Abstract
Yeast RNA polymerase (Pol) II consists of a 10-subunit core enzyme and the Rpb4/7 subcomplex, which is dispensable for catalytic activity and dissociates in vitro. To investigate whether Rpb4/7 is an integral part of DNA-associated Pol II in vivo, we used chromatin immunoprecipitation coupled to high resolution tiling microarray analysis. We show that the genome-wide occupancy profiles for Rpb7 and the core subunit Rpb3 are essentially identical. Thus, the complete Pol II associates with DNA in vivo, consistent with functional roles of Rpb4/7 throughout the transcription cycle.
Pol2 II is the enzyme responsible for mRNA synthesis during transcription of protein-coding genes in eukaryotic cells. The structure of the complete 12-subunit Pol II is known and consists of a 10-subunit core enzyme, which includes the active center, and the peripheral heterodimer of subunits Rpb4 and Rpb7 (Rpb4/7 subcomplex) (1–4). Whereas Rpb7 is essential for viability of the yeast Saccharomyces cerevisiae, Rpb4 is not, but it becomes essential at temperature extremes (5). In vitro, yeast Rpb4/7 is required for transcription initiation, can dissociate from Pol II, and is dispensable for catalytic RNA elongation (6). Rpb4/7 binds single-stranded nucleic acids and mediates a step during initiation subsequent to promoter DNA binding (7). These observations are consistent with the idea that Rpb4/7 is present during initiation at promoters but then dissociates from Pol II.
However, evidence has accumulated for additional functional roles of Rpb4/7 during transcription. Rpb7 remains associated with early elongation complexes (8) and binds Nrd1, a protein involved in RNA 3′-end processing (9). Rpb4/7 can be cross-linked to the transcribed region, and loss of Rpb4 decreases the association with 3′-processing factors and alters usage of the polyadenylation site at a tested gene (10). Thus, recent data suggest that Rpb4/7 is an integral part of the Pol II enzyme and is required not only for initiation but also for 3′-RNA processing at the end of transcription.
To investigate whether Rpb4/7 generally associates with Pol II in vivo, we carried out chromatin immunoprecipitation in yeast coupled to tiling microarray analysis at a technical resolution of 32 base pairs. We demonstrate that the occupancy profiles of Rpb7 and the Pol II core subunit Rpb3 are virtually identical, showing that the complete Pol II, including Rpb4/7, associates with DNA genome-wide. These data are consistent with functional roles of Rpb4/7 throughout the transcription cycle.
EXPERIMENTAL PROCEDURES
Yeast Strains—Experiments were performed with S. cerevisiae strains W303 wild type (MATa, ura3-1, trp1-1, his3-11,15, leu2-3,112, ade2-1, can1-100, GAL+) and W303 carrying a tandem affinity purification (TAP) tag sequence fused to the C terminus of the gene encoding Rpb3 (MATa, ura3-1, his3-11,15, leu2-3,112, ade2-1, can1-100, GAL+, RPB3-TAP::TRP1). In addition, S. cerevisiae S288C strains RPB3-TAP and RPB7-TAP (ATCC 201388:MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0; Open Biosystems) were used.
Production of Monoclonal Antibodies against Rpb4/7—Lou/C rats were immunized subcutaneously and intraperitoneally with a mixture of 50 μg of purified recombinant Rpb4/7-His fusion protein (2), 5 nmol of CpG oligonucleotide (ODN 2006; TIB Molbiol, Berlin, Germany), 500 μl of phosphate-buffered saline, and 500 μl of Incomplete Freund's Adjuvant (IFA). After a six-week interval a final boost without adjuvant was given 3 days before fusion of the rat spleen cells with the murine myeloma cell line P3X63-Ag8-653. Hybridoma supernatants were tested in an enzyme-linked immunosorbent assay using bacterial extracts from Escherichia coli expressing either the Rpb4/7 fusion protein or a His-tagged fusion protein. Antibodies B4 15H8 (rat IgG1) and B4 5F5 (rat IgG) recognized the protein specifically in Western blotting and were used for this study.
Chromatin Immunoprecipitation (ChIP)—ChIP was performed essentially as described (11). Briefly, chromatin was sheared with a Bioruptor™ UCD-200 (Diagenode) using 25 × 30 s cycles with 30 s breaks at an output of 200 W. Before proceeding with immunoprecipitation experiments 20 μl of chromatin solution was put aside and marked an Input sample. For immunoprecipitation of TAP-tagged Rpb3 and Rpb7, IgG-Sepharose™ 6 Fast Flow (GE Healthcare) was used. Precipitation was performed for 3 h at room temperature. For precipitation of Rpb4 and Rpb7 from W303 wild type cells, a mixture of protein A and protein G-Sepharose was incubated with rabbit anti-rat IgG antibodies for 1 h at room temperature and then incubated with rat monoclonal antibodies against Rbp4/7 for an additional 4–5 h at room temperature. These coupled beads were incubated overnight at 4 °C with chromatin solution.
DNA Amplification and Microarray Handling—DNA samples were amplified and reamplified with GenomePlex® Complete Whole Genome Amplification (WGA) kit and GenomePlex® WGA Reamplification kit (Sigma) using the Farnham Lab whole genome amplification protocol for ChIP chip. DNA quantity and quality control was performed with a ND-1000 Spectrophotometer (NanoDrop Technologies). Labeling, hybridization, array scanning, data extraction, and a preliminary data analysis were performed by imaGenes GmbH, as part of the NimbleGen ChIP chip service. For all of the experiments the NimbleGen S. cerevisiae ChIP Whole Genome Tiling Array (catalog number C4214-00-01) was used. ChIP chip datasets were derived from three or two biological replicates of S288C Rpb7-TAP or Rpb3-TAP strains, respectively, and from single experiments on W303 wild type and W303 Rpb3-TAP strains. All of the microarray measurements were performed with two-color technology using Cy3 and Cy5, i.e. the DNA bound to immunoprecipitated protein was labeled with one dye and genomic background DNA with another dye (MIAME-compliant microarray data submitted to the Gene Expression Omnibus data base under accession number GSE12060).
Bioinformatic Analysis—In all analyses, we worked with the logarithm of the fluorescent signal from the ChIP DNA divided by the signal from the genomic background. A standard background correction was performed on all such signals by subtracting their genome-wide average. We repeated the ChIP chip measurements for TAP-tagged Rpb3 and Rpb7 with exchanged dyes Cy5 and Cy3 and averaged over measurements to subtract out the strong, systematic, dye-related technical noise. This effectively gets rid of intensity-dependent saturation effects, as shown in the MA plots in supplemental Fig. S1. For Rpb3 we had two replica measurements with ChIP DNA labeled by Cy5 but only one with exchanged dyes. When averaging over the replicas, we weighted the Cy3-labeled signal doubly to cancel out systematic noise. To directly compare the Rpb3 and Rpb7 signals, we adjusted their relative scales, which can differ slightly from 1 because of technical effects (12). For this purpose, we performed a linear regression on the Rpb7 versus Rpb3 scatter plot (supplemental Fig. S2). This yielded a factor 1.06, by which we scaled up the Rpb7 signal. Linear regression is superior to the ratio of median absolute deviations or standard deviations as scale factor in the case of correlated signals, because it can be shown to minimize the standard deviation of the difference signal. This scale adjustment does not influence the estimated correlation coefficients (see Table 1).
TABLE 1.
Pearson correlation coefficients between occupancy profiles
|
Comparison |
Profile 1 |
Profile 2 |
Pearson correlation |
|||
|---|---|---|---|---|---|---|
| Subunit tag | Strain | Subunit tag | Strain | Unsmoothed | Smoothed | |
| Biological replicatesa | Rpb3-TAP | S288C | Rpb3-TAP | S288C | 0.88 | 0.92 |
| Rpb3 vs. Rpb7b | Rpb7-TAP | S288C | Rpb3-TAP | S288C | 0.91 | 0.93 |
| Different strainsa | Rpb3-TAP | S288C | Rpb3-TAP | W303 | 0.90 | 0.92 |
| TAP tag vs. antibodya | Rpb7-TAP | S288C | Rpb4/7 AB | W303 | 0.72 | 0.79 |
Profiles are single measurements with the ChIP DNA tagged with the Cy5 dye.
Profiles for Rpb3 and Rpb7 are averaged over three and two measurements, respectively, with one measurement in each case taken with interchanged fluorescent dyes. The averaging over replicas as well as the smoothing reduces noise and leads to higher correlation coefficients.
To correct for unspecific binding of proteins to the IgG beads during immunoprecipitation with the monoclonal antibody for Rpb4/7 (AB), we performed a mock experiment with empty IgG beads. However, random noise in the mock experiment was very strong, presumably because of the very low amount of nonspecifically bound DNA in the mock probe. Indeed, normalization of the AB-precipitated signal by the mock signal resulted in drastically increased noise, whereas positive effects of suppressing systematic errors were not apparent. In Fig. 1D we therefore show the AB-precipitated Rpb7 signal normalized by the genomic background instead of by the mock signal.
FIGURE 1.
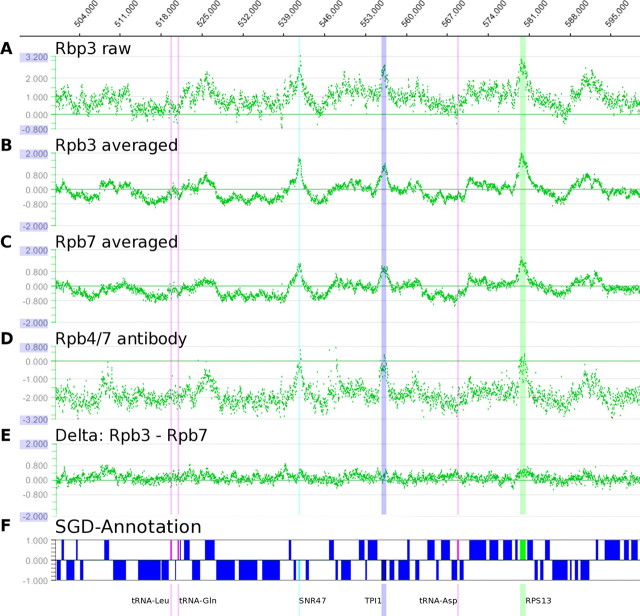
Genome-wide occupancy profiling of Pol II subunits. Profiles were obtained by chromatin immunoprecipitation coupled to high resolution tiling microarray analysis. A representative 100-kilobase pair sample on chromosome 4 (genomic positions 500,000–600,000) of the profile for the Rpb3 and Rpb7 subunits is depicted. Each green dot represents the signal for a single oligonucleotide probe on the tiling array, which has one probe every 32 bp. A, raw data trace for the TAP-tagged Rpb3 core subunit of Pol II. The signal intensity is the logarithm (base 2) of the fluorescence signal of Cy5-labeled Rpb3-bound DNA divided by the signal for Cy3-labeled genomic background DNA. B, average over three biological replicate traces for Rpb3, one of which with interchanged fluorescent dyes. C, average over two biological replicate traces for Rpb7, one with interchanged dyes. D, Pol II occupancy profile generated with a monoclonal antibody against Rpb4/7. E, the difference signal between averaged Rpb3 and Rpb7 occupancy profiles (B and C) fluctuates around a near-zero base line. F, annotation of genomic features according to the Saccharomyces Genome Data base. Genes encoding the intron-containing ribosomal protein RPS13 (green), the snoRNA SNR47 (cyan), and the housekeeping gene triose phosphate isomerase TPI1 show high Rpb3 occupancy at our growth conditions. Other protein-coding genes (blue) show lower Pol II occupancy. Three different tRNA genes (magenta) are transcribed by Pol III and show no increased Pol II occupancy.
The agreement between various pairs of genome-wide measurements of Rpb3 and Rpb7 occupancy was quantified by the Pearson correlation coefficient. The mitochondrial genome was omitted in this calculation, because, as expected, neither Rpb3 nor Rpb7 showed signs of binding to it. Because the Pearson correlation coefficient is sensitive to the presence of noise, we calculated the coefficients for the raw data and for smoothed traces. We used a local quadratic regression smoother with a Gaussian kernel (σ = 2; see Table 1), corresponding to averaging over approximately nine data points, respectively. We used smoothed curves for analyzing correlations between occupancy signals and genomic features (Fig. 2). For bioinformatics analysis of the ChIP chip data with respect to genome annotation, compare the legends to Figs. 1 and 2.
FIGURE 2.
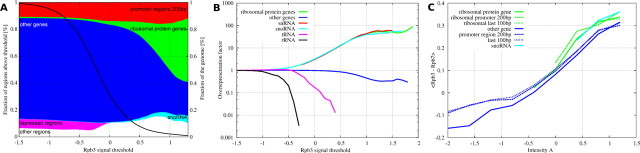
Distribution of genomic regions above a Pol II (Rpb3-TAP) occupancy threshold. A, the thick black trace shows the fraction of the genome within regions above the Rpb3 occupancy threshold. For example, ∼99% of the whole genome have occupancy signals above a threshold of –1.0, ∼30% have signals above 0 (this threshold corresponds to equal fluorescent signals from Rpb3 and the genomic background). At each threshold, the height of the colored areas indicates the fraction of above-threshold regions annotated with the given genomic feature. Nonribosomal protein genes (blue) make up almost 80% of the genome, but they constitute only 25% in regions with a Rpb3 occupancy above 1.0. Ribosomal protein genes (green) occupy only a small fraction of the genome, yet make up 35% of regions with an Rpb3 occupancy above 1.0. The fraction of all genes (ribosomal and nonribosomal) is approximately constant over a large range of thresholds, as is the fraction in promoter regions, defined as the 200 bp upstream of genes (red). The most overrepresented features in highly occupied regions are snoRNAs (cyan) and ribosomal genes (green). Repressed regions (magenta) summarize LTR (long terminal repeats) retrotransposons, transposable elements, and telomeres and are absent in regions above an occupancy threshold of 0. B, overrepresentation (“enhancement factor”) of selected genomic features in regions above the occupancy threshold. The colors are as in A except for the snRNAs (red). Other genes encompass intergenic regions of unknown function and more rarely occurring genomic features. C, dependence of the difference signal (Rpb3–Rpb7) on the average signal intensity A = (Rpb3 + Rpb7)/2. The colors are as in A and B. The solid traces refer to the coding region, dashed traces refer to promoter regions (i.e. 200 bp upstream of coding regions), and dotted traces refer to the last 100 bp of coding regions. Because the signals are calculated as logarithms (base 2) of the ChIP fluorescence divided by the genomic background fluorescence, a difference signal of 0.3 would correspond to a ratio of Rpb7 to Rpb3 occupancy of 2–0.30 = 0.81.
RESULTS AND DISCUSSION
Genome-wide ChIP of Pol II Subunits—To investigate whether the complete Pol II is the form of the enzyme that associates with the yeast genome or whether the complete Pol II and the core Pol II associate with different regions of the genome, we used ChIP coupled to occupancy profiling with high resolution tiling microarrays (ChIP chip). We used yeast strains that contained TAP tags at the C termini of the integral core subunit Rpb3 and the essential Rpb4/7 subunit Rpb7. Both subunit C termini are exposed in the Pol II structure (4), suggesting that the tag would not interfere with Pol II function in vivo.
Analysis and Quality of ChIP Chip Data—To investigate the reproducibility of the ChIP chip data, we carried out three biological replicate experiments with the S288C Rpb3-TAP strain. The reproducibility of the obtained data were very high, resulting in Pearson correlations between individual profiles of around 0.9 (Table 1). The Pearson coefficient is 1 for a perfect correlation and 0 for uncorrelated signals. Similarly, two biological replicates of S288C Rpb7-TAP profiles were obtained and showed very good agreement (Pearson correlation, 0.90; Table 1). Noise could be dramatically reduced using dual color microarray technology and replica measurements with interchanged dyes (13) (Fig. 1, A–C, and Table 1).
Occupancy Profiles for Rpb3 and Rpb7 Are Essentially Identical—We next compared the replica-averaged, noise-reduced profiles for Rpb3 and Rpb7. Differences in the two profiles would indicate that part of the genome associates with the complete Pol II and part of the genome associates with the core enzyme, and thus Rpb4/7 is dissociable in vivo. In contrast, a high similarity of the two profiles would indicate that Rpb4/7 is always part of the DNA-associated Pol II in vivo.
Comparison of the average, noise-reduced occupancy profiles for Rpb3-TAP and Rpb7-TAP showed that they are highly correlated throughout the genome (Fig. 1, B and C). There are only small differences between the profiles (Fig. 1E and supplemental Fig. S2). Weak systematic deviations from zero occur only for a small fraction of genomic locations with the highest signal intensity (supplemental Fig. S2; see discussion below). A Pearson correlation coefficient of 0.91 between the averaged Rpb3 and Rpb7 profiles compares favorably with the correlation obtained for biological replicates of the same profile (0.88; Table 1). Thus, statistically the agreement between the two average profiles of Rpb3 and Rpb7 was as good as the agreement between individual replicate profiles of the same strain. The near identity of the two profiles, within experimental errors, indicates that Rpb4/7 is present in DNA-associated Pol II genome-wide.
The Profiles Are Not Systematically Influenced by the Type of Yeast Strain—We next investigated whether the obtained occupancy profiles change when a different strain of yeast is used. We compared the Rpb3-TAP profile obtained with yeast strain S288C to a profile obtained under identical conditions with a W303 strain. The obtained profiles were highly similar, resulting in a Pearson correlation of 0.90 (Table 1), showing that the type of yeast strain did not influence the results.
The Profiles Are Not Influenced by the Affinity Tag—It is possible that the cellular function of Rpb7 is perturbed by its fusion to the TAP tag. To investigate this, we purified the recombinant stoichiometric Rpb4/7 subcomplex as described (2) and raised monoclonal antibodies against the pure subcomplex (“Experimental Procedures”). We then used this monoclonal antibody to measure an unbiased occupancy profile for Rpb4/7. The resulting occupancy profile contained more noise than the profiles obtained from TAP-tagged strains, but it did not show any systematic disagreement with the profile obtained from TAP-tagged strains (Fig. 1D), resulting in a Pearson correlation of 0.72 (0.79 for the smoothed traces; Table 1). Thus, the profiles obtained from TAP-tagged strains reflect those of unperturbed yeast cells and were not considerably influenced by the presence of the TAP tag.
Correlation of the Pol II Occupancy Profiles with Genome Features—The ChIP chip data contain a lot of information beyond the observation that Rpb4/7 is a component of DNA-associated Pol II in vivo. Analyses of Pol II occupancy profiles over a part of the yeast genome or at lower resolution were presented recently (14–16), but a comparison of these published profiles with our data is hampered because of the use of different experimental conditions and technological platforms.
To analyze which genomic features correlate with Rpb3 occupancy, we identified all genomic regions with a ChIP chip signal above an occupancy threshold that was varied continuously (Fig. 2A). We added up the base pairs within these regions belonging to the genomic features annotated in the Saccharomyces Genome Data base (version 11, March 2008). The colored areas in Fig. 2A indicate the fractions of the regions annotated by various genomic features at a chosen Pol II threshold. A threshold of zero corresponds to equal fluorescent signals from Rpb3 and the genomic background. The solid line shows the proportion of the genome above the threshold. Fig. 2B shows how much certain genomic features are overrepresented in regions with Pol II occupancy above a given threshold.
Pol II Distribution over the Genome—This analysis provides general insights into the global distribution of Pol II in growing yeast cells. First, most Pol II is bound to highly transcribed genes that encode small nuclear RNAs, small nucleolar RNAs, or ribosomal proteins (Fig. 2). Second, genomic repeat regions that are expected to be transcriptionally silenced show very low Pol II occupancy. Third, Pol III-transcribed genes that encode tRNAs do not show Pol II enrichment. Fourth, the fraction of all genes and promoter regions is approximately constant over a large range of Pol II occupancy thresholds (Fig. 2A). This supports previous reports that have found no evidence for wide-spread accumulation of Pol II by pausing at promoters in yeast, in contrast to higher eukaryotes (17). However, our findings are also consistent with other ChIP chip studies showing the preferential location of Pol II on promoter-bearing intragenic regions in yeast cells in stationary phase (15) because our data were obtained from yeast cells harvested in their logarithmic growth phase.
Persistent Presence of Rpb4/7—After this manuscript was submitted, a study comparing the genome-wide distribution of core Pol II and Rpb4/7 subcomplex was published (18). This study showed at lower resolution that Rpb3 and Rpb4 profiles are similar and generally agrees with our findings. The authors also note that Rpb4 can be underrepresented with respect to Rpb3, an effect that is more pronounced for shorter genes. However, we could not confirm that this is a significant or systematic effect, as demonstrated below.
The ratio of Rpb7 to Rpb3 does not fall below 0.78, and such deviations from 1 are limited to a very small fraction of the genome (Fig. 2C). We investigated whether these minor local deviations show systematic features and thus may be biologically significant. We analyzed the Rpb7-Rpb3 difference signal separately for several genomic categories, including ribosomal protein genes, nonribosomal protein genes, and snoRNA (Fig. 2C). This revealed that the difference signal increases with increasing occupancy on the x axis. The higher occupancy of Rpb3 versus Rpb7 for ribosomal proteins can be explained by the generally higher total Pol II occupancy on ribosomal protein genes. Rpb7 occupancy also did not depend on the position along the gene. The promoter regions and 5′ ends of genes show essentially the same difference signal as the coding regions. Thus, the ratio of Rpb3 to Rpb7 occupancy neither depends on the genomic feature type nor on the position along genes. Thus, there is no evidence for a functional significance of the minor local deviations between Rpb3 and Rpb7 levels.
Functional Roles of Rpb4/7—Our data suggest that Rpb4/7 is generally required throughout the transcription cycle in vivo and argue against models that propose the dissociation of Rpb4/7 during transcription. Our results are consistent with functional roles of Rpb4/7 during initiation (6, 7) as well as 3′-RNA processing (10). They are also consistent with microarray-based gene expression analysis of yeast strains lacking the gene for Rpb4, which suggested that Rpb4 is globally required for Pol II transcription (19, 20). They are further consistent with the observation that Rpb4 is essential in the fission yeast Schizosaccharomyces pombe and that Rpb4/7 is present in S. pombe Pol II in stoichiometric amounts during exponential cell growth (21).
Finally, our results are not inconsistent with the recent observations that Rpb4/7 shuttles between the nucleus and cytoplasm (22) and that Rpb4 and Rpb7 play roles in P-body function and mRNA decay (23), because it is possible that Rpb4/7 dissociates from core Pol II during transcription termination and that there is a cellular pool of free Rpb4/7. It also remains possible that Rpb4/7 dissociates under certain growth conditions.
Supplementary Material
Acknowledgments
We thank A. Thomma for assistance in the analysis and presentation of the microarray data and S. Jentsch, A. Mitterweger, T. Straub and P. Becker for advice. We thank J. Sydow, G. Damsma, E. Lehmann, A. Jawhari, and other members of the Cramer lab for help.
This work was supported by the Deutsche Forschungsgemeinschaft, the Sonderforschungsbereich SFB646, the Nanoscience Initiative Munich NIM, the Elitenetzwerk Bayern, the Transregio 5 Chromatin, and the Fonds der Chemischen Industrie. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
Footnotes
The abbreviations used are: Pol, polymerase; TAP, tandem affinity purification; ChIP, chromatin immunoprecipitation.
References
- 1.Cramer, P., Bushnell, D. A., and Kornberg, R. D. (2001) Science 292 1863–1876 [DOI] [PubMed] [Google Scholar]
- 2.Armache, K.-J., Kettenberger, H., and Cramer, P. (2003) Proc. Natl. Ac. Sc. U. S. A. 100 6964–6968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bushnell, D. A., and Kornberg, R. D. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 6969–6972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Armache, K.-J., Mitterweger, S., Meinhart, A., and Cramer, P. (2005) J. Biol. Chem. 280 7131–7134 [DOI] [PubMed] [Google Scholar]
- 5.Woychik, N. A., and Young, R. A. (1989) Mol. Cell Biol. 9 2854–2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edwards, A. M., Kane, C. M., Young, R. A., and Kornberg, R. D. (1991) J. Biol. Chem. 266 71–75 [PubMed] [Google Scholar]
- 7.Orlicky, S. M., Tran, P. T., Sayre, M. H., and Edwards, A. M. (2001) J. Biol. Chem. 276 10097–10102 [DOI] [PubMed] [Google Scholar]
- 8.Cojocaru, M., Jeronimo, C., Forget, D., Bouchard, A., Bergeron, D., Cote, P., Poirier, G. G., Greenblatt, J., and Coulombe, B. (2008) Biochem. J. 409 139–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mitsuzawa, H., Kanda, E., and Ishihama, A. (2003) Nucleic Acids Res. 31 4696–4701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Runner, V. M., Podolny, V., and Buratowski, S. (2008) Mol. Cell Biol. 28 1883–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aparicio, O., Geisberg, J. V., Sekinger, E., Yang, A., Moqtaderi, Z., and Struhl, K. (2005) Curr. Prot. Mol. Biol. 21 Units 21–23 [DOI] [PubMed] [Google Scholar]
- 12.Do, J. H., and Choi, D. K. (2006) Mol. Cell 22 254–261 [PubMed] [Google Scholar]
- 13.Yang, Y. H., and Speed, T. (2002) Nat. Rev. Genet. 3 579–588 [DOI] [PubMed] [Google Scholar]
- 14.Liu, C. L., Kaplan, T., Kim, M., Buratowski, S., Schreiber, S. L., Friedman, N., and Rando, O. J. (2005) PLoS Biol. 3 1753–1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Radonjic, M., Andrau, J. C., Lijnzaad, P., Kemmeren, P., Kockelkorn, T. T., van Leenen, D., van Berkum, N. L., and Holstege, F. C. (2005) Mol. Cell 18 171–183 [DOI] [PubMed] [Google Scholar]
- 16.Steinmetz, E. J., Warren, C. L., Kuehner, J. N., Panbehi, B., Ansari, A. Z., and Brow, D. A. (2006) Mol. Cell 24 735–746 [DOI] [PubMed] [Google Scholar]
- 17.Wade, J. T., and Struhl, K. (2008) Curr. Opin. Genet. Dev. 18 130–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verma-Gaur, J., Rao, S. N., Taya, T., and Sadhale, P. (2008) Eukaryot. Cell 7 1009–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miyao, T., Barnett, J. D., and Woychik, N. A. (2001) J. Biol. Chem. 276 46408–46413 [DOI] [PubMed] [Google Scholar]
- 20.Pillai, B., Verma, J., Abraham, A., Francis, P., Kumar, Y., Tatu, U., Brahmachari, S. K., and Sadhale, P. P. (2003) J. Biol. Chem. 278 3339–3346 [DOI] [PubMed] [Google Scholar]
- 21.Sakurai, H., Mitsuzawa, H., Kimura, M., and Ishihama, A. (1999) Mol. Cell Biol. 19 7511–7518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Selitrennik, M., Duek, L., Lotan, R., and Choder, M. (2006) Eukaryot. Cell 5 2092–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lotan, R., Bar-On, V. G., Harel-Sharvit, L., Duek, L., Melamed, D., and Choder, M. (2005) Genes Dev. 19 3004–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.