Abstract
The LIM-only protein FHL2 acts as a transcriptional modulator that positively or negatively regulates multiple signaling pathways. We recently reported that FHL2 cooperates with CREB-binding protein/p300 in the activation of β-catenin/T cell factor target gene cyclin D1. In this paper, we demonstrate that FHL2 is associated with the cyclin D1 promoter at the T cell factor/CRE site, providing evidence that cyclin D1 is a direct target of FHL2. We show that deficiency of FHL2 greatly reduces the proliferative capacity of spontaneously immortalized mouse fibroblasts, which is associated with decreased expression of cyclin D1 and p16INK4a, and hypophosphorylation of Rb. Reexpression of FHL2 in FHL2-null fibroblasts efficiently restores cyclin D1 levels and cell proliferative capacity, indicating that FHL2 is critical for cyclin D1 activation and cell growth. Moreover, ectopic cyclin D1 expression is sufficient to override growth inhibition of immortalized FHL2-null fibroblasts. Gene expression profiling revealed that FHL2 deficiency triggers a broad change of the cell cycle program that is associated with down-regulation of several G1/S and G2/M cyclins, E2F transcription factors, and DNA replication machinery, thus correlating with reduced cell proliferation. This change also involves down-regulation of the negative cell cycle regulators, particularly INK4 inhibitors, which could counteract the decreased expression of cyclins, allowing cells to grow. Our study illustrates that FHL2 can act on different aspects of the cell cycle program to finely regulate cell proliferation.
The LIM-only protein FHL2 is a member of the four-and-a-half LIM (FHL) family (1). Individual LIM domains consist of two zinc finger motifs rich in cysteine and histidine that serve as protein-binding interface for the assembly of multiprotein complexes. The zinc fingers of some transcriptional regulators can interact with DNA, but there is no evidence for DNA binding activity of a LIM domain. FHL2 interacts with multiple transcription factors, including the androgen receptor, AP1, CREB, PLZF, SKI, and β-catenin (2–8). It functions as either a coactivator or a corepressor, depending on cell type and promoter contexts (7, 9). Moreover, FHL2 can bind several transcription factors simultaneously and participates in the assembly of multiprotein complexes (10, 11).
FHL2 is found in both the cytoplasm and the nucleus (8, 12). In the cytoplasm, FHL2 interacts with integrins and focal adhesion kinase at focal adhesions (13, 14). Integrins bind extracellular matrix proteins and certain cell surface receptors, serving as sensors for both chemical and mechanical cues (15). FHL2 shuttles between focal adhesions and nuclei to relay the flow of genetic information at different execution points. Serum response factor, which regulates the expression of immediate early genes, directly controls FHL2 expression in a RhoA-dependent manner (16), and the timing of FHL2 induction is coordinated with that of the early response proteins Fos and Jun (3). Following stimulation with RhoA or serum, FHL2 is induced and translocated to the nucleus, thereby linking extracellular signals to gene expression programs (3, 17). Studies in various biological systems have shown that FHL2 plays diverse roles in many aspects of cell life. Mice deficient in FHL2 are viable (18) but display pathological phenotypes, including osteopenia, resulting from a decrease in the activity of osteoblasts (19), cardiac hypertrophy under β-adrenergic stimulation (20), and defects in skin wound healing (21).
We have previously demonstrated that FHL2 is a coactivator of β-catenin and that it cooperates with CREB5-binding protein/p300 to enhance transcription driven by the β-catenin·TCF complex β-catenin (8, 10, 22). We have shown that FHL2 and β-catenin synergistically increase the transcription of the β-catenin target gene cyclin D1. In addition, primary FHL2-/- mouse embryonic fibroblasts (MEFs) were found to express half the normal amount of cyclin D1 transcripts (10). Recent studies have shown that FHL2 antagonizes the p53-dependent antiproliferative effects of the transcription factor E4F1 (23) and regulates cell cycle-dependent p21 expression in breast cancer cells (24), thus linking FHL2 to cell cycle and proliferation.
In this study, we used MEFs derived from FHL2-null embryos to investigate the role of FHL2 in the regulation of cyclin D1 and cell proliferation. By chromatin immunoprecipitation (ChIP), we show that FHL2 is associated with the cyclin D1 promoter. We found that disruption of FHL2 decreased the proliferative capacity of murine fibroblasts. Finally, we used microarray to analyze gene expression at the whole genome level and show down-regulation of both positive and negative regulators of the cell cycle in FHL2-null cells. Overall, our results provide evidence for the function of FHL2 as an important regulator of cell cycle and proliferation.
EXPERIMENTAL PROCEDURES
MEF Generation and Establishment of Immortalized Cell Lines—Primary MEFs from wild type (WT) or FHL2-/- mice (18) were derived from embryos on day 13.5 or 16.5. Spontaneously immortalized cell lines were generated from primary MEFs using a 3T3 protocol. The growth curves of immortalized clones were obtained by seeding 2.5 × 104 cells into 12-well plates in triplicate. Cell numbers were determined every day for a total of 6 days.
Flow Cytometry Analysis—Cells were pulsed for 30 min with 10 μm BrdUrd (Sigma). After fixation in 70% ethanol, cells were denatured in 2 m HCl, neutralized with 0.1 m sodium borate, and then incubated with fluorescein isothiocyanate-conjugated anti-BrdUrd monoclonal antibody (556028; BD Pharmingen). DNA was stained with 10 μg/ml propidium iodide (Sigma) and treated with DNase-free RNase (Roche Applied Science). Cells were analyzed by flow cytometry with excitation at 488 nm and monitoring of BrdUrd-linked fluorescein isothiocyanate and propidium iodide at 514 and 575 nm, respectively, using appropriate filters.
Serum Stimulation and Immunoblotting—1.5 × 105 MEFs were seeded into 6-well plates and synchronized by incubation in 0.5% fetal bovine serum in Dulbecco's modified Eagle's medium for 96 h. Synchronized cells were stimulated by adding 10% fetal bovine serum in Dulbecco's modified Eagle's medium. Extracts were prepared at 0, 3, 9, 15, 18, 21, and 24 h post-stimulation and analyzed by immunoblotting as described previously (10). Antibodies against p16INK4a (sc-1207), p15INK4b (sc-613), p18INK4c (sc-1064), cyclin D1 (sc-450), cyclin D2 (sc-181), cyclin D3 (sc-182), cyclin A (sc-751), and cyclin E (sc-481) were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA); FHL2 (ab12327), p27Kip1 (ab6547), and phospho-Rb (ThrT821) (ab4787) from Abcam; Rb (554136) from BD Pharmingen; Phospho-Rb (Ser780) (9307) and (Ser795) (9301) from Cell Signaling; and actin (A5441) from Sigma.
Retroviral Vectors and Gene Transfer—pBabe-FHL2 was constructed by inserting full-length FHL2 cDNA in the pBabe-puro vector. pBabe puro cyclin D1 HA constructed by Dr. William Hahn was obtained from Addgene (Addgene plasmid 9050). Phoenix ecotropic virus packaging cells in 10-mm dishes were transfected by phosphate calcium precipitation with 10 μg of a retroviral plasmid. 24 h later, exponentially growing MEFs were infected with 5 ml of virus-containing supernatant supplemented with 8 μg/ml Polybrene/dish. This procedure was repeated three times every 3 h. Cells were selected in medium containing 4 μg/ml puromycin.
ChIP Assay—Synchronized MEFs were starved in Dulbecco's modified Eagle's medium with 0.5% fetal bovine serum for 96 h, followed by the addition of 10% serum for 15 h before harvest. Nonsynchronized or synchronized MEFs were used for ChIP assays with anti-FHL2 antibody, as described previously (25). The antibodies used were anti-FHL2 (5 μg; K0055-3; MBL) and irrelevant anti-HA antibody (Babco). The following primers were used to amplify the cyclin D1 promoter: F1 (5′-CAACGAAGCCAATCAAGAAGCT-3′) and R1 (5′-GAAAAGTAAATCGCTGCAAGTTATTAGTC-3′) for the AP1 site; F2 (5′-CTGCCCGGCTTTGATCTCT-3′) and R2 (5′-AGGACTTTGCAACTTCAACAAAACT-3′) for the TCF/CRE site. The primers used to amplify the negative control promoter of the cad (carbomoyltransferase-dihydroorotase) gene are 5′-GCCGTCGCAGTCGTGCT-3′ and ACCGACCCGTCCTCCAA-3′. Conventional PCR was carried out as described previously (25). Real time PCR was performed with F2 and R2 primers in a final volume of 10 μl in SYBR Green PCR Master Mix (Applied Biosystems). Accumulation of fluorescent products was monitored using the ABI PRISM 7900HT sequence detection system (Applied Biosystems). Each PCR generated only the expected specific amplicon, as shown by the melting temperature profiles of final products (dissociation curve automatically measured by the sequence detection system).
mRNA Analysis—Total RNA was extracted from WT and FHL2-/- MEFs with the RNeasy minikit (Qiagen). 1 μg of RNA was reverse-transcribed using random primers and the Superscript Reverse Transcriptase (Invitrogen). Real time PCR was performed using the following primers: for FHL2, 5′-GCGTGCCCTGCTATGAGAAG-3′ (sense) and 5′-AAGCGTTGCCCAGATAGCTG-3′ (antisense); for cyclin D1, 5′-CATCAAGTGTGACCCGGACTG-3′ (sense) and 5′-CCTCCTCCTCAGTGGCCTTG-3′ (antisense); for p15INK4b, 5′-ATTTGGGTGGGTGCAGTCA-3′ (sense) and 5′-GTTTCCCATTTAGCCTGTGGAT-3′ (antisense); for p18INK4c, 5′-ACCATCCCAGTCCTTCTGTCA-3′ (sense) and 5′-CCCCTTTCCTTTGCTCCTAATC-3′ (antisense); for p19INK4d, 5′-GGACGCTTCCGGTCTCACT-3′ (sense) and 5′-GCAGAATGTCCATGAGGTTCTG-3′ (antisense); for 18 S, 5′-GTAACCCGTTGAACCCCATT-3′ (sense) and 5′-CCATCCAATCGGTAGTAGCG-3′ (antisense). To quantitate expression of each gene in a given sample, the mean copy number of tested DNA from a triplicate determination was normalized to the mean copy number of 18 S RNA.
Microarray Analysis—Total RNA was extracted with the RNAeasy kit (Qiagen). The quality of RNA was monitored on Agilent RNA Nano LaChips (Agilent Technologies, Santa Clara, CA). Microarray hybridizations were performed on the Affymetrix mouse genome 430 2.0, containing probe sets for 39,000 transcripts. Three independent WT and FHL2-/- MEF clones were profiled via microarrays. For each array, the cell intensity files (*.CEL) were generated with GeneChipOperating software. Data analysis was performed using SPlus Array-Analyser software (Insightful). Data were processed using the RMA method. Statistical analysis to compare replicates arrays was done with the local pool error test. The p values (the probability that the variability in a gene behavior observed between classes could occur by chance) were adjusted using the Bonferroni algorithm.
Gene ontology and KEGG pathway annotation were performed by Onto-tools software (available on the World Wide Web). GSEA (26) was used to evaluate the correlation of a specific gene list with two different sample groups (genotypes). We used the signal/noise ratio as a statistic to compare specific and gene set permutations in order to evaluate statistical differences. Microarray data have been deposited in NCBI Gene Expression Omnibus (available on the World Wide Web). The GEO series accession number is GSE10902.
RESULTS
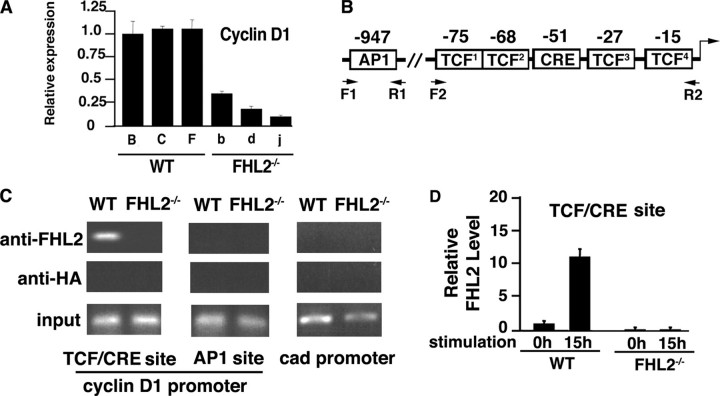
FHL2 Is Associated with the Cyclin D1 Promoter at the TCF/CRE Site—Our previous work has demonstrated that FHL2 cooperates with β-catenin in transcriptional activation of the cyclin D1 promoter in a luciferase reporter assay (10). To further study the role of FHL2 in the regulation of cyclin D1 and cell proliferation, we used MEFs derived from WT and FHL2-null embryos to generate WT and FHL2-/- MEF cell lines using the 3T3 protocol. Several independent immortalized clones were obtained in each genotype. Real time RT-PCR revealed strongly decreased cyclin D1 mRNA levels in all three FHL2-/- lines tested compared with WT cells (Fig. 1A). To determine whether cyclin D1 is a direct transcriptional target of FHL2, we carried out ChIP assays to examine the potential association of FHL2 with the cyclin D1 promoter. Since there is so far no evidence of DNA binding activity of LIM-only proteins, FHL2 could be associated with the cyclin D1 promoter through its interaction with transcription factors. We analyzed the proximal TCF/CRE region and the distal AP1 sites (Fig. 1B), because FHL2 is known to interact with β-catenin, CREB, and AP1 (3, 4, 7, 8). The proximal TCF/CRE region contains four putative TCF binding sites and one CRE element (Fig. 1B). A previous report (27) showed that these TCF sites were necessary for the activation of the cyclin D1 promoter by β-catenin. WT and FHL2-/- MEFs were first cross-linked with formaldehyde, and protein-DNA complexes were immunoprecipitated with either a FHL2 antibody or a HA antibody as control. Using chromatin precipitated with the FHL2 antibody as template, we were able to amplify the 79-bp region containing the TCF/CRE site in WT cells, whereas no amplification was obtained with chromatin precipitated from FHL2-/- cells (Fig. 1C). Amplification of chromatin immunoprecipitated with the control HA antibody did not give rise to any signal (Fig. 1C). Surprisingly, FHL2 was absent at the upstream region of the cyclin D1 promoter encompassing the AP1 site (Fig. 1C). No FHL2 binding signal was detected at the control promoter of the cad gene (Fig. 1C).
FIGURE 1.
FHL2 is associated with the cyclin D1 promoter. A, analysis of cyclin D1 transcripts in three independent WT and FHL2-/- immortalized clones by real time RT-PCR. Cyclin D1 expression was normalized to 18 S RNA. The ratio of the cyclin D1/18 S signal in WT clone B was arbitrarily set at 1. The average S.D. values and for three independent experiments are shown. Wild type clones are indicated by capital letters, and FHL2-/- clones are shown by lowercase letters. B, schematic representation of AP1 and TCF/CRE elements at the cyclin D1 promoter. C, association of FHL2 with the cyclin D1 promoter at the TCF/CRE site. WT and FHL2-/- MEFs were analyzed by ChIP. Antibodies against FHL2 and HA were used in parallel immunoprecipitations. Eluted DNA was amplified by PCR using cyclin D1 primers F1-R1 and F2-R2 indicated in B and control primers of the cad promoter (see “Experimental Procedures”). Data are representative of three independent experiments. D, graphic representation of the data of real time PCR for the TCF/CREB binding site in immortalized wild type and FHL2-/- MEFs. Calculation of the amount of immunoprecipitated DNA relative to that present in total input chromatin (percentage of total) is as follows: % total = 2ΔCT (where CT represents cycle threshold). ΔCT = CT (input) - CT (FHL2IP). The total percentage at the 0 h time point of serum stimulation in WT MEFs was arbitrarily set at 1. Two independent immortalized FHL2-/- clones were analyzed in two independent ChIP experiments.
Next, we assessed whether FHL2 binding to the cyclin D1 promoter is cell cycle-dependent by quantitative ChIP assays (28). After starvation during 96 h followed by stimulation with serum for 15 h, synchronized WT and FHL2-/- MEFs were cross-linked with formaldehyde, and protein-DNA complexes were immunoprecipitated with anti-FHL2 antibody. Quantitative data were obtained by real time PCR using F2 and R2 primers encompassing the TCF and CREB sites at the cyclin D1 promoter (Fig. 1B). As shown in Fig. 1D, quantitative ChIP analysis revealed a progressive association of FHL2 with the TCF/CRE target site in WT cells following serum stimulation, which is in accord with the observations that FHL2 can be induced by serum (3) (see Fig. 2A). Thus, these data indicate that FHL2 is directly involved in the control of the cyclin D1 promoter in a cell cycle-dependent manner.
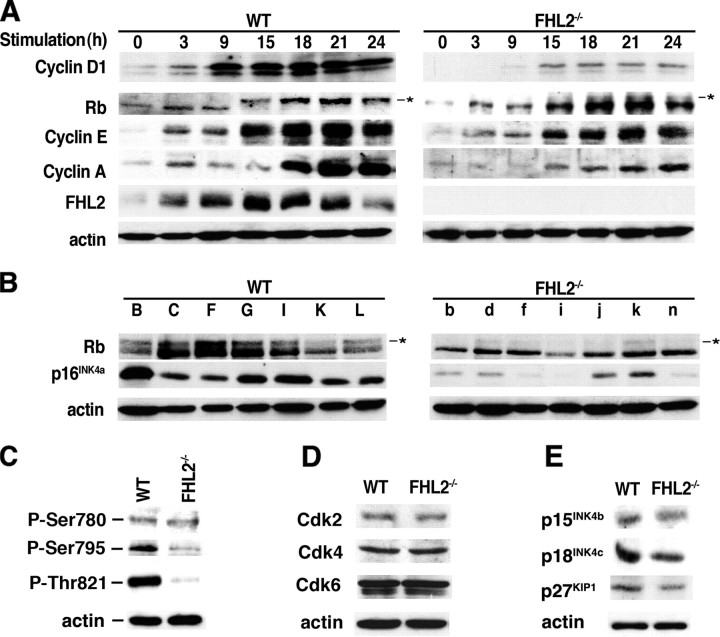
FIGURE 2.
Decreased expression of cell cycle regulators in FHL2-/- cells. A, immortalized WT clone B and FHL2-/- clone b were synchronized by serum starvation and stimulated by 10% serum. Cell lysates were prepared at the indicated time after stimulation and analyzed by immunoblotting. Similar results were obtained with the other two independent clones in each genotype. B, analysis of the expression of Rb and p16INK4a in seven immortalized clones for each genotype by immunoblotting. The asterisks in A and B show hyperphosphorylated Rb. C, lysates from WT and FHL2-/- cells were analyzed by immunoblotting with antiphosphospecific antibodies that recognize pRb phosphorylated at specific amino acid residues. D, expression of CDKs was not altered in FHL2-/- cells. Lysates from WT and FHL2-/- cells were analyzed by immunoblotting. E, reduced expression of Cdk inhibitors in FHL2-/- cells. Cells lysates were prepared from WT and FHL2-/- cells and analyzed by immunoblotting with the indicated antibodies.
Low Levels of Cyclin D1 Result in Hypophosphorylation of Rb and Reduced Proliferation of Immortalized FHL2-/- MEFs—Since cyclin D1 is considered as a critical cell cycle sensor of the extracellular mitogenic environment, we analyzed expression of cyclin D1 in FHL2-/- MEFs in response to serum stimulation after starving cells for 96 h followed by the addition of serum. Consistent with earlier observations (3), FHL2 was induced 3 h after the addition of serum, and its levels rapidly peaked at 15 h, thus confirming its behavior as an early response gene (Fig. 2A). Strikingly, the kinetics of induction of cyclin D1 in FHL2-/- MEFs was significantly delayed compared with WT cells, and the levels of the induction were greatly reduced (Fig. 2A). During the G1 phase of the cell division cycle, mitogenic stimulation triggers assembly of cyclin D-cyclin-dependent kinases 4 and 6 (Cdk) complexes, which collaborate with cyclin E-Cdk2 to phosphorylate Rb family members. Decreased cyclin D1 was associated with severely impaired phosphorylation of Rb in FHL2-/- MEFs (Fig. 2A, right). This post-translational modification is known to inactivate Rb transcriptional corepressor activity on E2F transcription factors and enables expression of E2F-dependent genes, including cyclins E and A. Since Rb was hypophosphorylated in FHL2-/- cells for up to 24 h after serum stimulation, the production of cyclins E and A was concomitantly decreased (Fig. 2A). Moreover, in cycling cells, the overall levels of Rb phosphorylation were very low in all seven FHL2-/- cell lines analyzed compared with WT clones (Fig. 2B), showing that immortalized FHL2-/- MEFs almost exclusively express an active hypophosphorylated form of Rb. Using antibodies against phosphorylated Rb at specific sites, we observed that the phosphorylation of Rb was affected at both cyclin D-Cdk and cyclin E-Cdk target sites (Fig. 2C). Analysis of Cdk expression showed similar levels of Cdk2, Cdk4, and Cdk6 between WT and FHL2-/- MEFs (Fig. 2D) but significant reduction of the Cdk inhibitor p16INK4a in FHL2-null cell lines (see Fig. 2B). However, p16INK4a gene deletion was excluded, because its expression was detected in all clones tested, including clone i after longer exposure (Fig. 2B). Decreased expression of p16INK4a in FHL2-/- cell lines was in sharp contrast to a previous report (29) that immortalized cells usually express high levels of functional p16INK4a, which was also observed in WT cells in this study (Fig. 2B). The levels of other Cdk inhibitors, including p15INK4b, p18INK4c, and p27Kip1, were also decreased, albeit in a lesser extent than p16INK4a (Fig. 2E).
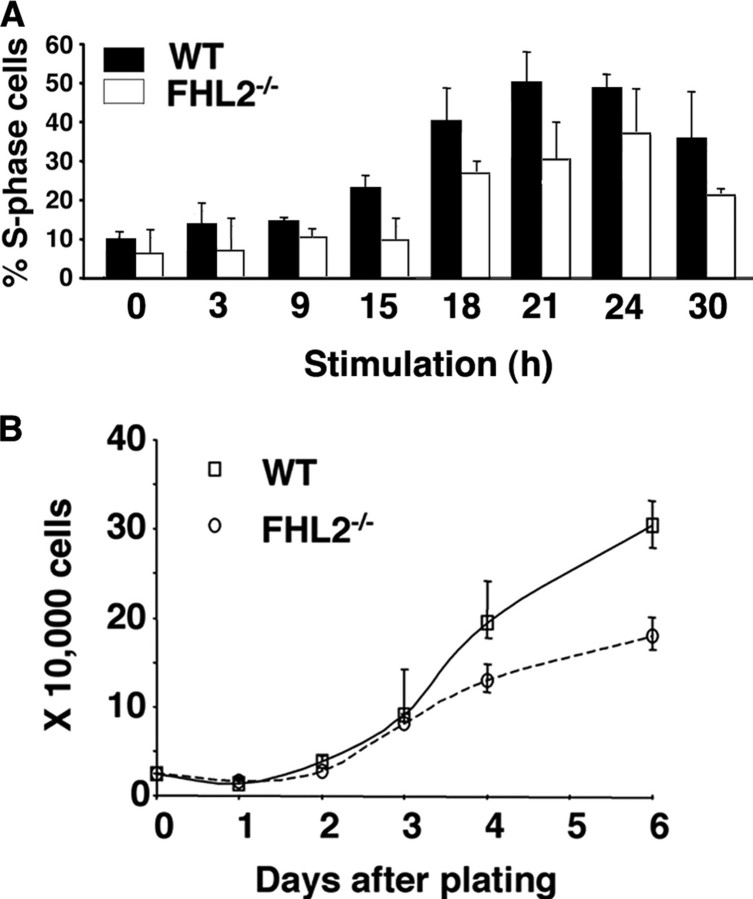
The critical role of cyclin D1 in the G1/S transition prompted us to investigate the effects of FHL2 deficiency on S phase entry and cell proliferation. We starved cells by serum deprivation for 96 h, followed by serum stimulation to assess the efficiency of S phase entry. Using flow cytometry analysis of BrdUrd-labeled cells, we found that the percentage of cells in S phase was consistently lower for FHL2-/- MEFs than for WT MEFs throughout the observation period (Fig. 3A). Additionally, FHL2-/- cells showed significantly lower rates of proliferation than WT MEFs (Fig. 3B). These data were confirmed in three independent cell lines.
FIGURE 3.
Cell proliferation in immortalized WT and FHL2-/- MEFs. A, kinetics of S phase entry in immortalized MEFs. After serum stimulation, cells were pulse-labeled with BrdUrd and harvested at the indicated time of stimulation. The percentage of cells in S phase at each time point was calculated and plotted based on flow cytometry results of three independent experiments. B, decreased cell proliferation in immortalized FHL2-/- cells. Equal numbers of spontaneously immortalized WT and FHL2-/- MEFs were plated in triplicate. Cell numbers were determined during 6 days. The data presented are the mean value ± S.D. obtained from three independent experiments. Data shown are representative of growth curves of three independent clones for each genotype.
These findings revealed that deficiency of FHL2 results in reduced proliferation potential associated with low levels of cyclin D1 and p16INK4a and hypophosphorylation of Rb.
Restoration of FHL2 Expression in FHL2-null Cells Is Sufficient to Increase Cell Proliferation—To test the specificity of FHL2 in the regulation of cyclin D1 expression and cell proliferation, we reintroduced FHL2 in primary FHL2-/- MEFs at early passage with a retroviral vector and assessed the effect of exogenous FHL2 expression on cyclin D1 and cell growth. Continuous passage of FHL2-restored MEFs readily gave rise to established 3T3 cell lines. By real time RT-PCR, the mRNA levels of cyclin D1 in FHL2-restored cells were 11-fold higher than those of FHL2-/- cells and comparable with those of WT cells, which was in sharp contrast to the control vector pBabe-infected cells (Fig. 4A, top). Accordingly, immunoblotting analysis showed abundant cyclin D1 protein in immortalized FHL2-restored cells (Fig. 4A, bottom). Of note, the mRNA levels of FHL2 in FHL2-restored cells were only half those in WT cells, but the cyclin D1 levels of both mRNA and protein were similar or even higher in FHL2-restored cells than WT cells, suggesting that activation of cyclin D1 is not stoichiometrically associated with FHL2 expression. Besides, we found that cyclins D2 and D3 were down-regulated in FHL2-/- cells (Fig. 4A, bottom). The levels of cyclins D2, D3, and E were efficiently restored after ectopic expression of FHL2 (Fig. 4A, bottom).
FIGURE 4.
Restoration of cyclin D1 and cell proliferation in FHL2-/- MEFs by re-expression of FHL2. A, up-regulation of cyclins D1, D2, D3, and E and Rb phosphorylation in FHL2-restored cells. Upper panel, mRNA levels of cyclin D1 and FHL2 in immortalized MEFs of different genotypes. Real time RT-PCR was carried out using either cyclin D1- or FHL2-specific primers on total RNA extracted from indicated cell lines. Their expression was normalized to 18 S RNA. The average and S.D. values for three independent experiments are shown. Data are representative of those from three independent clones for WT and FHL2-/- genotypes and three independent pools of FHL2-restored and pBabe-infected cells. The ratio of cyclin D1/18 S or FHL2/18 S in WT MEFs was arbitrarily set at 1. In the lower panel, cell lysates were analyzed by immunoblotting. The asterisk shows hyperphosphorylated Rb. B, growth curves of immortalized WT, FHL2-/-, pBabe-infected and pBabe-FHL2-infected FHL2-/- MEFs. The data presented are the mean ± S.D. obtained from three independent experiments. Data are representative of growth curves from two independent FHL2-infected MEF pools.
In FHL2-restored cells, Rb phosphorylation was efficiently reestablished (Fig. 4A, bottom). The proliferation rate of FHL2-restored cells was markedly increased, reaching similar levels as WT cells (Fig. 4B). Taken together, these results firm up the role of FHL2 in the regulation of cyclin D1 and cell proliferation.
Ectopic Expression of Cyclin D1 Is Sufficient to Confer Normal Cell Proliferation to FHL2-/- Cells—Next, we asked whether FHL2 is implicated in cell cycle control via its effect on cyclin D1. We ectopically expressed HA-tagged cyclin D1 in primary FHL2-/- MEFs with a retroviral vector and assessed the effect of cyclin D1 on cell proliferation. After continuous passages of cyclin D1-HA/FHL2-/- MEFs, immortalized clones emerged spontaneously. As shown in Fig. 5A, the cyclin D1 transgene in FHL2-/- MEFs was expressed at a level similar to that of endogenous cyclin D1 in WT cells, whereas in FHL2-/- MEFs infected with pBabe vector, cyclin D1 was maintained at low levels. Strikingly, expression of cyclins D2, D3, E, and A was efficiently restored, and the ratio of hyperphosphorylated over hypophosphorylated Rb was significantly augmented in cyclin D1-HA/FHL2-/- MEFs compared with FHL2-/- cells (Fig. 5A). Moreover, cell growth in cyclin D1-HA/FHL2-/- MEFs was restored, showing an even higher level than that of WT cells (Fig. 5B). Collectively, these data provide evidence that the effects of FHL2 on cell proliferation rely mainly upon cyclin D1.
FIGURE 5.
Ectopic expression of cyclin D1 in FHL2-/- MEFs. A, expression of cell cycle regulators in FHL2-/- MEFs infected with pBabecyclin D1-HA. Increased cyclin D1 expression in FHL2-/- MEFs after introduction of the cyclin D1-HA transgene was confirmed by immunoblotting with both anti-cyclin D1 and anti-HA antibodies. B, growth curves of immortalized wild type, FHL2-/-, pBabe-infected and pBabecyclin D1-HA-infected FHL2-/- MEFs. Equal numbers of cells were plated for triplicate at the beginning of the experiment. Cell numbers were determined every day for a total of 6 days. The data presented are the mean ± S.D. of values obtained from three independent experiments. The results shown are representative of those from two independent cyclin D1-HA-expressing MEF pools.
Microarray Analysis Shows Deregulation of Genes Associated with Cell Proliferation in FHL2-deficient Cells—To gain insight into the impact of FHL2 deficiency on gene expression at the whole genome level, we analyzed the gene expression profile of three independent FHL2-/- and three WT MEF clones using the Affymetrix mouse genome 430 2.0 microarray. Supervised class comparison using the local pool error test identified 998 genes differentially expressed at p < 0.05. The results of 97 genes that had a -fold change of 15 or more between the two genotypes were displayed based on the normalized intensity, with blue representing lower relative gene expression and red indicating higher expression (Fig. 6A). We verified the expression pattern of five top up-regulated and five top down-regulated genes using quantitative real time PCR normalizing to internal 18 S RNA in MEF cell lines. The PCR results confirmed the up- or down-regulation of these genes in FHL2-/- cells with excellent correlation between microarray and quantitative PCR (data not shown).
FIGURE 6.

A, expression profile of the top up-regulated and down-regulated genes at -fold change ≥15 that are differentially expressed in the FHL2-/- compared with WT clones. Wild type clones are indicated by capital letters, and FHL2-/- clones are shown by lowercase letters. Data are plotted as a heat map where red and blue correspond to high and low expression in log2-transformed scale. B, gene set enrichment analysis plot using a GenMAPP gene set list corresponding to the G1 to S cell cycle reactome (normalized enrichment score =-1.7, false discovery rate = 0.008, p < 0.0001). The enrichment score reflects the correlation of the gene set with FHL2-/- (KO) and WT genotypes. Significant results were also found in gene set enrichment analysis by using nine different cell cycle-related gene sets from the MIT data base (MSigDB). C, core group of genes of the G1 to S cell cycle reactome gene set that contributed to the enrichment score. The -fold change (KO/WT) corresponds to the mean value for those genes that have more than one probe set in the array.
In agreement with the data described above (Figs. 1A and 2), cyclins D1, D2, and E and Cdk inhibitors p16INK4a, p15INK4b, and p18INK4c were found significantly down-regulated in FHL2-/- cells. Pathway analysis using the Gene Ontology and KEGG data bases evidenced deregulation of genes involved in focal adhesion, extracellular matrix-receptor interaction, and response to external stimuli in cells deficient in FHL2, such as filamin, parvin, tenascin C, laminins (lama2, lama4, and lama5), thrombospondin 2, and procollagens (Col6a2, Col5a2, Col3a1, Col5a3, and Col2a1). This finding is in accord with previous reports that FHL2 interacts with integrins and focal adhesion kinase (13, 14), which are key components of focal adhesion (15). Overrepresentation of these functional modules is consistent with the view that FHL2 possesses sensor functions capable of detecting extracellular stimuli. Once receiving external signals, FHL2 can shuttle to the nucleus, where it regulates a large spectrum of genetic programs, including cell proliferation, in response to these signals.
Further evidence of alterations of cell cycle-associated genes in FHL2-/- cells was provided by gene set enrichment analysis, a computational method for assessing enrichment of a pre-defined gene list in one class compared with another (26). Using as a gene set the G1 to S cell cycle reactome (GenMAPP), we found that 50% of the 66 genes in this gene set were significantly down-regulated in FHL2-/- cells (Fig. 6, B and C). Specifically, besides cyclins D1, D2, and E, several critical G1/S regulators, including E2F transcription factors, were decreased in cells lacking FHL2. Moreover, cell cycle proteins involved in G2 to M transition appeared to be also affected, as demonstrated by the down-regulation of cyclin B1, Cdc25A, and Wee1 in FHL2-/- (Fig. 6C). Consistent with the lower rate of cell proliferation of FHL2-/- cells, several major components of the DNA replication machinery like Cdc45L, replication protein A subunits, Orc6L, and the minichromosome maintenance (Mcm) genes were down-regulated.
Remarkably, the most seemingly paradoxical effect of FHL2 deficiency was sustained repression of cell cycle negative regulators, including the four members of the INK4 family (p16INK4a, p15INK4b, p18INK4c, and p19INK4d), p57Kip2, and Wee1 (Fig. 6C). Real time RT-PCR analysis of p15INK4b, p18INK4c, and p19INK4d confirmed down-regulated expression of these genes in FHL2-/- cells (data not shown), in agreement with the immunoblotting analysis (Fig. 2E). The INK4 inhibitors and p57Kip2 specifically inhibit the activity of cyclin D-Cdk4/6 and cylin E-Cdk2 in G1/S, whereas Wee1 is an inhibitor of cyclin B-Cdk1 in G2/M. Reduced expression of Cdk inhibitors in FHL2-/- cells may rescue minimal activity of cyclin-Cdk complexes that is required for initiation of DNA synthesis and mitosis, thus maintaining the progression of the cell cycle at low levels. This FHL2-mediated regulation of cell proliferation seemingly involves not just cell cycle positive but also negative regulators controlling both G1/S and G2/M transitions. Taken together, the genome-wide analysis provides additional insight into FHL2-mediated regulation in cell proliferation and cell cycle proteins.
DISCUSSION
In this paper, biological analysis and genome-wide studies of gene expression show that deficiency of FHL2 in immortalized murine fibroblasts has a profound effect on expression of cell cycle proteins and cell proliferation. Spontaneously immortalized FHL2-/- MEFs express low levels of several G1/S and G2/M cyclins and E2F transcription factors, which are accompanied by hypophosphorylation of Rb and down-regulation of Cdk inhibitors. This unusual expression pattern with decreased levels of both cell cycle positive and negative regulators is associated with a reduced but not null proliferative capacity of FHL2-/- cells. These findings underscore an important function of FHL2 in regulating the expression of a large spectrum of genes implicated in cell division.
We demonstrate that cyclin D1 is directly regulated by FHL2 and represents a key effector of FHL2 in cell cycle control. Mitogen-induced signal transduction pathways promote the activation of cyclin D1 at many levels, including transcription, translation, and protein stability. Different lines of evidence indicate that FHL2 acts mostly at the transcriptional level. We provide evidence that FHL2 physically occupies the cyclin D1 promoter at the TCF/CRE site, which is in line with previous reports that FHL2 interacts directly with β-catenin and CREB transcription factors (4, 7, 8). FHL2 can regulate cyclin D1 by acting as a protein scaffold for the recruitment of the adaptor complexes, which in turn facilitates binding of general transcription factors. It has been shown that FHL2 is capable of binding the FOXO factors and deacetylase SIRT1 and can counteract the repression of FOXO1 on the cyclin D1 promoter by enhancing the interaction of FOXO1 with SIRT1 and the deacetylation of the transcription repressor (11). Thus, FHL2 may inhibit the activity of transcription repressors at the cyclin D1 promoter through its ability to interact with FOXO1 and SIRT1.
Alternatively, FHL2 may regulate the activity of the cyclin D1 promoter through its implication in acetylation of transcription factors and histones. We have previously shown that FHL2 increases β-catenin acetylation by p300 and that acetylation of β-catenin at residue Lys345 enhances the affinity of β-catenin for TCF4 (10, 22). Increasing acetylation of β-catenin might represent another mechanism by which FHL2 contributes to the activation of the cyclin D1 transcription. Likewise, FHL2 may be involved in acetylation of histones through its interaction with CREB-binding protein/p300. It is generally considered that histone acetylation is implicated in activation of transcription. Our preliminary results suggest that FHL2 deficiency is associated with a significant reduction of the occupancy of acetylated histone H3 at certain promoters, including the cyclin D1 promoter.6 Further study is needed to determine if FHL2 could regulate gene expression by participating in chromatin remodeling.
Finally, FHL2 may control cyclin D1 via indirect mechanisms by interfering in signaling pathways that modulate cyclin D1 activity. It has been shown that FHL2 can retain extracellular signal-regulated kinase 2 (ERK2) of the mitogen-activated protein kinase signaling pathways in cardiomyocytes, thus negatively regulating mitogen-activated protein kinase signaling (9). However, our microarray analysis showed that FHL2 deficiency in fibroblasts is associated with down-regulation of mitogen-activated protein kinases, which was confirmed by our preliminary results of the PathwayProfiler study (Upstate and Chemicon) and Western blot analyses.6 FHL2 is an early response gene (3), which argues that FHL2 possesses sensor functions that can relay mitogenic signals to nuclear effectors. Disruption of FHL2 may therefore partially block transduction of the signals that normally induce cyclin D1.
In contrast to immortalized cyclin D1-/- MEFs, which display a similar grow rate as WT MEFs (30), inactivation of FHL2 severely compromises proliferation of immortalized fibroblasts. This observation suggests that cyclin D1 is not the only target of FHL2. Indeed, gene expression profiling of FHL2-/- cells revealed deregulation of genes involved in focal adhesion, extracellular matrix-receptor interaction, response to external stimuli, and insulin-like growth factor signaling, demonstrating that FHL2 is in the front line of upstream signaling pathways emanating from diverse stimuli. These findings are also in agreement with the role of FHL2 in wound healing (21). Conversely, FHL2 deficiency in whole animals produces only limited abnormalities under various stimuli (19–21). FHL2 is thus dispensable for the proliferation of the majority of tissues in which, we speculate, other members of FHL family may provide substitutes for its functions.
The mechanisms involved in the down-regulation of other cell cycle-related genes by FHL2 can be both direct and indirect. FHL2 interacts with multiple transcription factors, which can directly regulate a large spectrum of gene expression program. On the other hand, cyclin D1 is such a critical regulator in cell division that its down-regulation in the absence of FHL2 can have an impact on the other cell cycle genes. Cyclin D-Cdk complexes play a crucial role in G1 progression by phosphorylating Rb family members, thus canceling their inhibitory functions on E2F transcription factors. This process controls an E2F-dependent transcriptional program that activates a large range of genes involved in cell division and DNA-replication machinery. Accordingly, as E2F targets, cyclin E, p107, Cdc25A, and Mcm genes were down-regulated in FHL2-/- cells. However, E2F target genes seemed still to be induced, since those cells are able to grow, albeit less rapidly than WT MEFs. In fact, in the massive decrease of G1/S cyclins and Rb phosphorylation in immortalized FHL2-/- cells, expression of Cdk inhibitors, in particular the INK4 family, was significantly reduced. This reduction may aim at rescuing a part of cyclin-Cdk activity, allowing minimal phosphorylation of Rb and perpetual growth of these cells. These findings provide a clear example of how FHL2 exerts its transcription modulator functions by acting on different aspects of multiple signaling pathways.
Acknowledgments
We thank C. Demeret and G. Soubigou for technical advice and C. Neuveut and D. Cougot for insightful discussion. We are grateful to K. Kean for comments on the manuscript.
This work was supported in part by the Association pour la Recherche sur le Cancer and the Comité de Paris of La Ligue contre le Cancer. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: CREB, cAMP-response element-binding protein; CRE, cAMP-response element; MEF, mouse embryo fibroblast; ChIP, chromatin immunoprecipitation; BrdUrd, bromodeoxyuridine; HA, hemagglutinin; RT, reverse transcription; WT, wild type; TCF, T cell factor.
Y. Nouët, F. Levillayer, M.-A. Buendia, and Y. Wei, unpublished data.
References
- 1.Johannessen, M., Moller, S., Hansen, T., Moens, U., and Van Ghelue, M. (2006) Cell Mol. Life Sci. 63 268-284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Muller, J. M., Isele, U., Metzger, E., Rempel, A., Moser, M., Pscherer, A., Breyer, T., Holubarsch, C., Buettner, R., and Schule, R. (2000) EMBO J. 19 359-369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morlon, A., and Sassone-Corsi, P. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 3977-3982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fimia, G. M., De Cesare, D., and Sassone-Corsi, P. (2000) Mol. Cell. Biol. 20 8613-8622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McLoughlin, P., Ehler, E., Carlile, G., Licht, J. D., and Schafer, B. W. (2002) J. Biol. Chem. 277 37045-37053 [DOI] [PubMed] [Google Scholar]
- 6.Chen, D., Xu, W., Bales, E., Colmenares, C., Conacci-Sorrell, M., Ishii, S., Stavnezer, E., Campisi, J., Fisher, D. E., Ben-Ze'ev, A., and Medrano, E. E. (2003) Cancer Res. 63 6626-6634 [PubMed] [Google Scholar]
- 7.Martin, B., Schneider, R., Janetzky, S., Waibler, Z., Pandur, P., Kuhl, M., Behrens, J., von der Mark, K., Starzinski-Powitz, A., and Wixler, V. (2002) J. Cell Biol. 159 113-122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wei, Y., Renard, C. A., Labalette, C., Wu, Y., Levy, L., Neuveut, C., Prieur, X., Flajolet, M., Prigent, S., and Buendia, M. A. (2003) J. Biol. Chem. 278 5188-5194 [DOI] [PubMed] [Google Scholar]
- 9.Purcell, N. H., Darwis, D., Bueno, O. F., Muller, J. M., Schule, R., and Molkentin, J. D. (2004) Mol. Cell. Biol. 24 1081-1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Labalette, C., Renard, C. A., Neuveut, C., Buendia, M. A., and Wei, Y. (2004) Mol. Cell. Biol. 24 10689-10702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang, Y., Hou, H., Haller, E. M., Nicosia, S. V., and Bai, W. (2005) EMBO J. 24 1021-1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scholl, F. A., McLoughlin, P., Ehler, E., de Giovanni, C., and Schafer, B. W. (2000) J. Cell Biol. 151 495-506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wixler, V., Geerts, D., Laplantine, E., Westhoff, D., Smyth, N., Aumailley, M., Sonnenberg, A., and Paulsson, M. (2000) J. Biol. Chem. 275 33669-33678 [DOI] [PubMed] [Google Scholar]
- 14.Gabriel, B., Mildenberger, S., Weisser, C. W., Metzger, E., Gitsch, G., Schule, R., and Muller, J. M. (2004) Anticancer Res. 24 921-927 [PubMed] [Google Scholar]
- 15.Hervy, M., Hoffman, L., and Beckerle, M. C. (2006) Curr. Opin. Cell Biol. 18 524-532 [DOI] [PubMed] [Google Scholar]
- 16.Philippar, U., Schratt, G., Dieterich, C., Muller, J. M., Galgoczy, P., Engel, F. B., Keating, M. T., Gertler, F., Schule, R., Vingron, M., and Nordheim, A. (2004) Mol. Cell 16 867-880 [DOI] [PubMed] [Google Scholar]
- 17.Muller, J. M., Metzger, E., Greschik, H., Bosserhoff, A. K., Mercep, L., Buettner, R., and Schule, R. (2002) EMBO J. 21 736-748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chu, P. H., Bardwell, W. M., Gu, Y., Ross, J., Jr., and Chen, J. (2000) Mol. Cell. Biol. 20 7460-7462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gunther, T., Poli, C., Muller, J. M., Catala-Lehnen, P., Schinke, T., Yin, N., Vomstein, S., Amling, M., and Schule, R. (2005) EMBO J. 24 3049-3056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kong, Y., Shelton, J. M., Rothermel, B., Li, X., Richardson, J. A., Bassel-Duby, R., and Williams, R. S. (2001) Circulation 103 2731-2738 [DOI] [PubMed] [Google Scholar]
- 21.Wixler, V., Hirner, S., Muller, J. M., Gullotti, L., Will, C., Kirfel, J., Gunther, T., Schneider, H., Bosserhoff, A., Schorle, H., Park, J., Schule, R., and Buettner, R. (2007) J. Cell Biol. 177 163-172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lévy, L., Wei, Y., Labalette, C., Wu, Y., Renard, C. A., Buendia, M. A., and Neuveut, C. (2004) Mol. Cell. Biol. 24 3404-3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paul, C., Lacroix, M., Iankova, I., Julien, E., Schafer, B. W., Labalette, C., Wei, Y., Cam, A. L., Cam, L. L., and Sardet, C. (2006) Oncogene 25 5475-5484 [DOI] [PubMed] [Google Scholar]
- 24.Martin, B. T., Kleiber, K., Wixler, V., Raab, M., Zimmer, B., Kaufmann, M., and Strebhardt, K. (2007) Cell Cycle 6 1779-1788 [DOI] [PubMed] [Google Scholar]
- 25.Cougot, D., Wu, Y., Cairo, S., Caramel, J., Renard, C. A., Levy, L., Buendia, M. A., and Neuveut, C. (2007) J. Biol. Chem. 282 4277-4287 [DOI] [PubMed] [Google Scholar]
- 26.Subramanian, A., Tamayo, P., Mootha, V. K., Mukherjee, S., Ebert, B. L., Gillette, M. A., Paulovich, A., Pomeroy, S. L., Golub, T. R., Lander, E. S., and Mesirov, J. P. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 15545-15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tetsu, O., and McCormick, F. (1999) Nature 398 422-426 [DOI] [PubMed] [Google Scholar]
- 28.Frank, S. R., Schroeder, M., Fernandez, P., Taubert, S., and Amati, B. (2001) Genes Dev. 15 2069-2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zindy, F., Quelle, D. E., Roussel, M. F., and Sherr, C. J. (1997) Oncogene 15 203-211 [DOI] [PubMed] [Google Scholar]
- 30.Yu, Q., Geng, Y., and Sicinski, P. (2001) Nature 411 1017-1021 [DOI] [PubMed] [Google Scholar]