Abstract
The functional capacity of the transcriptional regulatory CCAAT/enhancer-binding protein-β (C/EBPβ) is governed by protein interactions and post-translational protein modifications. In a proteome-wide interaction screen, the histone-lysine N-methyltransferase, H3 lysine 9-specific 3 (G9a), was found to directly interact with the C/EBPβ transactivation domain (TAD). Binding between G9a and C/EBPβ was confirmed by glutathione S-transferase pulldown and co-immunoprecipitation. Metabolic labeling showed that C/EBPβ is post-translationally modified by methylation in vivo. A conserved lysine residue in the C/EBPβ TAD served as a substrate for G9a-mediated methylation. G9a, but not a methyltransferase-defective G9a mutant, abrogated the transactivation potential of wild type C/EBPβ. A C/EBPβ TAD mutant that contained a lysine-to-alanine exchange was resistant to G9a-mediated inhibition. Moreover, the same mutation conferred super-activation of a chromatin-embedded, endogenous C/EBPβ target gene. Our data identify C/EBPβ as a direct substrate of G9a-mediated post-translational modification that alters the functional properties of C/EBPβ during gene regulation.
CCAAT/enhancer-binding protein-β (C/EBPβ)2 is a basic leucine zipper transcription factor that regulates tissue-specific gene expression, proliferation, and differentiation in a variety of cell types. C/EBPβ is an important factor for female reproduction (1), liver function and regeneration (2), acute phase response (3), as well as innate and adopted immune functions (4).
Although the locus encoding C/EBPβ lacks any intron, three protein isoforms are generated from a single mRNA transcript by alternative translation initiation, resulting in full-length and N-terminally truncated proteins termed LAP*, LAP, and LIP proteins, respectively (5, 6). The long C/EBPβ isoforms LAP* and LAP are transactivator proteins, and the small LIP isoform is thought of as a transcriptional dominant-negative inhibitor, although LAP may also function as a latent transcriptional repressor dependent on post-translational modifications (7, 8).
A number of post-translational modifications have been detected on C/EBPβ, including phosphorylation, acetylation, and SUMOylation. Of particular interest is a phosphoacceptor site in the central, regulatory part of the protein that is conserved throughout evolution (Thr-235 in human (9), Thr-188 in rat (10), and Thr-220 in chicken (11)). Phosphorylation at this site creates a conformational change that abrogates the inhibitory function of the central regulatory domain. This leads to a release of the N-terminal transactivation domain (TAD) (11, 12) and exchange of interacting Mediator components and activation of the repressed protein (13). Other phosphorylation sites and their corresponding kinases were also identified, including glycogen synthase kinase 3 (GSK-3) that phosphorylates Ser-184 in murine LAP (Ser-33 in LIP) (14); calcium/calmodulin-dependent protein kinase that phosphorylates Ser-276 (15); and protein kinase C that phosphorylates Ser-105 and Ser-240 and protein kinase A (PKA) that phosphorylates Ser-105, Ser-299, and Ser-240 in LAP (16–18); p90 ribosomal S kinase (p90rsk) that phosphorylates Ser-105 in rat C/EBPβ and Thr-217 in murine LAP (19); and Cdk2 and Cdc2 that phosphorylate Ser-64 and Thr-189 in murine C/EBPβ (20). Several of these phosphorylation events may also depend on each other (21) to modulate the transcriptional activity of C/EBPβ in response to external stimuli.
C/EBPβ, as well as other C/EBP proteins, is also modified by SUMOylation. SUMOylation of C/EBPα (22), C/EBPβ (23, 24), and C/EBPε (25) occurs at a highly conserved lysine residue in the regulatory domain. Mutation of the lysine acceptor site increases the activity of the respective transcription factor, suggesting that SUMOylation is involved in repressive C/EBP functions.
In addition, C/EBPβ can be acetylated at multiple lysine residues (26–28). The acetyltransferases p300 and p300/CREB-binding protein-associated factor (PCAF) catalyze acetylation of lysine 39, and mutation of lysine 39 has been shown to affect C/EBPβ-dependent reporter activity (26).
Acetylation is predominantly observed on N-terminal histone tails (29) and accompanied by methylation of lysine side chains (30). Methylation is involved in gene activation or gene repression depending on the state of methylation (mono-, di-, or trimethylation) and on its position on the histone tail (31). Lysine methylation of non-histone transcription factors and the concomitant alteration of their biological functions remain poorly investigated, with some notable exceptions, such as p53 (32–34). In support of a role of G9a to methylate non-histone targets, it was shown that G9a is automethylated, which raises the possibility of the enzymatic activity of G9a being utilized by other nuclear proteins (35). Such action by G9a is thought to impose an epigenetic signature both on histones and non-histone proteins alike (36).
We observed that C/EBPβ is also post-translationally methylated and set out to identify interacting proteins that may alter the C/EBPβ structure and its activity in gene regulation. Here we show that G9a binds to the TAD of C/EBPβ, methylates C/EBPβ on lysine 39, and alters the transcriptional capacity of C/EBPβ. Mutational analysis of C/EBPβ and of G9a suggests that methylation at lysine 39 obstructs the ability of C/EBPβ to activate gene expression.
EXPERIMENTAL PROCEDURES
Plasmids—Numbering of amino acid residues in C/EBPβ is based on the chicken sequence (EMBL accession number Z21646). The conserved regions and corresponding amino acids in C/EBPβ were previously published (11). Briefly, conserved region 1 (CR1) corresponds to AA 1–13, CR2 to AA 18–38, CR3 to AA 42–63, CR4 to AA 99–113, CR5 to AA 118–131, CR6 to AA 145–179, CR7 to AA 184–222, and CR8–9 (basic leucine zipper region) to AA 243–317. All mutations were introduced using the Stratagene QuikChange site-directed mutagenesis kit and confirmed by sequencing.
C/EBPβ-Gal4 constructs were previously published (11). Briefly, the Gal4 DNA binding domain (AA 1–147) was amplified from the pSG424 plasmid (37) by PCR, creating a NarI site (in-frame with the C/EBPβ NarI site in the DNA binding domain) at the 5′ end and an XbaI-translation stop codon at the 3′ end (5′ primer, GCACTAGTGAATTCAAGCTTACCATGGCGCTACTGTCTTCT; and 3′ primer, GCATCGATCTAGAGTGGATCCGACAGTCAACTGTCT). Appropriate C/EBPβ TAD constructs were ligated in-frame as HindIII-NarI fragments to the Gal4 DNA binding domain. Plasmids for full-length and mutant G9a have been published elsewhere (38).
Peptides—Peptides covering the C/EBPβ TAD were synthesized and are derived from the mouse sequence (Swiss-Prot P28033): peptide1, AA 1–21; peptide2, AA 22–55; peptide3, AA 49–81; and peptide4, AA 80–113.
GST Pulldown—Glutathione S-transferase (GST) fusion proteins were expressed in BL21(DE3) bacteria, lysed, and purified with glutathione-Sepharose 4B (GE Healthcare) using standard protocols. For pulldown assays, GST fusion proteins were bound to glutathione-Sepharose 4B and incubated with in vitro translated, [35S]methionine-labeled G9a.
Cell Culture—HEK-293 and HeLa cells were cultivated in Dulbecco's modified Eagle's medium (Invitrogen) containing 10% fetal bovine serum (Invitrogen) and 1% penicillin/streptomycin at 5% CO2 and 37 °C. U937 and Jurkat cells were cultivated in RPMI 1640 medium (Invitrogen) containing 10% fetal bovine serum and 1% penicillin/streptomycin at 5% CO2 and 37 °C. HD3 cells (39) were cultivated with 8% fetal bovine serum, 2% chicken serum (Invitrogen), and 1% penicillin/streptomycin at 5% CO2 and 37 °C. Phorbol 12-myristate 13-acetate (PMA) stimulation was performed for 1 h at a final concentration of 0.5 μm. Transient transfections were performed by the calcium phosphate method or with Metafectene reagent (Biontex).
Immunoprecipitation and Immunoblotting—The following antibodies were used: anti-C/EBPβ (Leutz lab or Santa Cruz Biotechnology (C-19, H-7)), anti-phospho-Thr-235 C/EBPβ (Cell Signaling Technology), anti-G9a (Walsh lab), anti-green fluorescent protein (GFP; Roche Applied Bioscience), anti-FLAG (Sigma), anti-H3K9me2 (Abcam), and anti-H3K9ac S10p (Abcam). HEK-293, U937, or Jurkat cells were lysed in 50 mm Tris·Cl (pH 8), 150 mm NaCl, 0.1% Nonidet P-40, 1 mm EDTA, 5 mm MgCl2, 50 μm ZnCl2, and protease inhibitor mixture (Roche Applied Bioscience). Samples were immunoprecipitated with appropriate antibodies for 2 h at 4 °C, and immunoprecipitates were collected on protein A- or G-Sepharose beads. Alternatively, anti-FLAG affinity matrix (Sigma) was used for 1 h. Immunoblots were developed and quantified using IRDye 700-coupled anti-mouse IgG or IRDye 800-coupled anti-rabbit IgG on the Odyssey infrared scanning system or by ECL (GE Healthcare).
Chromatin Immunoprecipitation—Chromatin immunoprecipitation was performed as described with minor changes (40). Briefly, U937 cells (2 × 107) were cross-linked with 1% formaldehyde, quenched with glycine (final concentration, 0.125 m), and lysed (10 mm Tris·Cl (pH 8), 10 mm NaCl, 0.2% Nonidet P-40, and protease inhibitor mixture). Cell debris was removed by centrifugation, nuclei were lysed (50 mm NaCl, 10 mm EDTA, 1% SDS, and protease inhibitor mixture), and DNA was sheared by sonication. After centrifugation the SDS concentration of the supernatant was reduced by adding 2 volumes of dilution buffer (20 mm Tris·Cl (pH 8), 2 mm EDTA, 150 mm NaCl, 1% Triton X-100, and protease inhibitor mixture). Immunoprecipitation (IP) was performed with one-fifth of the extract using 1–5 μg of the indicated antibody. One sample was spared for input control. Immunoprecipitates were collected with magnetic protein G beads (Dynabeads protein G-100, Invitrogen). Beads were washed repeatedly with IP buffer 1 (20 mm Tris·Cl (pH 8), 2 mm EDTA, 50 mm NaCl, 1% Triton X-100, and 0.1% SDS), IP buffer 2 (10 mm Tris·Cl (pH 8), 1 mm EDTA, 250 mm LiCl, 1% Nonidet P-40, and 1% sodium deoxycholic acid), and IP buffer 3 (20 mm Tris·Cl (pH 7.6) and 50 mm NaCl) and eluted with 150 μl of elution buffer (100 mm NaHCO3 and 1% SDS). Cross-links were reversed by incubation in 300 mm NaCl at 67 °C overnight. Nucleic acids were precipitated with isopropyl alcohol overnight, and precipitated DNA was analyzed by quantitative PCR.
Quantitative PCR—Quantitative PCR was performed using SYBR Green (Invitrogen) and a Light Cycler (Roche Applied Bioscience) according to the manufacturers' instructions. cDNA synthesis was performed with SuperScript reverse transcriptase (Invitrogen). Sequences of PCR primer pairs are published elsewhere (41).
In Vitro Methylation—pCMV-GFP-G9a or pCMV-GFP-G9aΔSET was transfected into HEK-293 cells. After 48 h the cells were lysed (50 mm Tris·Cl (pH 8), 150 mm NaCl, 0.1% Nonidet P-40, 1 mm EDTA, 5 mm MgCl2, 50 μm ZnCl2, and protease inhibitor mixture), and GFP fusion proteins were immunoprecipitated from lysates for 3 h with an anti-GFP antibody. Antigen-antibody complexes were bound by protein G-coated Dynabeads and used directly in the methyl transfer reaction. Each reaction was supplied with 1 μCi of S-adenosyl-l-[methyl-3H]methionine as a methyl donor and 1–5 μg of GST fusion protein or 1 μm of the corresponding peptide. Reactions were carried out for 3 h at 30 °C in assay buffer (50 mm Tris·Cl (pH 9), 5 mm MgCl2, and 4 mm dithiothreitol) (42). Methylated samples were either submitted to SDS-PAGE and detected by fluorography or spotted onto phosphocellulose membrane, and [3H]methyl incorporation was determined by scintillation counting.
In Vivo Methylation—Transiently transfected HEK-293 cells were labeled with 10 μCi/ml l-[methyl-3H]methionine (Amersham Biosciences) in the presence or absence of 20 μm homocysteine hydrolase inhibitor adenosine dialdehyde (Sigma) in methionine-free Dulbecco's modified Eagle's medium supplemented with the protein synthesis inhibitors cycloheximide (100 μg/ml; Sigma) and chloramphenicol (40 μg/ml; Sigma) as described previously (43). Inhibition of translation was monitored by labeling the cells with 20 μCi/ml [35S]methionine (MP Biomedicals). C/EBPβ was immunoprecipitated and separated by SDS-PAGE. For fluorography, gels were incubated with ENH3ANCE (Perkin Elmer Life Sciences), dried, and exposed to Eastman Kodak X-OMAT at –70 °C.
Gene Expression—The chicken myelomonocytic growth factor (cMGF) promoter-reporter construct pM82 or a Gal4 binding site derivative was used (44, 45). C/EBPβ expression vectors were transfected into HEK-293 cells together with a luciferase reporter in the absence and presence of coeffectors. Cells were lysed after 36–48 h (50 mm Tris·Cl (pH 7.5), 1 mm EDTA, 1 mm dithiothreitol, and 1% Triton), and relative reporter activities were determined in duplicate in a Berthold Lumat LB9501. Relative reporter activities were normalized to cotransfected (100 ng) pCMV-lacZ, as determined by β-galactosidase assay. Resident gene activation was determined by transfection of HD3 cells and Northern analysis exactly as described (11).
Macroarray Screening—A human colon protein expression library (RZPD, library 829) was screened using the C/EBPβ TAD (CR1–4) as a bait. The bait was generated as a GST-PKA-C/EBPβ fusion protein expressed in BL21(DE3). The GST-PKA-C/EBPβ CR1–4 fusion protein was affinity-purified with glutathione-Sepharose from lysates, and the GST moiety was cleaved off by thrombin protease. The PKA-C/EBPβ CR1–4 moiety was in vitro phosphorylated by recombinant PKA in kinase buffer (10 mm MgCl2, 20 mm MES, 2 mm EGTA, and 1 mm EDTA) in the presence of [γ-32P]ATP. The labeled protein was applied in blocking solution (3% skim milk in Tris-buffered saline/Tween) overnight to the expression library on polyvinylidene difluoride membrane. Interacting proteins were visualized by autoradiography. Identity of clones was determined according to the manufacturer's instructions.
RESULTS
C/EBPβ Interacts with the G9a Lysine Methyltransferase—A C/EBPβ bait protein for proteomic screening was constructed to identify proteins that interact with the TAD of C/EBPβ (supplemental Fig. 1). Briefly, the N terminus of C/EBPβ containing the entire TAD (consisting of CR1–4) (11) was cloned into the pGEX vector that contained or lacked a phosphorylation site for PKA (see “Experimental Procedures”). The resulting GST-C/EBPβ fusion proteins were expressed in bacteria, purified, and submitted to phosphorylation by PKA using [γ-32P]ATP. Only GST-C/EBPβ containing the PKA site was labeled (supplemental Fig. 1A). The GST moiety was cleaved off using thrombin protease, and the PKA-labeled C/EBPβ protein was used as a probe to screen a human colon protein expression library from Escherichia coli containing 18,432 independent clones, arrayed as duplicates. The histone-lysine N-methyltransferase, H3 lysine 9-specific 3 (G9a) (Unigene AB209433), was identified as an interacting protein (supplemental Fig. 1B). Interaction between G9a and C/EBPβ was confirmed by pull-down experiments with recombinant GST-C/EBPβ TAD and in vitro translated G9a, as shown in supplemental Fig. 1C.
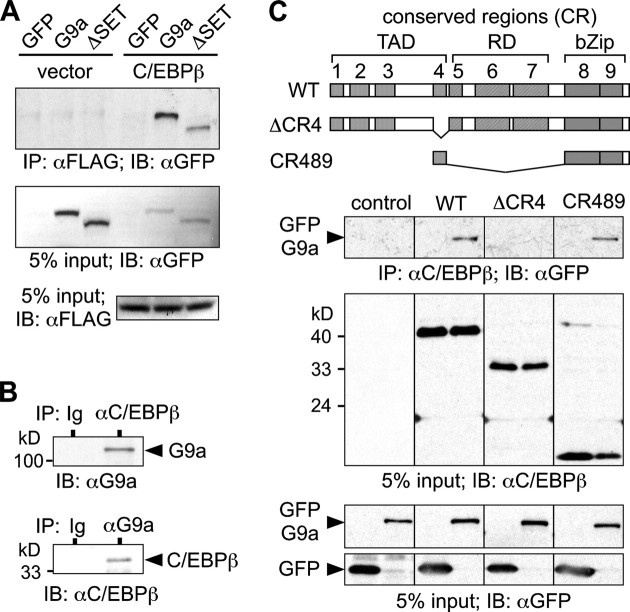
Co-immunoprecipitation of G9a and C/EBPβ showed that both G9a and ΔSET (a G9a mutant lacking the C-terminal catalytic domain) interact with C/EBPβ in eukaryotic cells (Fig. 1A). Interaction between endogenous C/EBPβ and G9a was confirmed in the T lymphocytic Jurkat cell line that expresses both proteins (Fig. 1B). Interaction between G9a and C/EBPβ depends on CR4, and CR4 was also found to be sufficient for the interaction, as shown in Fig. 1C.
FIGURE 1.
C/EBPβ and G9a interact in eukaryotic cells. A, empty vector or pcDNA3-C/EBPβ LAP*-FLAG was transfected together with pCMV-GFP, pCMV-GFP-G9a, and pCMV-GFP-G9aΔSET, as indicated, into HEK-293 cells and immunoprecipitated (IP) using anti-FLAG affinity matrix. Bound proteins were analyzed by immunoblotting (IB) using anti-GFP antibodies. Protein input was monitored with anti-FLAG or anti-GFP antibodies. B, Jurkat cells were lysed and immunoprecipitated with anti-C/EBPβ, anti-G9a, or Ig (control) antibodies as indicated. Bound proteins were analyzed by immunoblotting. C, structure of C/EBPβ, indicating TAD, regulatory domain (RD), and CR, is shown. pcDM8-C/EBPβ LAP*, pcDM8-C/EBPβ ΔCR4, pcDM8-C/EBPβ CR489, and pCMV-GFP-G9a constructs were transfected into HEK-293 cells and immunoprecipitated using anti-C/EBPβ antibody and protein A-Sepharose and revealed by immunoblotting, as indicated. WT, wild type; bZIP, basic leucine zipper.
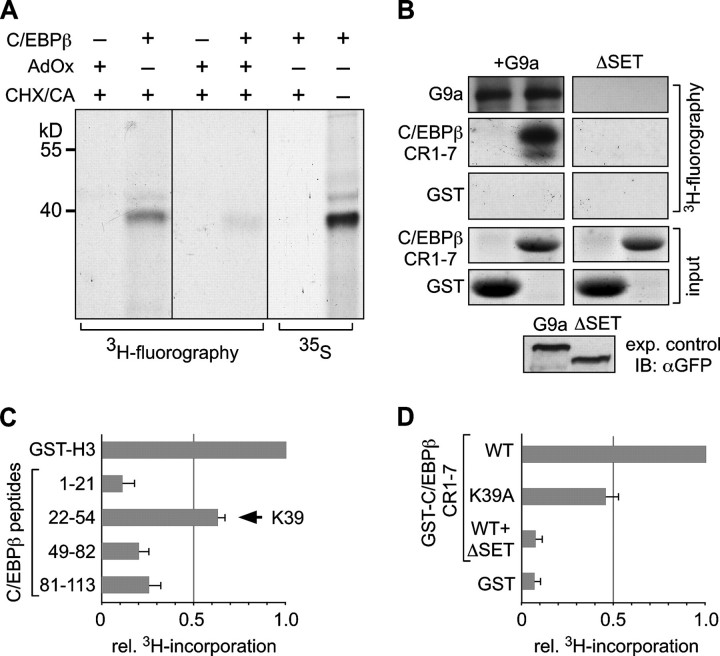
G9a Methylates the C/EBPβ TAD—Cells were labeled with l-[methyl-3H]methionine as a traceable methyl donor to examine whether C/EBPβ becomes methylated in vivo. The experiment was performed in the presence or absence of the homocysteine hydrolase inhibitor adenosine dialdehyde, which blocks generation of the cellular methyl donor S-adenosylmethionine (46), and in the presence of cycloheximide and chloramphenicol to block protein synthesis. Metabolic labeling with [35S]methionine was used to control inhibition of protein translation in the presence of cycloheximide/chloramphenicol (Fig. 2A, right panel). Immunoprecipitation of C/EBPβ and subsequent SDS-PAGE and fluorography revealed a [3H]methyl-labeled C/EBPβ product in the absence but not in the presence of adenosine dialdehyde (Fig. 2A, left and middle panels). These results suggest that C/EBPβ becomes post-translationally methylated in vivo.
FIGURE 2.
C/EBPβ is methylated at Lys-39 by G9a. A, HEK-293 cells were transfected with pcDM8-C/EBPβ and labeled with l-[methyl-3H]methionine 48 h after transfection in the absence or presence of adenosine dialdehyde (AdOx). Protein synthesis was blocked with cycloheximide (CHX) and chloramphenicol (CA). Inhibition of translation was monitored by labeling cells with [35S]methionine. After metabolic labeling, cells were lysed, and C/EBPβ was immunoprecipitated, separated by SDS-PAGE, and visualized by fluorography. B, in vitro methylation reactions were carried out with S-adenosyl-[methyl-3H]methionine as a [3H]methyl donor. G9a or G9aΔSET was expressed in HEK-293 cells and enriched with anti-GFP antibodies. Automethylation of GFP-G9a served as an internal control for the catalytic activity of the protein. 5 μg of GST or GST-C/EBPβ CR1–7 were used as a substrate. The reaction was performed for 3 h at 30 °C in assay buffer (see “Experimental Procedures”). Samples were separated by SDS-PAGE and visualized by autoradiography. GST protein input was visualized with Coomassie Brilliant Blue. GFP-G9a or GFP-G9aΔSET was detected by immunoblotting (IB) using anti-GFP antibodies in the expression control. C, in vitro methylation of C/EBPβ peptides by G9a is shown. The reactions were carried out as described in B with 1 μm of the corresponding peptides as substrates. Bars represent relative incorporation of S-adenosyl-l-[methyl-3H]methionine. Histone H3 was used as a positive control. Error bars represent the standard deviation from three independent determinations. D, in vitro methylation of recombinant C/EBPβ proteins by G9a is shown. Reactions were carried out as described in B with 5 μg of GST protein as a substrate. Bars represent relative incorporation of S-adenosyl-l-[methyl-3H]methionine. Histone H3 was used as a positive control. Error bars represent standard deviations from two independent determinations. WT, wild type.
Next, we asked whether G9a can methylate C/EBPβ. G9a or the ΔSET mutant was expressed as GFP fusion proteins in HEK-293 cells and immunoprecipitated from cell lysates. Affinity enriched G9a proteins were subsequently incubated with recombinant GST or GST-C/EBPβ CR1–7 in the presence of S-adenosyl-l-[methyl-3H]methionine. As shown in Fig. 2B, C/EBPβ CR1–7 was methylated by G9a but not by G9aΔSET. Automethylation of G9a served as an internal control for the activity of the enzyme (Fig. 2B) (36).
Comparison of the amino acid sequence of the C/EBPβ TAD between teleosts and mammals disclosed a single conserved lysine residue (Lys-39 in chick, mouse, and rat; Lys-43 in human) in the low complexity hinge region located between CR2 and CR3. Methylation assays with immunoprecipitated enzymes were repeated with peptides covering the C/EBPβ TAD to disclose a G9a methylation target site, and GST-H3 (N terminus) served as a positive control. A peptide that spanned C/EBPβ Lys-39 incorporated significantly more [3H]methyl label than other peptides that may or may not contain lysine residues (peptide 81–113 contains Lys-98, Lys-101, and Lys-102) (Fig. 2C). The data show that G9a specifically methylates peptide 22–55 that contains the conserved Lys-39. Next, in vitro methylation assays with various recombinant GST-C/EBPβ proteins and G9a were carried out (Fig. 2D). 3H-Methylation was significantly lower than in wild type when Lys-39 was replaced by alanine. These data suggest that Lys-39 of C/EBPβ serves as a target site for G9a methylation.
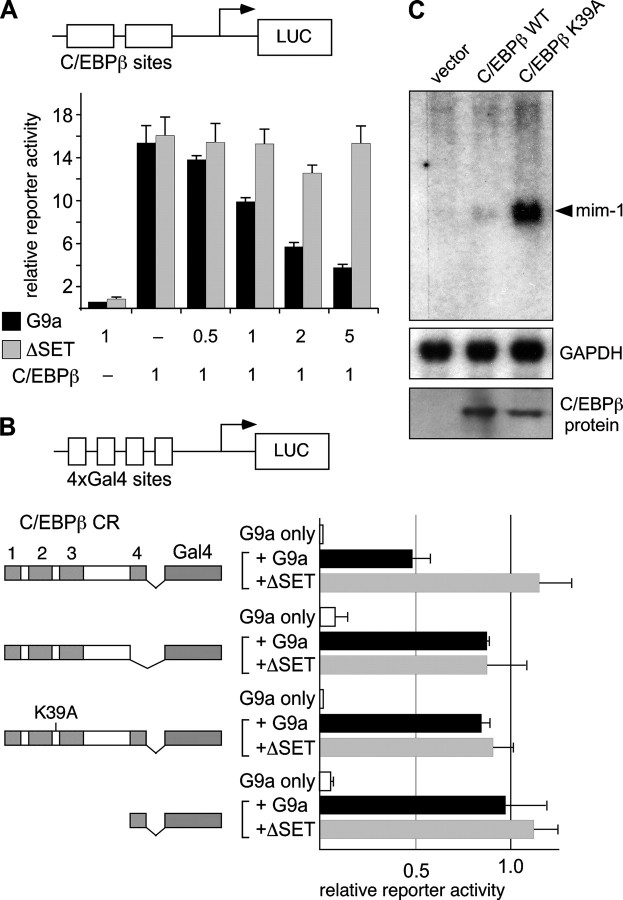
The G9a SET Domain Is Required for Repression of C/EBPβ Transactivation—The functional consequence of the interaction between C/EBPβ and G9a was addressed by reporter gene studies. G9a, but not G9aΔSET, diminished the transactivation potential of C/EBPβ in a concentration-dependent manner on the C/EBPβ-responsive cMGF reporter construct pM82 (Fig. 3, A and B). A chimeric construct with the complete C/EBPβ TAD and the heterologous DNA binding domain of Gal4 (11) was also repressed by G9a (Fig. 3B). The G9aΔSET variant failed to repress reporter activation, suggesting that the catalytic activity of G9a is important for repression. Repression by G9a depends on CR4, as its deletion overcame the sensitivity toward G9a (Fig. 3B). Interestingly, exchange of the conserved lysine at position 39 to alanine (K39A) also abrogated the sensitivity toward G9a. Taken together, these data suggest that both recruitment of G9a by CR4 and methylation of lysine 39 by G9a are prerequisites for suppression of the gene activation potential of C/EBPβ. The possibility that prevention of methylation at Lys-39 generates super-active C/EBPβ was examined by transfecting erythroid HD3 cells with wild type C/EBPβ or K39A and monitoring activation of the resident, chromatin-embedded myeloid mim-1 C/EBPβ target gene (11). As shown in Fig. 3C, the C/EBPβ-K39A mutant resulted in strongly enhanced activation of mim-1, suggesting that prevention of Lys-39 methylation causes C/EBPβ to become super-active.
FIGURE 3.
G9a acts as a repressor of C/EBPβ-dependent transcription. A, schematic representation of the cMGF promoter-pM82 reporter gene construct containing two C/EBPβ binding sites upstream of the luciferase (LUC) gene. G9a, G9aΔSET, and C/EBPβ were transfected into HEK-293 at the indicated plasmid concentrations. B, reporter construct as in A, with the C/EBPβ binding sites exchanged against four Gal4 binding sites (pM82-Gal4). G9a or G9aΔSET was transfected, together with reporter and Gal4 effector constructs (or without effector in the G9a-only controls). The ratio of transfected Gal4 effector constructs to G9a expression plasmids was 1:2. The reporter activity of the individual Gal4 constructs was set to 1 for comparison of G9a effects. Transfection efficiency was monitored by determining β-galactosidase activity of cotransfected pCMV-lacZ construct. Error bars in A and B represent to S.D. from two independent experiments. C, constructs, as indicated on the top, transfected into HD3 cells. RNA was extracted after 24 h and subjected to Northern hybridization using a probe to the mim-1 gene (arrowhead) or to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a control. A protein expression control (anti-C/EBPβ immunoblot from transfected cell lysates) is shown. WT, wild type.
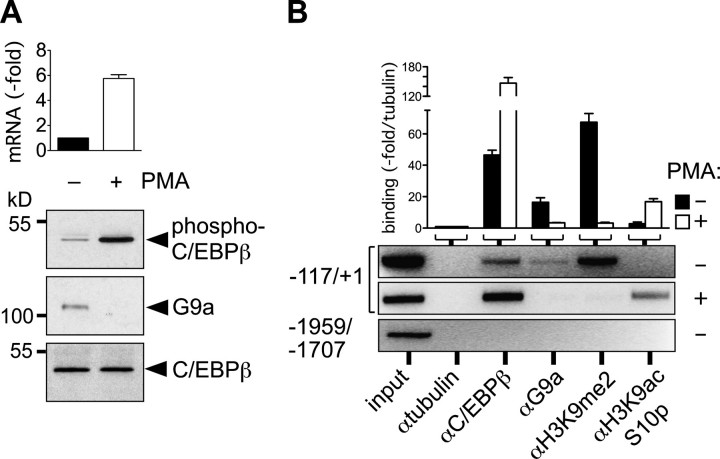
Association of C/EBPβ and G9a with the Promoter of the Neutrophil Elastase Gene (ELA2)—Previously, it has been shown that C/EBPβ may repress gene expression and that C/EBPβ can be activated by Ras signaling or PMA treatment through phosphorylation of an evolutionarily conserved mitogen-activated protein kinase (MAPK) site in its regulatory domain (9, 11, 12). Activation of C/EBPβ is accompanied by a conformational change that is allosterically transmitted to the TAD (13). We therefore examined physical and functional interactions between G9a and C/EBPβ under both conditions and on the resident human myeloid target gene hELA2 that can be activated by PMA in the U937 cell line (41, 47). As shown in Fig. 4A, PMA treatment of U937 cells enhances expression of the hELA2 gene (∼6-fold) and induces phosphorylation of C/EBPβ at Thr-235. Interaction between G9a and C/EBPβ was abrogated when C/EBPβ became hyperphosphorylated. This suggests that phosphorylation-dependent structural alterations in C/EBPβ that abrogate repression also abrogate interaction with G9a. Next, we examined G9a and C/EBPβ occupancy at the hELA2 promoter. A 117-bp fragment upstream of the TATA box has been characterized as a minimal promoter that is C/EBP-responsive (41). As shown in Fig. 4B, in unstimulated U937 cells both, C/EBPβ and G9a were associated with the minimal promoter (–117/+1), but not with an upstream sequence (–1959/–1707). In addition, “repressive” H3K9 dimethylation, a hallmark of G9a activity, was found at the promoter region, but no H3K9 acetylation and H3S10 phosphorylation could be detected (marks of transcriptionally active genes). PMA treatment abrogated both G9a association and H3K9 dimethylation and concomitantly enhanced promoter-bound C/EBPβ, H3K9 acetylation, and H3S10 phosphorylation (Fig. 4B). Altogether, these data suggest that C/EBPβ-G9a complexes reside at the ELA2 gene promoter and that G9a is shed from the promoter during PMA-induced gene activation.
FIGURE 4.
Interaction between C/EBPβ-G9a and hELA2 promoter occupancy. A, myeloid U937 cells were treated with or without PMA (2 h) and lysed, and sequential immunoprecipitation and immunoblotting were performed with anti-phospho-C/EBPβ (IP) and anti-C/EBPβ (H-7, immunoblot, upper panel), anti-C/EBPβ (C-19, IP) and anti-G9a (immunoblot, middle panel), or anti-C/EBPβ (C-19, IP) and anti-C/EBPβ (H-7, immunoblot; lower panel), respectively. hELA2 transcript was determined by quantitative PCR (bar graph) with and without PMA stimulation of U937 cells, as indicated. B, Chromatin immunoprecipitation assay of the hELA2 promoter (–117/+1; –1959/–1707 served as control; lower panel) from unstimulated (black bars and upper panel) or PMA-stimulated (white bars and middle panel) U937 cells. Antibodies were used as indicated. Quantitative PCR results are expressed as fold enrichment over anti-tubulin. Quantitative PCR conditions were 40 cycles at 95 °C for 30 s, 57 °C for 10 s, and 72 °C for 20 s. PCR products were analyzed on 1.8% agarose gels for visualization. Error bars in A and B represent the S.D. from two independent experiments.
DISCUSSION
The Lysine Methyltransferase G9a Interacts with C/EBPβ—G9a was isolated in a proteome-wide screen as an interaction partner of C/EBPβ, and the association was found to be evolutionarily conserved and independent of the catalytic SET domain of G9a. Beside the SET domain, G9a contains ankyrin repeats that may function as a protein-protein interaction motif. The G9a ankyrin repeats were recently shown to preferentially bind mono- and dimethylated lysines in histone H3 tails (48). Therefore, G9a represents a protein lysine methyltransferase that may “write” (with its SET domain) and “read” (with its ankyrin repeats) the same epigenetic mark. Whether methylation of C/EBPβ by G9a creates an additional binding site for G9a itself or other repressive histone-modifying complexes will require further analysis. It is also evident that only the long C/EBPβ isoforms LAP* and LAP may recruit G9a and serve repression (7, 8), because CR4 is absent in LIP.
An interesting observation in this context is that the repressive effect of G9a via C/EBPβ is observed only when methylation of Lys-39 may occur, although G9a binds to C/EBPβ independently of Lys-39. This observation may suggest that methylation of C/EBPβ by G9a creates a new binding site for a repressive protein complex or enhances interaction with C/EBPβ via “reading” methylated Lys-39 and thus exert repression, e.g. by enhancing histone H3 methylation in its vicinity. These notions are substantiated by the experimental results showing enhanced binding of wild type compared with G9aΔSET (Fig. 1A) and by chromatin immunoprecipitation of endogenous C/EBPβ and G9a proteins on the resident hELA2 gene in U937 cells (Fig. 4).
Methylation of Lys-39 Is a Novel Post-translational Modification of C/EBPβ—Data presented in this study identify lysine methylation at Lys-39 as a novel post-translational modification in C/EBPβ. The evolutionary conserved Lys-39 is part of the transactivation domain of C/EBPβ and is located in a low complexity hinge region between CR2 and CR3. Lysine methylation of nuclear proteins was first detected in histones, but evidence is accumulating that non-histone proteins are also subject to regulation by methylation of lysine side chains in a dynamic fashion, e.g. p53 was shown to be lysine-methylated in the C-terminal part of the protein by the histone methyltransferases Set8, Set9, and Smyd2 (32, 33, 49). Methylation of p53 may enhance or repress transcription in a target gene-dependent manner, whereas demethylation by LSD1 decreases its activity (49). Set9 also methylates TAF10 at a single lysine residue. This modification leads to a stronger association of the TATA box-binding protein (TBP)-associated factor with the RNA polymerase II (50). The retinoic acid receptor α was also found to be trimethylated in its ligand-binding domain. This modification appears to enhance the ability of retinoic acid receptor α to interact with co-factors like PCAF (51). Similarly, the lysine methylation of C/EBPβ could also alter the interaction with cofactors that still have to be identified.
G9a Catalyzes Lysine Methylation of C/EBPβ—Methylation of lysine 9 in histone H3 (H3K9) serves as a specific binding site for heterochromatin protein 1 (HP1) and is correlated with transcriptional gene silencing. Among others, Suv39h1 and G9a methylate H3K9 and repress transcription; however, Suv39h1 and G9a partition differently between heterochromatin and euchromatin, respectively. G9a rather appears to be the predominant euchromatic H3K9 methyltransferase in mammals as deletion of G9a in mice decreases H3K9 methylation in euchromatic regions (52). G9a cooperates with GLP as a heteromeric complex and is essential for maintaining normal methylation patterns of H3K9 throughout euchromatin (53), whereas the Suv39h class of enzymes maintains methylation of heterochromatin (54) and recruits HP1 (55). Biochemical studies showed that G9a is capable of mono-, di-, and trimethylating peptides that cover the N termini of H3 in vitro, although the final trimethylation step is rate-limiting (53, 56). Furthermore, G9a was shown to be automethylated in vitro, which did not alter the enzymatic activity, but increased the ability of G9a to bind HP1 (36). Accordingly, one may assume that repressive complexes can assemble on non-histone proteins after methylation by G9a.
G9aRepressesC/EBPβ—C/EBPβ-dependent reporter assays showed a concentration-dependent and SET-dependent repression of C/EBPβ activity by G9a. Interestingly, the murine equivalent of Lys-39 was previously found to be acetylated, and acetylation was suggested to enhance the transactivation potential of C/EBPβ (26). However, acetylation-dependent activation was deduced from mutational analysis that reduced transactivation in C/EBPβ K39A and K39R mutants on C/EBP-responsive sites (26). Currently, we cannot resolve the discrepancy between our data and the previous study, but one possibility is that post-translational modifications on C/EBPβ manifest in a promoter-specific manner. Notwithstanding the discrepancy, acetylation of the C/EBPβ TAD at Lys-39 could be associated with transcriptional activation and lysine methylation with repression, representing a site that is involved in tuning C/EBPβ activity dependent on the chromatin context.
Supplementary Material
Acknowledgments
We acknowledge Dr. Constantin Rüder for help with immunofluorescence microscopy and Karolin Friedrich for technical assistance.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
Footnotes
The abbreviations used are: C/EBPβ, CCAAT/enhancer-binding protein-β; GST, glutathione S-transferase; PCAF, p300/CREB-binding protein associated factor; AA, amino acid(s); PMA, phorbol 12-myristate 13-acetate; GFP, green fluorescent protein; IP, immunoprecipitation; MES, 4-morpholineethanesulfonic acid; TAD, transactivation domain; CR, conserved region; PKA, protein kinase A.
References
- 1.Sterneck, E., Tessarollo, L., and Johnson, P. F. (1997) Genes Dev. 11 2153–2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Greenbaum, L. E., Li, W., Cressman, D. E., Peng, Y., Ciliberto, G., Poli, V., and Taub, R. (1998) J. Clin. Investig. 102 996–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poli, V. (1998) J. Biol. Chem. 273 29279–29282 [DOI] [PubMed] [Google Scholar]
- 4.Tanaka, T., Akira, S., Yoshida, K., Umemoto, M., Yoneda, Y., Shirafuji, N., Fujiwara, H., Suematsu, S., Yoshida, N., and Kishimoto, T. (1995) Cell 80 353–361 [DOI] [PubMed] [Google Scholar]
- 5.Descombes, P., and Schibler, U. (1991) Cell 67 569–579 [DOI] [PubMed] [Google Scholar]
- 6.Calkhoven, C. F., Muller, C., and Leutz, A. (2000) Genes Dev. 14 1920–1932 [PMC free article] [PubMed] [Google Scholar]
- 7.Screpanti, I., Musiani, P., Bellavia, D., Cappelletti, M., Aiello, F. B., Maroder, M., Frati, L., Modesti, A., Gulino, A., and Poli, V. (1996) J. Exp. Med. 184 1561–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lamb, J., Ramaswamy, S., Ford, H. L., Contreras, B., Martinez, R. V., Kittrell, F. S., Zahnow, C. A., Patterson, N., Golub, T. R., and Ewen, M. E. (2003) Cell 114 323–334 [DOI] [PubMed] [Google Scholar]
- 9.Nakajima, T., Kinoshita, S., Sasagawa, T., Sasaki, K., Naruto, M., Kishimoto, T., and Akira, S. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 2207–2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanlon, M., Sturgill, T. W., and Sealy, L. (2001) J. Biol. Chem. 276 38449–38456 [DOI] [PubMed] [Google Scholar]
- 11.Kowenz-Leutz, E., Twamley, G., Ansieau, S., and Leutz, A. (1994) Genes Dev. 8 2781–2791 [DOI] [PubMed] [Google Scholar]
- 12.Williams, S. C., Baer, M., Dillner, A. J., and Johnson, P. F. (1995) EMBO J. 14 3170–3183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mo, X., Kowenz-Leutz, E., Xu, H., and Leutz, A. (2004) Mol. Cell 13 241–250 [DOI] [PubMed] [Google Scholar]
- 14.Piwien-Pilipuk, G., Van Mater, D., Ross, S. E., MacDougald, O. A., and Schwartz, J. (2001) J. Biol. Chem. 276 19664–19671 [DOI] [PubMed] [Google Scholar]
- 15.Wegner, M., Cao, Z., and Rosenfeld, M. G. (1992) Science 256 370–373 [DOI] [PubMed] [Google Scholar]
- 16.Chinery, R., Brockman, J. A., Dransfield, D. T., and Coffey, R. J. (1997) J. Biol. Chem. 272 30356–30361 [DOI] [PubMed] [Google Scholar]
- 17.Trautwein, C., van der Geer, P., Karin, M., Hunter, T., and Chojkier, M. (1994) J. Clin. Investig. 93 2554–2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trautwein, C., Caelles, C., van der Geer, P., Hunter, T., Karin, M., and Chojkier, M. (1993) Nature 364 544–547 [DOI] [PubMed] [Google Scholar]
- 19.Buck, M., Poli, V., van der Geer, P., Chojkier, M., and Hunter, T. (1999) Mol. Cell 4 1087–1092 [DOI] [PubMed] [Google Scholar]
- 20.Shuman, J. D., Sebastian, T., Kaldis, P., Copeland, T. D., Zhu, S., Smart, R. C., and Johnson, P. F. (2004) Mol. Cell. Biol. 24 7380–7391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, X., Kim, J. W., Gronborg, M., Urlaub, H., Lane, M. D., and Tang, Q. Q. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 11597–11602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Subramanian, L., Benson, M. D., and Iniguez-Lluhi, J. A. (2003) J. Biol. Chem. 278 9134–9141 [DOI] [PubMed] [Google Scholar]
- 23.Berberich-Siebelt, F., Berberich, I., Andrulis, M., Santner-Nanan, B., Jha, M. K., Klein-Hessling, S., Schimpl, A., and Serfling, E. (2006) J. Immunol. 176 4843–4851 [DOI] [PubMed] [Google Scholar]
- 24.Eaton, E. M., and Sealy, L. (2003) J. Biol. Chem. 278 33416–33421 [DOI] [PubMed] [Google Scholar]
- 25.Kim, J., Cantwell, C. A., Johnson, P. F., Pfarr, C. M., and Williams, S. C. (2002) J. Biol. Chem. 277 38037–38044 [DOI] [PubMed] [Google Scholar]
- 26.Cesena, T. I., Cardinaux, J. R., Kwok, R., and Schwartz, J. (2007) J. Biol. Chem. 282 956–967 [DOI] [PubMed] [Google Scholar]
- 27.Joo, M., Park, G. Y., Wright, J. G., Blackwell, T. S., Atchison, M. L., and Christman, J. W. (2004) J. Biol. Chem. 279 6658–6665 [DOI] [PubMed] [Google Scholar]
- 28.Xu, M., Nie, L., Kim, S. H., and Sun, X. H. (2003) EMBO J. 22 893–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Allfrey, V. G., Faulkner, R., and Mirsky, A. E. (1964) Proc. Natl. Acad. Sci. U. S. A. 51 786–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Comb, D. G., Sarkar, N., and Pinzino, C. J. (1966) J. Biol. Chem. 241 1857–1862 [PubMed] [Google Scholar]
- 31.Jenuwein, T., and Allis, C. D. (2001) Science 293 1074–1080 [DOI] [PubMed] [Google Scholar]
- 32.Chuikov, S., Kurash, J. K., Wilson, J. R., Xiao, B., Justin, N., Ivanov, G. S., McKinney, K., Tempst, P., Prives, C., Gamblin, S. J., Barlev, N. A., and Reinberg, D. (2004) Nature 432 353–360 [DOI] [PubMed] [Google Scholar]
- 33.Huang, J., Perez-Burgos, L., Placek, B. J., Sengupta, R., Richter, M., Dorsey, J. A., Kubicek, S., Opravil, S., Jenuwein, T., and Berger, S. L. (2006) Nature 444 629–632 [DOI] [PubMed] [Google Scholar]
- 34.Shi, X., Kachirskaia, I., Yamaguchi, H., West, L. E., Wen, H., Wang, E. W., Dutta, S., Appella, E., and Gozani, O. (2007) Mol. Cell 27 636–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sampath, S. C., Marazzi, I., Yap, K. L., Krutchinsky, A. N., Mecklenbrauker, I., Viale, A., Rudensky, E., Zhou, M. M., Chait, B. T., and Tarakhovsky, A. (2007) Mol. Cell 27 596–608 [DOI] [PubMed] [Google Scholar]
- 36.Chin, H. G., Esteve, P. O., Pradhan, M., Benner, J., Patnaik, D., Carey, M. F., and Pradhan, S. (2007) Nucleic Acids Res. 35 7313–7323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sadowski, I., and Ptashne, M. (1989) Nucleic Acids Res. 17 7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nishio, H., and Walsh, M. J. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 11257–11262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beug, H., Doederlein, G., Freudenstein, C., and Graf, T. (1982) J. Cell. Physiol. Suppl. 1 195–207 [DOI] [PubMed] [Google Scholar]
- 40.Weinmann, A. S., Bartley, S. M., Zhang, T., Zhang, M. Q., and Farnham, P. J. (2001) Mol. Cell. Biol. 21 6820–6832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lausen, J., Liu, S., Fliegauf, M., Lubbert, M., and Werner, M. H. (2006) Oncogene 25 1349–1357 [DOI] [PubMed] [Google Scholar]
- 42.Chin, H. G., Pradhan, M., Esteve, P. O., Patnaik, D., Evans, T. C., Jr., and Pradhan, S. (2005) Biochemistry 44 12998–13006 [DOI] [PubMed] [Google Scholar]
- 43.Liu, Q., and Dreyfuss, G. (1995) Mol. Cell. Biol. 15 2800–2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sterneck, E., Blattner, C., Graf, T., and Leutz, A. (1992) Mol. Cell. Biol. 12 1728–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sterneck, E., Muller, C., Katz, S., and Leutz, A. (1992) EMBO J. 11 115–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen, D. H., Wu, K. T., Hung, C. J., Hsieh, M., and Li, C. (2004) J. Biochem. (Tokyo) 136 371–376 [DOI] [PubMed] [Google Scholar]
- 47.Nuchprayoon, I., Simkevich, C. P., Luo, M., Friedman, A. D., and Rosmarin, A. G. (1997) Blood 89 4546–4554 [PubMed] [Google Scholar]
- 48.Collins, R. E., Northrop, J. P., Horton, J. R., Lee, D. Y., Zhang, X., Stallcup, M. R., and Cheng, X. (2008) Nat. Struct. Mol. Biol. 15 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang, J., Sengupta, R., Espejo, A. B., Lee, M. G., Dorsey, J. A., Richter, M., Opravil, S., Shiekhattar, R., Bedford, M. T., Jenuwein, T., and Berger, S. L. (2007) Nature 449 105–108 [DOI] [PubMed] [Google Scholar]
- 50.Kouskouti, A., Scheer, E., Staub, A., Tora, L., and Talianidis, I. (2004) Mol. Cell 14 175–182 [DOI] [PubMed] [Google Scholar]
- 51.Huq, M. M., Tsai, N. P., Khan, S. A., and Wei, L. N. (2007) Mol. Cell. Proteomics 6 677–688 [DOI] [PubMed] [Google Scholar]
- 52.Tachibana, M., Sugimoto, K., Nozaki, M., Ueda, J., Ohta, T., Ohki, M., Fukuda, M., Takeda, N., Niida, H., Kato, H., and Shinkai, Y. (2002) Genes Dev. 16 1779–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Collins, R. E., Tachibana, M., Tamaru, H., Smith, K. M., Jia, D., Zhang, X., Selker, E. U., Shinkai, Y., and Cheng, X. (2005) J. Biol. Chem. 280 5563–5570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rea, S., Eisenhaber, F., O'Carroll, D., Strahl, B. D., Sun, Z. W., Schmid, M., Opravil, S., Mechtler, K., Ponting, C. P., Allis, C. D., and Jenuwein, T. (2000) Nature 406 593–599 [DOI] [PubMed] [Google Scholar]
- 55.Stewart, M. D., Li, J., and Wong, J. (2005) Mol. Cell. Biol. 25 2525–2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Patnaik, D., Chin, H. G., Esteve, P. O., Benner, J., Jacobsen, S. E., and Pradhan, S. (2004) J. Biol. Chem. 279 53248–53258 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.