) Eur. J. Immunol. 37 2396–2399 [DOI] [PubMed] [Google Scholar]
- 19.Floess, S., Freyer, J., Siewert, C., Baron, U., Olek, S., Polansky, J., Schlawe, K., Chang, H. D., Bopp, T., Schmitt, E., Klein-Hessling, S., Serfling, E., Hamann, A., and Huehn, J. (2007) PLoS Biol. 5 e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roncarolo, M. G., Gregori, S., Battaglia, M., Bacchetta, R., Fleischhauer, K., and Levings, M. K. (2006) Immunol. Rev. 212 28–50 [DOI] [PubMed] [Google Scholar]
- 21.Chen, Y., Inobe, J., Kuchroo, V. K., Baron, J. L., Janeway, C. A., Jr., and Weiner, H. L. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 388–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seder, R. A., Marth, T., Sieve, M. C., Strober, W., Letterio, J. J., Roberts, A. B., and Kelsall, B. (1998) J. Immunol. 160 5719–5728 [PubMed] [Google Scholar]
- 23.Inobe, J., Slavin, A. J., Komagata, Y., Chen, Y., Liu, L., and Weiner, H. L. (1998) Eur. J. Immunol. 28 2780–2790 [DOI] [PubMed] [Google Scholar]
- 24.Nakamura, K., Kitani, A., and Strober, W. (2001) J. Exp. Med. 194 629–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsumura, Y., Kobayashi, T., Ichiyama, K., Yoshida, R., Hashimoto, M., Takimoto, T., Tanaka, K., Chinen, T., Shichita, T., Wyss-Coray, T., Sato, K., and Yoshimura, A. (2007) J. Immunol. 179 2170–2179 [DOI] [PubMed] [Google Scholar]
- 26.Takeyama, K., Kojima, R., Ohashi, R., Sato, T., Mano, H., Masushige, S., and Kato, S. (1996) Biochem. Biophys. Res. Commun. 222 395–400 [DOI] [PubMed] [Google Scholar]
- 27.Taniguchi, K., Kohno, R., Ayada, T., Kato, R., Ichiyama, K., Morisada, T., Oike, Y., Yonemitsu, Y., Maehara, Y., and Yoshimura, A. (2007) Mol. Cell. Biol. 27 4541–4550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kinjyo, I., Inoue, H., Hamano, S., Fukuyama, S., Yoshimura, T., Koga, K., Takaki, H., Himeno, K., Takaesu, G., Kobayashi, T., and Yoshimura, A. (2006) J. Exp. Med. 203 1021–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joo, A., Aburatani, H., Morii, E., Iba, H., and Yoshimura, A. (2004) Oncogene 23 726–734 [DOI] [PubMed] [Google Scholar]
- 30.Kim, H. P., and Leonard, W. J. (2007) J. Exp. Med. 204 1543–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park, I. K., Shultz, L. D., Letterio, J. J., and Gorham, J. D. (2005) J. Immunol. 175 5666–5674 [DOI] [PubMed] [Google Scholar]
- 32.Baron, U., Floess, S., Wieczorek, G., Baumann, K., Grutzkau, A., Dong, J., Thiel, A., Boeld, T. J., Hoffmann, P., Edinger, M., Turbachova, I., Hamann, A., Olek, S., and Huehn, J. (2007) Eur. J. Immunol. 37 2378–2389 [DOI] [PubMed] [Google Scholar]
- 33.Wang, R., Cherukuri, P., and Luo, J. (2005) J. Biol. Chem. 280 11528–11534 [DOI] [PubMed] [Google Scholar]
- 34.Hiroi, M., and Ohmori, Y. (2003) J. Biol. Chem. 278 651–660 [DOI] [PubMed] [Google Scholar]
- 35.Clarke, C. J., Taylor-Fishwick, D. A., Hales, A., Chernajovsky, Y., Sugamura, K., Feldmann, M., and Foxwell, B. M. (1995) Eur. J. Immunol. 25 2961–2966 [DOI] [PubMed] [Google Scholar]
- 36.te Velde, A. A., Huijbens, R. J., de Vries, J. E., and Figdor, C. G. (1990) J. Immunol. 144 3046–3051 [PubMed] [Google Scholar]
- 37.Bennett, B. L., Cruz, R., Lacson, R. G., and Manning, A. M. (1997) J. Biol. Chem. 272 10212–10219 [DOI] [PubMed] [Google Scholar]
- 38.Du, X., Tabeta, K., Mann, N., Crozat, K., Mudd, S., and Beutler, B. (2005) Eur. J. Immunol. 35 3414–3423 [DOI] [PubMed] [Google Scholar]
- 39.Nozaki, Y., Tamaki, C., Yamagata, T., Sugiyama, M., Ikoma, S., Kinoshita, K., and Funauchi, M. (2006) Rheumatology Int. 26 810–817 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.