Abstract
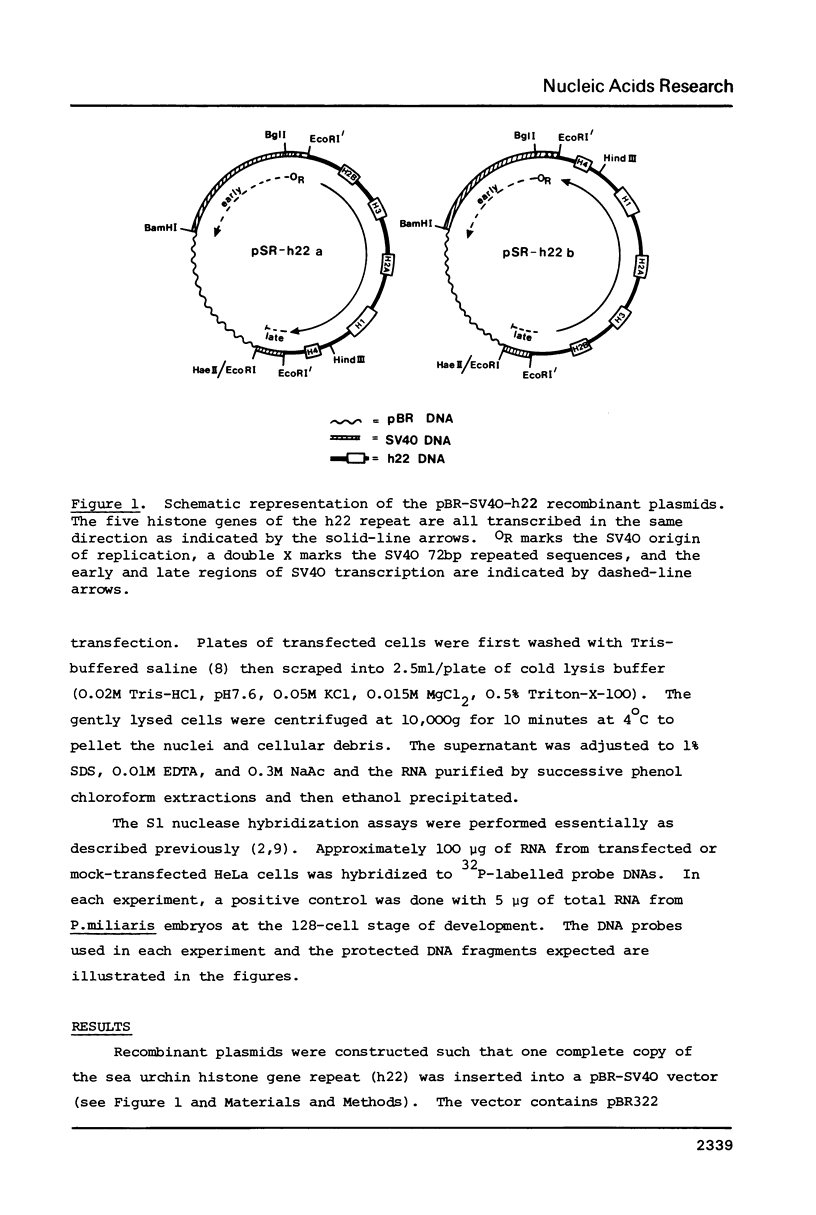
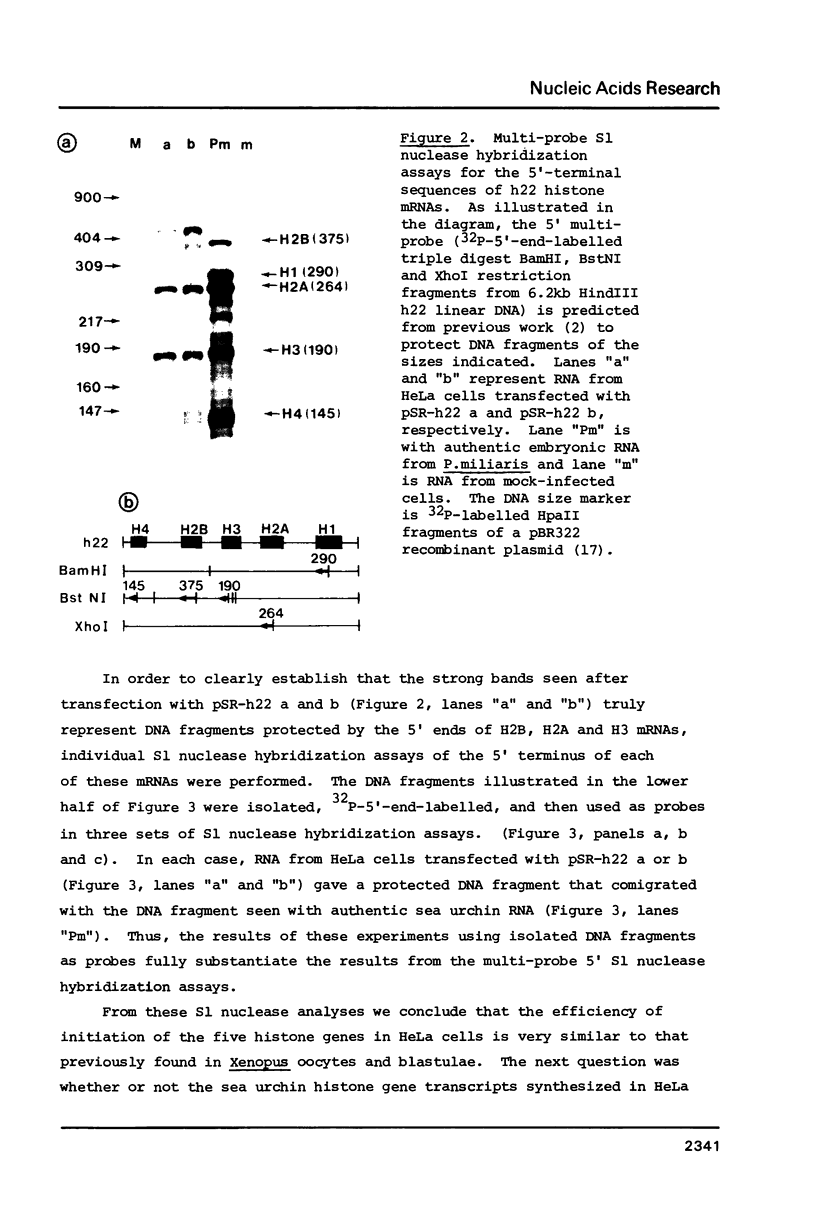
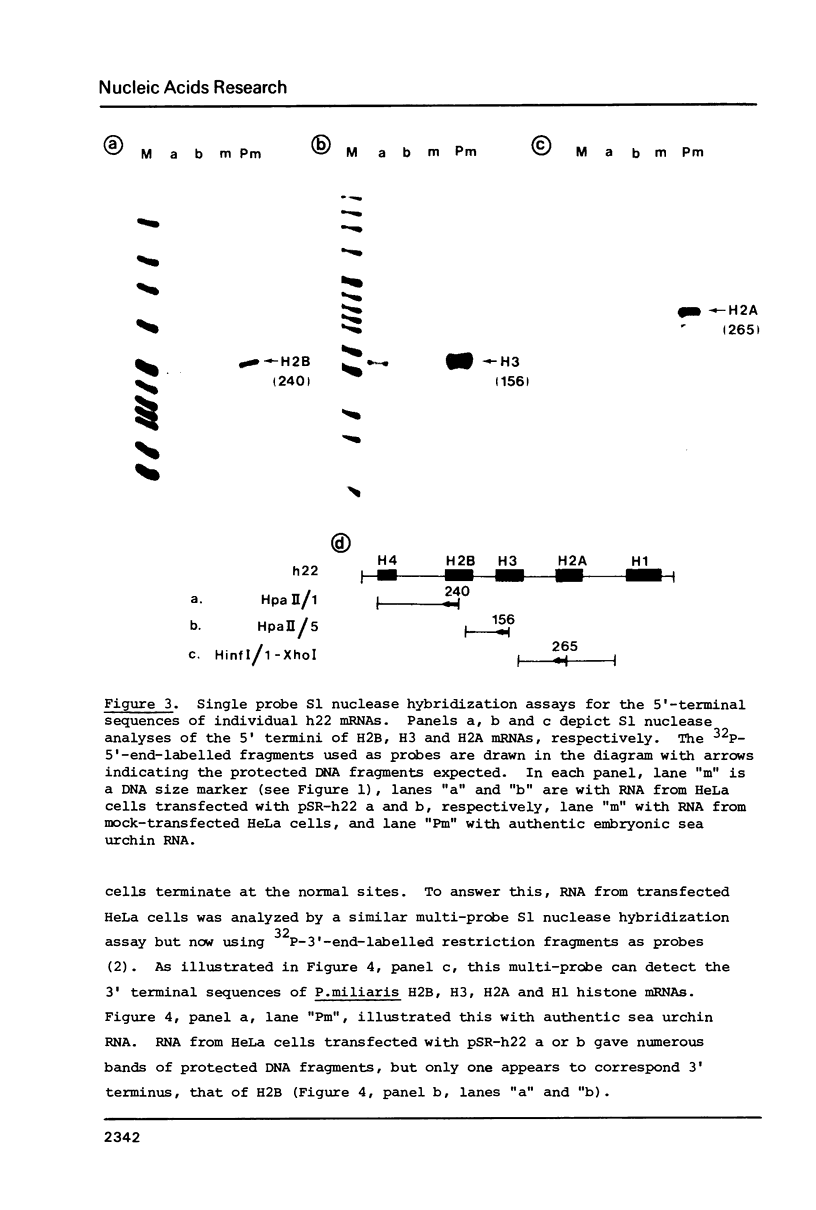
HeLa cells were transfected with recombinant DNAs containing the embryonic histone gene repeat of P.miliaris (h22) inserted in either orientation into a pBR-SV40 vector. After 2 to 3 days cytoplasmic RNA was analyzed for authentic sea urchin histone gene transcripts. The correct 5' termini of all five histone genes were detected, three (H2B, H2A and H3) at relatively high levels. In contrast, termination was largely aberrant with the correct 3' terminus of only the H2B mRNA found in significant amounts. The levels of histone gene transcription were dependent on the presence, but not the orientation, of SV40 DNA in the recombinant plasmid. The efficiency of initiation of transcription of the individual sea urchin histone genes in HeLa cells was very similar to that previously observed in Xenopus oocytes. The termination pattern, however, was quite different in that oocytes, all but the H3 gene terminate efficiently. The idiosyncrasies in termination efficiencies for these two heterologous transcription systems may reflect the presence of termination factors which are relatively species or even tissue specific and only some of which recognize the sea urchin "terminators" correctly.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Banerji J., Rusconi S., Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981 Dec;27(2 Pt 1):299–308. doi: 10.1016/0092-8674(81)90413-x. [DOI] [PubMed] [Google Scholar]
- Bendig M. M. Persistence and expression of histone genes injected into Xenopus eggs in early development. Nature. 1981 Jul 2;292(5818):65–67. doi: 10.1038/292065a0. [DOI] [PubMed] [Google Scholar]
- Benoist C., Chambon P. In vivo sequence requirements of the SV40 early promotor region. Nature. 1981 Mar 26;290(5804):304–310. doi: 10.1038/290304a0. [DOI] [PubMed] [Google Scholar]
- Birchmeier C., Grosschedl R., Birnstiel M. L. Generation of authentic 3' termini of an H2A mRNA in vivo is dependent on a short inverted DNA repeat and on spacer sequences. Cell. 1982 Apr;28(4):739–745. doi: 10.1016/0092-8674(82)90053-8. [DOI] [PubMed] [Google Scholar]
- Clarkson S. G., Smith H. O., Schaffner W., Gross K. W., Birnstiel M. L. Integration of eukaryotic genes for 5S RNA and histone proteins into a phage lambda receptor. Nucleic Acids Res. 1976 Oct;3(10):2617–2632. doi: 10.1093/nar/3.10.2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosschedl R., Birnstiel M. L. Delimitation of far upstream sequences required for maximal in vitro transcription of an H2A histone gene. Proc Natl Acad Sci U S A. 1982 Jan;79(2):297–301. doi: 10.1073/pnas.79.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss P., Dhar R., Khoury G. Simian virus 40 tandem repeated sequences as an element of the early promoter. Proc Natl Acad Sci U S A. 1981 Feb;78(2):943–947. doi: 10.1073/pnas.78.2.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentschel C. C., Birnstiel M. L. The organization and expression of histone gene families. Cell. 1981 Aug;25(2):301–313. doi: 10.1016/0092-8674(81)90048-9. [DOI] [PubMed] [Google Scholar]
- Hentschel C., Irminger J. C., Bucher P., Birnstiel M. L. Sea urchin histone mRNA termini are located in gene regions downstream from putative regulatory sequences. Nature. 1980 May 15;285(5761):147–151. doi: 10.1038/285147a0. [DOI] [PubMed] [Google Scholar]
- Hentschel C., Probst E., Birnstiel M. L. Transcriptional fidelity of histone genes injected into Xenopus oocyte nuclei. Nature. 1980 Nov 6;288(5786):100–102. doi: 10.1038/288100a0. [DOI] [PubMed] [Google Scholar]
- Hofstetter H., Kressman A., Birnstiel M. L. A split promoter for a eucaryotic tRNA gene. Cell. 1981 May;24(2):573–585. doi: 10.1016/0092-8674(81)90348-2. [DOI] [PubMed] [Google Scholar]
- Kimura G., Dulbecco R. Isolation and characterization of temperature-sensitive mutants of simian virus 40. Virology. 1972 Aug;49(2):394–403. doi: 10.1016/0042-6822(72)90492-8. [DOI] [PubMed] [Google Scholar]
- Mellado R. P., Delius H., Klein B., Murray K. Transcription of sea urchin histone genes in Escherichia coli. Nucleic Acids Res. 1981 Aug 25;9(16):3889–3906. doi: 10.1093/nar/9.16.3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertz J. E. Linear DNA does not form chromatin containing regularly spaced nucleosomes. Mol Cell Biol. 1982 Dec;2(12):1608–1618. doi: 10.1128/mcb.2.12.1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stunnenberg H. G., Birnstiel M. L. Bioassay for components regulating eukaryotic gene expression: a chromosomal factor involved in the generation of histone mRNA 3' termini. Proc Natl Acad Sci U S A. 1982 Oct;79(20):6201–6204. doi: 10.1073/pnas.79.20.6201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigler M., Pellicer A., Silverstein S., Axel R. Biochemical transfer of single-copy eucaryotic genes using total cellular DNA as donor. Cell. 1978 Jul;14(3):725–731. doi: 10.1016/0092-8674(78)90254-4. [DOI] [PubMed] [Google Scholar]
- Wyllie A. H., Laskey R. A., Finch J., Gurdon J. B. Selective DNA conservation and chromatin assembly after injection of SV40 DNA into Xenopus oocytes. Dev Biol. 1978 May;64(1):178–188. doi: 10.1016/0012-1606(78)90069-6. [DOI] [PubMed] [Google Scholar]