Abstract
The urokinase receptor, urokinase receptor (uPAR), is a glycosylphosphatidylinositol-anchored membrane protein engaged in pericellular proteolysis and cellular adhesion, migration, and modulation of cell morphology. A direct matrix adhesion is mediated through the binding of uPAR to vitronectin, and this event is followed by downstream effects including changes in the cytoskeletal organization. However, it remains unclear whether the adhesion through uPAR-vitronectin is the only event capable of initiating these morphological rearrangements or whether lateral interactions between uPAR and integrins can induce the same response. In this report, we show that both of these triggering mechanisms can be operative and that uPAR-dependent modulation of cell morphology can indeed occur independently of a direct vitronectin binding. Expression of wild-type uPAR on HEK293 cells led to pronounced vitronectin adhesion and cytoskeletal rearrangements, whereas a mutant uPAR, uPARW32A with defective vitronectin binding, failed to induce both phenomena. However, upon saturation of uPARW32A with the protease ligand, pro-uPA, or its receptor-binding domain, the ability to induce cytoskeletal rearrangements was restored, although this did not rescue the uPAR-vitronectin binding and adhesion capability. On the other hand, using other uPAR variants, we could show that uPAR-vitronectin adhesion is indeed capable and sufficient to induce the same morphological rearrangements. This was shown with cells expressing a different single-site mutant, uPARY57A, in the presence of a synthetic uPAR-binding peptide, as well as with wild-type uPAR, which underwent cytoskeletal rearrangements even when cultivated in uPA-deficient serum. Blocking of integrins with an Arg-Gly-Asp-containing peptide counteracted the matrix contacts necessary to initiate the uPAR-dependent cytoskeletal rearrangements, whereas inactivation of the Rac signaling pathway in all cases suppressed the occurrence of the same events.
The urokinase receptor (uPAR)3 is a glycosylphosphatidyl-inositol-anchored membrane protein that binds the plasminogen activator, uPA and the extracellular matrix protein, vitronectin (for review, see Ref. 1). uPAR plays several important roles in the interplay of cells with their surrounding matrix, including the governing of plasmin-dependent fibrinolysis and matrix degradation (2), the direct adhesion on vitronectin-containing matrices (3), and the modulation of morphology and adhesive properties of cells toward both vitronectin and other matrix proteins (4–6). Being expressed on tumor-associated stromal cells (7, 8) and, in some cancers, on the tumor cells themselves (9), uPAR is assumed to be important for cancer invasion, both through the localization of matrix degrading processes (10) and through the modulation of cell adhesion and migration (11). Therefore, a considerable research effort is devoted to the unraveling of the molecular and cellular interactions of uPAR.
Recent studies on the three-dimensional structure of uPAR (12, 13), combined with site-directed mutagenesis (14–17), have led to a detailed understanding of the structural basis for the binding of uPA and vitronectin to uPAR. In addition to these binding reactions, however, an increasing number of reports point to downstream effects of uPAR interactions that appear to include additional molecular partners, such as β1, β2, and β3 integrins (6, 18–21), G-protein-coupled receptors (22), or members of the epidermal growth factor receptor family (23), depending on the cell type in question.
Such interactions are likely to be involved in signaling effects that ultimately govern cell morphology and migration as well as proliferation (6, 24, 25), but their molecular basis is still much less well defined than that of the uPA and vitronectin binding reactions. Therefore, although direct interactions between uPAR and integrins have indeed been demonstrated (20, 26, 27), an unresolved question is whether the signaling reactions do actually depend on a defined molecular interplay between these membrane proteins and uPAR or whether they are secondary phenomena brought about, e.g. by uPAR-mediated matrix adhesion as proposed recently (16). In this report, we address an important aspect of this question by demonstrating a striking uPAR-dependent effect on the cytoskeleton that is independent of the uPAR-mediated cell adhesion.
EXPERIMENTAL PROCEDURES
Protein Reagents—The following recombinant proteins and synthetic reagents were produced as described previously: recombinant human full-length pro-uPA and its isolated amino-terminal fragment (ATF; residues 1–143) (15), the uPA growth factor domain (GFD; residues 1–48) (17), and the synthetic peptide antagonist, AE120 (28). The cyclic peptides, cyclo(Arg-Gly-Asp-d-Phe-Val) and cyclo(Arg-Ala-Asp-d-Phe-Val), were purchased from Peptides International, Louisville, KT. The following reagents were purchased from the commercial sources indicated: human vitronectin (BD Biosciences) and FITC-phalloidin (Invitrogen).
Transfection of HEK Cells with cDNAs Encoding uPAR Wild-type and Single-site Mutants or Dominant-negative Rac Construct—cDNA constructs encoding the human uPARWT full-length protein (signal peptide of 22 amino acid residues followed by residues 1–313; numbering referring to Ref. 29) and uPAR proteins with single site substitutions at position 32 (uPARW32A) and 57 (uPARY57A) were obtained by ligating the corresponding DNA constructs encoding soluble, truncated uPARs (14) into the full-length uPAR expression plasmid, pRc/CMV-uPAR (4), using appropriate restriction sites. HEK cells, (HEK293, purchased from ECACC) were transfected with the resulting uPAR expression vectors or with vector alone (mock-transfected) by electroporation followed by cloning by limiting dilution and characterization of transfectant clones by flow cytometry. Details for these procedures can be found in the supplemental materials. For transfection with a dominant-negative Rac construct, the plasmid pRK5-myc-Rac1-T17N (N17Rac) (4, 30) was used for electroporation. After transformation, the cells were immediately seeded on coverslips coated with vitronectin in the presence or absence of pro-uPA. Further details are described in the supplemental materials.
Adhesion Assays—Vitronectin adhesion assays with transfected cells were done in vitronectin-coated 96-well culture plates, using triplicate samples of 1 × 105 cells in PBS with 5 mm EDTA and 0.5% bovine serum albumin. After incubation and washing, adherent cells were quantified by thiazolyl blue tetrazolium bromide assay (31). The complete procedure is available in the supplemental materials.
Analyses of Cell Morphology and Cytoskeletal Rearrangements—Glass coverslips were placed in a 24-well culture plate and coated with 50 μl of a 5 μg/ml solution of vitronectin in PBS/calcium/magnesium (PBS with 0.9 mm CaCl2 and 0.5 mm MgCl2) followed by blocking with 2 mg/ml bovine serum albumin in PBS/calcium/magnesium. After washing the wells, transfected cells were seeded on the coverslips and grown for 2 days at 37 °C in 1 ml of serum-containing growth medium (see above), supplemented with 400 μg/ml Geneticin. In some experiments, this was followed by the addition of pro-uPA or other reagents, as indicated followed by culture for another 2 days with the cells still being subconfluent. In some experiments, cells were cultured for different time periods, as indicated. For examination of lamellipodia and cytoskeletal rearrangements, cells were fixed and permeabilized followed by staining with FITC-labeled phalloidin (see supplemental materials for details). The cells were then examined by fluorescence microscopy, using a Leica DM4000B fluorescence microscope with a Leica DFC-480 camera. Scoring of lamellipodia-positive cells (16) was done blindly in randomly selected fields from fluorescence microscopy of the FITC-phalloidin treated samples. For each cell type and each of the experimental conditions tested, five microscope fields were examined by four skilled researchers who were unaware of the sample identity. Each investigator assigned the number 0 (lamellipodia-negative), 0.5 (ambiguous), or 1 (lamellipodia-positive) to each field examined. The final score of each sample was defined as the cumulative number (0–20) obtained from all investigators for all fields from the sample in question after decoding the fields and samples. In specific experiments with uPA-free conditions, FCS was excluded from the cell culture medium and substituted with sterile serum from a uPA-deficient mouse (a kind gift from Dr. Leif R. Lund, the Finsen Laboratory). For studies with cells transfected with the dominant-negative Rac construct, quantification of lamellipodia-positive cells was done by counting of single cells; see supplemental materials.
RESULTS
Adhesion Properties and Morphology of HEK Cells Transfected with Mutated uPARs—Based on the biochemical mapping of the vitronectin binding determinant of soluble uPAR (15), we set out to study the importance of this binding reaction with cell-associated uPAR. Human embryonic kidney (HEK293) cells were transfected with vector alone (mock-transfected), with full-length human wild-type uPAR cDNA (uPARWT), or with uPAR cDNAs with targeted alanine substitutions at positions 32 (uPARW32A) or 57 (uPARY57A). Of these mutant proteins, when analyzed in their soluble form, uPARW32A has impaired vitronectin binding but normal binding of the protease ligand, uPA/pro-uPA (15). The same was shown to be the case for the corresponding cellular, membrane-bound uPAR mutant in a recent report (16), published during the course of this work. In contrast, uPARY57A has some reduction in the binding to uPA/pro-uPA (14) but, when complexed with this ligand, this mutant binds vitronectin normally (15).
Stable transfectant cell lines were isolated and shown to express comparable levels of cell surface uPAR by flow cytometric analysis (supplemental Fig. S1). The cells were then studied with respect to two properties, known from previous studies to be directly or indirectly dependent on the uPAR-vitronectin interaction, i.e. cellular adhesion on a vitronectin matrix (3, 6) and induction of cytoskeletal rearrangements (4). To separate the uPAR-mediated vitronectin adhesion from the integrin-mediated matrix interactions, cell adhesion assays were done as short term experiments in the presence of EDTA (4) (Fig. 1A). Under these conditions, adhesion was completely uPAR-dependent since efficient adhesion occurred with uPARWT transfectants, whereas no adhesion occurred with the mock-transfected control cells. The two mutant uPARs both failed to induce adhesion capability, in accordance with the notion that neither mutant binds to vitronectin under these conditions.
FIGURE 1.
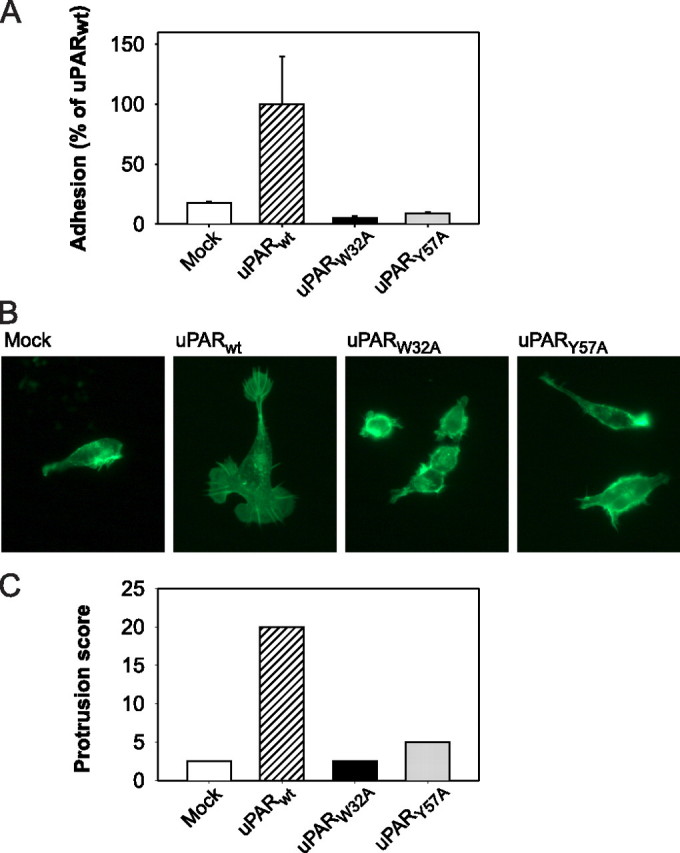
Vitronectin adhesion and cytoskeletal rearrangements in HEK cells expressing wild-type and mutated uPAR. A, adhesion of uPAR transfected cells on a reconstituted vitronectin matrix. Mock-transfected (Mock) cells or cells expressing uPARWT or uPAR mutant proteins were seeded in vitronectin-coated culture wells in the presence of EDTA and allowed to adhere during a 1-h incubation period at 37 °C. After washing, adherent cells were quantified by thiazolyl blue tetrazolium bromide assay. Each column represents the mean of a triple determination. The standard deviations are indicated. B, morphological changes and cytoskeletal rearrangements. Mock-transfected cells or cells expressing uPARWT or uPAR mutant proteins were cultured for 5 days on vitronectin-coated coverslips. The cells were then fixed and permeabilized followed by FITC-phalloidin staining and examination by fluorescence microscopy. Note the exclusive appearance of lamellipodia and cytoskeletal extensions in the uPARWT-transfected cells. C, quantification of fields with lamellipodia-positive cells. Cells were cultured, stained, and examined by fluorescence microscopy as in B. Each cell type was assigned an arbitrary designation, after which five randomly selected microscope fields for each cell type were scored blindly by four investigators for lamellipodia-positive cells (see “Experimental Procedures”). The cumulative score is represented for each sample.
Expression of uPARWT also induced marked changes in cell morphology. The uPARWT cells displayed pronounced protrusions and lamellipodia, which could be observed directly on live cells by phase contrast microscopy (result not shown), as well as by fluorescence microscopy after permeabilization and staining with FITC-phalloidin (Fig. 1B). These membranous structures were accompanied by marked cytoskeletal rearrangements, as also evident on the phalloidin-stained cells. Lamellipodia and cytoskeletal extensions were absent in the mock-transfected cells. This difference between lamellipodia-positive and -negative cell samples was highly striking (Fig. 1B) and was completely unambiguous as evident by the very consistent assignment of cell morphology status after unbiased scoring by four independent researchers (Fig. 1C; see “Experimental Procedures”). When analyzing the two uPAR mutants in the same system, neither of these was capable of inducing morphological change or cytoskeletal rearrangements (Fig. 1, B and C).
The Role of pro-uPA in the uPAR-induced Effects—Thus, in accordance with the results of Madsen et al. (16), transfection with wild-type uPAR led to efficient vitronectin adhesion and changes in cell morphology, whereas non-adhesive uPAR variants failed to induce cytoskeletal rearrangements. To further pursue this phenomenon, we examined the role of the protease ligand, pro-uPA, which can bind to uPAR simultaneously with the binding of vitronectin to the latter (3).
As seen in Fig. 2A, the addition of pro-uPA led to the expected pattern of cell adhesion on vitronectin in all cases. Thus, in accordance with the notion that pro-uPA stimulates the interaction between wild-type uPAR and vitronectin (32, 33), an even stronger adhesion was observed with the uPARWT-expressing cells upon the addition of the protease ligand, as compared with the same cells in the absence of pro-uPA. Furthermore, as expected, the addition of pro-uPA did not lead to adhesion of mock-transfected cells. Finally, a pronounced adhesion capability was induced by the addition of pro-uPA to the uPARY57A cells, whereas adhesion was in all cases negative with the uPARW32A cells. This was in complete agreement with our previous binding studies with isolated uPAR mutant proteins where uPARY57A was shown to bind vitronectin efficiently when saturated with pro-uPA, whereas no affinity for vitronectin could be induced in uPARW32A (15).
FIGURE 2.
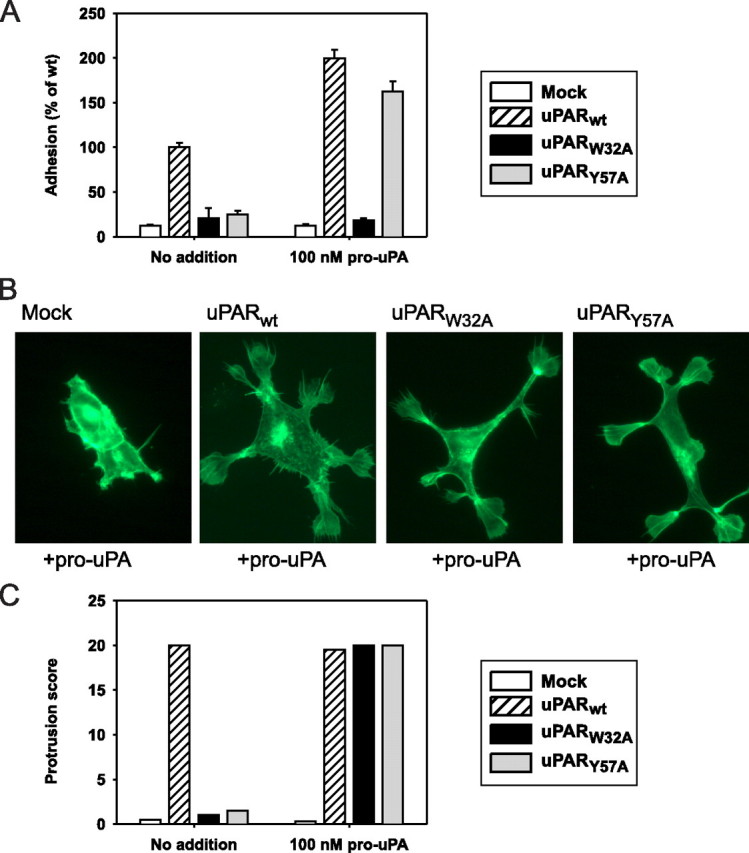
Effect of pro-uPA on vitronectin adhesion and cell morphology. Vitronectin adhesion and morphological properties of transfected HEK cells were analyzed as in Fig. 1, except that pro-uPA (100 nm) was added to the samples where indicated. Symbols designating the four transfectant cell types are indicated. A, cell adhesion on a vitronectin matrix in the presence or absence of pro-uPA. Mock, mock-transfected. B, cytoskeletal rearrangements as a consequence of the addition of pro-uPA during cell culture. After fixation, permeabilization, and FITC-phalloidin staining, cells were examined by fluorescence microscopy. C, occurrence of lamellipodia-positive cells in the presence or absence of pro-uPA. FITC-phalloidin-stained cell samples were scored blindly as described in the legend for Fig. 1 (panel C), after which the cumulative score for each type of sample was calculated.
Interestingly, however, under these conditions, studies on cell morphology revealed a clear distinction between cellular adhesive properties and the occurrence of cytoskeletal rearrangements. This distinction was evident in the case of the uPARW32A cells, which formed extensive cellular protrusions and lamellipodia in response to pro-uPA (Fig. 2, B and C), although the same uPAR mutant was non-adhesive in the vitronectin adhesion assay as noted above. The other transfectant cell types, however, displayed the same pro-uPA dependence as found in the adhesion assay. Thus, for the uPARY57A cells, lamellipodia were formed only with cells grown in the presence of pro-uPA (Fig. 2, B and C), whereas uPARWT cells were in all cases lamellipodia-positive (Fig. 2C). These cytoskeletal rearrangements were not a result of long term cell culture or high concentrations of pro-uPA because the same outcome was obtained after overnight cell culture with only 10 nm pro-uPA (supplemental Fig. S2A). In conclusion, the uPAR-dependent mechanism for modulation of cell morphology was found to follow the capability for uPAR-mediated vitronectin adhesion in some but not in all cases. The protease ligand could induce uPAR-dependent cytoskeletal rearrangements in uPARW32A transfectants without conferring vitronectin affinity.
The Effect of pro-uPA Is Exerted by the Growth Factor Domain and Is Sensitive to Blocking of Integrins—Pro-uPA includes a receptor-binding GFD, a Kringle domain, and a catalytic domain (34), and uPA itself has been shown to have some affinity for vitronectin (35). Thus, it was important to learn whether the effect of pro-uPA in the phenomena studied in this work required the complete protein or whether only part of the molecule was needed. Therefore, we tested the effect of the isolated receptor-binding units in the same phenomena as studied above. As shown in Fig. 3, A and B, the effects of pro-uPA were in all cases reproduced not only by ATF (including the Kringle and the GFD) but also by the isolated GFD, comprising just 48 amino acid residues. Thus, ATF and GFD both induced a pronounced adhesive capability in the uPARY57A cells but failed to induce any adhesion of the uPARW32A cells (Fig. 3A). In the morphology study, both reagents successfully rescued the lamellipodia phenotype of the uPARW32A as well as the uPARY57A cells, just like the effect exerted by the complete pro-uPA protein (Fig. 3B and supplemental Fig. S2B). In conclusion, the GFD of uPA retains all of the structural features required to elicit these cellular effects.
FIGURE 3.
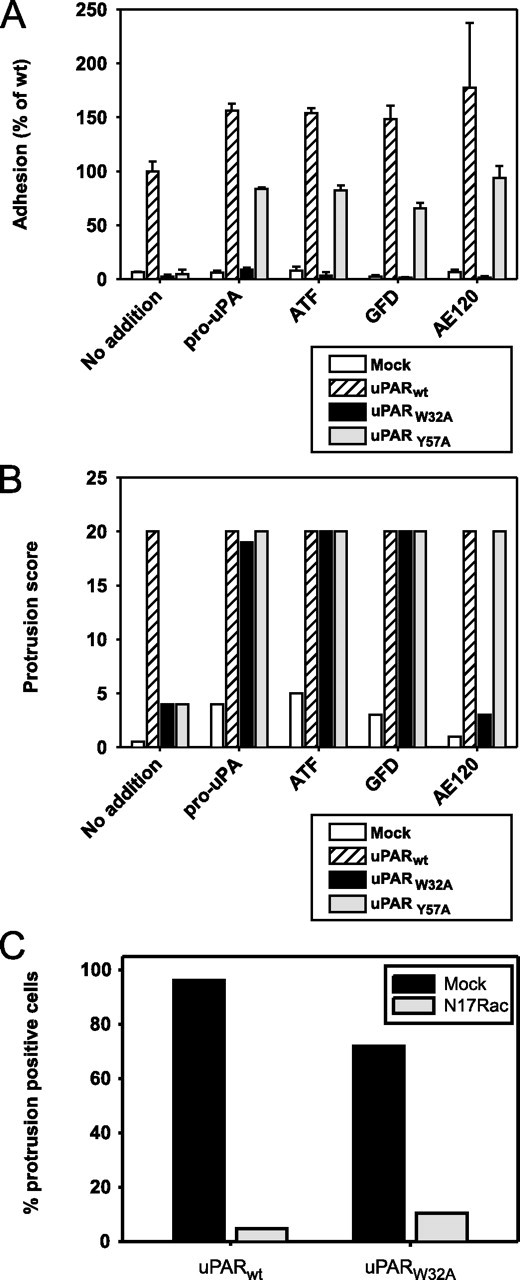
Dependence on different uPAR ligands and the Rac pathway. A and B, adhesion (A) and lamellipodia-positive cells (B), analyzed as in Fig. 2, A and C, but in the presence of the indicated ligands. Ligands were added at final concentrations of 100 nm (pro-uPA, ATF, and GFD) or 1 μm (synthetic peptide AE120), respectively. Mock, cells transfected with vector alone. C, inactivation of the Rac pathway. Cells were transfected with the dominant-negative Rac expression plasmid, N17Rac, or with vector alone, as indicated, and cultured in the presence of 20 nm pro-uPA. After fixation, permeabilization, and FITC-phalloidin staining, randomly selected microscope fields representing at least 50 cells were scored for the percentage of protrusion-positive cells.
A non-natural, uPAR-binding peptide, designated AE120, has been developed by phage display technology and combinatorial chemistry and has been shown to interact with uPAR with a high affinity (28). X-ray crystallography of the complex of uPAR and a truncated version of this peptide has shown that it binds directly in the uPA-binding pocket of uPAR (12), but the conformation of the complex is slightly different from the complex between uPAR and ATF in terms of the interdomain orientation and the flexible regions of the receptor (13, 36). Therefore, we included AE120 in these studies to learn whether ligation of uPAR with this peptide would mimic the effects of the biological ligand, pro-uPA/GFD. Interestingly, as seen in Fig. 3, A and B, this was the case for some properties but not for others. AE120 was indeed able to induce adhesion and cytoskeletal rearrangements of uPARY57A cells. However, unlike pro-uPA, ATF, and GFD, the peptide failed to induce cytoskeletal rearrangements (as well as adhesion) of uPARW32A cells; see also supplemental Fig. S2B.
Under some experimental conditions, uPAR has been found to stimulate the vitronectin binding capability of neighboring integrins (6), without this involving a direct uPAR-vitronectin interaction. To investigate whether this phenomenon was important for the uPAR-dependent morphological changes in our assay system, we studied the effect of a cyclic RGD peptide that efficiently blocks integrin-vitronectin interactions (37). In the presence of this peptide, cells expressing uPARW32A failed to grow adherently on the vitronectin matrix, thus precluding the washing steps needed for fixation and phalloidin staining. However, the cell shape and the formation of lamellipodia could indeed be evaluated directly by phase contrast microscopy of non-fixed cells (supplemental Fig. S3). It was evident that the RGD peptide had a strong impact on the morphology and the matrix contact formation of the uPAR-transfected cells. In the case of uPARWT cells, both in the presence and in the absence of pro-uPA, this peptide prevented the morphological changes previously observed in all cases with wild-type uPAR. In contrast to the characteristic flattened cell morphology with large lamellipodia, shown above, uPARWT cells in the presence of the RGD peptide displayed a rounded shape with much smaller protrusions (supplemental Fig. S3, A and E). In the uPARW32A cells, where cytoskeletal rearrangements and lamellipodia would otherwise be induced by the addition of the protease ligand, pro-uPA failed to induce these alterations when the RGD peptide was present (supplemental Fig. S3B). If an RAD control peptide was used instead of the RGD peptide, uPARWT cells were in all cases lamellipodia-positive (supplemental Fig. S3, C and G), and the uPARW32A cells regained the ability to form lamellipodia in response to pro-uPA (supplemental Fig. S3D), just like the situation found when no peptide had been added. Thus, the blocking of integrins with an RGD-containing peptide had an impact on cell-matrix contact and cell morphology irrespective of which uPAR variant was expressed by the cells.
The uPAR-dependent Cytoskeletal Rearrangements Proceed through a Rac-dependent Pathway—In fibroblast-like cells, expression of wild-type uPAR also leads to cytoskeletal rearrangements, with this effect proceeding through a signaling pathway involving the Rho GTPase, Rac (4). Therefore, we wanted to study whether this pathway is also operative in the present system. Consequently, we studied the effect of transfecting the uPAR-expressing HEK cells with a dominant-negative Rac expression plasmid (N17Rac), which has been positively shown to inactivate the Rac pathway (4). After transfection with N17Rac or vector alone, cells were seeded on vitronectin-coated coverslips in the presence or absence of pro-uPA and studied with respect to cell morphology as above. Initial transfection studies with a green fluorescent protein-encoding plasmid under the same conditions showed that a 100% transfection efficiency could not be obtained (result not shown), indicating that all microscopy fields would include a certain number of cells without knock-down of Rac activity. Therefore, rather than scoring whole microscopy fields, quantification in this particular experiment was done by counting of single cells with cytoskeletal rearrangements (Fig. 3C).
Transfection with vector alone had no effect on the subsequent behavior of the cells, with the large majority of uPARWT and uPARW32A cells being lamellipodia-positive in the presence of pro-uPA (Fig. 3C, black columns) and with uPARWT cells also being positive without protease ligand (results not shown). In contrast, transfection with the dominant-negative Rac construct strongly suppressed the cytoskeletal rearrangements in all cases (Fig. 3C, gray columns).
Efficient Binding of uPAR to Vitronectin Supersedes the Need for uPA—The results shown in Fig. 2, B and C, indicated that although the addition of pro-uPA is necessary for inducing morphological rearrangements in cells that express the uPAR mutants with defective vitronectin binding, this addition of pro-uPA is not needed for the modulation of cell morphology in the case of wild-type uPAR. However, although HEK cells themselves do not express uPA (3),4 the cells were cultured in the presence of fetal calf serum, which contributes a low concentration of bovine pro-uPA. Therefore, these experiments did not address the question whether a completely uPA-independent function of uPAR exists in these systems. To answer this question, we set up a new cell morphology study, substituting the FCS with serum from a uPA-deficient mouse (38). Using medium supplemented with this serum, we analyzed the properties of mock-transfected and uPAR-transfected cells and compared the outcome with that obtained with FCS-containing medium (supplemental Fig. S4).
In contrast to the situation observed with FCS, a slight appearance of cellular protrusions was observed in some mock-transfected cells when cultured with the mouse serum (supplemental Fig. S4). However, although the background for this phenomenon is not known, these protrusions were much smaller than those observed in the previous experiments with uPAR-transfected cells and were only observed in a minority of the microscopy fields when scoring the morphology status (supplemental Fig. S4B). Otherwise, the result of this experiment was strikingly similar to that obtained with cells cultured in FCS. Mock-transfected cells and cells transfected with uPARW32A behaved identically and showed a very low formation of protrusions, whereas transfection with the uPARWT encoding DNA led to strong cytoskeletal rearrangements and lamellipodia, indistinguishable from those observed in the previous experiments (supplemental Fig. S4, A and B).
In conclusion, cells that adhere efficiently to vitronectin through a direct interaction with uPAR are capable of cytoskeletal rearrangements, even without a need for uPA. In the absence of a direct uPAR-vitronectin interaction, very similar uPAR-dependent rearrangements can be brought about by an alternative mechanism that requires uPA binding.
DISCUSSION
The integrated role of uPAR in pericellular proteolysis, cell adhesion, and signal transduction is the result of several molecular interactions. Although the binding of uPAR to vitronectin and to uPA has been rigorously analyzed as structurally defined protein interactions (13–15, 17), the existence of lateral interactions with other cell surface proteins has been inferred mostly from observations of molecular function. Many of these latter interactions seem to include matrix-binding integrins (for a review, see Ref. 39). Thus, the function of various integrins is assumed to be regulated through a molecular interplay with uPAR, with this leading to signal transduction and modulation of cell morphology. However, since the uPAR-dependent phenomena often include changes in cellular adhesion, an alternative possibility has been opened in that the uPAR-dependent signaling events may be just a consequence of the physical adhesion process. In a recent report, Madsen et al. (16) formulated the hypothesis that the uPAR-mediated formation of contacts with the vitronectin matrix facilitates the engagement of various signal-transducing membrane proteins with their respective matrix ligands, which, in turn, leads to signal transduction and cytoskeletal rearrangements. According to that model, these latter effects are indeed induced by the uPAR-vitronectin adhesion, but in molecular terms, they are being conducted through other receptors that are not necessarily associated with uPAR.
Our work serves to solve part of this controversy. It shows that the morphological response in this system can be triggered through more than one mechanism, depending on the inherent vitronectin adhesion capability of uPAR as well as the availability of the protease ligand, pro-uPA. Using a mutant uPAR (uPARW32A) that is defective in vitronectin binding, we succeeded in finding conditions where the formation of cytoskeletal rearrangements occurs independently of a direct uPAR-mediated adhesion. Thus, the ligation of uPARW32A with pro-uPA or its receptor-binding derivatives enabled the formation of pronounced cellular protrusions and lamellipodia in the absence of a uPAR-mediated vitronectin adhesion. A different mutant uPAR, uPARY57A, could be modulated with respect to vitronectin adhesion through the addition of pro-uPA or its derivatives in concentrations, leading to receptor saturation (14). With this mutant, the formation of cytoskeletal rearrangements in all aspects followed the adhesion ability. Thus, with uPARY57A cells, both the adhesion and the formation of lamellipodia could also be induced by the synthetic peptide antagonist, AE120, which failed to induce morphological alterations in the case of the uPARW32A mutant that has an equally high affinity for the peptide (28).
Therefore, we suggest that uPAR facilitates the occurrence of cytoskeletal rearrangements, both as a consequence of direct vitronectin binding/adhesion and as a result of uPA-dependent, lateral interactions with integrins on the cell surface (see further below). This would raise the intriguing question as to how two different interactions of uPAR, occurring at different sites on the molecular surface (13, 15, 17), can lead to the same cellular outcome. A likely explanation, however, can be suggested based on the assumption that the morphologic response is in both cases a consequence of the formation of physical cell-matrix contacts, which engages signaling receptors. The formation of these adhesive contacts can be brought about by the direct binding of uPAR to vitronectin, but it can also be initiated when uPAR stimulates the vitronectin binding of neighboring integrins (6). Although the former reaction may occur even in the total absence of uPA as shown in our experiments with cells in uPA-deficient serum (supplemental Fig. S4), the latter process is uPA-dependent (Fig. 2 and supplemental Fig.S3). Importantly, there is no conflict between this model and the lack of adhesion of some mutant uPAR transfectants in our adhesion assays. Unlike the study by Madsen et al. (16), we performed our adhesion assays in the presence of EDTA to isolate the direct uPAR-mediated adhesion event, thus excluding the contribution from integrins (4). Under cell culture conditions, however, the integrin-mediated adherence can occur even with transfectants that do not bind vitronectin directly through uPAR. This was shown directly with the uPARW32A transfectants, which grew adherently under standard conditions but failed to do so upon integrin blocking with an RGD peptide, a condition that also prevented the cytoskeletal rearrangements induced by pro-uPA. Although the detailed signaling mechanisms involved in the cytoskeletal rearrangements are outside the scope of the present work, we showed that they include a Rac-dependent pathway, just like the situation previously documented thoroughly with fibroblast-like cells (4).
Interestingly, GFD was sufficient to induce these morphological effects, thus excluding a need for a “bridging” function of the rest of the uPA molecule in the formation of lateral molecular contacts. Nevertheless, as noted above, the mere engagement of the uPA-binding cleft of uPAR, as obtained by saturation with the peptide antagonist AE120, did not lead to the same outcome. This opens the possibility that, in the induction of lateral interactions with integrins, an important conformational factor may be the shift of the interdomain organization of uPAR that occurs upon binding of GFD but not AE120 (36). It cannot be excluded, however, that the small exposed surface of the uPAR-bound GFD (13) could also play a role in these interactions.
Altogether, we have shown that expression of uPAR leads to pronounced changes in the adhesive and morphological properties of HEK cells and that these effects are modulated by both of the extracellular uPAR ligands, i.e. uPA and vitronectin. Importantly, the effect of each of these ligands on cell morphology can be exerted in the absence of an interaction between uPAR and the other ligand.
Supplementary Material
Acknowledgments
We thank Dr. Leif Lund for the generous gift of uPA-deficient mouse serum. The excellent technical assistance of Suzanne K. Møller, Jette Christiansen, Anette L. Poulsen, and Katharina H. Stegmann is gratefully acknowledged.
This work was supported by European Union contract LSHC-CT-2003-503297 and by grants from the Danish Cancer Society, the Danish Cancer Research Foundation, and the Danish Medical Research Council. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental text and supplemental figures.
Footnotes
The abbreviations used are: uPA, urokinase plasminogen activator; uPAR, uPA receptor; PBS, phosphate-buffered saline; FCS, fetal calf serum; FITC, fluorescein isothiocyanate; ATF, amino-terminal fragment of uPA; GFD, growth factor domain of uPA; WT, wild type.
T. Hillig, L. H. Engelholm, and N. Behrendt, unpublished results.
References
- 1.Behrendt, N. (2004) Biol. Chem. 385 103-136 [DOI] [PubMed] [Google Scholar]
- 2.Bugge, T. H., Flick, M. J., Danton, M. J., Daugherty, C. C., Rømer, J., Danø, K., Carmeliet, P., Collen, D., and Degen, J. L. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 5899-5904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wei, Y., Waltz, D. A., Rao, N., Drummond, R. J., Rosenberg, S., and Chapman, H. A. (1994) J. Biol. Chem. 269 32380-32388 [PubMed] [Google Scholar]
- 4.Kjøller, L., and Hall, A. (2001) J. Cell Biol. 152 1145-1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kjøller, L. (2002) Biol. Chem. 383 5-1911928822 [Google Scholar]
- 6.Wei, Y., Lukashev, M., Simon, D. I., Bodary, S. C., Rosenberg, S., Doyle, M. V., and Chapman, H. A. (1996) Science. 273 1551-1555 [DOI] [PubMed] [Google Scholar]
- 7.Nielsen, B. S., Rank, F., Illemann, M., Lund, L. R., and Danø, K. (2007) Int. J. Cancer. 120 2086-2095 [DOI] [PubMed] [Google Scholar]
- 8.Pyke, C., Ralfkiaer, E., Rønne, E., Høyer-Hansen, G., Kirkeby, L., and Danø, K. (1994) Histopathology (Oxf.) 24 131-138 [DOI] [PubMed] [Google Scholar]
- 9.Rømer, J., Pyke, C., Lund, L. R., Ralfkiaer, E., and Danø, K. (2001) J. Investig. Dermatol. 116 353-358 [DOI] [PubMed] [Google Scholar]
- 10.Danø, K., Behrendt, N., Høyer-Hansen, G., Johnsen, M., Lund, L. R., Ploug, M., and Rømer, J. (2005) Thromb. Haemostasis 93 676-681 [DOI] [PubMed] [Google Scholar]
- 11.Blasi, F., and Carmeliet, P. (2002) Nat. Rev. Mol. Cell. Biol. 3 932-943 [DOI] [PubMed] [Google Scholar]
- 12.Llinas, P., Le Du, M. H., Gårdsvoll, H., Danø, K., Ploug, M., Gilquin, B., Stura, E. A., and Menez, A. (2005) EMBO J. 24 1655-1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huai, Q., Mazar, A. P., Kuo, A., Parry, G. C., Shaw, D. E., Callahan, J., Li, Y., Yuan, C., Bian, C., Chen, L., Furie, B., Furie, B. C., Cines, D. B., and Huang, M. (2006) Science 311 656-659 [DOI] [PubMed] [Google Scholar]
- 14.Gårdsvoll, H., Danø, K., and Ploug, M. (1999) J. Biol. Chem. 274 37995-38003 [DOI] [PubMed] [Google Scholar]
- 15.Gårdsvoll, H., and Ploug, M. (2007) J. Biol. Chem. 282 13561-13572 [DOI] [PubMed] [Google Scholar]
- 16.Madsen, C. D., Ferraris, G. M., Andolfo, A., Cunningham, O., and Sidenius, N. (2007) J. Cell Biol. 177 927-939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gårdsvoll, H., Gilquin, B., Le Du, M. H., Menez, A., Jorgensen, T. J., and Ploug, M. (2006) J. Biol. Chem. 281 19260-19272 [DOI] [PubMed] [Google Scholar]
- 18.Xue, W., Kindzelskii, A. L., Todd, R. F., III, and Petty, H. R. (1994) J. Immunol. 152 4630-4640 [PubMed] [Google Scholar]
- 19.Xue, W., Mizukami, I., Todd, R. F., III, and Petty, H. R. (1997) Cancer Res. 57 1682-1689 [PubMed] [Google Scholar]
- 20.Pluskota, E., Soloviev, D. A., and Plow, E. F. (2003) Blood 101 1582-1590 [DOI] [PubMed] [Google Scholar]
- 21.Chaurasia, P., Aguirre-Ghiso, J. A., Liang, O. D., Gårdsvoll, H., Ploug, M., and Ossowski, L. (2006) J. Biol. Chem. 281 14852-14863 [DOI] [PubMed] [Google Scholar]
- 22.Resnati, M., Pallavicini, I., Wang, J. M., Oppenheim, J., Serhan, C. N., Romano, M., and Blasi, F. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 1359-1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jo, M., Thomas, K. S., Wu, L., and Gonias, S. L. (2003) J. Biol. Chem. 278 46692-46698 [DOI] [PubMed] [Google Scholar]
- 24.Nguyen, D. H., Hussaini, I. M., and Gonias, S. L. (1998) J. Biol. Chem. 273 8502-8507 [DOI] [PubMed] [Google Scholar]
- 25.Aguirre Ghiso, J. A., Kovalski, K., and Ossowski, L. (1999) J. Cell Biol. 147 89-104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei, Y., Eble, J. A., Wang, Z., Kreidberg, J. A., and Chapman, H. A. (2001) Mol. Biol. Cell 12 2975-2986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carriero, M. V., Franco, P., Gargiulo, L., Vocca, I., Cito, L., Fontana, L., Iaccarino, C., Del, P. G., Guardiola, J., and Stoppelli, M. P. (2002) Biol. Chem. 383 107-113 [DOI] [PubMed] [Google Scholar]
- 28.Ploug, M., Ostergaard, S., Gårdsvoll, H., Kovalski, K., Holst-Hansen, C., Holm, A., Ossowski, L., and Danø, K. (2001) Biochemistry 40 12157-12168 [DOI] [PubMed] [Google Scholar]
- 29.Behrendt, N., Ploug, M., Patthy, L., Houen, G., Blasi, F., and Danø, K. (1991) J. Biol. Chem. 266 7842-7847 [PubMed] [Google Scholar]
- 30.Caron, E., and Hall, A. (1998) Science 282 1717-1721 [DOI] [PubMed] [Google Scholar]
- 31.Liu, S., Bugge, T. H., and Leppla, S. H. (2001) J. Biol. Chem. 276 17976-17984 [DOI] [PubMed] [Google Scholar]
- 32.Kanse, S. M., Kost, C., Wilhelm, O. G., Andreasen, P. A., and Preissner, K. T. (1996) Exp. Cell Res. 224 344-353 [DOI] [PubMed] [Google Scholar]
- 33.Høyer-Hansen, G., Behrendt, N., Ploug, M., Danø, K., and Preissner, K. T. (1997) FEBS Lett. 420 79-85 [DOI] [PubMed] [Google Scholar]
- 34.Appella, E., Robinson, E. A., Ullrich, S. J., Stoppelli, M. P., Corti, A., Cassani, G., and Blasi, F. (1987) J. Biol. Chem. 262 4437-4440 [PubMed] [Google Scholar]
- 35.Moser, T. L., Enghild, J. J., Pizzo, S. V., and Stack, M. S. (1995) Biochem. J. 307 867-873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yuan, C., and Huang, M. (2007) CMLS Cell. Mol. Life Sci. 64 1033-1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aumailley, M., Gurrath, M., Muller, G., Calvete, J., Timpl, R., and Kessler, H. (1991) FEBS Lett. 291 50-54 [DOI] [PubMed] [Google Scholar]
- 38.Lund, L. R., Green, K. A., Stoop, A. A., Ploug, M., Almholt, K., Lilla, J., Nielsen, B. S., Christensen, I. J., Craik, C. S., Werb, Z., Danø, K., and Rømer, J. (2006) EMBO J. 25 2686-2697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chapman, H. A., and Wei, Y. (2001) Thromb. Haemostasis 86 124-129 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


