Abstract
Here, we report the identification, cloning, and functional characterization of three Caenorhabditis elegans G protein-coupled pigment dispersing factor (PDF) receptors, which we designated as Ce_PDFR-1a, -b, and -c. They represent three splice isoforms of the same gene (C13B9.4), which share a high degree of similarity with the Drosophila PDF receptor and are distantly related to the mammalian vasoactive intestinal peptide receptors (VPAC2) and calcitonin receptors. In a reverse pharmacological screen, three bioactive C. elegans neuropeptides, which were recently identified as the Drosophila PDF orthologues, were able to activate these receptors in a dose-dependent manner with nanomolar potency (isoforms a and b). Integrated green fluorescent protein reporter constructs reveal the expression of these PDF receptors in all body wall muscle cells and many head and tail neurons involved in the integration of environmental stimuli and the control of locomotion. Using a custom data analysis system, we demonstrate the involvement of this newly discovered neuropeptide signaling system in the regulation of locomotor behavior. Overexpression of PDF-2 phenocopies the locomotor defects of a PDF-1 null mutant, suggesting that they elicit opposite effects on locomotion through the identified PDF receptors. Our findings strengthen the hypothesis that the PDF signaling system, which imposes the circadian clock rhythm on behavior in Drosophila, has been functionally conserved throughout the protostomian evolutionary lineage.
The neuropeptide pigment dispersing factor (PDF)3 was initially discovered in crustaceans (as pigment-dispersing hormone), where it drives a daily rhythm of color changes (1). Thereafter, highly conserved PDF peptides were identified in many species of insects and recently also in nematodes (2).4 PDF is a crucial component of the insect circadian clock and has been characterized as a putative output factor, controlling daily rhythms in locomotor activity (3, 4). In 2005, three independent studies identified CG13758, a class B peptide G protein-coupled receptor (GPCR), as the receptor for PDF in Drosophila melanogaster (PDFR) (5–7). It is related to the mammalian VIP receptor (VPAC2) and to the calcitonin receptor, both of which are expressed in the mammalian master clock. Fly PDFR mutants and flies lacking PDF both exhibit severe deficits in free-running locomotor rhythms (4–6). Recently, 3 endogenous PDF-like neuropeptides were discovered in the free-living nematode model organism C. elegans.4 They are expressed mainly in neurons involved in chemosensation, mechanosensation, oxygen sensing, and locomotion. Circadian analysis revealed that at least two of these peptides (e.g. PDF-1a and -b) are involved in the control of daily locomotor rhythms in C. elegans.4 Mutants lacking PDF-1 mimic the behavioral phenotype of Drosophila PDF mutants with respect to free-running locomotor rhythms. This led us to the hypothesis that the PDF signaling system, which imposes the clock rhythm on behavior, may be functionally conserved during evolution, at least in invertebrates.
To fortify this hypothesis, we set out to find and characterize the cognate receptors of the C. elegans PDF neuropeptides. In this study we report the identification of 3 orphan G protein-coupled PDF receptor isoforms and unveil the PDF peptides as their endogenous matching ligands by means of a reverse pharmacological approach. The PDF receptors are distantly related to the mammalian VPAC2 and calcitonin receptors and show expression in body wall muscles and neuronal cells that play a role in the integration of environmental stimuli and the control of locomotion. Our functional analysis of the PDF signaling system reveals that overexpression of PDF-2 phenocopies the locomotor defects of the PDF-1 null mutant, suggesting that they elicit opposite effects on locomotion through these PDF receptors.
EXPERIMENTAL PROCEDURES
Strains and Media—All C. elegans strains were grown at 20 °C on nematode growth medium agar seeded with Escherichia coli OP50 bacteria. The strains used were Bristol N2; BC11358, C13B9.4::gfp, and FX1996, T07E3.6(tm1996). Mutants were backcrossed at least two times to a wild-type Bristol N2 background prior to phenotypic analysis.
Molecular Cloning—The open reading frame of Ce_pdfr-1c (C13B9.4c) was obtained by reverse transcriptase-PCR. mRNA, extracted from mixed stage C. elegans N2 (QuickPrep Micro mRNA Purification Kit, Amersham Biosciences), was used as a template for cDNA synthesis (SuperScript First-Strand Synthesis System for RT-PCR, Invitrogen). The full-length cDNA of Ce_pdfr-1c was then amplified by means of PCR, using gene-specific oligonucleotide primers (Sigma) based on the predicted cDNA sequence (Wormbase). The open reading frames of Ce_pdfr-1a (C13B9.4a) and Ce_pdfr-1b (C13B9.4b) were amplified from EST clones yk1101h12 and yk1404c05, respectively. Because the 5′ end of splice isoforms a, b, and c and the 3′ end of isoforms a and b, are identical, the following primers were used: forward primers a, b, and c, 5′-CACCATGGCGGATGCCACGTCACC-3′; reverse primers a and b, 5′-TTATGGAGATTTTGTGAGCGATTGG-3′; reverse primer c, 5′-AATTTATTCTTTGTTTTCTACTCTTCATAC-3′. A partial Kozak sequence (CACC) was also incorporated immediately preceding the authentic initiation codon, to optimize initiation of translation. The resulting PCR product of each receptor isoform was cloned directly into the eukaryotic expression vector pcDNA3.1D (pcDNA3.1 Directional TOPO Expression Kit, Invitrogen) and sequenced. Each isoform was confirmed to be identical to the Wormbase predicted cDNA sequence.
Cell Culture and Transfections—Chinese hamster ovary cells (CHO-K1), stably overexpressing the mitochondrially targeted apo-aequorin (mtAEQ) and the human Gα16 subunit were used for Ca2+ measurements and cultured in Ham's F-12 medium (Biowhittaker/Cambrex) containing 10% fetal bovine serum, 100 IU/ml of penicillin/streptomycin, 250 μg/ml Zeocin, and 2.5 μg/ml Fungizone (Amphoterin B). Human embryonic kidney cells (HEK293), used in the CRE-luciferase reporter assay, were maintained in Dulbecco's modified Eagle's medium (with l-glutamine, 4,500 mg/liter d-glucose, 110 mg/liter sodium pyruvate) supplemented with 10% fetal bovine serum and 100 IU/ml of a penicillin/streptomycin solution. Both cell lines were split every 3 days (1:10 and 1:5, respectively) and grown at 37 °C in a humidified atmosphere of 5% CO2 in air. CHO/mtAEQ/Gα16 and HEK293 cells were transiently transfected with the receptor cDNA constructs using the FuGENE 6 reagent (Roche), according to the manufacturer's instructions. HEK293 cells were also co-transfected with a multimerized cyclic AMP response element (CRE) luciferase reporter construct (pCRE(6X)-Luc) in a 1:5 (luciferase reporter:pdfr) ratio.
Bioluminescence Assay—Intracellular calcium was monitored as previously described (7). Briefly, CHO/mtAEQ/Gα16 cells expressing the receptor were collected 2 days post-transfection in bovine serum albumin medium (Dulbecco's modified Eagle's medium/Ham's F-12 with 15 mm Hepes, without phenol red, supplemented with 0.1% bovine serum albumin) and loaded with 5 μm coelenterazine h (Invitrogen) for 4 h to reconstitute the holoenzyme aequorin. After a 10-fold dilution, cells (25,000/well) were exposed to potential peptide ligands, i.e. HPLC fractions (1/20) or synthetic peptides reconstituted in bovine serum albumin medium. The calcium response was recorded for 30 s on a Microlumat Plus, LB96V microplate luminometer (EG&G Berthold, Germany). Triton X-100 (0.1%) was used as a positive control, bovine serum albumin medium as a negative control, and 1 μm ATP was used to check the functional response. Cells transfected with an empty pcDNA3.1D vector were used as negative control.
CRE-Luciferase Reporter Assay—Twenty-four hours post-transfection, HEK293 cells (50,000/well) were exposed to potential peptide ligands for 4 h in serum-free medium supplemented with 200 μm 3-isobutyl-1-methylxanthine. Forskolin (Sigma) was added at 10 μm to test for Gαi activity. The luciferase activity was quantified with a LucLite Kit (PerkinElmer Life Sciences), and luminescence was measured on a Microlumat Plus, LB96V microplate luminometer in quadruplicate. EC50 values were calculated from dose-response curves, constructed using a computerized nonlinear regression analysis, with a sigmoidal dose-response equation (SigmaPlot 8.0).
Peptides—Based on in silico predictions and in-house peptidomics data (8), a library of 156 synthetic C. elegans peptides was composed and custom-synthesized by Sigma Genosys. See supplemental Table S1 for the entire list. All peptides were initially tested at a concentration of 10 μm. Receptor activating peptides were purified further using reversed phase HPLC and quantified with the bicinchoninic acid (BCA) protein assay.
Peptide Extraction and HPLC Fractionation—Peptide extraction from four 650-ml liquid cultures (Fernbach flasks) of mixed stage wild-type N2 worms was performed as previously described (8). The resulting aqueous phase was desalted by solid phase extraction (MegabondElute, Varian). The peptides eluted by 0–60% CH3CN in 0.1% trifluoroacetic acid were further fractionated by reversed-phase HPLC on a Delta-pack C18 (2× (25 × 100 mm)) column (Waters) with a solvent flow rate of 12 ml/min. After injection, a wash step of 10 min using 0.1% aqueous trifluoroacetic acid was initiated, followed by a linear gradient over 80 min to a final concentration of 60% CH3CN in 0.1% aqueous trifluoroacetic acid. 12-ml fractions were collected automatically every minute starting from the beginning of the gradient.
Transgenes—A stable transgenic line containing an integrated promoter::gfp construct of C13B9.4 (BC11358) was kindly provided by David Baillie. GFP expression was visualized using the LSM510 multiphoton confocal microscope (Zeiss) and cells were identified using a combination of their position and morphology (Nomarski DIC imaging, Axio Imager Z1, Zeiss). Overexpression of the Ce_PDF genes was accomplished by introducing extra copies of the respective wild-type genes as transgenes. The genomic DNA, including the predicted promoter, the open reading frame, and the 3′-untranslated region sequence for each gene was amplified by PCR, purified, and microinjected (100 ng/μl) in wild-type C. elegans together with 30 ng/μl elt-2::gfp as a selection marker. In the rescue experiment, pdf-1(tm1996) mutants were injected with 50 or 5 ng/μl of the overexpression construct for pdf-1 and 30 ng/μl of elt-2::gfp. Transgenic (GFP+) animals from three independent lines were selected and assayed for rescue of the pdf-1(tm1996) mutant phenotype. Rescue phenotypes were compared with those of nontransgenic (GFP-) siblings of wild-type N2. All PCR primers used are listed in supplemental Table S3.
Locomotion Assay—L4 staged hermaphrodites were picked 16 h prior to behavioral analysis. Worms were tracked around midday for 2 min at 22 °C on nematode growth medium plates containing a thin lawn of freshly grown (8 h) OP50 bacteria. The monitoring system consists of a Leica MZ-16F stereomicroscope equipped with a digital camera (DFC320, Leica Microsystems GmbH, Germany), which was controlled by image and video handling software (Leica Application Suite). The data were compressed and integrated into MPEG format for feature extraction. Time lapse image processing was performed using eaZYX-IMAGING software (Maia Scientific, Geel, Belgium). Worms were segmented from background using linear scale space detection of elongated structures (9, 10). To quantify the centroid velocity and the frequency of reversals and directional changes, segmented objects were first skeletonized and then the center of gravity coordinates was calculated for each worm in each frame of the sequence. To allow for fast visual inspection of nematode behavior in a video sequence, all the binary image analysis results (worm skeletons) from each sequence were overlaid in a single image using the data reduction tools in the eaZYX-IMAGING software. All strains tested were cultivated and analyzed the same way and a similar numbers of tracking assays (n ≥ 12) was carried out. The statistical significance of behavioral assays was determined using the two-tailed Student's t test.
RESULTS
Identification and Cloning of the C. elegans PDF Receptors—A BLAST analysis using the amino acid sequence of the recently characterized D. melanogaster PDF receptor (5–6, 11) (CG13758) as a query revealed 3 potential orthologues in the C. elegans genome (CE30860, CE37087, and CE37088) with 36/54, 35/53, and 36/54% sequence identity and % similarity, respectively. These 3 G protein-coupled receptors, hereafter named PDFR-1a, -b, and -c, represent differentially spliced isoforms of the same gene, C13B9.4 (a, b, and c) (Fig. 1b and supplemental Fig. S2). Sequence-specific primers were used to amplify the open reading frame of pdfr-1c by reverse transcriptase-PCR. The open reading frames of pdfr-1a (C13B9.4a) and pdfr-1b (C13B9.4b) were amplified from C. elegans EST clones yk1101h12 and yk1404c05, respectively. The resulting PCR products (a, 1,641 bp; b, 1,611 bp; and c, 1,626 bp) were cloned directly into the eukaryotic expression vector pcDNA3.1D and sequenced. Each splice isoform was confirmed to be identical to the predicted cDNA sequence (Wormbase) (EF141316, EF141317, and EF141318). The deduced proteins of pdfr-1a, -b, and -c, composed of 546, 536, and 541 amino acids, respectively, all contain seven putative transmembrane domains (TMPRED_form). The amino-terminal extracellular region contains six conserved cysteine residues, two tryptophan residues, and one aspartate, whereas one cysteine residue appears to be conserved in extracellular loops E1 and E2 (Fig. 1b). This is typical for the class B (secretin) GPCRs (12). Interestingly, the extracellular NH2-terminal domain of PDFR-1b differs from that of isoforms a and c due to alternative splicing of the pdfr-1 gene. PDFR-1c, on the other hand, contains a carboxyl terminus that is different from that of the other isoforms (Fig. 1b and supplemental Fig. S2).
FIGURE 1.
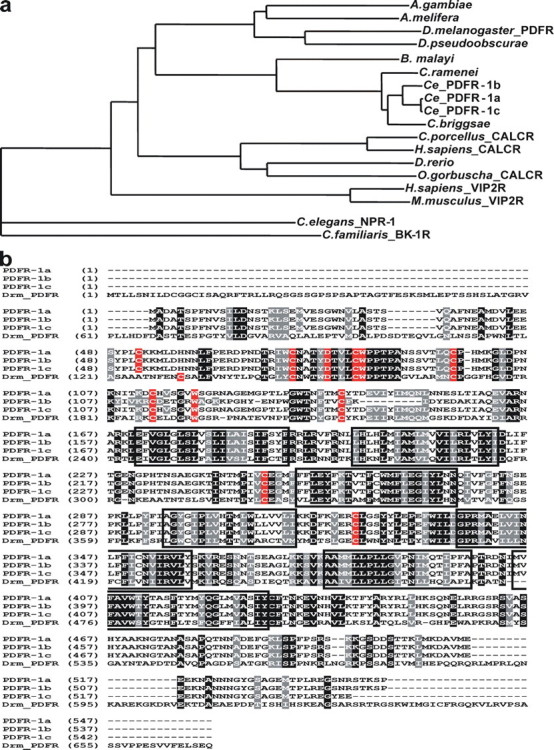
Sequence comparison of Ce_PDFR-1a, -b, and -c with the most closely related invertebrate and vertebrate orthologues. a, phylogenetic relationship of the C. elegans PDF receptors (Ce_PDFR-1a, -b, and -c; C13B9.4a,b,c) and related receptors (AlignX software, Vector NTI Advance 10, Invitrogen). C. elegans NPR-1 (AAA93419) and the C. familiaris bradykinin receptor B1 (AAN16466) were used as an outgroup. b, amino acid sequence alignment of Ce_PDFR-1a, -b, and -c with the PDF receptor of D. melanogaster (Drm_PDFR; CG13758). Identical amino acids are highlighted in black, similar amino acids in gray. Membrane-spanning regions are boxed and cysteine, tryptophan, and aspartate residues conserved in class B GPCRs are highlighted in red.
A phylogenetic analysis of the C. elegans PDF receptors was performed with various related GPCRs (Fig. 1), selected based on BLASTP and tBLASTn analysis. C. elegans NPR-1 (AAA93419) and the Canis familiaris bradykinin receptor B1 (AAN16466) were used as an outgroup. The phylogenetic tree shows a clear clustering of nematode PDF receptor-like receptors, insect PDF receptor-like receptors, and vertebrate calcitonin receptors. The C. elegans PDF receptors are almost identical to the putative GPCRs of the nematodes Caenorhabditis briggsae (78%; CBG1 6586), Caenorhabditis ramenei (92%; Car_Cr01.s ctg4.wum.45.1), and Brugia malayi (63%; 14136. m00015) (Fig. 1), indicating that the receptor is highly conserved in nematodes. In addition, PDFR-1a, -b, and -c are very closely related to the PDF receptor of D. melanogaster (CG13758) and the orphan insect receptors ENSAPMT0000000 7039 (Apis melifera), Q29FR1 (Drosophila pseudoobscurae), and Q7PQE3 (Anopheles gambiae). They are also related, but to a lesser extent than the insect orthologues, to the vertebrate calcitonin GPCRs O08893 (Cavia porcellus), Q8AXU4 (Oncorhynchus gorbuscha), P30988 (Homo sapiens), and Q68EK2 (Danio rerio, putative orphan receptor). The vertebrate VIP2 receptors CAA64474 (Homo sapiens) and NP_033537 (Mus musculus) seem to share less similarity with the nematode PDF receptors.
Identification of the C. elegans PDF Receptor Ligands—Following successful cloning and the construction of CHO cell lines (stably overexpressing Gα16 and apo-aequorin) transiently expressing PDFR-1a, -b, or -c, the cells were challenged with a library of 156 synthetic C. elegans peptides, consisting of neuropeptides belonging to the established FMRFamide-like peptide (FLP) and neuropeptide-like protein (NLP) families of peptides (supplemental Table S1). Only synthetic analogues of the recently identified C. elegans PDF neuropeptides PDF-1a (SNAELINGLIGMDLGKLSAVamide), PDF-1b (SNAELINGLLSMNLNKLSGAamide), and PDF-2 (NLP-37, NNAEVVNHILKNFGALDRLGDVamide)4 were able to activate both receptors PDFR-1a and PDFR-1b in the calcium bioluminescence assay (supplemental Fig. S1, a and b). This activity was not seen in cells transfected with the empty pcDNA3.1D vector. The pdfr-1 receptor gene (C13B9.4) and the neuropeptide precursor gene pdf-1, which encodes two of its activating ligands, both reside on chromosome III, only 245.9 kb (42 genes) from each other. The linking peptides encoded by pdf-1 were not active. Challenging the cells with 10 μm of a synthetic D. melanogaster PDF peptide (CG6496-PA) (Table 1) also resulted in activation of receptors PDFR-1a and PDFR-1b (data not shown). This supports our hypothesis that PDFR-1a and -b are indeed the orthologues of the Drosophila PDF receptor. To verify whether the Ce_PDFs indeed constitute the endogenous cognate ligands for these receptors, we also challenged the cells with HPLC fractions of a whole body peptide extract of mixed stage C. elegans and checked whether any other endogenous peptides were able to activate these receptors. Only fractions 53 and 55, which contain the 3 endogenous PDF peptides (as previously described in Ref. 1), were able to elicit a calcium response, but only in cells expressing receptors PDFR-1a and -b (supplemental Fig. S1a). Surprisingly, PDFR-1c did not react to any of the tested peptide ligands in the calcium bioluminescence assay.
TABLE 1.
|
Peptide |
Sequence |
Ca2+ assay |
CRE-luciferase assay |
||||
|---|---|---|---|---|---|---|---|
| PDFR-1a | PDFR-1b | PDFR-1c | PDFR-1a | PDFR-1b | PDFR-1c | ||
| Ce_PDF-1a | SNAELINGLIGMDLGKLSAVamide | +a | 127.4c | –b | 385.5 ± 5.1 | 6.99 ± 0.03 | 10,600 ± 400 |
| Ce_PDF-1b | SNAELINGLLSMNLNKLSGAamide | + | 360.7 | – | 5370 ± 50 | 15.56 ± 0.04 | 1,900 ± 40 |
| Ce_PDF-2 | NNAEVVNHILKNFGALDRLGDVamide | + | 33.8 | – | 114.5 ± 4.1 | 0.59 ± 0.03 | 7,300 ± 30 |
| Drm_PDF (10 μm) | NSELINSLLSLPKNMNDAamide | + | + | – | + | + | + |
| VIP (PHM-27) | YLESLMamide | – | – | – | – | – | – |
| VIP (PHV-42) | VSSNISEDPVPV | – | – | – | – | – | – |
| PACAP27 | SYSRYRKQMAVKKYLAAVLamide | – | – | – | – | – | – |
| PACAP38 | YLAAVLGKRYKQRVKNKamide | – | – | – | – | – | – |
Plus sign, receptor activation observed with a single dose of 10 μm
Minus sign, no receptor activation observed with a single dose of 10 μm
EC50 values for each receptor-ligand couple (mean ± S.E.) as indicated when available
C-terminal fragments of the human vasoactive intestinal peptide (VIP) (YLESLM amide (PHM-27) and VSSNISEDPVPV (PHV-42)) and pituitary adenylate cyclase activating peptides (SYSRYRKQMAVKKYLAAVL amide (PACAP27) and YLAAVLGKRYKQRVKNK amide (PACAP38)) were also tested (Table 1). None of these peptides, however, was able to activate the PDFR-1 receptors.
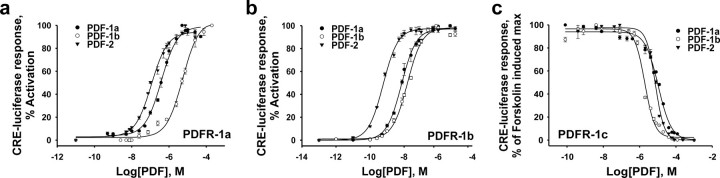
Pharmacological Characterization of the C. elegans PDF Receptors—To analyze the G protein signaling pathway, all three Ce_PDFs were tested for their ability to elicit a Ca2+ response in the calcium assay, using CHO/mtAEQ cells expressing PDFR-1a or -b, but lacking Gα16. The observed absence of activity suggests that these receptors do not signal through Gαq. Class B GPCRs, like calcitonin, VIP, and PACAP receptors and also the Drosophila PDF receptor, typically signal through adenylate cyclase (11, 12), regulating intracellular concentrations of cAMP. To test the involvement of adenylate cyclase in the PDFR-1 signal transduction pathway, HEK293 cells were transfected with receptor cDNA (PDFR-1a, -b, or -c) and a multimerized CRE-luciferase reporter gene, and subsequently assayed for luciferase activity 24 h post-transfection with peptides Ce_PDF-1a, Ce_PDF-1b, and Ce_PDF-2. This resulted in a clear and dose-dependent increase of CRE-luciferase activity for PDFR-1a and PDFR-1b (Fig. 2, a and b), suggesting that they signal through Gαs. Interestingly, in the CRE-luciferase reporter assay, PDFR-1c could be activated by all three PDF peptides (although not as potently as PDFR-1a and -b). The C. elegans PDF peptides are able to inhibit forskolin-induced cAMP formation in cells expressing PDFR-1c, resulting in a dose-dependent decrease of luciferase activity (Fig. 2c). These data suggest that PDFR-1c might signal through a Gαi/o type of G protein. These CRE-luciferase responses were all mediated through the PDFR-1 receptors because no response was obtained with mock cells or cells expressing the empty pcDNA3.1 plasmid.
FIGURE 2.
Dose-dependent CRE-luciferase responses to PDF-1a, PDF-1b, and PDF-2 of HEK293 cells expressing the PDR-1 receptors. a–c, dose-response curves for PDF-1a, PDF-1b, and PDF-2 on receptors PDR-1a, PDR-1b, and PDR-1c, respectively. Each data point represents the mean ± S.E. of at least two independent experiments carried out in quadruplicate.
Analysis of pdfr-1 Expression Pattern—To examine the spatial expression of pdfr-1 in C. elegans, GFP expression of a stable transgenic line containing an integrated promoter::gfp construct of C13B9.4 (BC11358, kindly provided by David Baillie) was visualized using an LSM510 multiphoton confocal microscope (Zeiss). Fluorescent signals from PDFR-1::GFP could be observed in hermaphrodites throughout post-embryonic life. pdfr-1::gfp transgenic lines showed consistent GFP expression in all 95 body wall muscle cells and less frequently in muscle arms. Expression was also observed in the mechanosensory neuron pairs PLM, ALM, FLP, OLQD, and OLQV, the chemosensory neuron pairs PHA and PHB, the ring motor neurons RMED and RMEV, and the pharyngeal interneuron pair I1 (Fig. 3, a and b). In addition, fluorescent signals were observed in 2 vulva cells and several additional head and tail neurons that could not be unequivocally identified. The expression pattern of pdfr-1 could be confirmed by immunohistochemistry using anti-PDFR-1 polyclonal antibodies (raised in rabbit against synthetic C. elegans PDFR-1 fragment 54–67 (KMLDHNNLFPERDP), Sigma; data not shown). In males, a similar expression pattern was observed in the head and body wall muscle cells. Other neuronal cells, like the sensory neuron R3, that has a possible role in male mating behavior (13), showed GFP fluorescence in the male tail (Fig. 3, c and d) (see also supplemental Table S2 for an overview and corresponding references). Because the pdfr-1::gfp transgenic animals expressed a transcriptional fusion construct of C13B9.4, we could not differentiate between splice isoforms. For clarity, we also created three-dimensional movies from confocal Z-stack projections of pdfr-1::gfp transgenic animals (see supplementary Movies online).
FIGURE 3.
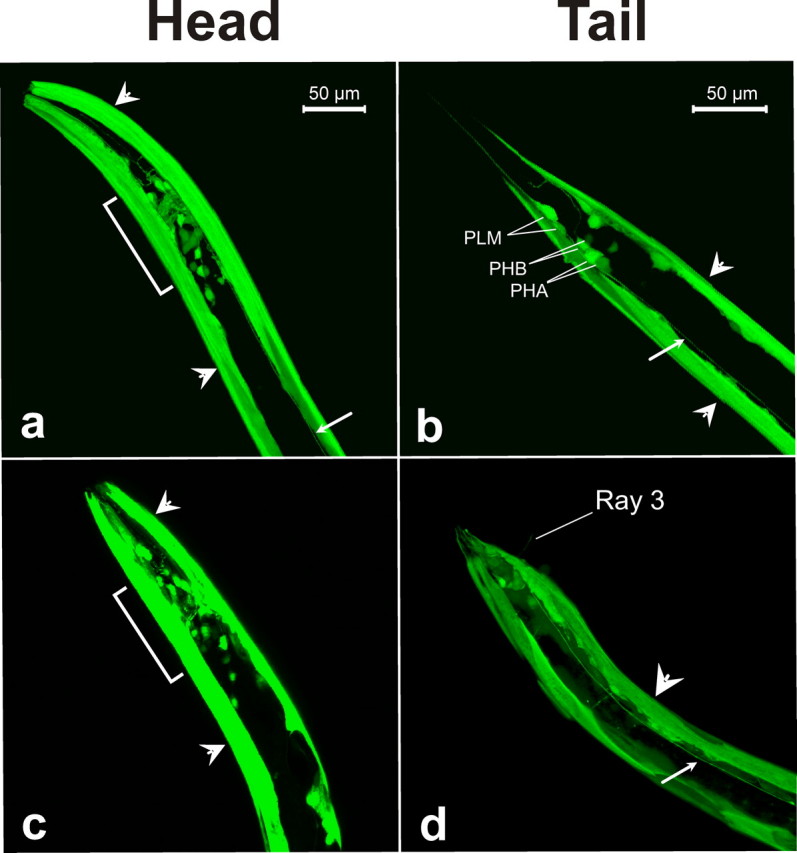
Expression pattern of pdfr-1. Confocal projections of transgenic wild-type N2 worms expressing GFP under the control of the pdfr-1 promoter sequence. Only the anterior head and posterior tail regions of the worm are shown. a and b, hermaphrodite head and tail. c and d, male head and tail. Brackets indicate neurons of the head, small arrows indicate the ventral nerve cord and large arrowheads indicate body wall muscle cells. The position and identity of fluorescent cell bodies in the tail are indicated.
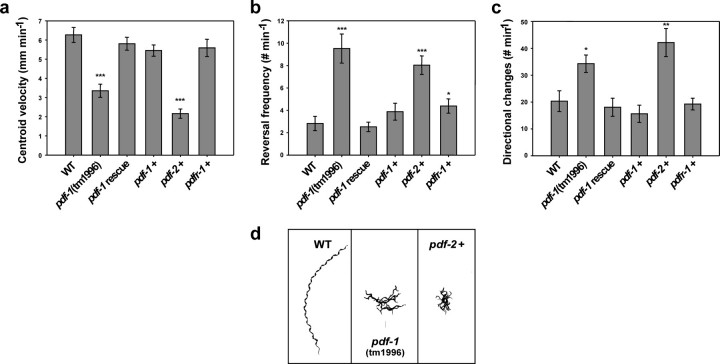
Functional Characterization of the C. elegans PDF Signaling System—In D. melanogaster, mutations in pdf or the PDF receptor result in an aberrant locomotor behavior (4, 11). Also C. elegans PDF-1 mutants display an abnormal free-running locomotor rhythm.4 To study the function of the C. elegans PDF system in detail, we analyzed the locomotor behavior of a pdf-1 deletion mutant (FX1996, pdf-1(tm1996)) and multicopy overexpression lines of pdf-1, pdf-2, and pdfr-1. Worms were recorded for 2 min and the data analyzed with eaZYX-IMAGING software to quantify various locomotion parameters, including centroid velocity (change in centroid position (forward + backward) over time), frequency of reversals, and frequency of directional changes. pdf-1 mutants displayed three distinct locomotion defects when compared with wild-type N2 worms. They display a significant decrease in centroid velocity, which is a measure for the moving speed of the worm (Fig. 4a). Their movements are almost 50% slower than that of wild-type animals (p = 2.4160E-5). A 3-fold increase in reversal frequency was also observed (p = scored as any shift from forward to backward movement. pdf-1 mutants display a frequent shift from forward to backward movement, in contrast to wild-type animals (in which forward movement is occasionally alternated with backward movements). In addition, these mutants change direction more often, resulting in an increase of 75% in the frequency of directional changes (p = 0.0275) (Fig. 4c). Due to these locomotor defects, the net forward movement (radial distance from origin) is drastically reduced when compared with wild-type animals (Fig. 4d). All pdf-1(null) locomotion defects were fully suppressed to normal locomotion (rescued) when pdf-1 genomic DNA (both 50 and 5 ng/μl concentrations) was re-introduced into mutants by microinjection (Fig. 4, a–c), confirming that the deletion mutation in pdf-1 (the absence of PDF-1a,b peptides) specifically caused these defects. Overexpression of pdf-1 from its own control sequences did not result in any significant (p < 0.05) locomotor defects, when compared with wild-type N2 (Fig. 4, a–c).
FIGURE 4.
Analysis of defective locomotion of pdf-1 mutants and transgenic lines overexpressing pdfr-1 (pdfr-1 +), pdf-1 (pdf-1 +), and pdf-2 (pdf-2 +). a, centroid velocity is decreased in pdf-1 mutants (pdf-1(tm1996)) and worms overexpressing pdf-2 (pdf-2 +). b, increased frequency of reversals in pdf-1 mutants and worms overexpressing pdf-2. c, increased frequency of directional changes in pdf-1 mutants and worms overexpressing pdf-2. d, rope images of moving worms (2 min). Error bars indicate S.E. (n ≥ 12). *, p < 0.05; **, p < 0.005; ***, p < 0.0005.
Interestingly, high-copy expression of pdf-2 resulted in a locomotor phenotype identical to one of the pdf-1 null mutants. Animals overexpressing pdf-2 display a 3-fold decrease in centroid velocity (p = 3.6100E-8), when compared with wild-type animals (Fig. 4a). They also show a 184% increase in reversal frequency (p = 6.9910E-5) (Fig. 4b) and a 2-fold increase in the frequency of directional changes (p = 2.0941E-3) (Fig. 4c). Compared with pdf-1 null mutants, the locomotor defects in centroid velocity and frequency of directional changes are even more pronounced in animals overexpressing pdf-2. Hence, the net forward movement (radial distance from origin) of these animals is even more reduced (Fig. 4d). pdfr-1 overexpression did not significantly alter the centroid velocity, nor the frequency of directional changes (Fig. 4, a and b). It did, however, result in a 55% increase (p = 0.0491) in reversal frequency, when compared with wild-type animals.
DISCUSSION
G protein-coupled receptors represent the largest and most important group of therapeutic targets in medicine. Although C. elegans is an important model organism in developmental biology and neurobiology, information on the GPCR signaling systems in this nematode is scarce (14). Only 6 FLP-activating G protein-coupled receptors have been characterized so far (7, 15–19). For the remaining peptides (>230), most of which belong to the NLP class of peptides (157), the corresponding receptors are currently unknown.
By using a reverse pharmacology approach, we have identified and coupled three PDF receptor isoforms in C. elegans to their matching peptide ligands. These are the first class B receptors to be deorphanized in C. elegans and also the first receptors having NLP-type peptides (non-FLP) as cognate ligands. So far, PDF neuropeptide signaling had only been reported in arthropods. In this paper we identify C13B9.4 as the C. elegans orthologue of the recently identified D. melanogaster PDF receptor (CG13758) (5, 6, 11). C13B9.4 represents one of six class B (secretin) GPCR genes predicted in C. elegans and it encodes three different splice isoforms, Ce_PDFR-1a, -b, and -c, which are closely related to insect PDF receptor-like receptors, and the vertebrate calcitonin and VIP receptors. Ca2+ and CRE-luciferase reporter screening assays reveal that the Ce_PDF-like neuropeptides4 are the cognate ligands of PDFR-1a, -b, and -c. Overall we find that both PDF and its receptor seem to be well conserved between insects and nematodes. This type of co-evolution between neuropeptide receptors and their ligands has been rarely observed in C. elegans and emphasizes the functional importance of PDF signaling in nematodes.
All three PDF peptides were able to activate each of the PDFR isoforms in the CRE-luciferase reporter assay, but with a significant difference in potency. The lack of PDFR-1c activation observed in the Ca2+ assay might be caused by a defective membrane trafficking of PDFR-1c in CHO cells. PDFR-1b shows the highest PDF affinity of all receptors and PDFR-1c the lowest. PDF-2 is the most potent activator of PDFR-1a and PDFR-1b, whereas PDF-1b is the least potent ligand. There is, however, a serious (200-fold) difference in PDF affinity between receptor isoforms a and b, which can be explained by the difference in the extracellular NH2-terminal domains. The calcium screening experiments indicate that PDFR-1a and -b do not signal through Gαq. CRE-luciferase activity measurements, in particular, suggest that these receptors couple to a Gαs type of G protein, which is in agreement with the findings for PDFR in Drosophila (11). The PDFR-1c isoform, however, seems to signal through a Gαi/o type of G protein. In addition, PDF-1b is able to activate this receptor isoform more potently than PDF-1a and PDF-2. The PDF-1a and PDF-2 activity of PDFR-1c seems very low when compared with PDFR-1a, although they share identical extracellular NH2-terminal domains. This might reflect the coupling to different G proteins as PDFR-1c differs at the carboxyl terminus or it might be due to difficulties in functional expression in the heterologous assay, or the necessity of receptor activity-modifying proteins or oligomerization (20).
Spatial and temporal expression patterns of the individual PDFR-1 isoforms might be different from the observed pattern because the latter reflects the expression pattern of all three isoforms together. They are expressed throughout post-embryonic life in at least 11 types of neurons, five of which are involved in mechanosensation, two in chemosensation, and two in locomotion. Interestingly, they also show strong expression in all body wall muscle cells. The Drosophila PDFR is expressed in several subgroups of clock neurons and some non-clock neuronal cells (5, 11). In contrast with our findings, however, PDFR expression in muscle cells has so far not been reported in arthropods. The PDFR-1 expression in body wall muscles suggests that PDF-1 and PDF-2 may also act directly on muscle cells to modulate their activity. Only a few PDFR-1 expressing cells make direct contact with the cells expressing PDF, supporting the idea that the Ce_PDFs may act as local neuromodulators.4
Circadian analysis already revealed that PDF-1 peptides are involved in the control of daily locomotor rhythms in C. elegans.4 Mutants lacking PDF-1 mimic the behavioral phenotype of Drosophila PDF mutants with respect to free-running locomotor rhythms. Our further characterization of the Ce_PDF system indicates that PDF-1 mutants display a severely altered locomotor behavior, as rendered by a decreased moving speed, an increased reversal frequency and an increased frequency of directional changes. Also overexpression of PDFR-1 resulted in an increase in reversal frequency. Simmer et al. (21) already reported an abnormal locomotion phenotype for C13B9.4 (pdfr-1) using the RNA interference hypersensitive strain rrf-3(pk1426) (WBRNAi00027171). This is consistent with the observed PDF/PDFR spatial expression patterns as almost all neurons that show PDF and/or PDFR expression in C. elegans play a role in the sensing and integration of environmental stimuli or in the control of locomotion.4 Movement in C. elegans is directed by sensory inputs reflecting the internal state of the animal and its environment (22). Interestingly, overexpression of pdf-2 resulted in a locomotor phenotype identical to one of the pdf-1(null) mutants. Although we have already screened over 30 predicted neuropeptide GPCRs (out of 50–60 ones predicted (23)) with synthetic PDFs, we cannot rule out the possibility that PDF-1 and PDF-2 might also interact with other receptors in vivo. Even though there is no PDFR mutant available, our present data strongly suggest that the PDF-1 peptides act through PDFR-1 to stimulate forward movement and/or to inhibit backward movement and that the PDF-2 peptides act through PDFR-1 to inhibit forward movement and/or to stimulate backward movement. PDF-1 and PDF-2 peptides may thus elicit opposite effects in vivo, both activating PDFR-1 isoforms to modulate forward and backward locomotion. We hypothesize that modulation of circadian locomotor behavior through PDF signaling might therefore depend on a state of equilibrium between the levels of PDF-1 and PDF-2, and that these levels have to be fine-tuned. In this way, spatial and/or temporal differences in expression levels of PDF-1, PDF-2, and PDFR-1 isoforms could provide a basic mechanism in the regulation of the observed rhythmic locomotor behavior in C. elegans (24, 25).4
The presence of PDFR-1 in many mechanosensory neurons (ALM, FLP, OLQD, OLQV, and PLM) suggests that PDF signaling might also be involved in modulating touch sensitivity. These neurons all provide input to the command (inter)neurons (PVC, AVB, AVD, and AVA), which drive forward and backward locomotion. In C. elegans, touch initiates and modulates several behaviors, including locomotion (26), egg laying, feeding (27, 28), defecation (29), and mating (13), which all display an ultradian or circadian rhythm (25, 30).4 In 2000, Hendricks et al. (31) reported that rest in Drosophila, like sleep in mammals, features a decrease in responsiveness to mechanical stimuli. Whether this is also the case in C. elegans and is modulated by the PDF system remains to be determined.
The identification and functional characterization of the PDF receptors in nematodes strengthens the hypothesis that the PDF system, which imposes the clock rhythm on behavior in Drosophila, has been conserved throughout the protostomian evolutionary lineage. Several studies have shown that parasitic nematodes display biological activity rhythms that are adjusted to the circadian rhythm of their host or carrier/vector (32–33). GPCRs like the PDF receptors, which are involved in control of locomotion, make interesting targets for the development of novel, more selective and environmentally friendly antihelminthic drugs. As many species of plant and animal parasitic nematodes directly or indirectly affect billions of people worldwide (34), it is not difficult to comprehend the importance of a better understanding of the circadian clock in nematodes.
Supplementary Material
Acknowledgments
We thank the Genome BC C. elegans Gene Expression Consortium, which is funded by Genome Canada and Genome British Columbia; the Caenorhabditis Genetics Center, which is funded by the NIH National Centre for Research Resources (NCRR), and the C. elegans Knock-out consortium for providing the C. elegans strains. We are grateful to Y. Kohara for EST clones, A. Fire for plasmid pPD95.75, P. Taghert for the pCRE(6x)-LUC construct, and R. Tsien for the mCherry plasmid. We acknowledge the Cell Imaging Core of the K. U. Leuven Impulse for financing heavy equipment and for access to the multiphoton fluorescence microscope. We also thank L. Vandenbosch, S. Van Soest, J. Gijbels, M. Vandereecken, and J. Puttemans for excellent technical assistance. We are especially grateful to A. De Loof for critical reading and support.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBank™/EBI Data Bank with accession number(s) EF141316 (pdfr-1a), EF141317 (pdfr-1b), and EF141318 (pdfr-1c).
This work was supported in part by Research Foundation, Flanders, Grants (FWO) G.0270.04, G.O580.06, and G.0434.07. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S3, Figs. S1 and S2, and Movies 1–3.
Supported by a Ph.D. scholarship from Research Foundation, Flandersi (FWO)
Footnotes
The abbreviations used are: PDF, pigment dispersing factor; GPCR, G protein-coupled receptor; CHO, Chinese hamster ovary; HEK, human embryonic kidney; CRE, cyclic AMP response element; NLP, neuropeptide-like protein; FLP, FMRFamide-like peptide; VIP, vasoactive intestinal peptide; HPLC, high pressure liquid chromatography; GFP, green fluorescent protein.
T. Janssen, S. J. Husson, M. Lindemans, K. Verstraelen, E. Meelkop, L. Temmerman, R. Rademakers, I. Mertens, M. Nitabach, G. Jansen, and L. Schoofs, submitted for publication.
References
- 1.Rao, K. R., and Riehm, J. P. (1993) Ann. N. Y. Acad. Sci. 680 78-88 [DOI] [PubMed] [Google Scholar]
- 2.Sato, S., Chuman, Y., Matsushima, A., Tominaga, Y., Shimohigashi, Y., and Shimohigashi, M. (2002) Zoolog. Sci. 19 821-828 [DOI] [PubMed] [Google Scholar]
- 3.Helfrich-Forster, C., Tauber, M., Park, J. H., Muhlig-Versen, M., Schneuwly, S., and Hofbauer, A. (2000) J. Neurosci. 20 3339-3353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Renn, S. C., Park, J. H., Rosbash, M., Hall, J. C., and Taghert, P. H. (1999) Cell 99 791-802 [DOI] [PubMed] [Google Scholar]
- 5.Hyun, S., Lee, Y., Hong, S. T., Bang, S., Paik, D., Kang, J., Shin, J., Lee, J., Jeon, K., Hwang, S., Bae, E., and Kim, J. (2005) Neuron 48 267-278 [DOI] [PubMed] [Google Scholar]
- 6.Lear, B. C., Merrill, C. E., Lin, J. M., Schroeder, A., Zhang, L., and Allada, R. (2005) Neuron 48 221-227 [DOI] [PubMed] [Google Scholar]
- 7.Mertens, I., Meeusen, T., Janssen, T., Nachman, R., and Schoofs, L. (2005) Biochem. Biophys. Res. Commun. 330 967-974 [DOI] [PubMed] [Google Scholar]
- 8.Husson, S. J., Clynen, E., Baggerman, G., De Loof, A., and Schoofs, L. (2005) Biochem. Biophys. Res. Commun. 335 76-86 [DOI] [PubMed] [Google Scholar]
- 9.Geusebroek, J. M., Smeulders, A. W. M., and Geerts, H. (2001) Int. J. Comput. Vision. 43 99-111 [Google Scholar]
- 10.Lindeberg, T. (1994) Scale-Space Theory in Computer Vision, Kluwer Academic Publishers, Boston
- 11.Mertens, I., Vandingenen, A., Johnson, E. C., Shafer, O. T., Li, W., Trigg, J. S., De Loof, A., Schoofs, L., and Taghert, P. H. (2005) Neuron 48 213-219 [DOI] [PubMed] [Google Scholar]
- 12.Harmar, A. J. (2001) Genome Biol. 2 REVIEWS3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu, K. S., and Sternberg, P. W. (1995) Neuron 14 79-89 [DOI] [PubMed] [Google Scholar]
- 14.Husson, S. J., Mertens, I., Janssen, T., Lindemans, M., and Schoofs, L. (2007) Prog. Neurobiol. 82 33-55 [DOI] [PubMed] [Google Scholar]
- 15.Kubiak, T. M., Larsen, M. J., Zantello, M. R., Bowman, J. W., Nulf, S. C., and Lowery, D. E. (2003) J. Biol. Chem. 278 42115-42120 [DOI] [PubMed] [Google Scholar]
- 16.Kubiak, T. M., Larsen, M. J., Nulf, S. C., Zantello, M. R., Burton, K. J., Bowman, J. W., Modric, T., and Lowery, D. E. (2003) J. Biol. Chem. 278 33724-33729 [DOI] [PubMed] [Google Scholar]
- 17.Mertens, I., Vandingenen, A., Meeusen, T., Janssen, T., Luyten, W., Nachman, R. J., De Loof, A., and Schoofs, L. (2004) FEBS Lett. 573 55-60 [DOI] [PubMed] [Google Scholar]
- 18.Mertens, I., Clinckspoor, I., Janssen, T., Nachman, R., and Schoofs, L. (2006) Peptides 27 1291-1296 [DOI] [PubMed] [Google Scholar]
- 19.Rogers, C., Reale, V., Kim, K., Chatwin, H., Li, C., Evans, P., and de Bono, M. (2003) Nat. Neurosci. 6 1178-1185 [DOI] [PubMed] [Google Scholar]
- 20.Conner, A. C., Simms, J., Hay, D. L., Mahmoud, K., Howitt, S. G., Wheatley, M., and Poyner, D. R. (2004) Biochem. Soc. Trans. 32 843-846 [DOI] [PubMed] [Google Scholar]
- 21.Simmer, F., Moorman, C., van der Linden, A. M., Kuijk, E., van den Berghe, P. V., Kamath, R. S., Fraser, A. G., Ahringer, J., and Plasterk, R. H. (2003) PLoS Biol. 1 E12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao, B., Khare, P., Feldman, L., and Dent, J. A. (2003) J. Neurosci. 23 5319-5328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keating, C. D., Kriek, N., Daniels, M., Ashcroft, N. R., Hopper, N. A., Siney, E. J., Holden-Dye, L., and Burke, J. F. (2003) Curr. Biol. 13 1715-1720 [DOI] [PubMed] [Google Scholar]
- 24.Hasegawa, K., Saigusa, T., and Tamai, Y. (2005) Chronobiol. Int. 22 1-19 [DOI] [PubMed] [Google Scholar]
- 25.Saigusa, T., Ishizaki, S., Watabiki, S., Ishii, N., Tanakadate, A., Tamai, Y., and Hasegawa, K. (2002) Curr. Biol. 12 R46-R47 [DOI] [PubMed] [Google Scholar]
- 26.Wicks, S. R., and Rankin, C. H. (1995) J. Neurosci. 15 2434-2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chalfie, M., Sulston, J. E., White, J. G., Southgate, E., Thomson, J. N., and Brenner, S. (1985) J. Neurosci. 5 956-964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keane, J., and Avery, L. (2003) Genetics 164 153-162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomas, J. H. (1990) Genetics 124 855-872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iwasaki, K., Liu, D. W., and Thomas, J. H. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 10317-10321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hendricks, J. C., Finn, S. M., Panckeri, K. A., Chavkin, J., Williams, J. A., Sehgal, A., and Pack, A. I. (2000) Neuron 25 129-138 [DOI] [PubMed] [Google Scholar]
- 32.Brown, E. D., Macdonald, D. W., Tew, T. E., and Todd, I. A. (1994) J. Helminthol. 68 105-108 [DOI] [PubMed] [Google Scholar]
- 33.Pichon, G., and Treuil, J. P. (2004) C. R. Biol. 327 1087-1094 [DOI] [PubMed] [Google Scholar]
- 34.Parkinson, J., Mitreva, M., Whitton, C., Thomson, M., Daub, J., Martin, J., Schmid, R., Hall, N., Barrell, B., Waterston, R. H., McCarter, J. P., and Blaxter, M. L. (2004) Nat. Genet. 36 1259-1267 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.