Abstract
AAK-2 is one of two α isoforms of the AMP-activated protein kinase in Caenorhabditis elegans and is involved in life span maintenance, stress responses, and germ cell cycle arrest upon dauer entry. We found that AAK-2 was phosphorylated at threonine 243 in response to paraquat treatment and that this phosphorylation depends on PAR-4, the C. elegans LKB1 homologue. Both aak-2 mutation and par-4 knockdown increased the sensitivity of C. elegans worms to paraquat, and the double deficiency did not further increase sensitivity, indicating that aak-2 and par-4 act in a linear pathway. Both mutations also slowed body bending during locomotion and failed to reduce head oscillation in response to anterior touch. Consistent with this abnormal motility and behavioral response, expression of the AAK-2::green fluorescent protein fusion protein was observed in the ventral cord, some neurons, body wall muscle, pharynx, vulva, somatic gonad, and excretory cell. Our study suggests that AMPK can influence the behavior of C. elegans worms in addition to its well known function in metabolic control.
AMP-activated protein kinase (AMPK)3 is activated by low glucose intake, extensive physical exercise, and various stresses such as oxidative stress, hypoxia, ischemia, and heat shock (1). AMPK activates catabolic enzymes and inhibits anabolic enzymes such as acetyl-CoA carboxylase and 3-hydroxy-3-methylglutaryl-CoA reductase. In addition to its role as a key metabolic switch, it inhibits protein translation and cell growth by inhibiting the target of rapamycin (TOR) pathway through phosphorylating TSC2 (2). AMPK is a therapeutic target that is activated by the anti-diabetic drug metformin and by resveratrol, a health supplement derived from grapes (3–5).
AMPK is composed of three subunits: the α catalytic subunit and the β and γ regulatory subunits. In mammals there are two α, two β, and three γ isoforms. In α2 knock-out mice, insulin secretion is attenuated and resistance to insulin is induced in peripheral tissues (6). In contrast, α1 knock-out mice do not show a phenotype. In Drosophila melanogaster, a deletion of the single AMPK α gene results in lethality, with severe abnormalities in cell polarity and mitosis (7). The role of AMPK in epithelial cell polarity has also been observed in a Madin-Darby canine kidney cell line (8). Caenorhabditis elegans encodes two catalytic subunits, namely aak-1 and aak-2. Although no phenotype was reported for aak-1, an aak-2 mutation was found to result in reduced life span and hypersensitivity to heat shock and to a mitochondrial poison (9). However, both AAK-1 and AAK-2 are needed in conjunction with DAF-18(PTEN) to inhibit germ line cell proliferation during dauer development (10). The AAK kinase-mediated inhibition of germ cell proliferation is reminiscent of the observation that AMPK induces G1 arrest by phosphorylating p53 in mouse embryonic fibroblasts after exposure to a low glucose level (11).
Mammalian LKB1 kinase, a deficiency in which is associated with Peutz-Jeghers syndrome, a disease that shows increased susceptibility to certain cancers (12), and its Drosophila homologue are the major upstream Ser/Thr kinases acting on AMPK (7, 8, 13–16). In addition, the β isoform of Ca2+/calmodulindependent protein kinase kinase β also phosphorylates and activates AMPK in mammalian cells (17, 18). In this study, we demonstrate that the C. elegans homologue of LKB1, PAR-4, is the upstream kinase of the AAK-2 AMPK α subunit and that PAR-4 and AAK-2 deficiency sensitizes worms to oxidative stress. In addition, par-4 depletion and aak-2 mutations reduce worm motility and affect behavioral response. In agreement with these phenotypes, AAK-2 is expressed in neurons and muscle cells.
EXPERIMENTAL PROCEDURES
Strains and Culture Conditions—Wild-type N2, aak-2(ok524), aak-2(rr48), par-4(it57), and par-4(it47) C. elegans strains were obtained from the Caenorhabditis Genetics Center (Minneapolis, MN). The aak-2(gt33) deletion mutant was generated by random mutagenesis using UV-activated trimethylpsoralen (19). The ok524 allele has a 408-bp deletion (from nucleotide 180 of Exon 3 to nucleotide 118 of Intron 3), and the gt33 allele has a 606-bp deletion (from nucleotide 22 of Exon 3 to nucleotide 160 of Intron 3) (see Fig. 1). The rr48 allele has a substitution mutation, H208Y, in Exon 3 (see Fig. 1) and is a semidominant-negative mutation (10). Wild-type N2 and aak-2 mutant worms were maintained at 20 °C according to standard procedures except for experiments where a growth temperature of 25 °C is specified. Temperature-sensitive par-4 mutants were maintained at 16 °C and shifted to 25 °C at the L1 stage when needed. The aak-1 expressed sequence tag clone, yk1173f11, was a generous gift from Dr. Yuji Kohara (National Institute of Genetics, Japan).
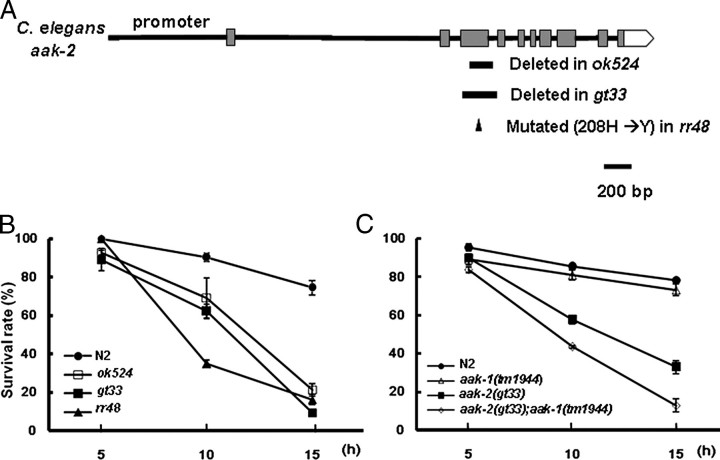
FIGURE 1.
aak-1 and aak-2 affect paraquat sensitivity of adult C. elegans worms. A, a schematic representation of the aak-2 gene (T01C8.1a in the WormBase Database) in wild-type and mutant strains. B, survival of paraquat-treated wild-type N2 and aak-2 mutant C. elegans worms. Young adult C. elegans worms were incubated in 100 mm paraquat solution, and survival was scored under a stereomicroscope at 5-h intervals. Fifty worms of each strain were treated with paraquat, and the experiment was performed three times. p > 0.05 at 10- and 15-h time points between any pair of aak-2 alleles, except for rr48 at the 10-h time point, where the p value was <0.003 compared with the two other alleles. C, paraquat sensitivities of single and double deficient strains for aak-2 and aak-1. p > 0.15 between N2 and aak-1 at 10 and 15 h, and p < 0.03 between aak-1(tm1944); aak-2(gt33) and aak-2(gt33) at 10 and 15 h. Error bars are S.E.
Mutant Out-cross—The aak-2(ok524) and aak-2(gt33) strains were out-crossed eight and three times, respectively, with the wild-type N2 strain. The aak-2(rr48), par-4(it57), and par-4(it47) strains were used as received from the C. elegans Genetics Center and have been previously out-crossed once for aak-2 and more than three times for both par-4 alleles. aak-1(tm1944) with a deletion of 618 bp was obtained from the National Bioresource Project (Japan), out-crossed three times with N2, and then crossed with aak-2(gt33) to generate the double mutant. We also made the aak-2(ok524);par-4(it57) double mutant.
Bacterially Mediated RNAi—A par-4 cDNA fragment was amplified by polymerase chain reaction from C. elegans cDNA using primers 5′-CCCGGGATGGATGCTCCGTCGACATC and 5′-TCTAGACTAAGCACTATCGGTACGAGAAC. cDNA was prepared using a Power cDNA synthesis kit (iNtRON Biotechnology) after oligo(dT) priming of total RNA isolated from C. elegans worms using TRIzol reagent (Invitrogen). The par-4 cDNA fragment was cloned into pGEM ®-T vector (Promega), and the 1.5-kb-long BamHI-BamHI fragment was inserted into the pPD129.36 vector, which contains convergent T7 polymerase promoters separated by a multi-cloning site. The recombinant DNA construct was transformed into Escherichia coli HT115(DE3) (20, 21). Transformants were grown for 18 h in LB medium containing 50 μg/ml ampicillin and seeded onto NGM plates supplemented with 50 μg/ml ampicillin and 1 mm isopropyl β-d-thiogalactopyranoside. RNAi was performed at 25 °C by placing L1 larvae on plates seeded with E. coli HT115(DE3) cells expressing double-stranded RNA of par-4.
Survival of C. elegans Worms in Paraquat—To collect synchronized eggs, gravid wild-type and mutant worms were placed on NGM agar plates seeded with E. coli OP50, allowed to lay eggs for 3–6 h, and removed from the plates. C. elegans L1 larvae were placed on NGM plates covered with an E. coli OP50 lawn and allowed to grow at 25 °C to the young adult stage. For par-4 knockdown, wild-type N2 (or aak-2 mutant) worms were fed E. coli HT115(DE3) cells expressing cognate double-stranded RNA. The young adult worms were transferred to an M9 solution containing 100 mm paraquat (1,1′-dimethyl-4,4′-bipyridinium dichloride, Sigma-Aldrich) and incubated at 25 °C. Dead worms were counted every 5 h up to the 20-h time point. Fifty worms from each strain were treated with paraquat, and the experiment was performed three times. Sensitivity to hydrogen peroxide (5 mm) was measured in the same way as for paraquat.
Western Blot Analysis—Wild-type N2, aak-2(gt33), aak-2(ok524), and the temperature-sensitive mutants par-4(it57) and par-4(it47) were grown at 25 °C from the L1 stage until the second adult day. Adult worms were washed from six NGM plates (55-mm diameter) using phosphate-buffered saline with 0.1% Tween 20 and resuspended in the same buffer containing 100 mm paraquat, followed by incubation at 25 °C for 2 h. After removing the supernatant, the worms were mixed with 2 volumes of 2× sample loading buffer (200 mm Tris-Cl, pH 8.0, 500 mm NaCl, 0.1 mm EDTA, 0.1% Triton X-100, and 0.4 mm phenylmethylsulfonyl fluoride) and boiled for 10 min. The worm lysate was electrophoresed on an 8% SDS-polyacrylamide gel and electroblotted onto a nitrocellulose membrane (Schleicher & Schüll). The membrane was incubated in a blocking solution containing anti-phospho-AMPK anti-rabbit antibody (1:300 dilution; Cell Signaling Technology) or anti-α-tubulin mouse antibody (1:5000 dilution; Developmental Studies Hybridoma Bank) at 4 °C overnight, followed by incubation with anti-mouse or anti-rabbit horseradish peroxidase antibody (1:5000 dilution; Jackson ImmunoResearch) for 1 h at 25°C. The ECL Western blotting analysis system (Amersham Biosciences) was used to detect the secondary antibodies on the membrane. Luminescence of the blot was captured using an LAS-3000 imaging system (Fujifilm).
GFP-tagged AAK-2 Expression in C. elegans—The genomic aak-2 DNA fragment including the 2.0-kb upstream sequence and the transcribed region proximal to the stop codon was amplified from C. elegans genomic DNA using primers 5′-CTGCAGACCAAACGACCACTTCAAAAATTA and 5′-GGATCCAACGAGCCAGTGTTCCAATCAA. The 8.8-kb genomic aak-2 DNA fragment was cloned between the PstI and BamHI sites of the pPD95.75 plasmid DNA (from Dr. Andrew Fire, Stanford University). The recombinant plasmid DNA (20 μg/ml) and pRF4 plasmid DNA (50 μg/ml) were coinjected into adult N2 worms. Rolling and green fluorescent worms were selected and observed by fluorescence microscopy (DMR HC, Leica). To localize GFP expression in head neurons, transgenic animals were incubated in M9 buffer containing 10 μg/ml DiI (Molecular Probes) for 2 h at room temperature and were then observed with a fluorescence microscope (AxioPhot2, Zeiss). To derive aak-2 expression in neurons, full-length aak-2 cDNA was fused to the 2.0-kb-long unc-119 promoter region and shuffled into pPD95.79 to obtain a C-terminal fusion with GFP. The resulting construct was first transformed into N2 and then aak-2(gt33) worms by crossing.
Behavioral Assays—To measure the frequency of body bending during locomotion, well fed worms on the first adult day (n > 30) were placed on NGM plates without food, and the number of sinusoidal curves made during locomotion was scored for 20 s (22). Foraging behavior was scored according to the procedure of Kindt et al. (23). Briefly, worms on the first day of adulthood (n > 60) were placed on NGM plates seeded with an E. coli OP50 lawn, and while moving forward, worms were touched behind the posterior pharynx bulb with a human eyelash. Touching induces backward movement and suppresses head oscillation in most wild-type worms. When at least two consecutive body bends occurred without foraging during the backward movement, foraging was scored as suppressed, as described by Kindt et al. (23). Foraging and body bending assays were repeated twice for each animal.
RESULTS
AAK-2 Mutants Are Hypersensitive to Oxidative Stress—Given that aak-2 worms show a reduced life span (9), a phenotype often correlated with oxidative stress, we directly tested the effect of oxidative stress on the survival of wild-type and aak-2 mutant worms. We used three different alleles of aak-2. The gt33 allele, which contains the largest deletion, was generated in this study (Fig. 1A). When survival was measured after treating young adults with paraquat to generate superoxide anions in mitochondria (24), all three aak-2 mutant worms showed hypersensitivity to paraquat (Fig. 1B). The finding that the rr48 missense mutant was more sensitive to paraquat at the 10-h time point as compared with the two deletion mutants might be due to a previously reported partial dominant-negative effect of this point mutation likely affecting AAK-1 function (10). To test whether aak-2 deficiency also confers hypersensitivity to another oxidative stress-inducing regimen, we compared the sensitivity of wild-type and gt33 worms to hydrogen peroxide, a molecule known to generate hydroxyl radicals (supplemental Fig. S1). In contrast to aak-2 alleles, paraquat hypersensitivity was not observed in the aak-1(tm1944) deletion mutant or upon RNAi-mediated knockdown of aak-1 (data not shown). However, an aak-1(tm1944);aak-2(gt33) double mutant showed enhanced paraquat sensitivity (Fig. 1C), suggesting that there might be a partial redundancy between aak-2 and aak-1 and that aak-1 might be able to partially compensate for the absence of aak-2. A similar observation of distinct but overlapping functions of the two isoforms was previously made in relation to the inhibition of germ cell proliferation during dauer development (10).
The LKB1 Homologue PAR-4 Phosphorylates AAK-2, and par-4 Deficiency Causes Hypersensitivity to Oxidative Stress—To determine whether the C. elegans LKB1 homologue PAR-4 (25) is the upstream kinase of AAK-2, we examined the phosphorylation status of AAK-2. The Western blot analysis of cell extracts was carried out using an antibody against phospho-Thr172 of human AMPK α, a phosphorylation site that is embedded in a sequence conserved between human and worm AAK-2 (Fig. 2A). We observed a specific band at the predicted size of phospho-Thr243 of the AAK-2 protein (corresponding to Thr172 of the human AMPK α subunit) upon Western blotting wild-type extracts of paraquat-treated worms. This signal was reduced to close to background levels in untreated worm extracts and was absent from paraquat-treated aak-2(gt33) and aak-2(ok524) worms (data not shown), indicating that the antibody specifically detects phosphorylated AAK-2. The band slightly below pAAK-2, with a molecular mass of ∼68 kDa, is unspecific. To further show that the antibody specifically detects phospho-Thr243 of AAK-2, we also blotted extracts from transgenic worms containing GFP-tagged AAK-2 and confirmed a band that was dramatically enhanced in extracts from paraquat-treated worms at the predicted molecular mass of the AAK-2::GFP fusion (Fig. 2A). Endogenous AAK-2 in the transgenic worms did not show any clear-cut phosphorylation, likely because of competition between the endogenous and GFP-fused AAK-2 for a limited level of upstream kinase. To test whether AAK-2 phosphorylation depended on PAR-4, we took advantage of par-4 temperature-sensitive mutants. In the par-4(it57) (and also in the par-4(it47) mutant, data not shown), the paraquat treatment-dependent phosphorylation was significantly reduced at the non-permissive temperature (25 °C), suggesting that the LKB1 homologue functions as an upstream kinase of AAK-2 (Fig. 2B). Similarly, the phosphorylation of the AAK-2::GFP fusion protein after paraquat treatment was greatly suppressed by par-4 knockdown (Fig. 2B), further confirming a crucial role of PAR-4 in the AAK-2 phosphorylation.
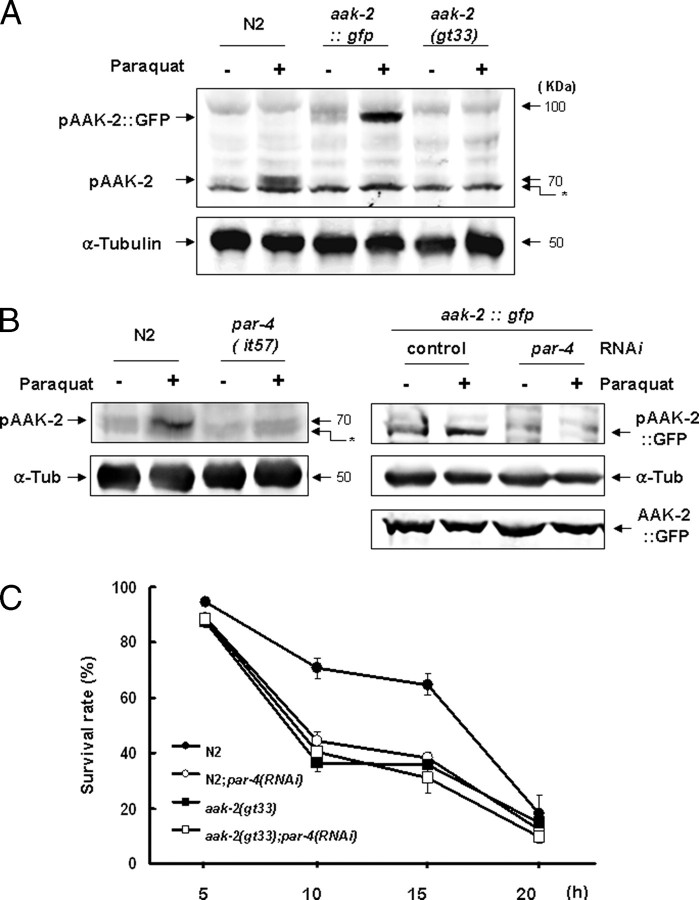
FIGURE 2.
Genetic interaction of aak-2 and par-4 in AAK-2 phosphorylation and worm survival after paraquat treatment. A, phosphorylation of AKK-2 after paraquat treatment of wild-type N2, an aak-2::gfp-overexpressing line, and aak-2(gt33). First-day adult worms were exposed to 100 mm paraquat for 2 h, and worm extracts were analyzed by Western blotting using antibodies to phospho-AMPK α and α-tubulin as a loading control. Arrows indicate the positions of molecular mass standard proteins of 70 and 50 kDa. The band marked with an asterisk is present even in the aak-2 deletion mutant gt-33, indicating that it is unrelated to aak-2. B, phosphorylation of AAK-2 after paraquat treatment in N2 and par-4 mutant strains and of AAK-2::GFP with or without par-4 RNAi. Anti-GFP rabbit antibody (Santa Cruz Biotechnology) was used to probe AAK-2::GFP. α-Tub, α-tubulin. C, survival of C. elegans worms with single or double deficiency of aak-2 and par-4. C. elegans worms were incubated in 100 mm paraquat, and survival was scored at 5-h intervals. p > 0.05 for all pairs of single and double deficient strains at 10 and 15 h. Fifty worms were treated with paraquat for each strain, and the experiment was performed three times. Error bars are S.E.
Given that AAK-2 phosphorylation depends on par-4, we wished to genetically determine whether par-4 might act in the same genetic pathway mediating resistance to oxidative stress as aak-2 and assessed whether par-4 depletion might confer paraquat hypersensitivity. We found that RNAi depletion of par-4 conferred paraquat hypersensitivity and that aak-2(gt33); par-4(RNAi) worms were no more sensitive to paraquat than aak-2(gt33) worms, suggesting that par-4 and aak-2 function in the same pathway (Fig. 2C). Unexpectedly, the par-4(it57) mutant at 25 °C, in contrast to par-4(RNAi), was not significantly hypersensitive to paraquat (supplemental Fig. S2). This difference between the par-4 mutation and the knockdown may be due to an only partially functional inactivation of par-4(it57) at the restrictive temperature. The double mutant aak-2(gt33);par-4(it57) was slightly more hypersensitive than aak-2(gt33) (supplemental Fig. S2).
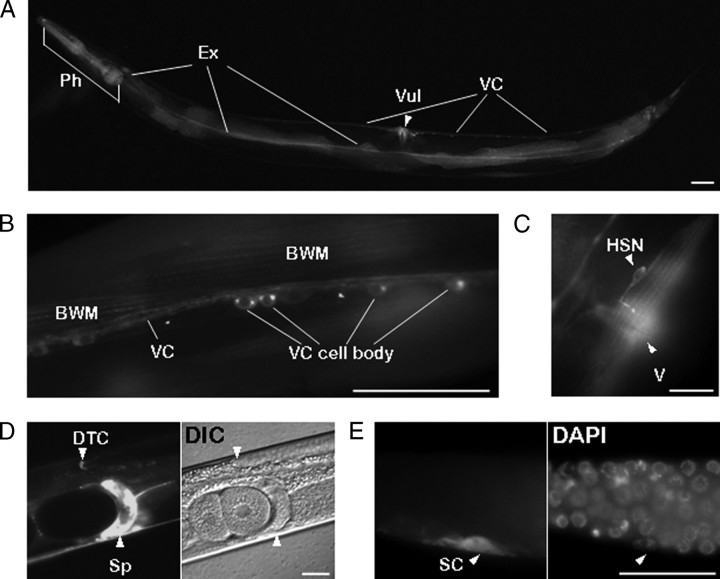
GFP-tagged AAK-2 Is Expressed in Several Tissues, Including Ventral Cord and Body Wall Muscle—In the transgenic C. elegans line expressing the AAK-2::GFP fusion protein, expression was observed in the pharynx, the ventral cord, other neurons including the hermaphrodite-specific neuron (HSN), body wall muscles, the vulva, the excretory canal, and weakly in the intestine (Fig. 3). A similar expression pattern was also reported in a global analysis of expression patterns that included an aak-2 promoter::gfp fusion (BC10615 and 10616 strains in WormBase) (26). In ventral cord motor neurons, the protein was found in the axons and the cytoplasm of the cell bodies, where it accumulated in two foci (Fig. 3, A and B). The expression in the pharynx, ventral cord motor neurons, and body wall muscle cells suggests that AAK-2 plays important roles in cells that are associated with movement. GFP expression was also seen in the distal tip cells, spermatheca, and sheath cells (Fig. 3, D and E). The GFP expression in distal tip cells, which produce a signal for germ cell proliferation, may be associated with the role of AAK-2 in inhibiting germ line proliferation in dauers (10). The observation that aak-2 mutants have partial egg-laying defects (data not shown) could be explained by AAK-2 expression in hermaphrodite-specific neurons (Fig. 3C). In addition, AAK-2 is expressed in vulva cells (Fig. 3A).
FIGURE 3.
Expression of the AAK-2::GFP fusion protein in C. elegans. The genomic DNA region including the 2.0 kb upstream of the start ATG and coding sequences of aak-2 was fused to GFP, and a transgenic C. elegans line was generated. A, green fluorescence can be seen in the pharynx (Ph), excretory cell (Ex), vulva (Vul), and ventral cord (VC) of a whole adult worm. Green fluorescence was also present in body wall muscle (BWM) and in the cell bodies of the ventral cord (B), HSN neuron and vulva (V)(C), distal tip cell (DTC) and spermatheca (Sp)(D), and sheath cell (SC)(E). Scale bar, 50 μm. DIC, differential interference contrast; DAPI, 4′,6-diamidino-2-phenylindole.
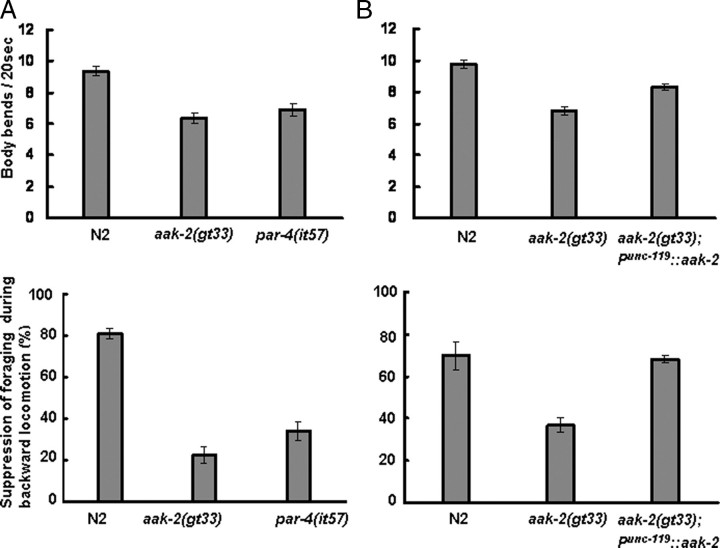
aak-2 and par-4 Deficiencies Result in Slow Body Bending and Abnormal Modulation of Head Oscillation—Because strong expression of the AAK-2::GFP fusion protein was observed in ventral cord motor neurons and at a lower level in body wall muscle, we compared the frequency of body bending in wild-type, aak-2(gt33), and par-4(it57) strains during locomotion (Fig. 4A). The frequency of body bending was reduced by 30% in both mutants (p < 0.001), suggesting that the two proteins modulate neural and/or muscular activities. C. elegans worms oscillate their heads while foraging, but in response to gentle anterior touch, they generally move backwards and cease head oscillation. Both mutants also showed a reduced (p < 0.001) ability to suppress head oscillations when touched on the anterior side (Fig. 4A). The fact that body bending and suppression of foraging during backward movement were affected to the same extent (p = 0.19 and 0.30, respectively) in the two mutants agrees with the argument that the two affected genes function in the same genetic pathway. The tyraminergic neuron RIM and the backward locomotion command neurons AVA and AVD are involved in the suppression of head oscillations in response to anterior touch (27). In the aak-2::gfp-overexpressing line, GFP was observed in the RIM neuron as well as in neighboring head neurons (supplemental Fig. S3A), but we could not detect any expression in the AVA or AVD neuron. Other behaviors such as pharyngeal pumping, egg laying, and directional reversal of locomotion were unaffected in the two mutants (data not shown).
FIGURE 4.
Locomotion and foraging behavior of wild-type N2, aak-2, and par-4 strains and the rescue of aak-2 mutation by neuronal expression of AAK-2::GFP. First-day adult worms (n > 30) were placed on NGM plates, and the number of body bends occurring within 20 s was counted. To examine foraging behavior, first-day adult forward-moving worms (n > 60) were touched with a human eyelash behind the posterior pharynx bulb. When no head oscillation occurred during backward movement with at least two consecutive body bending cycles, foraging was scored as suppressed. A, wild-type N2, aak-2(gt33), and par-4(it57) strains. B, effects of Punc-119::aak-2::gfp overexpression in the aak-2(gt33) background. The p value between N2 and the gt33;Punc-119::aak-2::gfp was 0 in the upper graph and 0.83 in the lower graph. Error bars are S.E.
To test whether the abnormal behavior of the aak-2 mutants in Fig. 4A was due to neuronal defect, we expressed AAK-2::GFP in neurons using the pan-neuronal unc-119 promoter (supplemental Fig. S3) (28). We observed that the attenuated body bending of aak-2(gt33) worms was partially reversed and that the failure to suppress foraging was completely reversed (Fig. 4B). This suggests that expression of AAK-2 in neurons is essential for the modulation of foraging behavior during backward movement. The incomplete rescue of body bending implies that normal rates of body bending might also be controlled by AAK-2 expressed in non-neuronal tissues. We also observed that paraquat hypersensitivity of aak-2(gt33) worms was partially rescued by the pan-neuronal expression of AAK-2::GFP (data not shown).
DISCUSSION
In mammals, LKB1 is the major upstream kinase of AMPK (8, 13, 14). In addition, activity of Ca2+/calmodulin-dependent protein kinase kinase β on AMPK was observed in the absence of LKB1 (18). In Drosophila, an LKB1 homologue was also demonstrated to be an upstream kinase of AMPK, which is essential for normal development and survival (7, 15). In C. elegans, the LKB1 homologue PAR-4 has been proposed to function upstream of AAK-1 but in parallel with AAK-2 in suppressing germ cell proliferation in dauers (10). However, we found in the present study that PAR-4 is the major upstream kinase of AAK-2, at least in the response to oxidative stress. We could not determine whether PAR-4 is also the upstream kinase of AAK-1 because the peptide sequence neighboring the phosphorylation site Thr172 of mammalian AMPK α is not conserved in AAK-1. AAK-2 contributes to the resistance of C. elegans to heat shock and the mitochondrial poison sodium azide as well as to the extension of life span (9). Recently, overexpression of a constitutively active AMPK Y2 subunit was found to increase C. elegans life span as well as resistance to paraquat (29). Both of these effects were dependent on DAF-16, a FOXO homologue, suggesting that activation of AAK-2 in response to oxidative stress is one mechanism leading to a longer life span (29). In addition, AAK-2 was found to increase mitochondrial respiration and oxidative stress under glucose restriction, thereby leading to the extension of life span (30).
We found that AAK-2 protein was restricted to the pharynx, ventral cord, body wall muscle, vulva, excretory cell, and somatic gonad, which is in contrast to the ubiquitous expression of vertebrate AMPK (31). Such tissue- and cell type-specific expression suggests a more important role of AAK-2 in the neural and muscular systems than in the hypodermis. In the ventral cord, the protein was detected in the cytoplasm of axons and cell bodies but not within nuclei. In mammals, two isoforms of the AMPK catalytic subunit α are localized in the cytoplasm, and a significant fraction of the α2 isoform is also present in nuclei (32). Thus, AAK-2 in C. elegans differs from its mammalian homologue in terms of tissue- and cell type-specific localization as well as in its exclusive restriction to the cytoplasm. A crucial role of AMPK in protecting neurons under stress was demonstrated in the rat brain, where AMPK phosphorylates the R2 subunit of γ-aminobutyric acid type B receptors, leading to slow synaptic inhibition (33). In accord with the regulatory role of mammalian AMPK in neuronal synapses, we found that AAK-2 modulates body bending and foraging behavior via its expression in neurons. In summary, one of the C. elegans AMPK α subunits, AAK-2, and the upstream kinase PAR-4 protect the animal from oxidative stress and modulate its motility and behavioral responses.
Supplementary Material
Acknowledgments
C. elegans N2 aak-2(ok524), aak-2(rr48), par-4(it57), and par-4(it47) strains were obtained from the Caenorhabditis Genetics Center, which is supported by the National Center for Research Resources. The aak-1(tm1944) strain was kindly provided by the National Bioresource Project, and the expressed sequence tag clone yk1173f11 of aak-1 was from Dr. Yuji Kohara. We thank Dr. Andrew Fire for the pPD95.75 vector, Sebastian Greiss for helping with the generation of the disruption library, and Dr. Robert Johnsen (Simon Fraser University) for the BC10615 C. elegans strain. Hyun-Ok Song in the laboratory of Dr. Joohong Ahnn (Hanyang University) kindly provided the plasmid containing the unc-119 promoter, and Kyuhyung Kim (Brandeis University) and Junsu Kang in the laboratory of Dr. Junho Lee (Seoul National University) helped with the identification of fluorescent neurons. We thank Dr. Kee Yang Chung (Yonsei College of Medicine) for introducing us to AMPK.
This work was supported by Korea Research Foundation Grant KRF-2005-041-C00305 funded by the Korean Government (Ministry of Education and Human Resources Development) (to H.-S. K.) and by a Cancer Research United Kingdom Career Development Award fellowship (to A. G.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
Footnotes
The abbreviations used are: AMPK, AMP-activated protein kinase; RNAi, RNA interference; NGM, nematode growth medium; GFP, green fluorescent protein.
References
- 1.Towler, M. C., and Hardie, D. G. (2007) Circ. Res. 100 328–341 [DOI] [PubMed] [Google Scholar]
- 2.Motoshima, H., Goldstein, B. J., Igata, M., and Araki, E. (2006) J. Physiol. 574 63–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carling, D. (2005) Biochimie (Paris) 87 87–91 [DOI] [PubMed] [Google Scholar]
- 4.Hardie, D. G. (2004) J. Cell Sci. 117 5479–5487 [DOI] [PubMed] [Google Scholar]
- 5.Dasgupta, B., and Milbrandt, J. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 7217–7222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Viollet, B., Andreelli, F., Jørgensen, S. B., Perrin, C., Geloen, A., Flamez, D., Mu, J., Lenzner, C., Baud, O., Bennoun, M., Gomas, E., Nicolas, G., Wojtaszewski, J. F., Kahn, A., Carling, D., et al. (2003) J. Clin. Investig. 111 91–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee, J. H., Koh, H. J., Kim, M. J., Kim, Y. S., Lee, S. Y., Karess, R. E., Lee, S. H., Shong, M. H., Kim, J. M., Kim, J. S., and Chung, J. K. (2007) Nature 447 1017–1020 [DOI] [PubMed] [Google Scholar]
- 8.Zheng, B., and Cantley, L. C. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 819–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Apfeld, J., O'Connor, G., McDonagh, T., DiStefano, P. S., and Curtis, R. (2004) Genes Dev. 18 3004–3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Narbonne, P., and Roy, R. (2006) Development (Camb.) 133 611–619 [DOI] [PubMed] [Google Scholar]
- 11.Jones, R. G., Plas, D. R., Kubek, S., Buzzai, M., Mu, J., Xu, Y., Birnbaum, M. J., and Thompson, C. B. (2005) Mol. Cell 18 283–293 [DOI] [PubMed] [Google Scholar]
- 12.Hemminki, A., Markie, D., Tomlinson, I., Avizienyte, E., Roth, S., Loukola, A., Bignell, G., Warren, W., Aminoff, M., Höglund, P., Järvinen, H., Kristo, P., Pelin, K., Ridanpää, M., Salovaaro, R., et al. (1998) Nature 391 184–187 [DOI] [PubMed] [Google Scholar]
- 13.Hawley, S. A., Boudeau, J., Reid, J. L., Mustard, K. J., Udd, L., Makela, T. P., Alessi, D. R., and Hardie, D. G. (2003) J. Biol. (Bronx N. Y.) 2 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shaw, R. J., Kosmatka, M., Bardeesy, N., Hurley, R. L., Witters, L. A., DePinho, R. A., and Cantley, L. C. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 3329–3335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mirouse, V., Swick, L. L., Kazgan, N., St. Johnston, D., and Brenman, J. E. (2007) J. Cell Biol. 177 387–392 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Woods, A., Johnstone, S. R., Dickerson, K., Leiper, F. C., Fryer, L. G., Neumann, D., Schlattner, U., Wallimann, T., Carlson, M., and Carling, D. (2003) Curr. Biol. 13 2004–2008 [DOI] [PubMed] [Google Scholar]
- 17.Hurley, R. L., Anderson, K. A., Franzone, J. M., Kemp, B. E., Means, A. R., and Witters, L. A. (2005) J. Biol. Chem. 280 29060–29066 [DOI] [PubMed] [Google Scholar]
- 18.Woods, A., Dickerson, K., Heath, R., Hong, S. P., Momcilovic, M., Johnstone, S. R., Carlson, M., and Carling, D. (2005) Cell Metab. 2 21–33 [DOI] [PubMed] [Google Scholar]
- 19.Jansen, G., Hazendonk, E., Thijssen, K. L., and Plasterk, R. H. (1997) Nat. Genet. 17 119–121 [DOI] [PubMed] [Google Scholar]
- 20.Timmons, L., and Fire, A. (1998) Nature 395 854. [DOI] [PubMed] [Google Scholar]
- 21.Kamath, R. S., Martinez-Campos, M., Zipperlen, P., Fraser, A. G., and Ahringer, J. (2001) Genome Biol. 2 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsalik, E. L., and Hobert, O. (2003) J. Neurobiol. 56 178–197 [DOI] [PubMed] [Google Scholar]
- 23.Kindt, K. S., Viswanath, V., Macpherson, L., Quast, K., Hu, H., Patapoutian, A., and Schafer, W. R. (2007) Nat. Neurosci. 10 568–577 [DOI] [PubMed] [Google Scholar]
- 24.Castello, P. R., Drechsel, D. A., and Patel, M. (2007) J. Biol. Chem. 282 14186–14193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watts, J. L., Morton, D. G., Bestman, J., and Kemphues, K. J. (2000) Development (Camb.) 127 1467–1475 [DOI] [PubMed] [Google Scholar]
- 26.Hunt-Newbury, R., Viveiros, R., Johnsen, R., Mah, A., Anastas, D., Fang, L., Halfnight, E., Lee, D., Lin, J., Lorch, A., McKay, S., Okada, H. M., Pan, J., Schulz, A. K., Tu, D., et al. (2007) PLoS Biol. 5 e237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alkema, M. J., Hunter-Ensor, M., Ringstad, N., and Horvitz, H. R. (2005) Neuron 46 247–260 [DOI] [PubMed] [Google Scholar]
- 28.Maduro, M., and Pilgrim, D. (1995) Genetics 141 977–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greer, E. L., Dowlatshahi, D., Banko, M. R., Villen, J., Hoang, K., Blanchard, D., Gygi, S. P., and Brunet, A. (2007) Curr. Biol. 17 1646–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schulz, T. J., Zarse, K., Voigt, A., Urban, N., Birringer, M., and Ristow, M. (2007) Cell Metab. 6 280–293 [DOI] [PubMed] [Google Scholar]
- 31.Proszkowiec-Weglarz, M., Richards, M. P., Ramachandran, R., and McMurtry, J. P. (2006) Comp. Biochem. Physiol. B 143 92–106 [DOI] [PubMed] [Google Scholar]
- 32.Salt, I., Celler, J. W., Hawley, S. A., Prescott, A., Woods, A., Carling, D., and Hardie, D. G. (1998) Biochem. J. 334 177–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuramoto, N., Wilkins, M. E., Fairfax, B. P., Revilla-Sanchez, R., Terunuma, M., Tamaki, K., Lemata, M., Warren, N., Couve, A., Calver, A., Horvath, Z., Freeman, K., Carling, D., Huang, L., Gonzales, C., Cooper, E., Smart, T. G., Pangalos, M. N., and Moss, S. J. (2007) Neuron 53 233–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.