Abstract
Mulberry latex contains extremely high concentrations of alkaloidal sugar mimic glycosidase inhibitors, such as 1,4-dideoxy-1,4-imino-d-arabinitol (d-AB1) and 1-deoxynojirimycin (DNJ). Although these compounds do not harm the silkworm, Bombyx mori, a mulberry specialist, they are highly toxic to insects that do not normally feed on mulberry leaves. d-AB1 and DNJ are strong inhibitors of α-glucosidases (EC 3.2.1.20); however, they do not affect the activity ofβ-fructofuranosidases (EC 3.2.1.26). Althoughα-glucosidase genes are found in a wide range of organisms, β-fructofuranosidase genes have not been identified in any animals so far. In this study, we report the identification and characterization of β-fructofuranosidase genes (BmSuc1 and BmSuc2) from B. mori. The BmSuc1 gene was highly expressed in the midgut and silk gland, whereas the expression of BmSuc2 gene was not detected. BmSuc1 encodes a functional β-fructofuranosidase, whose enzymatic activity was not inhibited by DNJ or d-AB1. We also showed that BmSUC1 protein localized within the midgut goblet cell cavities. Collectively, our data clearly demonstrated that BmSuc1 serves as a sugar-digesting enzyme in the silkworm physiology. This anomalous presence of the β-fructofuranosidase gene in the B. mori genome may partly explain why the silkworm can circumvent the mulberry's defense system.
Certain plants have evolved defense mechanisms against insect predation through the production of defense compounds (1–3). This is reflected by the limited number of insects capable of feeding on such plants. In particular, plant latex often contains a wide variety of toxic compounds, such as alkaloids and proteases, which play an important role in a plant's defense mechanism against insect herbivory (2, 4–6).
Mulberry latex contains extremely high concentrations of alkaloidal sugar mimic glycosidase inhibitors, such as 1,4-dideoxy-1,4-imino-d-arabinitol (d-AB1),2 1-deoxynojirimycin (DNJ), and 1,4-dideoxy-1,4-imino-d-ribitol (2, 7). These sugar mimic alkaloids are not toxic to larvae of the silkworm Bombyx mori (family Bombycidae), which feed only on mulberry leaves and have been reared on them for thousands of years (2). However, these compounds are highly toxic to other caterpillars, such as the eri-silkworm Samia cynthia ricini (family Saturniidae) and cabbage moth Mamestra brassicae (family Noctuidae), for which mulberry trees are not the host plant in natural conditions (2). This indicates that the silkworm has evolved an unknown mechanism to circumvent the toxic effects of such sugar mimic alkaloids, thus enabling it to feed and grow well on mulberry leaves (2, 8).
Sucrases are digestive enzymes that hydrolyze α-glucosyl (α-glucosidase, EC 3.2.1.20) or β-fructosyl residue (β-fructofuranosidase, EC 3.2.1.26) of the substrate. d-AB1 and DNJ are strong inhibitors of α-glucosidases; however, they do not exhibit inhibitory activity against β-fructofuranosidases (7). Although α-glucosidases are found in many types of organisms, including bacteria, fungi, plants, and animals, it has been generally assumed that β-fructofuranosidases do not exist in animals (9). However, several reports have suggested that β-fructofuranosidases are present anomalously in certain species of insects (10–13), including the silkworm. In the silkworm, sucrases in larval midgut tissue showed a kinetic property that is characteristic of β-fructofuranosidase (11, 12), and more importantly, silkworm sucrases were shown to be less affected by sugar mimic alkaloids than those of other organisms (7, 8).
Although there has been no direct experimental evidence that the β-fructofuranosidase gene is encoded in the genome of animals, these observations lead us to hypothesize that the β-fructofuranosidase gene is encoded in the silkworm genome and could play an important role in the silkworm's ability to feed on mulberry leaves. By using β-fructofuranosidases as digestive enzymes, the silkworm could possibly avoid the toxic effects of d-AB1 and DNJ that are present in extremely high concentrations in the mulberry latex. As the first step toward understanding the mechanism by which the silkworm circumvents the mulberry's defense system, we have cloned and functionally characterized the β-fructofuranosidase genes from the silkworm, B. mori.
We successfully isolated two β-fructofuranosidase genes from the silkworm. This is the first report of isolation of β-fructofuranosidase genes from an animal species. One of them (BmSuc1) appears to encode a functional enzyme, whereas the expression of the other (BmSuc2) was not detected. We then expressed the recombinant BmSUC1 protein and investigated its enzymatic properties. We further examined the expression and localization of BmSUC1 protein. Our data clearly indicated that the functional β-fructofuranosidase gene is actually encoded in the silkworm genome, and the product serves as a digestive enzyme in the silkworm.
EXPERIMENTAL PROCEDURES
Insects and Cell Line—The B. mori strains p50T and N4 were reared on an artificial diet in a conditioned insect rearing room (25 °C, 12-h light/12-h dark photoperiod) (14). The Spodoptera frugiperda cell line (Sf9) was maintained in TC-100 medium containing 10% fetal bovine serum as described previously (15).
Cloning of β-Fructofuranosidase Genes—Expressed sequence tags (ESTs) (16) and genome sequences (17, 18) showing homology to β-fructofuranosidase genes were investigated using the BLAST program. By sequencing and assembling them, we obtained two nonredundant genes (BmSuc1 and BmSuc2) that putatively encode for full-length β-fructofuranosidases.
Genomic DNA Blot and RNA Blot Analysis—Genomic DNA blot and RNA blot experiments were performed as described previously (14). Briefly, 5 μg of genomic DNA fully digested with restriction enzymes or total RNA was electrophoresed and transferred onto a nylon membrane. Labeling, hybridization, and signal detection were performed using the digoxigenin (DIG) labeling and detection system (Roche Applied Science) following the supplier's instructions. DNA probes were amplified by PCR using the EST clone mg-0575 (for BmSuc1) (16) or genomic DNA (for BmSuc2) as templates. Primers used for PCR are listed in Table 1.
TABLE 1.
List of primers
| Primer | Sequence (5′ to 3′) | Purpose |
|---|---|---|
| BmSuc1S-F | AATCCAGTCCTCTCCTACGTGC | Probe synthesis for BmSuc1 |
| BmSuc1S-R | TCCGGTCTGATACGTGTTCTTG | |
| BmSuc2S-F | ACGTGCAACTGTGACTCTCCTG | Probe synthesis for BmSuc2 |
| BmSuc2S-R | CTGATGCCTCCTGTTAGGGAAG | |
| pBacSUC1-F | CCTAGGCCTAGTTATGTTCGCCTGGAGCACACCGGTG | Construction of His-tagged BmSUC1 |
| pBacSUC1-R | CCTAGGCCTTCAATGGTGATGGTGATGATGACCAGCGGGTACACTTCTTCTCAATCG | |
| BmSuc1RT-F | GGGCTGGAATTCTTTACGACC | RT-PCR of BmSuc1 |
| BmSuc1RT-R | GCTGGATGAATGACCCTAACG |
Phylogenetic Analysis—Sequence homologies were analyzed by the BLAST program for public DNA/protein data bases. The amino acid sequences were aligned with the ClustalX program (19), and phylogenetic trees were constructed by neighbor-joining methods using the MEGA3 program (20) as previously described (15).
Expression and Purification of BmSUC1—The recombinant Autographa californica nucleopolyhedrovirus (AcMNPV) was constructed using a Bac-to-Bac system (Invitrogen). The coding region of BmSuc1 with a His tag sequence at the C terminus was PCR-amplified with primers pBacSUC1-F and pBacSUC1-R (Table 1), using the EST clone mg-0757 as a template. The PCR product was cloned into the cloning site (StuI) of the pFastBac1 vector (Invitrogen). Virus propagation and infection were performed as described previously (15). Monolayers of Sf9 cells cultured in a 150-mm dish were infected with the virus at a multiplicity of infection of 5. After 72 h, the culture medium was centrifuged at 20,000 × g for 5 min, and the supernatant was collected. Protein purification was performed using nickel chromatography. The medium was then concentrated from 300 to 20 ml using Centriprep YM-30 (Amicon), dialyzed against a binding buffer (20 mm sodium phosphate, pH 7.4, 500 mm NaCl, and 10 mm imidazole) overnight at 4 °C, and loaded onto a HiTrap column (Amersham Biosciences). After washing with 5 ml of a wash buffer (20 mm sodium phosphate, pH 7.4, 500 mm NaCl, and 40 mm imidazole), elution was performed using an elution buffer (20 mm sodium phosphate, pH 7.4, 500 mm NaCl, and 200 mm imidazole). The eluate was dialyzed against a 20 mm sodium phosphate buffer (pH 7.4) and stored at 4 °C or -80 °C until use.
Enzyme Assay—Enzyme activity was assayed by two methods using glucose as a standard substrate. Glucose released in the reaction was detected by the glucose oxidase-peroxidase method, and the reducing sugars were determined by the Somogyi-Nelson method. A 100-μl reaction containing 1 μg of purified protein, 100 mm substrate, and 10 mm sodium phosphate buffer (pH 7.0), was incubated for 1–20 min at 30 °C. After incubation, the reaction was stopped by boiling for 5 min. To measure the glucose liberated, a glucose test kit (Wako) was used as indicated by the supplier's protocol. The reducing sugars that were liberated during the reaction were estimated by the Somogyi-Nelson method. To establish the effect of pH on the sucrose hydrolytic activity, a 100-μl reaction containing 1 μg of purified protein, 100 mm sucrose, and 20 mm Britton-Robinson's wide range buffer (pH 4.0–11.0) was incubated for 1–20 min at 30 °C. The glucose liberated was measured using a glucose test kit (Wako). To see the effect of DNJ, an inhibitor of α-glucosidase, the sucrose hydrolytic activity was determined by a procedure identical to that described above except for the addition of DNJ (Sigma) to the reaction. For comparison, α-glucosidase from a bacterium Bacillus stearothermophilus (Sigma) was also assayed.
RT-PCR Analysis—Total RNA from the tissues was extracted using TRIZOL reagent (Invitrogen). cDNAs were obtained using the Superscript II reverse transcriptase (Invitrogen), following the manufacturer's instructions. PCR conditions were as follows: 94 °C for 2 min followed by 30 cycles of 94 °C for 30 s; 55 °C for 30 s; and 72 °C for 1 min. Primers used for PCR are listed in Table 1. PCR products were analyzed on a 1.0% agarose gel.
SDS-PAGE and Immunoblot Analysis—SDS-PAGE and immunoblot were performed as described previously (15). An antiserum against recombinant BmSUC1 was raised in rabbits (Operon Biotechnology). Protein samples were extracted from B. mori tissues as follows. Frozen tissues were homogenized in a 10 mm phosphate buffer (pH 7.0) containing the Complete protein inhibitor mixture (Roche Applied Science) using liquid nitrogen and centrifuged at 20,000 × g for 10 min at 4 °C, and the supernatants were collected. The primary antibody was the penta-His antibody (Qiagen) at a dilution of 1:2,000 or the anti-BmSUC1 antiserum at a dilution of 1:4,000. Signals were detected with the Immobilon Western Kit (Millipore) using the LAS1000 Plus imaging system (Fuji Film).
Immunohistochemistry—Immunohistochemistry was carried out as described previously (15). Sections of the middle part of the midgut were obtained from 3-day-old fifth instar of strain p50T. Sections of silk glands were obtained from the same developmental stage of strain N4, since this strain expresses carotenoid-binding protein (CBP) (21), which was used for control experiments. The primary antibody was anti-BmSUC1 at a dilution of 1:200, and the secondary antibody was an AlexaFluor488- or AlexaFluor546-labeled goat anti-rabbit IgG F(ab)2 fragment (Invitrogen) at a dilution of 1:200. The slides were counterstained with a 4′,6-diamidino-2-phenylindole dihydrochloride solution (DAPI; Wako) or AlexaFluor594-conjugated phalloidin (Molecular Probes, Inc., Eugene, OR). The fluorescence was observed under a fluorescence microscope (Olympus BX51) and photographed. For control experiments, antisera against soluble and membrane-bound alkaline phosphatase of B. mori (sALP and mALP) (22), and CBP (21) were also used at a dilution of 1:200, following the procedure described above.
RESULTS
Cloning of β-Fructofuranosidase Genes from B. mori—We performed homology searches against the whole genome data of B. mori and found two genes that show significant homology to bacterial β-fructofuranosidase (BmSuc1 and BmSuc2) (Fig. 1). BmSuc1 was found also in the silkworm EST data base (e.g. clone mg-0575) (16), whereas BmSuc2 was not.
FIGURE 1.

Amino acid sequences of BmSUC1 and BmSUC2. Amino acid sequences were compared with the sequence of β-fructofuranosidase from the bacterium B. licheniformis ATCC 14580 (GenBank™ accession number AAU25612). Sequences were aligned with the ClustalX program (19). Using the Boxshade program, identical and similar residues, in more than two sequences, have been highlighted in black and gray, respectively. Putative signal peptides are boxed, and the active sites of the yeast invertase (23) are underlined.
The BmSuc1 gene contained an open reading frame of 1464 bp encoding 488 amino acid residues with a predicted molecular mass of 55.9 kDa. The BmSuc2 gene contained an open reading frame of 1518 bp encoding 506 amino acid residues with a predicted molecular mass of 58.0 kDa. There was no intron in the coding region of either of the genes.
These genes were predicted to encode proteins that exhibited high homology to bacterial β-fructofuranosidases (Fig. 1). BmSUC1 and BmSUC2 showed 45 and 39% identity (in amino acids) to Bacillus licheniformis β-fructofuranosidase (GenBank™ accession number AAU25612), respectively. The active site of invertase (Asp-23 in yeast invertase) (23) was conserved in BmSUC1 (Asp-63) but not in BmSUC2 (His-54) (Fig. 1), suggesting that BmSUC2 might not possess β-fructofuranosidase activity. The N-terminal sequence of both predicted polypeptides consisted of a putative signal peptide that contained hydrophobic residues. Signal P prediction (24) positioned the putative cleavage site between residues 21 and 22 for BmSUC1 and between residues 18 and 19 for BmSUC2 (Fig. 1).
Genomic DNA Blot and Phylogenetic Analysis—To eliminate the possibility that β-fructofuranosidase genes were obtained because of the contamination of the microbial DNA or RNA, genomic DNA blot analysis was performed (data not shown). Genomic DNA of the B. mori p50T strain was digested with selected restriction enzymes, blotted onto a nylon membrane, and then probed with DIG-labeled PCR products of either BmSuc1 or BmSuc2. A single band was detected in each case, and the molecular size of the signal was different between them, suggesting that both BmSuc1 and BmSuc2 are single copy genes located on the silkworm genome (data not shown).
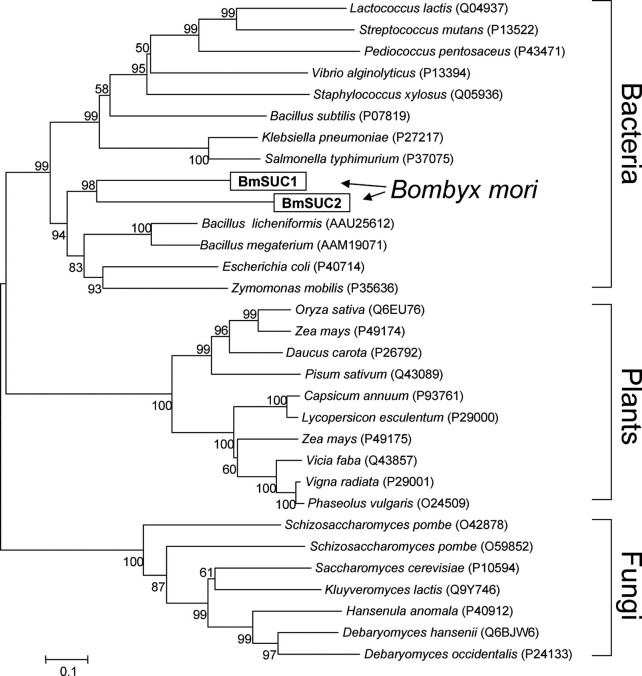
To investigate the evolutionary relationship between the BmSuc1, BmSuc2, and other β-fructofuranosidase genes, phylogenetic analysis was performed (Fig. 2). We searched proteins that show homology to BmSUC1 and BmSUC2 from public data bases with the BLAST program. The BLAST search retrieved many bacterial, fungal, and plant sucrases with high sequence homology. Their amino acid sequences were aligned with the ClustalX program (19), and a phylogenetic tree was constructed with the neighbor-joining method (Fig. 2). BmSUC1 and BmSUC2 were shown to form a monophyletic clade, suggesting that they may have been derived from a gene duplication event in an ancestral insect. Interestingly, these two genes were shown to belong to bacterial lineages. This may suggest a horizontal gene transfer from bacteria to an ancestral insect, although the phylogeny showed a deep division between bacterial and silkworm β-fructofuranosidases.
FIGURE 2.
Phylogenetic analysis of β-fructofuranosidases. Amino acid sequences of β-fructofuranosidases were aligned by the ClustalX program, and the phylogenetic tree was constructed by the neighbor-joining method using the MEGA3 program package (20). Bootstrap values after 1,000 replications are shown. GenBank™ or SwissProt accession numbers of sequences are shown in parentheses.
Expression and Purification of Recombinant BmSUC1 Protein—To characterize the enzymatic property, recombinant His-tagged BmSUC1 protein was expressed using a baculovirus expression system (Fig. 3). Since the expression of BmSuc2 was not detected in our RT-PCR analysis, only BmSuc1 was analyzed further.
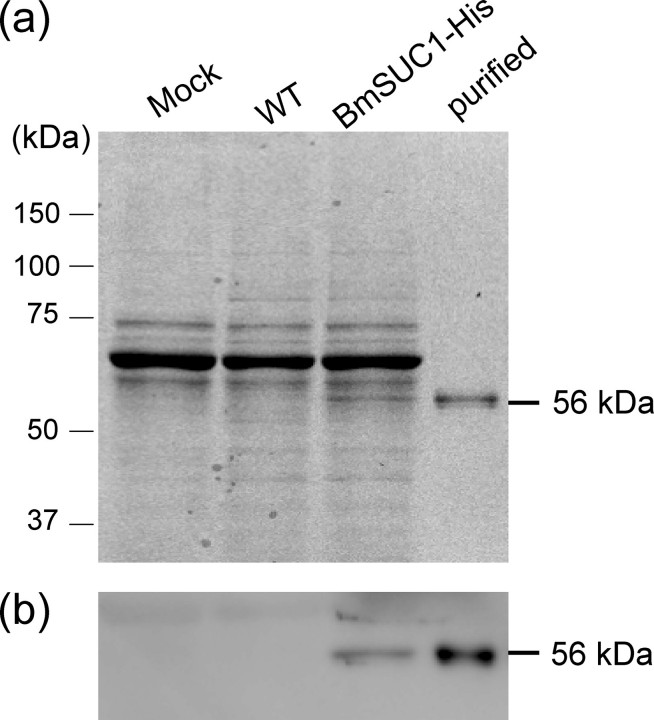
FIGURE 3.
Expression and purification of the His-tagged BmSUC1 protein. Recombinant BmSUC1 protein with the His tag at the C terminus was expressed using a baculovirus expression system. BmSUC1 was purified from the medium of virus-infected cells by nickel chromatography. Protein samples were analyzed by SDS-PAGE and stained with CBB (a), and the same samples were analyzed by immunoblot with the anti-His antibody (b). The molecular masses of the protein standards are shown on the left, and the estimated molecular mass of BmSUC1 is shown on the right. The following are the protein samples used. Mock, the medium from mock-infected cells; WT, the medium from parental AcMNPV-infected cells; BmSUC1-His, the medium from cells infected with the recombinant AcMNPV that expressed BmSUC1 protein; purified, the purified BmSUC1 protein.
The BmSUC1 protein of ∼56 kDa, which was consistent with the predicted molecular mass, was purified by nickel-chelating chromatography from the culture medium of virus-infected cells (Fig. 3a). The presence of the His tag sequence at the C terminus was confirmed by immunoblot analysis (Fig. 3b). The purified protein was used for further examination of the enzymatic properties.
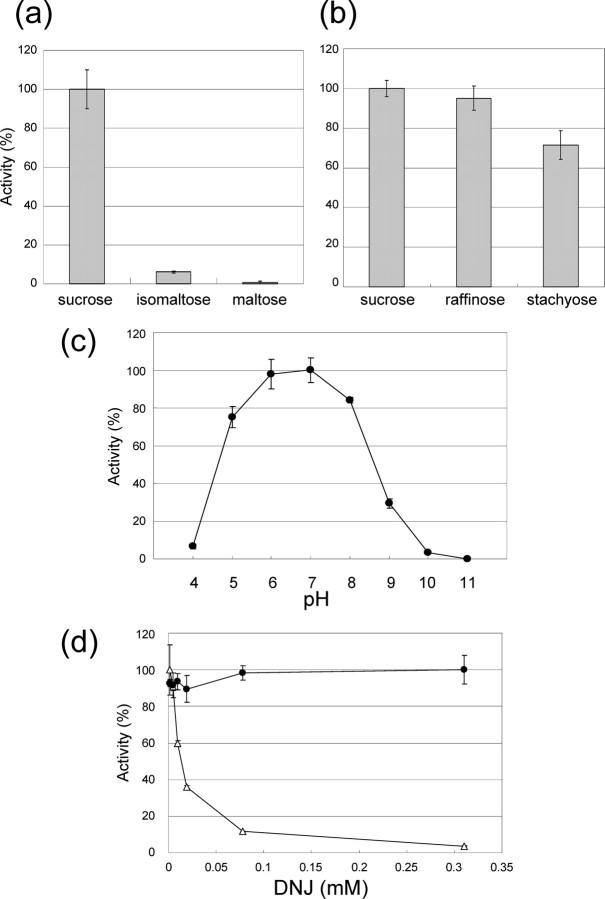
Enzyme Assay of BmSUC1—The substrate specificity of BmSUC1 was assayed using several sugar substrates (sucrose, isomaltose, maltose, raffinose, or stachyose) (Fig. 4). BmSUC1 was active on sucrose, raffinose, and stachyose, which possess the nonreducing β-d-fructofuranoside residues (Fig. 4b). However, BmSUC1 was not active on isomaltose or maltose, which lacks the β-fructofuranoside residue (Fig. 4a). This result clearly indicates that the BmSuc1 gene encodes a β-fructofuranosidase (EC 3.2.1.26) that hydrolyzed the β-fructosyl residue of the substrate. The pH profile of BmSUC1 was determined using sucrose as the substrate (Fig. 4c). The assay was performed in a pH range of 4.0–11.0, and the condition of pH 7.0 was shown to be optimal.
FIGURE 4.
Enzymatic properties of recombinant BmSUC1 protein. BmSUC1 protein was incubated with selected substrates (sucrose, isomaltose, maltose, raffinose, or stachose), and the substrate specificity of BmSUC1 was measured. a, the liberated glucose was measured by the glucose oxidase method; b, the reducing sugars released were estimated by the Somogyi-Nelson method. Bars, mean ± S.D. (n = 3). c, pH-activity relationship of the recombinant BmSUC1, determined using sucrose as a substrate. The points indicate the mean ± S.D. (n = 3). d, inhibitory assay of DNJ on the recombinant BmSUC1 (filled circle) and the α-glucosidase (open triangle) from a bacterium (Sigma), using sucrose as a substrate. The same unit (nmol of glucose produced/min) of the BmSUC1 and bacterial α-glucosidase was used for the analysis. The points indicate the mean ± S.D. (n = 3).
Next, we analyzed the inhibitory activities of DNJ on BmSUC1 activity (Fig. 4d). For comparison, α-glucosidase from bacterium Bacillus stearothermophilus was also used in the assay. As expected, DNJ strongly inhibited the α-glucosidase with an IC50 value of 10 μm. In contrast, BmSUC1 activity was not inhibited by DNJ (Fig. 4d), even at a final concentration of 1200 μm (IC50 > 1200 μm; data not shown).
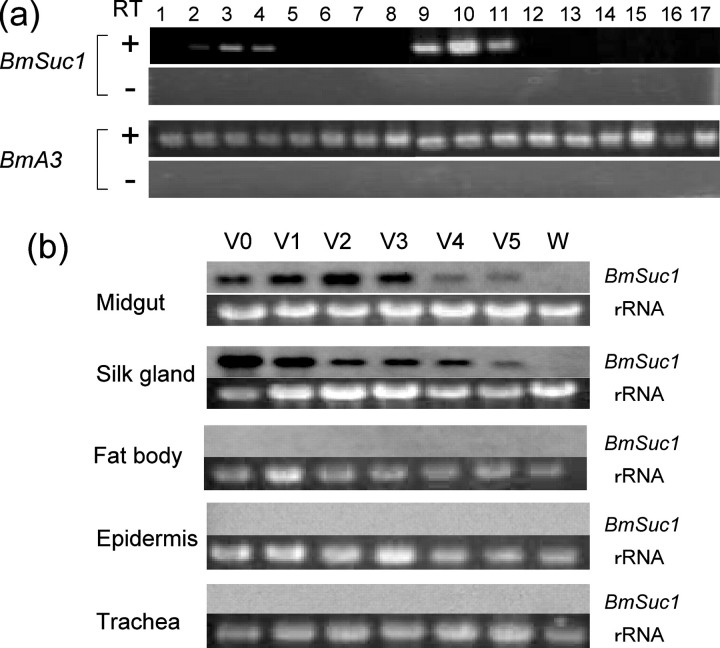
Expression Profiles of BmSuc1 and BmSuc2 Genes—The expression profile of BmSuc1 mRNA was examined by RT-PCR (Fig. 5a). On the 3rd day of the fifth instar larvae, BmSuc1 mRNA was expressed in midgut and, interestingly, also in the silk gland. Signals were detected throughout the midgut (i.e. anterior, middle, and posterior parts of the midgut) but were not detected in the foregut and hindgut of B. mori. In the silk glands, signals were detected in the anterior silk gland and anterior and middle part of the middle silk gland (MSG) but were not detected in the posterior part of MSG and in the posterior silk gland. Signals were not detected in other tissues, such as testis, fat bodies, epidermis, trachea, or malphigian tubules.
FIGURE 5.
Expression profiles of BmSuc1 mRNA. a, RT-PCR analysis of BmSuc1 expression. Total RNA from the 3rd day of the fifth instar larvae were used in the RT-PCR analysis. The B. mori actin 3 (BmA3) gene was used as the control. Template cDNAs used for RT-PCR experiments were synthesized with (+) or without (-) reverse transcriptase. Tissues used for analysis were as follows. Lane 1, foregut; lane 2, anterior part of the midgut; lane 3, middle part of the midgut; lane 4, posterior part of the midgut; lane 5, anterior part of the intestine; lane 6, posterior part of the intestine; lane 7, rectum; lane 8, testis; lane 9, anterior silk gland; lane 10, anterior part of the MSG; lane 11, middle part of the MSG; lane 12, posterior part of the MSG; lane 13, posterior silk gland; lane 14, malphigian tubules; lane 15, fat bodies; lane 16, trachea; lane 17, epidermis. b, RNA blot analysis of BmSuc1. Total RNA (5 μg) of the midgut, silk gland, fat bodies, epidermis, and trachea from day 0–5 of fifth instar larvae (V0–V5, respectively) and from larvae at wandering stage (W; day 6) were analyzed. rRNA stained with ethidium bromide was used as a loading control.
We then performed RNA blot analysis to further examine the temporal expression of BmSuc1 during the fifth instar (Fig. 5b). Consistent with the result of RT-PCR analysis, signals were detected specifically in the midgut and silk gland but not in fat bodies, epidermis, or trachea. In the midgut and silk gland, signals were detected during the feeding stage (Fig. 5, V0–V5) but disappeared by the wandering stage (W), just prior to the commencement of spinning. These results indicate that the expression of BmSuc1 is temporally and spatially controlled in the silkworm, presumably to utilize the gene product as a digestive enzyme. Investigation of the expression of BmSuc2 mRNA revealed that it was not expressed in any of the tissues examined in this study (data not shown).
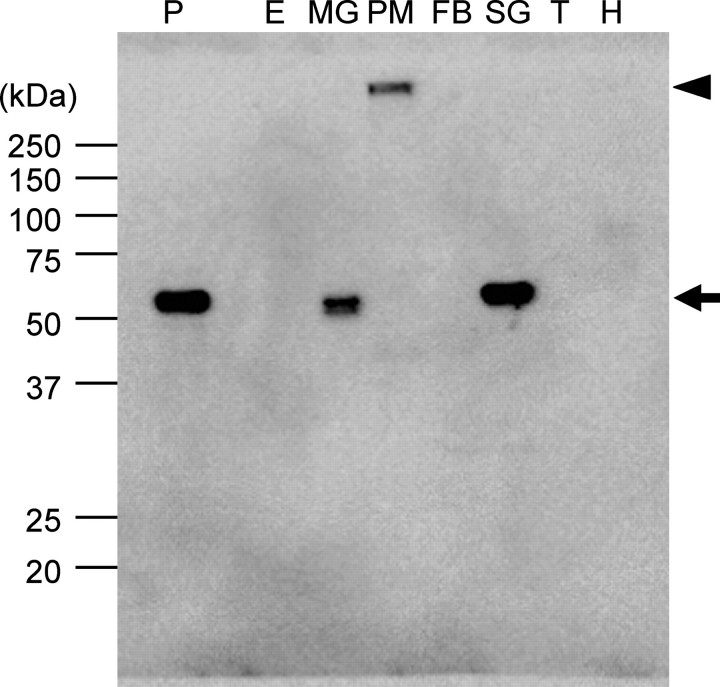
Expression and Localization of BmSUC1 Protein—We further investigated the expression and localization of the BmSUC1 protein on the 3rd day of the fifth instar larvae (Figs. 6, 7, 8). Our immunoblot analysis revealed that BmSUC1 is highly expressed in the midgut and silk gland but not in the epidermis, fat bodies, trachea, or hemolymph (Fig. 6). We found that the signal was also detected from the supernatant of homogenized peritrophic membrane, but its molecular mass (>250 kDa) was much higher than expected (56 kDa). This may suggest that the BmSUC1 protein might exist as a complex with other enzymes or components in the peritrophic membrane, which is composed of a network of chitin and proteins and therefore difficult to solubilize using the conventional method.
FIGURE 6.
Immunoblot analysis of BmSUC1 protein. Immunoblot analysis using anti-BmSUC1 antiserum was performed. Protein samples were isolated from the epidermis (E), midgut (MG), soluble fraction (supernatant of the homogenate) of peritrophic membrane (PM), fat bodies (FB), silk gland (SG), trachea (T), and hemolymph (H) from the 3-day-old fifth instar larvae. 2 μg of protein was loaded in each lane. As a positive control, the medium from cells infected with AcMNPV that expressed BmSUC1 protein was also loaded (P). The size and position of the protein standards are indicated on the left. BmSUC1 protein signals are shown by an arrow. An arrowhead shows the signal from putative BmSUC1 protein from the peritrophic membrane, which did not enter the running gel.
FIGURE 7.
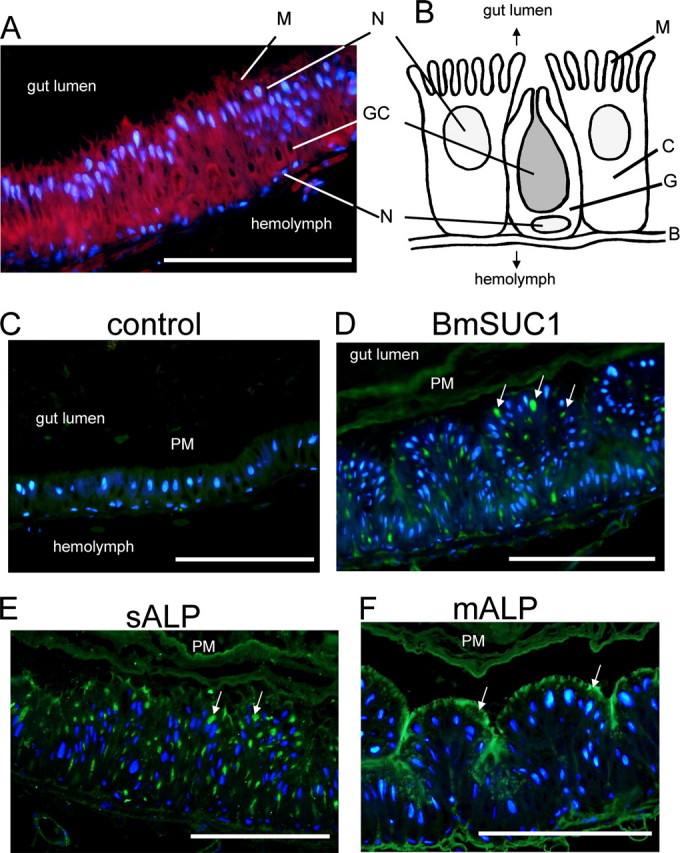
Immunohistochemical analysis of BmSUC1 protein in the midgut. A, midgut cells stained with DAPI (blue) and AlexaFluor594-conjugated phalloidin (red). B, schematic representation of midgut cells of B. mori. C, columnar cell; G, goblet cell; GC, goblet cell cavity; N, nucleus; B, basal lamina; M, microvilli. C, control experiments using preimmune serum. D, immunofluorescence visualization of BmSUC1. Sections were incubated with anti-BmSUC1 antibody followed by the secondary antibody labeled with AlexaFluor488 (green) and counterstained with DAPI. Strong signals were detected in the goblet cell cavities. E and F, control experiments using antisera against sALP (green) and mALP (green), respectively. Note that localization of BmSUC1 is closely similar to that of sALP, which was shown to localize within goblet cell cavities. Arrows, localization of the protein; bar, 200 μm.
FIGURE 8.
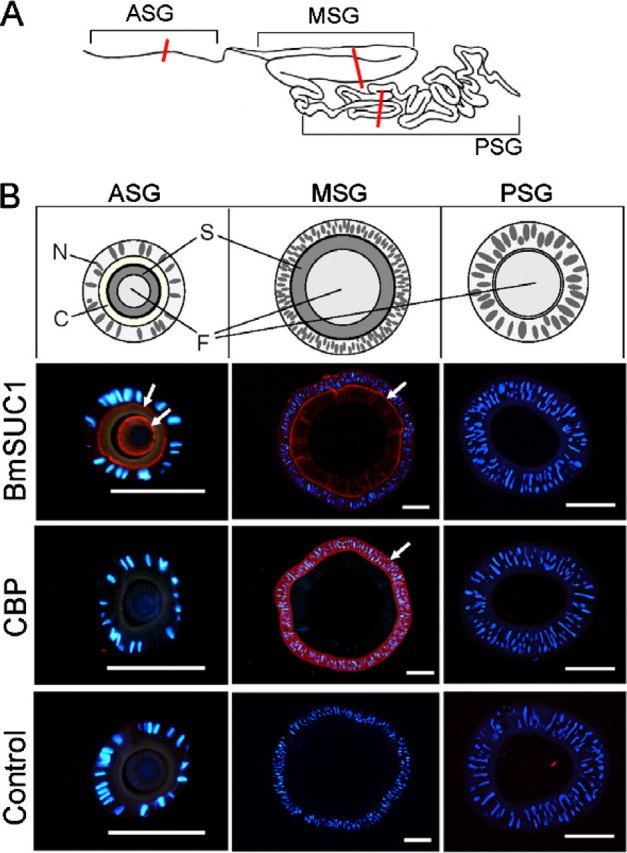
Immunohistochemical analysis of BmSUC1 protein in the silk gland. A, schematic representation of the silk gland of the silkworm. Sections were obtained from the middle part of the anterior silk gland (ASG), MSG, and posterior silk gland (PSG), as indicated by red lines. B, immunofluorescence visualization of BmSUC1 and CBP. Slides were incubated with anti-BmSUC1 or anti-CBP antiserum followed by the secondary antibody labeled with AlexaFluor546 (red) and counterstained with DAPI (blue). Control experiments were also preformed using preimmune serum. Arrows, localization of the protein; bar, 200 μm. Schematic representations of sections are also shown (top). N, nucleus; C, cuticle layer; S, sericin layer; F, fibroin layer.
Next, we performed immunohistochemical studies to investigate the localization of the BmSUC1 protein in the midgut (Fig. 7) and silk gland (Fig. 8). In the midgut from the 3rd day of the fifth instar, strong signals of BmSUC1 were observed in the goblet cell cavities (Fig. 7D), and weak signals were detected in the peritrophic membrane. The signals were not observed when the slides were incubated with preimmune serum (Fig. 7C). To compare this localization with other midgut proteins whose localization has been already demonstrated, we also examined the localization of sALP and mALP (22). As reported previously (22), sALP localized predominantly in the goblet cell cavities (Fig. 7E), and mALP localized in the apical microvilli of the midgut cells (Fig. 7F), demonstrating that the localization of BmSUC1 is closely similar to that of sALP but is quite different from that of mALP.
In the silk gland, signals of BmSUC1 were observed in the anterior and middle silk gland but not in the posterior silk gland (Fig. 8), consistent with the result of RT-PCR analysis (Fig. 5a). In the middle silk gland, BmSUC1 was shown to localize predominantly in the microvilli of the silk gland cells and distal surface of the sericin layer. In the anterior silk gland, strong signals of BmSUC1 were observed in the microvilli of the cells and in the sericin layer. This localization was distinct from that of CBP, which is expressed only in the middle silk gland and localizes within these cells (Fig. 8) (21). These results suggest that, in the silk gland, the BmSUC1 protein is secreted from anterior and middle silk gland cells and is incorporated into the sericin layer.
DISCUSSION
In this report, we have identified two β-fructofuranosidase genes from the silkworm, B. mori. It was earlier presumed that β-fructofuranosidase genes exist only in plants, bacteria, and fungi, not having ever been found in any animals (9). Although several reports have suggested the possible existence of β-fructofuranosidases in some insect species (10–13), no direct evidence of its existence in animals (i.e. cloning of the gene) could be obtained. The present study clearly demonstrates that β-fructofuranosidase genes are encoded in an animal genome.
We have identified two β-fructofuranosidase genes (BmSuc1 and BmSuc2) from the silkworm (Fig. 1). Sequence analysis suggests that the product of BmSuc2 might not possess β-fructofuranosidase activity, and its transcript was not detected by RT-PCR in our experiments, suggesting that BmSuc2 may not be functional.
Interestingly, these two genes exhibit high sequence homology to the bacterial β-fructofuranosidases. Phylogenetic analysis suggests a horizontal gene transfer from bacteria to an ancestral insect (Fig. 2). To establish this, however, we need to procure additional evidence by investigating the β-fructofuranosidase gene distribution across broad taxa of insects.
To measure the enzymatic activity of BmSUC1, we produced the recombinant protein using the baculovirus expression system (Figs. 3 and 4). Our enzymological studies clearly indicated that the BmSuc1 gene encoded the functional β-fructofuranosidase (EC 3.2.1.26).
We further investigated the expression profile of the BmSuc1 mRNA (Fig. 5). RT-PCR and RNA blot analysis showed that the BmSuc1 gene was expressed specifically in the midgut and silk gland. Although the biological significance of the expression of BmSuc1 in the silk gland is unclear at this point, our data clearly suggest that BmSUC1 plays a role as a digestive enzyme in the silkworm.
Immunohistochemical studies of BmSUC1 were conducted to determine the localization of the BmSUC1 protein (Figs. 7 and 8). In the larval midgut, the BmSUC1 signal was detected predominantly in the goblet cell cavities. This result is surprising, since it has long been presumed that the primary role of the midgut goblet cells is the active transport of potassium from the hemolymph into the gut lumen. Similarly, another sugar-digestive enzyme, trehalase-1 of the silkworm, which hydrolyzes the insect blood sugar trehalose into glucose, was also found to localize within the midgut goblet cell cavities (25). Considering these findings, we speculate that the midgut goblet cells have been overlooked for their physiological roles in the food digestion, although it is not clear whether both enzymes hydrolyze their substrates within the goblet cell cavities or they are released from the cavity. Further studies are required to elucidate how the digestive enzymes within midgut goblet cell cavities function in the process of digestion and/or absorption of nutrients.
The silkworm has a pair of salivary glands arising from the mandibular segment, in addition to the labial silk glands (26). The silk glands of B. mori have been considered as evolutionary homologues of the Drosophila salivary glands because they share many similarities, such as the segment identity (labial) (26, 27). It has been widely accepted that, in Lepidoptera, the labial silk glands became specialized for silk synthesis, and the mandibular salivary glands function in the digestion process (26, 27). Therefore, It was intriguing to find that the BmSuc1 gene is highly expressed in the silk gland of the silkworm. We also showed that the BmSUC1 protein predominantly localized in the apical microvilli of silk gland cells and in the distal surface of the sericin layer (Fig. 8). This suggests that the protein is secreted into the lumen of the silk gland and is incorporated into the sericin, which is the outer layer of silk fiber. Consistent with this, previous reports have also described the existence of sucrose-digesting enzymes in the liquid silk and cocoon of the silkworm (28, 29). Furthermore, we also found that a β-glucosidase gene of B. mori (EST clone F1mgP23A02f) (16) was expressed in the anterior silk gland.3 Their physiological roles should be clarified in future studies.
It has been demonstrated that mulberry latex contains a large amount of sugar mimic alkaloids, such as DNJ and d-AB1. These compounds are not toxic to B. mori, a mulberry specialist, although they are highly toxic to caterpillars, such as S. cynthia ricini, a generalist herbivore (2). This suggests that the silkworm has evolved some adaptive mechanisms to circumvent the toxic effects of such sugar mimic alkaloids to be able to feed on mulberry leaves. Recently, Hirayama et al. (8) suggested that the sucrase and trehalase are the main targets of sugar mimic alkaloids in the mulberry latex. They also showed that sucrase and trehalase of B. mori are highly insensitive to sugar mimic alkaloids, whereas those of S. cynthia are highly sensitive (8). These results strongly indicate that B. mori larvae evolved enzymatic adaptations against the mulberry's defense system and further suggest that sucrase and/or trehalase of B. mori are the key enzymes responsible for its resistance to sugar mimic alkaloids. In the present study, we identified β-fructofuranosidase genes from the silkworm and demonstrated that the enzyme was highly insensitive to DNJ and served as a sugar-digesting enzyme in the silkworm physiology. Our data suggest that the BmSUC1 plays a critical role in the silkworm's ability to avoid the toxic effects of sugar mimic alkaloids present in the mulberry latex.
Toward understanding the adaptive mechanism of B. mori to sugar mimic alkaloids, further studies are required to investigate whether β-fructofuranosidase genes are encoded in other insects. If the β-fructofuranosidase genes exist only in the B. mori genome, the tolerance to sugar mimic alkaloids contained in mulberry latex could be simply explained by the presence of the β-fructofuranosidase gene. If the genes are present in other insects that do not feed on mulberry leaves, such as S. cynthia, comparative studies on their enzymatic properties, expressions, and localizations would provide further clues into the silkworm's ability to circumvent the mulberry's defense system. Since previous reports have described several insect sucrases that exhibited a kinetic property characteristic of a β-fructofuranosidase (10, 13), it is probable that β-fructofuranosidase genes exist in other insects too. We are currently conducting further experiments to identify and characterize the β-fructofuranosidase genes from other insects.
As well as β-fructofuranosidase genes in this study, some animals are known to encode “anomalous” genes that have been believed to be absent in animal kingdom. For example, termites encode their own cellulase genes, which are assumed to facilitate the digestion of wood cellulose (30), and ascidians encode cellulose synthase genes (31, 32), which explains why tunicates are the only animals that can perform cellulose biosynthesis. As new animal genomes are sequenced and as the nucleotide/protein data bases expand, the list of such genes will increase. Isolation and characterization of these genes are particularly important in comparative genomics, since these genes may reflect unique physiology, ecology, and evolutionary history of organisms that encode them. The β-fructofuranosidase gene of the silkworm would be an intriguing example of such “anomalous” genes, since this gene may be a central player in the evolutionary arms race between mulberry trees and the silkworm, a mulberry specialist.
Acknowledgments
We thank Naoko Omuro for technical assistance. We greatly appreciate the gift of B. mori strain N4 and anti-CBP antiserum from Dr. Kozo Tsuchida (National Institute of Infectious Diseases, Japan) and anti-sALP and anti-mALP antisera from Dr. Masanobu Itoh (Kyoto Institute of Technology, Japan).
The nucleotide sequence(s) reported in this paper has been submitted to the GenBank™/EBI Data Bank with accession number(s) AB366559 and AB366560.
This work was supported by Ministry of Education, Culture, Sports, Science and Technology Grant 17018007 (to T. S.), a grant from Ministry of Agriculture, Forestry and Fisheries-National Institute of Agrobiological Sciences (Agrigenome Research Program), a grant from Japan Science and Technology Agency (Professional Program for Agricultural Bioinformatics), and Japan Society for the Promotion of Science Research Fellowship for Young Scientists 16·11613 (to T. D.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: d-AB1, 1,4-dideoxy-1,4-imino-d-arabinitol; DNJ, 1-deoxynojirimycin; DAPI, 4′,6-diamidino-2-phenylindole dihydrochloride; MSG, middle silk gland; sALP, soluble alkaline phosphatase; mALP, membrane-bound alkaline phosphatase; CBP, carotenoid-binding protein; DIG, digoxigenin; EST, expressed sequence tag; RT, reverse transcription.
T. Shimada, T. Daimon, T. Taguchi, and K. Mita, unpublished data.
References
- 1.Zagrobelny, M., Bak, S., Rasmussen, A. V., Jorgensen, B., Naumann, C. M., and Lindberg Moller, B. (2004) Phytochemistry 65 293-306 [DOI] [PubMed] [Google Scholar]
- 2.Konno, K., Ono, H., Nakamura, M., Tateishi, K., Hirayama, C., Tamura, Y., Hattori, M., Koyama, A., and Kohno, K. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 1337-1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nishida, R. (2002) Annu. Rev. Entomol. 47 57-92 [DOI] [PubMed] [Google Scholar]
- 4.Dussorurd, D. E. (1993) in Caterpillars: Ecological and Evolutionary Constraints on Foraging (Stamp, N. E., Casey, T. M., ed) pp. 92-131, Chapman & Hall, New York
- 5.Dussourd, D. E., and Eisner, T. (1987) Science 237 898-901 [DOI] [PubMed] [Google Scholar]
- 6.Konno, K., Hirayama, C., Nakamura, M., Tateishi, K., Tamura, Y., Hattori, M., and Kohno, K. (2004) Plant J. 37 370-378 [DOI] [PubMed] [Google Scholar]
- 7.Asano, N., Yamashita, T., Yasuda, K., Ikeda, K., Kizu, H., Kameda, Y., Kato, A., Nash, R. J., Lee, H. S., and Ryu, K. S. (2001) J. Agric. Food Chem. 49 4208-4213 [DOI] [PubMed] [Google Scholar]
- 8.Hirayama, C., Konno, K., Wasano, N., and Nakamura, M. (2007) Insect. Biochem. Mol. Biol. 37 1348-1358 [DOI] [PubMed] [Google Scholar]
- 9.Krasikov, V. V., Karelov, D. V., and Firsov, L. M. (2001) Biochemistry (Mosc.) 66 267-281 [DOI] [PubMed] [Google Scholar]
- 10.Santos, C. D., and Terra, W. R. (1986) Insect Biochem. 16 819-824 [Google Scholar]
- 11.Sumida, M., Yuan, X. L., and Matsubara, F. (1994) Comp. Biochem. Physiol. B Biochem. Mol. Biol. 107 273-284 [Google Scholar]
- 12.Sumida, M., Yuan, X. L., and Matsubara, F. (1994) Comp. Biochem. Physiol. A Biochem. Mol. Biol. 108 255-264 [Google Scholar]
- 13.Carneiro, C. N., Isejima, E. M., Samuels, R. I., and Silva, C. P. (2004) J. Insect Physiol. 50 1093-1101 [DOI] [PubMed] [Google Scholar]
- 14.Daimon, T., Hamada, K., Mita, K., Okano, K., Suzuki, M. G., Kobayashi, M., and Shimada, T. (2003) Insect Biochem. Mol. Biol. 33 749-759 [DOI] [PubMed] [Google Scholar]
- 15.Daimon, T., Katsuma, S., Iwanaga, M., Kang, W., and Shimada, T. (2005) Insect Biochem. Mol. Biol. 35 1112-1123 [DOI] [PubMed] [Google Scholar]
- 16.Mita, K., Morimyo, M., Okano, K., Koike, Y., Nohata, J., Kawasaki, H., Kadono-Okuda, K., Yamamoto, K., Suzuki, M. G., Shimada, T., Goldsmith, M. R., and Maeda, S. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 14121-14126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mita, K., Kasahara, M., Sasaki, S., Nagayasu, Y., Yamada, T., Kanamori, H., Namiki, N., Kitagawa, M., Yamashita, H., Yasukochi, Y., Kadono-Okuda, K., Yamamoto, K., Ajimura, M., Ravikumar, G., Shimomura, M., Nagamura, Y., Shin, I. T., Abe, H., Shimada, T., Morishita, S., and Sasaki, T. (2004) DNA Res. 11 27-35 [DOI] [PubMed] [Google Scholar]
- 18.Xia, Q., Zhou, Z., Lu, C., Cheng, D., Dai, F., Li, B., Zhao, P., Zha, X., Cheng, T., Chai, C., Pan, G., Xu, J., Liu, C., Lin, Y., Qian, J., Hou, Y., Wu, Z., Li, G., Pan, M., Li, C., Shen, Y., Lan, X., Yuan, L., Li, T., Xu, H., Yang, G., Wan, Y., Zhu, Y., Yu, M., Shen, W., Wu, D., Xiang, Z., Yu, J., Wang, J., Li, R., Shi, J., Li, H., Su, J., Wang, X., Zhang, Z., Wu, Q., Li, J., Zhang, Q., Wei, N., Sun, H., Dong, L., Liu, D., Zhao, S., Zhao, X., Meng, Q., Lan, F., Huang, X., Li, Y., Fang, L., Li, D., Sun, Y., Yang, Z., Huang, Y., Xi, Y., Qi, Q., He, D., Huang, H., Zhang, X., Wang, Z., Li, W., Cao, Y., Yu, Y., Yu, H., Ye, J., Chen, H., Zhou, Y., Liu, B., Ji, H., Li, S., Ni, P., Zhang, J., Zhang, Y., Zheng, H., Mao, B., Wang, W., Ye, C., Wong, G. K., and Yang, H. (2004) Science 306 1937-194015591204 [Google Scholar]
- 19.Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (1997) Nucleic Acids Res. 25 4876-4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar, S., Tamura, K., and Nei, M. (2004) Brief Bioinform. 5 150-163 [DOI] [PubMed] [Google Scholar]
- 21.Tsuchida, K., Jouni, Z. E., Gardetto, J., Kobayashi, Y., Tabunoki, H., Azuma, M., Sugiyama, H., Takada, N., Maekawa, H., Banno, Y., Fujii, H., Iwano, H., and Wells, M. A. (2004) J. Insect Physiol. 50 363-372 [DOI] [PubMed] [Google Scholar]
- 22.Azuma, M., and Eguchi, M. (1989) J. Exp. Zool. 251 108-112 [Google Scholar]
- 23.Reddy, A., and Maley, F. (1996) J. Biol. Chem. 271 13953-13957 [DOI] [PubMed] [Google Scholar]
- 24.Emanuelsson, O., Brunak, S., von Heijne, G., and Nielsen, H. (2007) Nat. Protoc. 2 953-971 [DOI] [PubMed] [Google Scholar]
- 25.Mitsumasu, K., Azuma, M., Niimi, T., Yamashita, O., and Yaginuma, T. (2005) Insect Mol. Biol. 14 501-508 [DOI] [PubMed] [Google Scholar]
- 26.Toyama, K. (1909) The Studies on Bombyx mori Eggs, Murayama-sha Press, Tokyo
- 27.Parthasarathy, R., and Gopinathan, K. P. (2005) Gene Expr. Patterns 5 323-339 [DOI] [PubMed] [Google Scholar]
- 28.Shimada, S., and Hayashiya, K. (1975) Insect Biochem. 5 357-369 [Google Scholar]
- 29.Shimada, S., and Hayashiya, K. (1975) Insect Biochem. 5 653-657 [Google Scholar]
- 30.Watanabe, H., Noda, H., Tokuda, G., and Lo, N. (1998) Nature 394 330-331 [DOI] [PubMed] [Google Scholar]
- 31.Nakashima, K., Yamada, L., Satou, Y., Azuma, J., and Satoh, N. (2004) Dev. Genes Evol. 214 81-88 [DOI] [PubMed] [Google Scholar]
- 32.Sasakura, Y., Nakashima, K., Awazu, S., Matsuoka, T., Nakayama, A., Azuma, J., and Satoh, N. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 15134-15139 [DOI] [PMC free article] [PubMed] [Google Scholar]