Abstract
MazG is a nucleoside triphosphate pyrophosphohydrolase that hydrolyzes all canonical nucleoside triphosphates. The mazG gene located downstream from the chromosomal mazEF “addiction module,” that mediated programmed cell death in Escherichia coli. MazG activity is inhibited by the MazEF complex both in vivo and in vitro. Enzymatic activity of MazG in vivo affects the cellular level of guanosine 3′,5′-bispyrophosphate (ppGpp), synthesized by RelA under amino acid starvation. The reduction of ppGpp, caused by MazG, may extend the period of cell survival under nutritional stress. Here we describe the first crystal structure of active MazG from E. coli, which is composed of two similarly folded globular domains in tandem. Among the two putative catalytic domains, only the C-terminal domain has well ordered active sites and exhibits an NTPase activity. The MazG-ATP complex structure and subsequent mutagenesis studies explain the peculiar active site environment accommodating all eight canonical NTPs as substrates. In vivo nutrient starvation experiments show that the C terminus NTPase activity is responsible for the regulation of bacterial cell survival under nutritional stress.
MazG is a nucleoside triphosphate pyrophosphohydrolase that hydrolyzes all canonical nucleoside triphosphates into their corresponding nucleoside monophosphates and PPi and interacts with the gene product of era (1). Although mazG is highly conserved among bacteria, it appears not to be essential to the bacterial viability, because an Escherichia coli strain lacking mazG still survived under laboratory conditions (1). It was proposed that MazG might have a “housecleaning” function by hydrolyzing noncanonical NTPs, such as 8-oxo-dGTP, dUTP, dITP, and 2-oxo-dATP, whose incorporation into the nascent DNA leads to the increased mutagenesis and DNA damage (2, 3). A recent report suggested that a physiological function of MazG is related to the cellular addiction system, mazEF, which is located downstream from the relA gene (4). Addiction systems are specified by the labile antitoxin (MazE in this case), which is rapidly degraded by a specific protease, and its cognate stable toxin (MazF) that, in the absence of antitoxin, acts on its target, causing immediate cell growth inhibition and, after a prolonged period, death (5–7). Thus, the cells are addicted to the constant presence of the antitoxin (8).
mazG is transcribed from the same polycistronic mRNA of mazEF, and its activity is inhibited by the MazEF complex both in vivo and in vitro (4). Activation of MazG causes reduction of (p)ppGpp cellular level, a product of enzymatic activity of ribosome-bound RelA synthetase under amino acid starvation (9). (p)ppGpp induces the stringent response through alteration of genome-wide transcriptional profiles (10). Comparison of the survival ability of wild-type E. coli and its ΔmazG mutant under amino acid starvation allows us to suggest that MazG has a function of delaying the “point of no return” (4, 11) (i.e. under nutritional stress, MazG can extend the period of cell survival until final switching on the mechanism of programmed cell death).
Despite the substantial involvement of MazG in the stringent response, molecular background of MazG activity is still obscure. Nevertheless, the crystal structures of five its single-domain analogs were solved: Vibrio sp. iMazG (12), Sulfolobus solfataricus MazG (Protein Data Bank code 1vmg; Joint Center for Structural Genomics), Mus musculus RS21-C6 (13), Chromobacterium violaceum MazG (Protein Data Bank code 2a7w; Northeast Structural Genomics Consortium), and Bacillus subtilis YpjD (Protein Data Bank code 2gta; Northeast Structural Genomics Consortium). Studies on phylogenetic distribution of MazG domains suggest that the most abundant form of MazG, to which MazG of E. coli belongs, occurs as a protein with two sequence-related domains in tandem (12). Sequence homology searches showed that in all bacterial genomes currently defined, the gene encoding the subfamily of tandem domain MazG occurs in conjunction with the relA gene (e.g. in all five Vibrio genomes available, in Deinococcus radiodurans, and, of course, in E. coli (12). The biological role of such tandem domain MazG proteins appears to be to regulate cellular levels of ppGpp (4, 14).
Here, we report the first crystal structure of tandem domain MazG from E. coli (EcMazG) at 2.0 Å resolution, which forms a dimer through the interaction of two repeated MazG-like domains. Two domains have similarly folded globular structures; however, NTPase activity was observed only at the C-terminal domain. Crystal structure of an ATP-bound form and its subsequent site-directed mutagenesis study revealed the key residues for the magnesium coordination, substrate stability, and enzyme catalysis. Starvation experiments suggest that MazG prevents cell death under conditions of nutrient starvation using the C-terminal NTPase activity.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification of MazG from E. coli—The gene coding for full-length EcMazG (residues 1–263) was amplified from the chromosomal DNA of E. coli strain by PCR with the primers as shown in supplemental Table 1. The PCR product was then subcloned into pPROEX HTa (Invitrogen) with His6 at the N terminus and a recombinant TEV protease cleavage site. The resulting expression vector, pPROEX HTa: EcmazG, was transformed into E. coli B834 strain and was grown in an LB medium containing 100 μg/ml ampicillin at 37 °C to A600 of 0.6. After induction with 1.0 mm IPTG2 for a further 20 h at 22 °C, the culture was harvested by centrifugation at 5,000 × g for 15 min at 4 °C. Cell pellet was resuspended in ice-cold buffer A (50 mm Tris-HCl, pH 8.0, 5 mm β-mercaptoethanol) and disrupted by ultrasonication. The cell debris was removed by centrifugation at 11,000 × g for 1 h, and lysate was bound to Ni2+-nitrilotriacetic acid-agarose (Qiagen). After washing with buffer A containing 10 mm imidazole, the bound proteins were eluted with 300 mm imidazole in buffer A. The His6 tag was released from the EcMazG by incubating with recombinant TEV protease (Invitrogen). Further purification was done by applying the sequential chromatographic steps of HiTrap Q ion exchange and Superdex 200 size exclusion. The purified protein with three-residue cloning artifact (Gly-His-Met) at the N terminus was concentrated to 20 mg/ml in 50 mm Tris-HCl, pH 8.0, and stored at -80 °C for crystallization trials. SDS-PAGE analysis of the purified protein showed a single band of ∼30.0 kDa that corresponds to the calculated molecular weight of an EcMazG monomer. The SeMet-substituted protein was expressed in a minimal medium supplemented with SeMet and purified under the same conditions as the wild-type protein.
Crystallization, Data Collection, and Structure Determination—The initial crystallizing conditions were obtained from Sparse Matrix Screening (15). Suitable crystals for diffraction appeared around 0.1 m trisodium citrate (pH 5.5), 0.2 m ammonium acetate, and 20% polyethylene glycol 4000 at 22 °C within 2 days and reached up to their maximal sizes in about 7 days with dimensions of ∼0.4 × 0.3 × 0.3 mm. For data collection, 30% (w/v) glycerol was added to the crystallizing precipitant as a cryoprotectant, and the crystals were immediately placed in a -173 °C nitrogen gas stream. X-ray diffraction data of native crystals were collected at a resolution of 2.0 Å at the 6C1 beamline of the Pohang Accelerator Laboratory using a QUANTUM 210 CCD detector (ADSC, San Diego, CA). For the ATP-bound EcMazG structure, crystals of EcMazG K257A mutant were obtained from the same crystallization conditions as for wild-type EcMazG, supplemented with 25 mm MgCl2, and soaked with ATP to a final concentration of 2.5 mm for 1 h. The data were then indexed, integrated, and scaled using the HKL2000 suite (16). The crystals belonged to the space group P212121, with the unit cell parameters a = 62.78, b = 67.42, c = 140.075 Å. With two EcMazG molecules in the asymmetric unit, the crystal volume per unit of protein mass was 2.53 Å3 Da-1, corresponding to a solvent content of 51% (17). The SeMet-substituted protein crystals were obtained from the same crystallizing conditions as for the native protein crystal. The single wave-length anomalous dispersion data with the SeMet crystal were collected at the 6C1 beamline of the Pohang Accelerator Laboratory at a wavelength of 0.97953 Å. Fourteen selenium atoms of the expected 16 in the asymmetric unit were identified at 2.9 Å resolution using the program SOLVE (18). The electron density was improved by density modification using the program RESOLVE (19), resulting in 52% of the cloned residues being automatically built. Further model building was performed manually using the program WinCoot (20), and the refinement was performed with CCP4 refmac5 (21) and CNS (22). The data statistics are summarized in Table 1. The refined models were deposited in the Protein Data Bank (wild-type EcMazG, Protein Data Bank entry 3cra; EcMazG complexed with ATP, Protein Data Bank entry 3crc).
TABLE 1.
Data collection and refinement statistics Values in parentheses are for the highest resolution shell. PAL, Pohang Accelerator Laboratory.
| Selenium peak | EcMazG (native) | EcMazG (ATP) | |
|---|---|---|---|
| Beamline | 6C1(MXII), PAL | 6C1(MXII), PAL | 6C1(MXII), PAL |
| Wavelength (Å) | 0.97953 | 1.23985 | 1.23985 |
| Space group | P212121 | P212121 | P212121 |
| Cell parameters, a, b, c (Å) | 63.39, 68.17, 140.30 | 62.78, 67.42, 140.07 | 63.23, 66.91, 140.70 |
| Resolution limits (Å) | 50-2.48 (2.57-2.48) | 50-2.0 (2.07-2.0) | 50.0-3.0 (3.11-3.0) |
| Total reflections | 291,271 | 172,203 | 48,461 |
| Unique reflections | 21,953 | 41,095 | 12,574 |
| Completeness (%) | 99.8 (98.1) | 93.7 (84.0) | 95.5 (93.7) |
| Rmergea (%) | 7.6 (66.2) | 3.8 (32.4) | 7.6 (19.9) |
| I/σ (I) | 26.2 (2.4) | 37.1 (3.1) | 16.6 (3.7) |
| Figure of merit,b SOLVE/RESOLVE | 0.33/0.74 | ||
| Rfactorc (%) | 23.5 | 22.1 | |
| Rfreed (%) | 26.6 | 30.1 | |
| No. of atoms, protein/water | 3787/233 | 3634/125 | |
| Root mean square deviation, bonds (Å)/angles (degrees) | 0.006/0.89 | 0.007/1.13 | |
| Geometry (most favored) (%) | 94.4 | 92.0 | |
| Geometry (additionally allowed) (%) | 5.2 | 7.1 |
Rmerge = Σhkl Σj|Ij – 〈I〉|/ΣhklΣjIj, where 〈I〉 is the mean intensity of reflection hkl
Figure of merit = |ΣP(α)eiα/ΣP(α)|, where P(α) is the phase probability distribution and α is the phase (50.0-2.9 Å)
Rfactor = Σhkl∥Fo| – |Fc∥/Σhkl|Fo|, where Fo and Fc are the observed and calculated structure factor amplitude, respectively, for reflections hkl included in the refinement
Rfree is the same as Rfactor but calculated over a randomly selected fraction (7%) of reflection data not included in the refinement
Preparations of Other Proteins Used in This Study—The point mutants used in the study were constructed by site-directed mutagenesis, and the PCR primer sequences are summarized in supplemental Table 1. For the preparation of EcMazG deletion mutants, genes coding for N-terminal (residues from Met1 to Ala127) and C-terminal domains (residues from Ser138 to Leu263) of EcMazG were amplified from the pPROEX HTa: EcmazG by PCR. The PCR product was then subcloned into pPROEX HTa (Invitrogen) with His6 at the N terminus and a recombinant TEV protease cleavage site. Expression and purification of point and deletion mutants of EcMazG were performed with the same protocol as described above.
NTPase Activity Assay—ATP and dTTP were used for representing substrates of purine-based ribonucleoside triphosphate and pyrimidine-based deoxyribonucleoside triphosphate, respectively. The NTPase activity of wild-type and mutant EcMazG was assayed by detecting hydrolyzed nucleoside monophosphate by TLC. The assay was carried out in 20 μl of reaction buffer (40 mm Tris-HCl, pH 8.0, 2.5 mm MgCl2, and 1 mm DTT) containing ATP or dTTP to a final concentration of 1 mm. An appropriate protein to the final concentration of 20 μm was added in order to initiate the reaction and incubated at 37 °C for 10 min. The reaction was terminated by the addition of equal volume of stop buffer (2% SDS and 20 mm EDTA), and 2 μl of terminated reaction mixture was spotted on polyethyleneimine-cellulose TLC plate, which was then developed in 0.75 m KH2PO4, pH 3.3. For detection of hydrolyzed products, spots were visualized by UV shadowing.
Cell Growth Rate Measurement and Nutrient Starvation Experiment—E. coli strain BL21 ΔmazG was obtained by insertion of the kan+ coding gene into the mazG using a suicide vector, pCVD422 (23), and the strain was used for cell growth rate measurement. The strain was transformed with expression plasmids producing wild-type EcMazG and its mutants. Transformants were grown overnight at 37 °C in LB-Amp/Kan medium. Overnight culture was inoculated onto fresh medium, and 1 mm IPTG was added to induce protein expression at A600 = 0.2. Samples were collected at the appropriate time, and cell growth was monitored by measuring the A600. For viable cell counts, samples were collected at the appropriate time and washed with M9 medium. After serial dilutions, 100 μl of cultures was spotted on LB-Amp/Kan plates. Viable cell growth was then monitored by measuring colony-forming units. In order to test the effect of nutrient starvation on cell growth, E. coli strain BL21 ΔmazG transformed with an appropriate expression plasmid was grown on M9 minimal medium with supplements (0.2% glucose, all amino acids (20 μg/ml each), 0.05 mm thiamine, 2 mm MgSO4, 0.1 mm CaCl2) to A600 = 0.2, and 1 mm IPTG was added. Thirty min after induction, serine hydroxamate (SH) was added to the final concentration of 1 mg/ml, and cells were grown for 5 h in nutrient starvation conditions. Optical density and viable cell count measurement was performed as described above.
Primer Extension Experiments—E. coli MC4100 ΔmazEFG relA+ strain was transformed with pSK10Δ6-pef plasmid carrying lacZ gene under mazEFG promoter (24). The transformants were grown in M9 minimal medium, supplemented with 100 μg/ml ampicillin, 0.2% glucose, and a mixture of all amino acids at 37 °C to an A600 of 0.2. At this point, cell culture was divided into two flasks, and one of them was treated with 1 mg/ml SH. Samples for primer extension analysis were taken just prior to the division and 5, 10, and 20 min after it. RNA extraction was carried out using the RNeasy Mini kit (Qiagen). Primer extension experiments were carried out with avian myeloblastosis virus reverse transcriptase (U.S. Biochemical Corp.) according to Gafny et al. (25). The oligonucleotide primer used for the pSK10Δ6-pef construct was “-40 M13 forward” (Amersham Biosciences) from the lacZ gene to the transcription start sites of the mazEFG promoter. The primer was end-labeled by polynucleotide kinase (New England Biolabs, Inc.) with [γ-32P]ATP (Amersham Biosciences) and purified on the Sephadex G-50 columns (Roche Applied Science). Reaction products were resolved on a 6% sequencing gel. A DNA sequencing reaction was performed with the same plasmid and primer and run on the gel parallel to the primer extension reaction. To quantify the RNA levels, the gels were analyzed, and the bands were quantified using the Fujix BAS100 PhosphorImager.
RESULTS
Overall Fold of E. coli MazG—EcMazG is mostly composed of α-helices. A monomer contains 10 α-helices and can be subdivided into two domains, the N- and C-terminal domains. Each domain consists of five α-helices (α1–α5 for the N-terminal and α6–α10 for the C-terminal domain) with a linker (Ala124–Pro140) between them (Fig. 1A). Intertwining of two respective domains generates two new globular structures in the dimeric EcMazG structure, which will be referred to hereafter as ND and CD for the N- and C-terminal domains, respectively (Fig. 1B). Hydrophobic interactions between α2 and α3 helices or α7 and α8 helices mediate protein dimerization (supplemental Fig. 1). Two sets of two α-helices (α2 and α3 of the ND and α7 and α8 of the CD) form a four-helical bundle in both ND and CD. Both domains have heart-shaped “rigid core” regions, which are formed by four α-helices (α1–α4 and α6–α9 for the ND and CD, respectively). In ND, two α-helices of α1 and α4 are located perpendicular to the four-helical bundle and stabilized mostly by hydrophobic interactions through a series of nonpolar residues from α1, α3, and α4. The CD is similarly folded as that of the ND using its corresponding α-helices (Fig. 1B). The last helix α5 or α10 of the respective ND or CD is located in parallel with the four-helical bundle of each domain. Although the flexible α5 in the ND is traced only in one molecule, the α10 in the CD is tightly bound to the extended region of α9, which forms an “additional region” (Fig. 1B).
FIGURE 1.
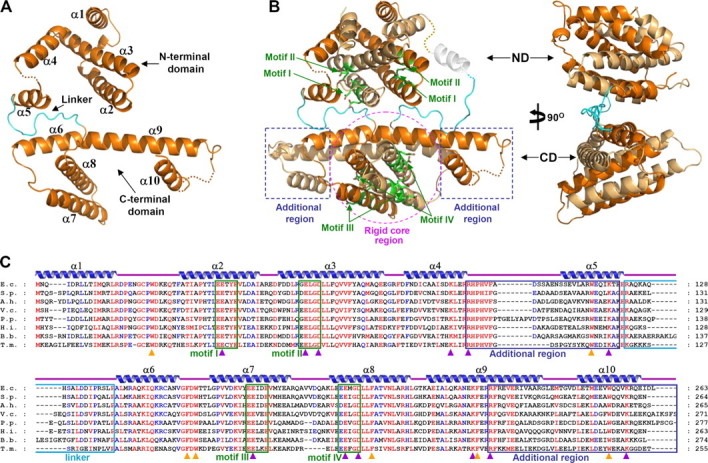
Overall shape of EcMazG. A, a monomeric EcMazG structure. EcMazG is composed of two similarly folded MazG-like domains, the N- and the C-terminal domains, and these domains are connected by a linker (cyan). The domains and secondary structure elements are labeled. B, overall shape of an EcMazG dimer. The N-terminal (ND) and the C-terminal (CD) domains are indicated by arrows. Two polypeptides are differentiated by colors of orange and light orange. A rigid core region of the CD is indicated by a purple circle, and additional regions are by blue rectangles. EEXX(E/D) motifs are presented by green stick models, and labeled by their corresponding numbers. C, alignment of amino acid sequences of tandem domain MazGs. Secondary structure elements are shown appropriately. Identical and highly conserved residues are presented in red and blue characters, respectively. Four EEXX(E/D) motifs, two on each domain, are indicated by green rectangles. Two additional regions and a linker are indicated by blue and cyan rectangles, respectively. Residues involved in the magnesium coordination and enzyme catalysis are marked with purple rectangles, and those involved in hydrophobic cavity formation for the binding of NTP bases are marked with orange rectangles. E.c., E. coli; S.p., Serratia proteamaculans; A.h., Aeromonas hydrophila; V.c., Vibrio cholerae; P.p., Pseudomonas putida; H.i., Hemophilus influenzae; B.b., Brucella abortus biovar; T.m., Thermotoga maritima.
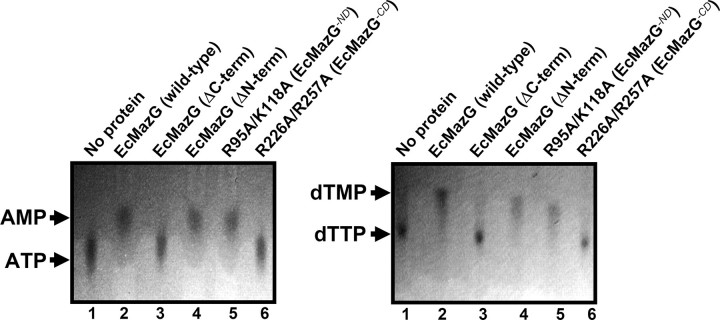
CD of EcMazG Has an NTPase Activity—The crystal structure of EcMazG shows four potential active sites at an EcMazG dimer, two each from the ND and the CD, all of which contain the EEXX(E/D) motif: Glu38, Glu39, Glu42, Glu58, and Asp61 from the α2 and α3 in the ND domain, Glu171, Glu172, Glu175, Glu192, Glu193, and Asp196 from α7 and α8 in the CD domain (Fig. 1, B and C). In both domains, several conserved positively charged residues (Lys91, Arg95, and Lys119 in the ND and Lys222, Arg226, and Lys257 in the CD) are located close to the EEXX(E/D) motif. To find out the domain(s) that has an NTPase activity, we performed NTPase assays using the wild-type EcMazG and several point and deletion mutants. Among all eight canonical nucleotides (ATP, TTP, GTP, CTP, dTTP, dGTP, dCTP, and UTP), ATP and dTTP were used as representatives of purine-based ribonucleoside triphosphate and pyrimidine-based deoxyribonucleoside triphosphate, respectively. Wild-type EcMazG showed a strong nucleotide pyrophosphohydrolysis activity against ATP and dTTP. Although the mutations on the ND (R95A/K118A) do not affect the nucleotide pyrophosphohydrolysis activity, those on the CD (R226A/K257A) caused loss of the activity against both ATP and dTTP (Fig. 2). The structure-based deletion mutants further confirmed these data; contrary to a C-terminal deletion mutant (Met1–Ala127) with no nucleotide pyrophosphohydrolysis activity, the N-terminal deletion mutant (Ser138–Leu263) showed the same amount of enzymatic activity as that of the wild-type protein (Fig. 2).
FIGURE 2.
NTPase activity of EcMazG. ATP and dTTP were used as representing substrates for purine-based ribonucleoside triphosphates and pyrimidine-based deoxyribonucleoside triphosphates, respectively. Hydrolysis experiments were carried out using wild-type and point/deletion mutant EcMazG. The hydrolysis products were analyzed by polyethyleneimine-cellulose thin layer chromatography. Lane 1 for both TLC assays indicates reaction without protein, and lanes 2–6 indicate reactions using EcMazG of wild type, ND alone (residues from Met1 to Ala127), CD alone (residues from Ser138 to Leu263), R95A/K118A mutation, and R226A/K257A mutation, respectively. Detailed reaction procedures are described under “Experimental Procedures.”
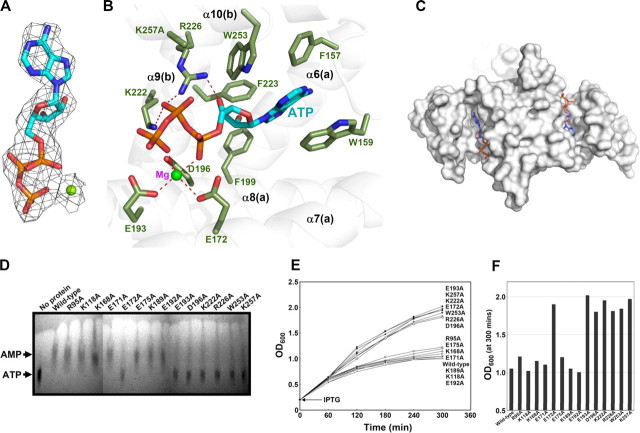
The Active Site of CD—The active site of the CD domain was revealed by the EcMazG-ATP complex structure and the subsequent mutation studies. To prevent the hydrolysis of the ATP molecule during crystallization, the K118A/K257A mutant of EcMazG was used for the complex structure with ATP. There was no structural difference between the apo- and the ATP-bound form of the enzyme. Only one ATP molecule was bound at one of the two C-terminal active sites with no ATP molecule at the other active site, in which the side chain of Met241 from the symmetry-related molecule occupies, and as expected, no ATP molecule was found at the N-terminal active sites. At the active site of the CD, one magnesium ion was coordinated by three acidic residues of Glu172, Glu193, and Asp196 from the two EEXX(E/D) motifs with two hydroxyl groups of α- and γ-phosphates as the remnant ligands. β-Phosphate interacts with the side chains of Lys222 and Arg226 and with two water molecules. The ribose ring is stabilized by forming a hydrogen bond between the 3′-hydroxyl group of the ring and the side chain of Arg226. The adenine base was located in a hydrophobic pocket constituted by a series of conserved aromatic residues, Phe157, Trp159, Phe199, Phe223, and Trp253 (Fig. 3, A–C).
FIGURE 3.
The active site of the EcMazG C-terminal domain. A, diagram showing the ATP molecule and a magnesium ion bound at the active site of the EcMazG CD. The 2Fo - Fc electron density (black mesh) is contoured at 1.0 σ. Carbons, oxygens, nitrogens, and phosphates are shown as green, red, blue, and orange, respectively. B, diagram showing the active site of the EcMazG CD bound with an ATP molecule. Secondary structure elements from two different polypeptides are labeled a and b. The ATP molecule is presented as a ball-and-stick model. Residues involved in the magnesium coordination, enzyme catalysis, and hydrophobic cavity formation for adenine base binding are labeled and presented as a light green stick model. Ionic and hydrogen bonds are shown with red dotted lines. A magnesium ion is presented with as a green ball. C, surface-fill model of EcMazG bound with ATP. ATP molecules bound at the C-terminal active sites are shown with a stick model. Only one ATP molecule was bound at one (right side) of the two C-terminal active sites with no ATP molecule at the other active site (left side), which the side chain of Met241 from the symmetry-related molecule occupies, and an ATP molecule was modeled in the diagram. D, NTPase activity assays. Hydrolysis of ATP to AMP was carried out using wild type and 14 point mutants of EcMazG. An EcMazG mutant used for each reaction was identified at the top of the TLC. Detailed reaction procedures are described under “Experimental Procedures.” E and F, toxic effect of mutant EcMazG on cell growth. E. coli strain BL21ΔmazG was transformed with plasmids expressing an appropriate protein. The transformants were grown to an A600 of 0.2, and 1 mm IPTG was added for induction. Cell growth was then measured at A600. A600 at 300 min after the IPTG induction on E was redrawn and differentiated on F.
Based on the structural features of the active site and amino acid sequence conservation analysis (Figs. 1C and 3B), 14 point mutants were constructed, including R95A, K118A, K168A, E171A, E172A, E175A, K189A, E192A, E193A, D196A, K222A, R226A, W253A, and K257A, and their NTPase activity was checked. The ATP pyrophosphohydrolysis activity was detected for the mutants of R95A, K118A, K168A, E171A, E175A, K189A, and E192A but not for those of E172A, E193A, D196A, K222A, R226A, W253A, and K257A (Fig. 3D). In order to test the inhibitory effect of MazG overexpression on cell growth rate in E. coli, the activities of the mutants were further tested in vivo. Like wild-type EcMazG, the overexpression of the mutants of R95A, K118A, K168A, E171A, E175A, K189A, or E192A significantly retarded the E. coli growth. However, the strain transformed with the mutants of E172A, E193A, D196A, K222A, R226A, W253A, or K257A has grown normally (Fig. 3, E and F).
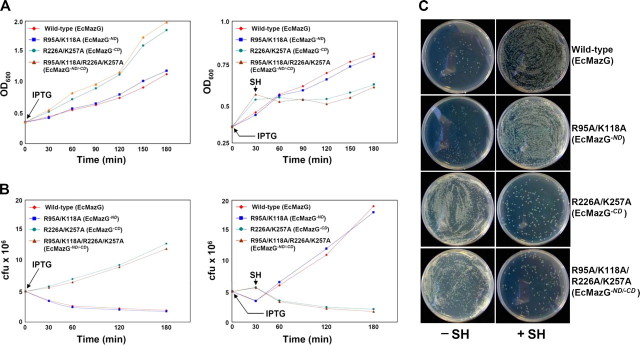
The Physiological Effect of EcMazG Mutants at Nutritional Stress—In order to identify the domain that is responsible for MazG-dependent cellular response to nutrition starvation and to test how the abolishment of NTPase activity of the CD domain influences the function, amino acid starvation experiments were performed using various constructs of EcMazG. mazG-deleted E. coli strain BL21 was transformed with four expression plasmids: the wild-type EcMazG and three of its mutants: R95A/K118A (MazG-ND), R226A/K257A (MazG-CD), and R95A/K118A/R226A/K257A (MazG-ND/-CD), which represent EcMazG proteins lacking activity in the ND, the CD, and both the ND and CD, respectively. Serine hydroxamate (SH), a serine analog, at a concentration of 1 mg/ml was used for the induction of the stringent response (4). In accordance with the data described above (Fig. 3, E and F), whereas the growth of cells expressing the wild-type or MazG-ND mutant was arrested under normal conditions, cells expressing MazG-CD or MazG-ND/-CD mutants grow without growth arrest. However, the induction of amino acid starvation by SH addition reverses the patterns of cell growth; cells expressing wild-type or MazG-ND mutant revert to their normal growth, whereas cells expressing MazG-CD or MazG-ND/-CD mutants show growth arrest (Fig. 4A). The effect of SH induction on the cell growth was also measured by monitoring colony-forming units of the same set of constructs used above, which also showed the same tendency as the optical density cell measurements (Fig. 4, B and C).
FIGURE 4.
Effects of EcMazG mutants on cell growth by the induction of stringent response. A, E. coli strain BL21ΔmazG was transformed with plasmids expressing wild-type EcMazG, EcMazG-ND (R95A/K118A), EcMazG-CD (R226A/K257A), and EcMazG-ND/-CD (R95A/K118A/R226A/K257A). The transformants were grown to an A600 of 0.2, and 1 mm IPTG was added for induction. Thirty min later, SH was added to a final concentration of 1 mg/ml for the induction of stringent response. Cell growth was then measured at A600 and compared with those with no SH addition. B, effect of EcMazG mutants on cell growth by the induction of stringent response was also measured by monitoring viable cell counts. C, colony formations after exposure to SH. Experiments were performed with the same procedures as in A. Cells exposed to SH were grown for 5 h. Treated cells were harvested and washed with M9 medium to remove SH. After 1 × 104 times dilution, 100 μl of cell suspensions were plated onto LB-Amp plates. Cells grown with the SH addition were also plated for negative controls.
DISCUSSION
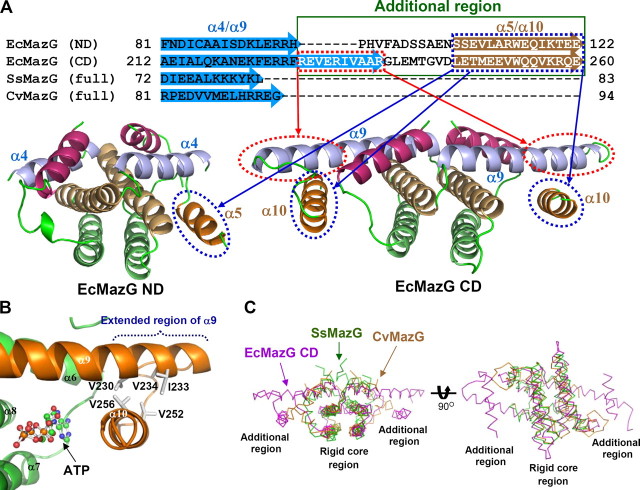
Unique Structural Features of the CD—The crystal structure of EcMazG reveals that the protein is composed of two similarly folded MazG-like domains in tandem. Point and deletion mutagenesis studies showed that only the CD domain exhibited an NTPase activity, which might be explained by the structural differences between the potential active sites in two domains. The well organized active sites of the CD are derived from the tightly formed “additional region,” in which α10 helix is stabilized on the extended region of the α9 through hydrophobic interactions among Val230, Ile233, Val234, Val252, and Val256 (Fig. 5, A and B). On the other hand, the lack of the corresponding region at the short α4 induces the last helix α5 flexible in ND (Fig. 5A). The extended region, seven amino acids, of the α9 helix serves as a rigid supporter for the α10 helix, and they together form an additional region, where the α10 helix provides key residues for substrate binding and enzyme catalysis, such as Trp263 and Lys257 (Figs. 3B and 5A). Moreover, among five conserved aromatic residues (Phe157, Trp159, Phe199, Phe223, and Trp253) involved in the formation of the nucleotide base binding pocket at the C-terminal active site, only two residues (Trp23 and Trp115) are conserved in the N-terminal active site, implying different nucleotide base binding pocket environments between the active sites of these two domains (Fig. 1C). The S. solfataricus MazG (Protein Data Bank code 1vmg) and C. violaceum MazG (Protein Data Bank code 2a7w) are short versions of the MazG-like proteins, which are composed of one MazG-like domain with 83 and 108 amino acid residues, respectively. They are composed of only four α-helices and well superimposed with the rigid core region of both ND and CD of EcMazG, resulting in no additional region (Fig. 5, A and C). It was suggested that this type of one-domain MazG proteins has a “housecleaning” function by degrading noncanonical NTPs, whose incorporation into the nascent DNA leads to increased mutagenesis and DNA damage (12). Until now, we could not identify the enzyme activity and physiological function of the ND domain of EcMazG. Our study reveals that the ND domain of EcMazG does not exhibit an NTPase activity (Fig. 2); however, the domain contains structural features responsible for nucleotide pyrophosphohydrolysis activity, such as EEXX(E/D) motifs and key basic catalytic residues as well (Fig. 1). These observations suggest that the ND domain of EcMazG might have a “housecleaning” function by hydrolyzing noncanonical nucleotides (2, 3).
FIGURE 5.
Unique structural features of the CD of EcMazG. A, amino acid sequences composing the fourth and fifth α-helices of MazG-like proteins were aligned. An additional region is identified by a green box. The extended region of α9 in the CD of EcMazG is indicated by a red box and circle. α5 and α10 from the ND and the CD of EcMazG, respectively, are indicated by a blue box and circle. B, hydrophobic interactions to form a rigid additional region of the CD. An extended region of the α9 is indicated, and five nonpolar residues involved in the hydrophobic interactions between α9 and α10 are presented with a gray stick model. C, EcMazG CD was superimposed with SsMazG and CvMazG, both of which lack additional region and cover only the rigid core region of the EcMazG CD. The right image is 90° vertically rotated from the left image.
Enzymatic Activities of the CD—Our crystal structure reveals that all four potential active sites in a dimeric EcMazG contain EEXX(E/D) motifs, which are frequently found in enzymes, requiring magnesium or manganese ions for their activities (26–31). Campylobacter jejuni dUTPase coordinates a magnesium ion with acidic residues of Glu46, Glu49, Glu74, and Asp77 and hydrolyzes the substrate using basic residues of Lys175, Arg182, and Lys194 (31). The substrate binding mode and enzymatic reaction mechanism of the C-terminal NTPase activity might be deduced from the crystal structure of EcMazG-ATP complex and site-directed mutagenesis studies (Fig. 3), which are similar to those of C. jejuni dUTPase. However, there are a few differences between the active sites of these two enzymes. First, although the active site of EcMazG contains the conserved EEXX(E/D) motifs, C. jejuni dUTPase has EXX(E/D) motifs. Second, the EEXX(E/D) motifs and three basic residues are provided from two independent polypeptides in EcMazG but from the same polypeptide in C. jejuni dUTPase. Third, the active site of C. jejuni dUTPase facilitates only a uracil ring by providing several hydrogen bonders with the base and the ribose ring, whereas no specific interaction was observed between the adenine ring of ATP and EcMazG at the active site, which is able to accommodate all four bases. Nonpolar aromatic residues constitute a hydrophobic pocket for bases in EcMazG without any interaction to the 2′-hydroxyl group of the ribose ring, explaining how both oxy- and deoxyribonucleotides could be the substrates of EcMazG (Fig. 3B).
EcMazG functions as a regulator of cellular response to starvation by lowering the cellular concentration of (p)ppGpp, which can be mediated through direct degradation of (p)ppGpp and/or indirectly through degradation of NTPs, substrates for (p)ppGpp synthesis by RelA (4). Our work provides detailed information on the NTPase activity of EcMazG. Although we tested ppGpp hydrolysis activity of this protein, a very low hydrolysis rate of ppGpp was detected for prolonged incubation time (data not shown). However, several structural features of the active site of the EcMazG might provide clues for the potential (p)ppGpp hydrolysis activity. As described previously, the active site of EcMazG contains six conserved acidic residues (Glu171, Glu172, Glu175, Glu192, Glu193, and Asp196), and only three of them (Glu172, Glu193, and Asp196) are involved in the magnesium coordination for NTPase activity. The remaining three acidic residues (Glu172, Glu193, and Asp196) might assist the binding of phosphates from (p)ppGpp. Beside the adenine binding pocket, another is constructed by conserved residues of Phe199, Ala219, Asn220, and Phe223, which is near the EEXX(E/D) motifs and three basic catalytic residues (Lys222, Arg226, and Lys257). A modeled structure of EcMazG with ppGpp shows that an α-phosphate bonded to a 3′-carbon of a ppGpp ribose ring is located at a similar position of the β-phosphate of the ATP, implying that three basic catalytic residues (Lys222, Arg226, and Lys257) for the ATP hydrolysis might be applied for the ppGpp hydrolysis as well (supplemental Fig. 2).
Regulation of Bacterial Response to Amino Acid Starvation by NTPase Activity of the CD—In E. coli, mazG is transcribed in the same polycistronic mRNA with mazEF, and MazG has a function of limiting the deleterious activity of MazF toxin (4). Overexpression of MazG in the E. coli ΔmazG strain actually lowers the cellular (p)ppGpp concentration and recovers cellular growth in nutrient starvation conditions, which explains the regulatory role of MazG under amino acid starvation stress (4). Enzymatic activity of MazG is inhibited by MazEF complex at the logarithmic phase of growth. Under stress conditions, causing repression of the mazEFG promoter, quick degradation of labile antitoxin MazE leads to activation of both MazF toxin and MazG regulator. The toxic effect of MazF originates from its endoribonuclease activity (32). Prolonged duration of mazEFG promoter repression causes complete exhaustion of MazE antitoxin, and, in the absence of improvement of environmental conditions, cells pass the “point of no return” and die (11, 33).
Mutational analysis of in vivo MazG NTPase activity confirms its inclusion in regulatory processes. The overexpressions of the wild-type MazG and MazG-ND have a deleterious effect on cell growth; those of MazG-CD and MazG-ND/-CD, however, did not influence cell growth under normal growth conditions (-SH), indicating that the NTPase activity of the CD has a toxic effect on the cell growth by degrading, at least, NTPs that are essential necessities in the cellular metabolism (Fig. 4). Under the nutrient starvation conditions (+SH), however, cell growth patterns were reversed. Cells expressing wild-type MazG and MazG-ND reverted their growth, and those expressing MazG-CD and MazG-ND/-CD are strongly inhibited (Fig. 4). As described above, the ND domain of EcMazG does not exhibit an NTPase activity and rather has a housecleaning function. Therefore, we suggest that the NTPase activity of the MazG CD domain is responsible for its regulatory function in amino acid starvation conditions.
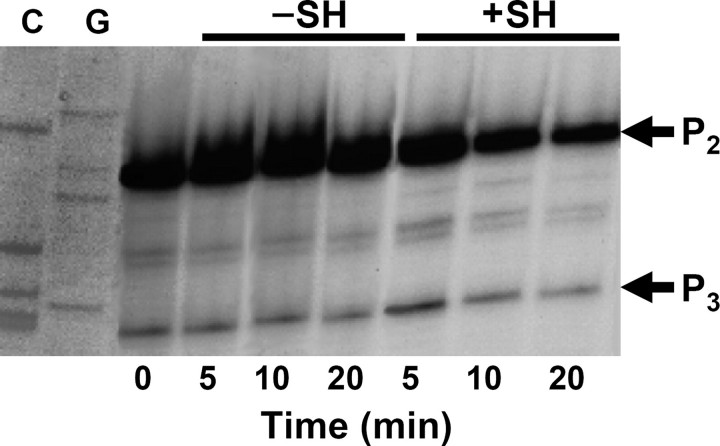
In order to assess the substantial involvement of the mazEFG operon in bacterial programmed cell death, we wanted to confirm the transcription inhibition of the mazEFG operon under amino acid starvation conditions. For that purpose, we have constructed a mazEFG-lacZ fusion and performed primer extension experiment under amino acid starvation conditions, resulting in 56.2 and 23.7% repression of P2 and P3 mazEFG promoters, respectively (Fig. 6). Thus, the function of MazG is to extend the waiting period under amino acid starvation stress through lowering the (p)ppGpp levels (4).
FIGURE 6.
mazEFG promoters are inhibited under SH-induced starvation conditions. E. coli strain MC4100 ΔmazEFG relA+ was transformed with the pSK10Δ6-pef plasmid (24). At A600 of 0.2, bacterial culture was divided into two flasks, one of which was supplemented with 1 mg/ml SH. At the indicated time points, samples were taken for total RNA isolation and primer extension reaction (see “Experimental Procedures”). C and G, sequence reactions of the same plasmid carried out with the terminators ddCTP and ddGTP, respectively.
A Proposed Inhibition Mechanism of MazG by MazEF Complex—MazEF inhibited the activity of MazG in vivo, and the addition of MazEF protein inhibited the NTPase activity of MazG by 70% in vitro (4). Because a C-terminal NTPase activity of MazG is responsible for its regulatory function, this activity might be inhibited by MazEF protein in normal growth conditions. Contrary to SsMazG and CvMazG that form a tetramer through the hydrophobic patches on the bottom face, a negatively charged patch is presented at the CD of EcMazG (supplemental Fig. 3). As indicated by size exclusion chromatography and crystal structure, EcMazG exists as a dimer, not as a tetramer. Because of a rigid additional region that hinders the substrate access from the side, the active site of the CD of EcMazG is accessible only from the bottom of the enzyme (Fig. 5, B and C, and supplemental Fig. 3). Interestingly, MazEF complex contains a positively charged valley that is constituted by several basic residues from both MazE and MazF (6). Therefore, the inhibition mode of the MazG CD by MazEF could be proposed based on the observed structural features of MazG and MazEF complex, which has a negatively charged patch and has a positively charged valley, respectively (supplemental Fig. 4). Taken together, in normal cell growth conditions, MazG might directly bind MazEF through charge compensation. The interaction between them would block the active sites of the MazG CD. In nutrient starvation conditions, ClpPA degrades MazE, and the constant cellular level of MazEF complex formation cannot be sustained (34). The lack of the MazEF complex would invoke the active sites of the MazG CD constantly open to achieve the NTPase activity for delaying cell death. Further crystallographic and biochemical studies might be needed for more detailed understanding of the inhibition mechanism of MazG by MazEF complex.
In summary, we present a crystal structure of EcMazG with two tandem MazG-like domains. A CD, but not an ND, of EcMazG showed an NTPase activity. The key residues and the enzymatic mechanism of the NTP pyrophosphohydrolase activity were revealed by the EcMazG-ATP complex structure and the mutagenesis analysis. The study also explains how the protein could accommodate all eight canonical NTPs as substrates. The cell growth measurements at normal and amino acid stress conditions revealed that the CD is responsible for the regulatory function using its NTPase activity. This work provides important structural and biochemical clues to understand the role of MazG in the E. coli survival under nutrient starvation.
Supplementary Material
The atomic coordinates and structure factors (codes 3cra and 3crc) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was funded by the 21C Frontier Microbial Genomics and Application Center Program, Ministry of Science and Technology, Republic of Korea. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. 1–4.
Footnotes
The abbreviations used are: IPTG, isopropyl 1-thio-β-d-galactopyranoside; SeMet, selenomethionine; DTT, dithiothreitol; SH, serine hydroxamate; ND, N-terminal domain; CT, C-terminal domain.
References
- 1.Zhang, J., and Inouye, M. (2002) J. Bacteriol. 184 5323-5329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moroz, O. V., Murzin, A. G., Makarova, K. S., Koonin, E. V., Wilson, W. S., and Galperin, M. Y. (2005) J. Mol. Biol. 347 243-255 [DOI] [PubMed] [Google Scholar]
- 3.Galperin, M. Y., Moroz, L. V., Wilson, K. S., and Murzin, A. G. (2006) Mol. Microbiol. 59 5-19 [DOI] [PubMed] [Google Scholar]
- 4.Gross, M., Marianovsky, I., and Glaser, G. (2006) Mol. Microbiol. 59 590-601 [DOI] [PubMed] [Google Scholar]
- 5.Engelberg-Kulka, H., and Glaser, G. (1999) Annu. Rev. Microbiol. 53 43-70 [DOI] [PubMed] [Google Scholar]
- 6.Kamada, K., Hanaoka, F., and Berley, S. K. (2003) Mol. Cell 11 875-884 [DOI] [PubMed] [Google Scholar]
- 7.Artsimovitch, I., Patlan, V., Sekine, S., Vassylyeva, M. N., Hosaka, T., Ochi, K., Yokoyama, S., and Vassylyev, D. G. (2004) Cell 117 299-310 [DOI] [PubMed] [Google Scholar]
- 8.Yarmolinsky, M. B. (1995) Science 267 836-837 [DOI] [PubMed] [Google Scholar]
- 9.Hogg, T., Mechold, U., Malke, H., Cashel, M., and Hilgenfeld, R. (2004) Cell 117 57-68 [DOI] [PubMed] [Google Scholar]
- 10.Durfee, T., Hansen, A. M., Zhi, H., Blattner, F. R., and Jin, D. J. (2008) J. Bacteriol. 190 1084-1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amitai, S., Yassin, Y., and Engelberg-Kulka, H. (2004) J. Bacteriol. 186 8295-8300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robinson, A., Guilfoyle, A. P., Harrop, S. J., Boucher, Y., Stokes, H. W., Curmi, P. N., and Mabbutt, B. C. (2007) Mol. Microbiol. 66 610-621 [DOI] [PubMed] [Google Scholar]
- 13.Wu, B., Liu, Y., Zhao, Q., Liao, S., Zhang, J., Bartlam, M., Chen, W., and Rao, Z. (2007) J. Mol. Biol. 369 1405-1412 [DOI] [PubMed] [Google Scholar]
- 14.Braeken, K., Moris, M., Daniels, R., Vanderleyden, J., and Michiels, J. (2006) Trends Microbiol. 14 45-54 [DOI] [PubMed] [Google Scholar]
- 15.Jancarik, J., and Kim, S. H. (1991) J. Appl. Crystallogr. 24 409-411 [Google Scholar]
- 16.Otwinowski, Z., and Minor, W. (1997) Methods Enzymol. 276 307-326 [DOI] [PubMed] [Google Scholar]
- 17.Matthews, B. W. (1968) J. Mol. Biol. 33 491-497 [DOI] [PubMed] [Google Scholar]
- 18.Terwilliger, T. C., and Berendzen, J. (1999) Acta Crystallogr. Sect. D Biol. Crystallogr. 55 849-861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Terwilliger, T. C. (2000) Acta Crystallogr. Sect. D Biol. Crystallogr. 56 965-972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Emsley, P., and Cowtan, K. (2004) Acta Crystallogr. Sect. D Biol. Crystallogr. 60 2126-2132 [DOI] [PubMed] [Google Scholar]
- 21.Murshudov, G. N., Vagin, A. A., and Dodson, E. J. (1997) Acta Crystallogr. Sect. D Biol. Crystallogr. 53 240-255 [DOI] [PubMed] [Google Scholar]
- 22.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T., and Warren, G. L. (1998) Acta Crystallogr. Sect. D Biol. Crystallogr. 54 905-921 [DOI] [PubMed] [Google Scholar]
- 23.Philippe, N., Alcaraz, J. P., Coursange, E., Geiselmann, J., and Schneider, D. (2004) Plasmid 51 246-255 [DOI] [PubMed] [Google Scholar]
- 24.Marianovsky, I., Aizenman, E., Engelberg-Kulka, H., and Glaser, G. (2001) J. Biol. Chem. 276. 5975-5984 [DOI] [PubMed] [Google Scholar]
- 25.Gafny, R., Hyman, H. C., Razin, S., and Glaser, G. (1988) Nucleic Acids Res. 16 61-76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith, D. W. E., and Ames, B. N. (1965) J. Biol. Chem. 240 3056-3063 [PubMed] [Google Scholar]
- 27.Kwon, J. H., Chun, J. Y., Lee, H. S., Cheon, C. I., Song, E. S., Min, K. H., and Lee, M. S. (2000) Can. J. Microbiol. 46 848-855 [PubMed] [Google Scholar]
- 28.Guo, R. T., Kuo, C. J., Chou, C. C., Ko, T. P., Shr, H. L., Liang, P. H., and Wang, A. H. (2004) J. Biol. Chem. 279 4903-4912 [DOI] [PubMed] [Google Scholar]
- 29.Chang, T. H., Guo, R. T., Ko, C. C., Wang, A. H., and Liang, P. H. (2006) J. Biol. Chem. 281 14991-15000 [DOI] [PubMed] [Google Scholar]
- 30.Peters, R. J., and Croteau, R. B. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 580-584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moroz, O. V., Harkiolaki, M., Galperin, M. Y., Vagin, A. A., Gonzalez-Pacanowska, D., and Wilson, K. S. (2004) J. Mol. Biol. 342 1583-1597 [DOI] [PubMed] [Google Scholar]
- 32.Zhang, Y., Zhang, J., Hoeflich, K. P., Ikura, M., Qing, G., and Inouye, M. (2003) Mol. Cell 12 913-923 [DOI] [PubMed] [Google Scholar]
- 33.Kolodkin-Gal, I., and Engelberg-Kulka, H. (2006) J. Bacteriol. 188 3420-3423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aizenman, E., Engelberg-Kulka, H., and Glaser, G. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 6059-6063 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.