Abstract
Increasing evidence indicates that bone morphogenetic proteins (BMPs) are crucial for cardiac induction, specification, and development. Although signaling of BMPs is tightly regulated through soluble BMP-binding proteins, how they regulate BMP signaling during cardiac differentiation remains unknown. To identify molecules responsible for BMP signaling during early cardiomyocyte differentiation of P19 cells, cDNA subtraction was performed. We found a bimodal expression of the BMP-binding protein Crossveinless-2 (Cv2) during cardiomyocyte differentiation; Cv2 is temporally expressed earlier than cardiac transcription factors such as Nkx2.5 and Tbx5 and acts as a suppressor for BMP signaling in P19 cells. We established a P19 clonal cell line harboring a cardiac alpha-myosin heavy chain promoter-driven enhanced green fluorescent protein gene to monitor cardiac differentiation by flow cytometry. Treatment with BMP2 during the first 2 days of differentiation suppressed cardiomyocyte differentiation through activation of down-stream targets Smad1/5/8 protein and Id1 gene, whereas treatment with Cv2 conversely inhibited Smad1/5/8 activation and Id1 expression, leading to increased generation of cardiac cells. RNA interference-mediated knockdown (KD) of endogenous Cv2 showed increased Smad1/5/8 activation and impaired cardiomyocyte differentiation. Expression of cardiac mesoderm markers was reduced, whereas expression of Id1 and endoderm markers such as Sox7, Hnf4, and E-cadherin was induced in Cv2-kinase dead cells. These phenotypes were rescued by the addition of Cv2 protein to the culture media during the first 2 days of differentiation or co-culture with parental cells. These data suggest that Cv2 may specify cardiac mesodermal lineage through inhibition of BMP signaling at early stage of cardiogenesis.
Heart development during embryogenesis is a multistep process that involves cardiac induction of mesodermal progenitor cells into the cardiac lineage. Although the genetic blueprint for cardiac differentiation and development is rapidly being elucidated (1-3), there is still uncertainty about cardiac-inducing factors which might be involved in cardiogenic induction and specification.
Increasing evidence indicates that BMPs2 are crucial for cardiac induction, specification, and development (2-7). Conventional deletion of BMP receptor 1a in mice showed no formation of mesoderm and heart (8). Studies with an in vitro cardiac differentiation model demonstrated an essential role of BMP signals in cardiomyogenesis (9). In contrast, functional disruption of the different BMPs in mice displayed a role in late but not in early cardiogenesis (10). For example, although deletion of BMP2 in the heart resulted in abnormal heart development, specification of cardiac mesoderm occurred normally (11). Explant cultures in chicken revealed that activation of BMP signaling inhibits cardiogenesis at early developmental stages (7). Until now, whether activation or suppression of BMP signaling is required during early cardiogenesis has not been resolved. It is also not known how the inhibitory effect of BMP signaling on early cardiogenesis is regulated.
BMPs belong to the transforming growth factor-β superfamily of secreted growth factors. The BMP family signals have been shown to play multiple roles in the control of embryogenesis, including cell-type specification, maturation, and dorsoventral axis determination (12). These pleiotropic functions of BMPs implicate a need for tight regulation of their activities. One way by which this is achieved is via soluble BMP-binding proteins, which tightly regulate BMP signaling (12, 13). During gastrulation of early embryogenesis, collaboration with BMPs and their binding proteins specifies the patterning of mesoderm by limiting the spatial extent of each other (13, 14).
In Drosophila, genetic analyses have identified a five cysteine-rich (CR) domain-containing molecule, Crossveinless-2 (Cv2), which promotes decapentaplegic (homologous to vertebrate BMP) activity in wings (15). Cv2 transcripts emerge first at gastrulation and are detected both in the precardiac mesoderm and in the posterior primitive streak in mouse embryos (16), suggesting the developmental role of Cv2 in cardiogenesis. Cv2 loss-of-function studies demonstrated the essential role of Cv2 as a pro-BMP factor in development (17, 18). However, either antagonistic or agonistic effects of Cv2 on BMP signal have been reported in vertebrates (17-22). Thus, the molecular mechanism by which Cv2 regulates BMP signal remains unclear.
In the present study we established a novel P19 embryonal carcinoma (P19)-derived clonal cell line EN8 cells harboring a cardiac α-myosin heavy chain (αMHC) promoter-driven enhanced green fluorescent protein (EGFP) gene to monitor the generation of cardiac cells by flow cytometric analysis (Fig. 1) and found a unique requirement for Cv2 in cardiac lineage decision through the temporal suppression of BMP activity during the first 2 days of differentiation. These findings imply that cardiac fate decision requires the temporal suppression of BMP signals at the early stages of development.
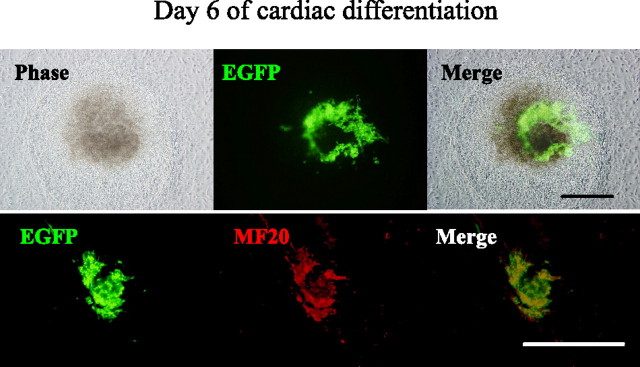
FIGURE 1.
Cardiac differentiation of P19EN8 cells harboring αMHC promoter-driven EGFP gene. Cells showing bright fluorescence matching beating areas have immunoreactivities for the primary antibody against sarcomeric myosin MF20 after 6 days of cardiac differentiation with DMSO. Scale bars equal 250 μm in panels.
EXPERIMENTAL PROCEDURES
Cells, Differentiation, and Transfection—Maintenance of P19 cells and dimethyl sulfoxide (DMSO)-induced differentiation were achieved as described previously (23). Briefly, cell aggregates were formed in hanging drops of 1000 cells in 50 μl of medium with 1% DMSO for the first 4 days. Aggregates were plated 2 days later onto tissue culture grade surfaces and maintained in DMSO-free medium. P19 EN8 cells were developed by transfecting the αMHC-green fluorescent protein vector (24) into P19Cl6 cells. P19Cl6 cells stably expressing FLAG-tagged Cv2 cDNA were also isolated. The BMP-responsive luciferase reporter (25) was from Dr. T. Katagiri (Saitama Medical School). pRL-TK was from Promega. Epitope-tag Cv2 was generated by amplifying Cv2 lacking the signal peptide and by subcloning the fragment in-frame into the pSec-Tag2 vector (Invitrogen). To develop the Cv2 knockdown cell and control cell lines, mouse Cv2 short hairpin RNA and control construct were developed by subcloning the target sequence (GCATAATGTGTGTGTGTTTGA) and GC%-matched scramble sequence into the pBA-puro vector (Takara, Japan), respectively.
cDNA Subtraction—Poly (A)-RNA (1 μg) from undifferentiated (driver) or differentiated P 19 cells at day 6 (tester) was used for double-stranded cDNA synthesis. The tester cDNA was digested with EcoRV and HincII. The driver cDNA library was amplified with the biotin-14-dCTP (Invitrogen) using the cDNA PCR library kit (Takara). Digested tester cDNA and biotin-labeled driver cDNA library were dissolved in 20 μl of a hybridization solution (0.5 mol/liter NaCl, 50 mmol/liter Tris, pH 7.5, 0.15% SDS, 40% formamide). After hybridizing for 48 h at 42 °C, the mixture was extracted, precipitated, and dissolved in 10 μl of H2O. After the mixture was incubated with 1.2 mg of streptavidin paramagnetic particles (Promega) in 100 μl of binding solution (10 mmol/liter Tris, pH 7.5, 1 mmol/liter EDTA, 100 mmol/liter NaCl), the supernatant was precipitated. Part of the products was ligated into pCR4Blunt-TOPO plasmids (Invitrogen). Purified plasmids went through sequence analysis, and the obtained data were compared with the GenBank™ databases with the BLAST program.
RT-PCR and Real-time PCR—T, Gata4, E-cadherin, Foxa2, NeuroD1, Pax6, Hnf4, Sox7, and cytokeratin 18 (CK) primer sequences can be found in Tada et al. (26). The PCR cycling conditions were as follows: 1 cycle of 94 °C for 4 min; 30 cycles of 94 °C for 30 s, 55 °C for 30 s, 72 °C for 1 min; 1 cycle of 72 °C for 30 s. Primers used for the PCR are as follows (forward and reverse): Cv2, 5-GGGTAAAATTCTCAACAGGA-3 and 5-CCACCAATCAAGTCATCACG-3; Mesp1, 5-CTGGCCATCCGCTACATTGG-3 and 5-CGTTGCATTGTCCCCTCCAC-3; Nkx2.5, 5-CAGTGGAGCTGGACAAAGCC-3 and 5-TAGCGACGGTTCTGGAACCA-3; Tbx5, 5-GCAGGGCCTGAGTACCTCTT-3 and 5-GGCTGATGGGCCACTGAGGT-3; Bmp2, 5-CGGGAACAGATACAGGAAGC-3 and 5-GCAAGGGGAAAAGGACACTC-3; Bmp4, 5-TGTGAGGAGTTTCCATCACG-3 and 5-TTATTCTTCTTCCTGGACCG-3; Inhibitor of DNA binding/differentiation 1 (Id1), 5-GGTACCGTACAACCTTTCTCCAACTTC-3 and 5-GGCTGGAGTCCATCTGGTCCCTCAGTGC-3. For quantitative analysis of gene expression levels, real-time PCRs were done using the ABI Prism 7700 sequence detection system. Data were normalized to glyceraldehyde-3-phosphate dehydrogenase. Primers used for the real-time PCR were as follows: T, Mm00436877_m1; Mesp1, Mm00801883_g1; Nkx2.5, Mm00657783_m1; Tbx5, Mm00803521_m1; and αMHC, Mm00440354_m1 (Applied Biosystems).
Whole-mount in Situ Analysis—Whole-mount in situ hybridization was performed using digoxigenin-UTP-labeled RNA probes according to manufacturers' protocols. Probes used were cDNAs for mouse Cv2. All experiments were performed in accordance with local institutional guidelines for animal experiments.
Flow Cytometry and Fluorescent-assisted Cell Sorting—Cells were prepared into a single-cell suspension by treatment with trypsin/EDTA. Flow cytometric analysis was performed with a fluorescent-assisted cell-sorting (FACS) machine (FACSCalibur, BD Biosciences). Sorted cells were collected by FACS Aria (BD Biosciences).
Western Blotting and Immunoprecipitation—Standard Western blot methods were used. FLAG-tagged secreted proteins were obtained by transient transfection. For immunoprecipitation analysis, an aliquot of the supernatant was incubated with anti-FLAG M2 antibody overnight at 4 °C. The immune complexes were collected with Protein G plus-agarose beads (Promega). For the in vitro receptor-ligand assays, BMP was incubated for 2 h at 4 °C with BMPRIa-Fc protein (R&D Systems) and then incubated with Cv2 (R&D Systems) for 2 h at 4 °C. The protein-A-agarose was preblocked with 1 mg/ml bovine serum albumin and then used to pull down the BMPR1A-Fc.
Data Analysis—Results are expressed as the mean ± S.D. Statistical significance was determined by Student's t test or one-way analysis of variance. p < 0.05 was used to determine a significant difference.
RESULTS
Identification of Genes Expressed during Early Cardiomyocyte Differentiation in P19 Cells—P19 cells are pluripotent stem cells that can mimic in vitro the first stages of cellular differentiation, which occur during normal mouse embryogenesis (23). To identify molecules responsible for BMP signaling during early cardiomyocyte differentiation of P19 cells, cDNA subtraction was employed. This analysis (performed by searching GenBank™ (NCBI) using the BLAST program) resulted in the identification of six distinct products including three genes that encode partial cDNA sequences of secreted proteins (Table 1). They are Wnt-3a, Sparc, and Cv2. Except for Cv2, these candidate genes have been reported on in detail elsewhere (27, 28). Among them, BMP-binding protein Cv2 was selected for further analysis based on the established role of BMPs in the regulation of cardiogenesis. A full-length Cv2 cDNA was obtained by using 5′ and 3′ rapid amplification of cDNA end technique. RT-PCR using cDNA from differentiated P19 cells at day 6 confirmed a substantial increase in Cv2 mRNA induced by DMSO (1%) but not retinoic acid (0.3 μmol/liter) nor a no-treatment control (Fig. 2A), suggesting that Cv2 is a DMSO-inducible factor in cardiomyocyte differentiation of P19 cells.
TABLE 1.
List of genes expressed during early cardiomyocyte differentiation in P19 cells identified by cDNA subtraction analysis
| Gene symbol | Gene name | NCBI GenBank™ |
|---|---|---|
| Cv2/Bmper | Crossveinless-2/BMP binding endothelial regulator | AF454954/NM028472 |
| Mapkap1 | Mitogen-activated kinase-associated protein 1 | NM177345 |
| Igf2bp3 | Insulin-like growth factor 2 mRNA-binding protein 3 | NM023670 |
| Sparc | Secreted acidic cysteine-rich glycoprotein/osteonectin | NM009242 |
| Fkhl18 | Forkhead-like 18 | NM010226 |
| Wnt3a | Wingless-related MMTV integration site 3A | NM009522 |
FIGURE 2.
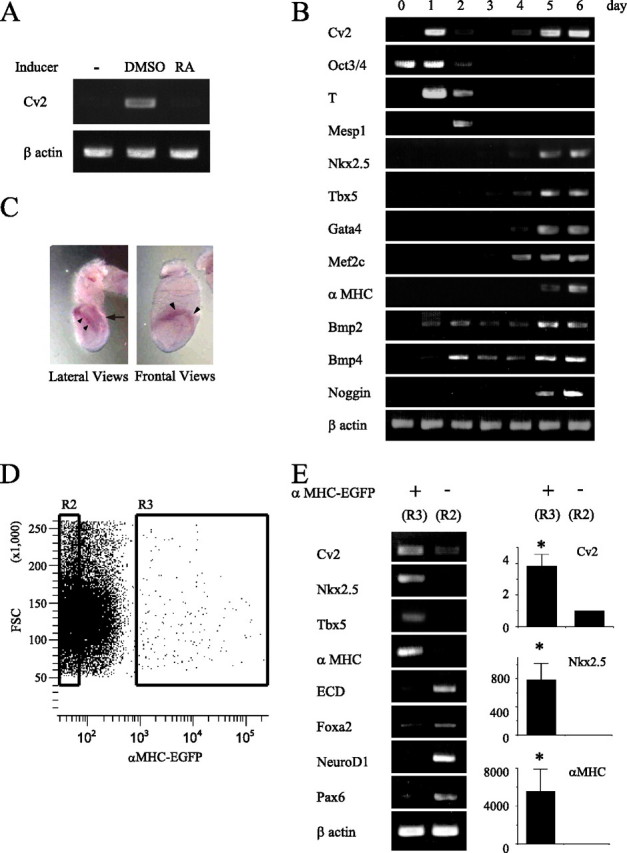
Gene expression during an in vitro cardiac differentiation model of P19 cells. A, RT-PCR analysis confirmed Cv2 expression in differentiating P19 cells (day 6) induced by DMSO but not retinoic acid (RA) or no-treatment control. B, kinetic analysis of gene expression in differentiating P19 cells showing that the progressive loss of an undifferentiating cell marker and the sequential acquisition of transcripts indicative of specific stages of embryonic development. RNA was isolated from undifferentiated P19 cells (day 0), aggregates harvested at daily interval (days 1-4), and adherent cultures (days 5 and 6). C, whole-mount in situ hybridization with Cv2 probes at 7.5 post-coitum. Lateral view and frontal view are shown. Cv2 transcripts were detected in the cardiac crescent (arrowhead) and the posterior primitive streak (arrow). D, purification of EGFP-positive cells (R3) and negative cells (R2) by FACS at day 7. FSC, forward scatter. E, RT-PCR and real-time PCR analysis of various lineage markers in each population. EGFP-expressing cells are cells differentiating into cardiac myocytes (n = 3; *, p < 0.05, R2 versus R3).
Bimodal Expression of Cv2 during Cardiomyocyte Differentiation—RT-PCR analysis of differentiating P19 cells as aggregates demonstrated the progressive down-regulation of a stem cell marker such as Oct3/4 accompanied by the sequential acquisition of mesodermal markers of T, Mesp1, Nkx2.5, and Tbx5 (Fig. 2B). Expression of T and Mesp1 peaked at day 1 and day 2, respectively. Transcription of Nkx2.5, Tbx5, Gata4, and Mef2c was also up-regulated from day 4, and αMHC was expressed from day 5. The fractions of aggregates with beating activity at day 6 are 80% in culture with DMSO and 0% in culture without DMSO (data not shown). Bmp2 and Bmp4 were expressed from day 1 and day 2, respectively. Noggin, the specific BMP antagonist, was expressed from day 5, consistent with the previous findings (29). These data indicate that our in vitro cardiac differentiation model of P19 cells reproduces the natural course of differentiation as described in the literature (23, 25), and this model is able to follow the Cv2 expression during the primary steps of cardiomyocyte differentiation. The kinetics of Cv2 expression showed a bimodal pattern; Cv2 is initially expressed earlier than cardiac transcription factors such as Nkx2.5 and Tbx5, and the second expression was up-regulated from day 4. This initial increase of Cv2 would correspond to just before or during gastrulation in intact embryos, in comparison with expression of T as a marker of primitive streak mesoderm (30). Because cells that are fated to become cardiac mesoderm are specified during gastrulation (31), this initial expression of Cv2 may be involved in the specification of cardiac lineage via modulating BMP activity. Whole-mount in situ hybridization revealed that at 7.5 days postcoitum Cv2 transcripts were predominantly found on the cardiac mesoderm (cardiac crescent) and the posterior primitive streak (Fig. 2C) as described previously (16).
Differentiating Cardiomyocytes Autologously Express Cv2—Next we analyzed Cv2 expression in differentiating P19EN8 cells (Fig. 2, D and E), because Cv2 may be expressed in a lineage-restricted manner. The EGFP-positive (differentiating cardiomyocytes) and EGFP-negative sorted populations were isolated by FACS, and gene expression was profiled by RT-PCR. Cv2 transcripts were determined to be 4-fold higher in the EGFP-positive population compared with the EGFP-negative population. These results indicate that Cv2 re-expresses autologously in the population of differentiating cardiac lineage.
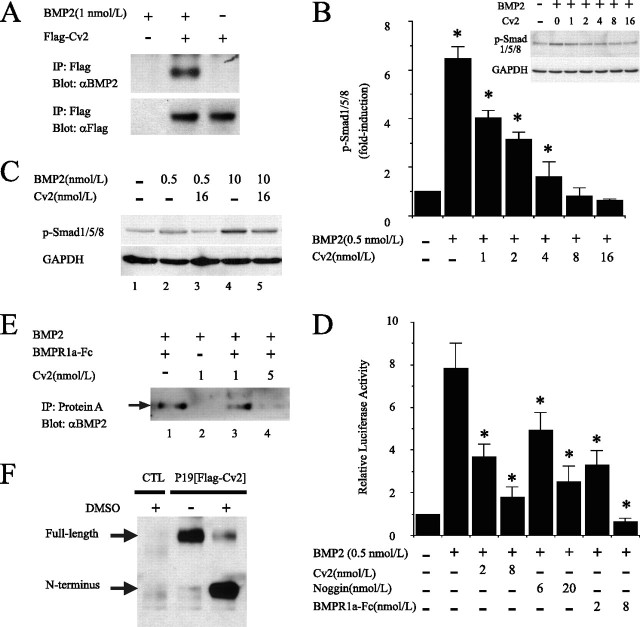
Cv2 Antagonizes BMP2 Signaling by Blocking Receptor Binding—First, we confirmed the BMP binding to Cv2 (Fig. 3A) using the FLAG-epitope-tagged construct of Cv2, in agreement with previous studies (19-22). This binding was inhibited in the presence of 10-fold molar excess of BMP4, but not activin A, transforming growth factor-β1 and epidermal growth factor (data not shown). Next, to examine whether Cv2 can antagonize BMP activity, we performed two independent experiments; the phosphorylation state of Smad1/5/8 proteins and the activation of a reporter gene under the control of a BMP-responsive sequence (Fig. 3, B and C). In Western blots, increasing amounts of Cv2 inhibited BMP-induced phosphorylated Smad1/5/8 (pSmad1/5/8) in a dose-dependent manner with 50% inhibition (IC50) at 2 nmol/liter (Fig. 3B). Although pSmad1/5/8 induced by 0.5 nmol/liter BMP2 was completely inhibited by the addition of more than 8 nmol/liter Cv2 (Fig. 3, B and C, lane 3), an excess of BMP2 (10 nmol/liter) was able to quench the antagonistic effect of Cv2 (16 nmol/liter) on BMP2 action (Fig. 3C, lane 5). Cv2 (16 nmol/liter) failed to affect pSmad2 induced by 1 nmol/liter transforming growth factor-β1 or 1 n mol/liter activin (data not shown). In luciferase assay of P19 cells transfected with the BMP-responsive reporter (Fig. 3D), Cv2 as well as Noggin and BMPRIa-Fc protein (a synthetic BMP antagonist) inhibited BMP-induced reporter expression in a dose-dependent manner. These results suggest Cv2 antagonism of BMP signals in P19 cells, consistent with the findings of others (19-21). To further address the molecular mechanism by which Cv2 antagonizes BMP2 signaling, we examined the effect of Cv2 on the binding of BMP2 to its receptor. The BMPRIa-Fc protein was immobilized on protein A-agarose, and bound BMP2 was analyzed. Increasing amounts of Cv2 (1 and 5 nmol/liter) caused a decrease in the amount of BMP2 bound to 1 nmol/liter BMPRIa-Fc protein (Fig. 3E, lane 3 and 4). We also observed an increase in the cleaved form of Cv2 in concert with cardiac differentiation using P19 cells stably expressing FLAG-tagged Cv2 cDNA (Fig. 3F). Together, these results strengthen the notion that Cv2 functionally interacts with BMP signaling by blocking interaction of BMP2 with its cognate receptor.
FIGURE 3.
Cv2 binds BMP2, and antagonizes BMP2 signaling by inhibiting receptor binding. A, representative Western blot (IB) analysis of BMP2 bound to full-length Cv2 after immunoprecipitation (IP). B, Cv2 inhibited pSmad1/5/8 by BMP2 (0.5 nmol/liter) in a dose-dependent manner in P19 cells (n = 3; *, p < 0.05 versus 0 nmol/liter Cv2). C, representative Western blot showing that inhibition of pSmad1/5/8 in P19 cells by Cv2 at low, but not high, BMP2 concentrations. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. D, reporter analysis of P19 cells transfected with BRE reporter (n = 3-4; *, p < 0.05 versus 0.5 nmol/liter BMP2). E, pulldown assay with protein A beads shows Cv2 inhibition of BMP2 (1 nmol/liter) binding to BMPRIa-Fc (1 nmol/liter). F, representative Western blot analysis of conditioned medium of P19 cells stably expressing N terminus FLAG-tagged Cv2 (designated as P19[FLAG-Cv2]) before and after cardiac differentiation. CTL, control.
Distinct Roles of BMP2 during Early and Late Cardiomyocyte Differentiation—BMP2 is an important signaling molecule for cardiac differentiation and development (2-7). To confirm this evidence, we treated differentiating P19 cells with BMP2 in a dose-dependent manner (Fig. 4A) and then evaluated cardiomyocyte differentiation at day 6 (Fig. 4B) and at day 7 (Fig. 4C). Contrary to our expectations, exposure to BMP2 during the first 2 days of differentiation suppressed the generation of cardiac cells (green fluorescent protein-positive cells), whereas exposure to BMP2 from day 5 to day 6 enhanced the generation of cardiac cells, as compared with the no-treatment control. The same observation applied to BMP4 (data not shown). These results suggest that at the early time frame from day 1 to day 2 of differentiation, corresponding to prior to or during gastrulation in development, BMP2 acts as an anti-cardiogenic factor. Our results agree with previous studies in chick embryos that BMPs inhibited cardiogenesis when applied at an early stage of gastrulation (7).
FIGURE 4.
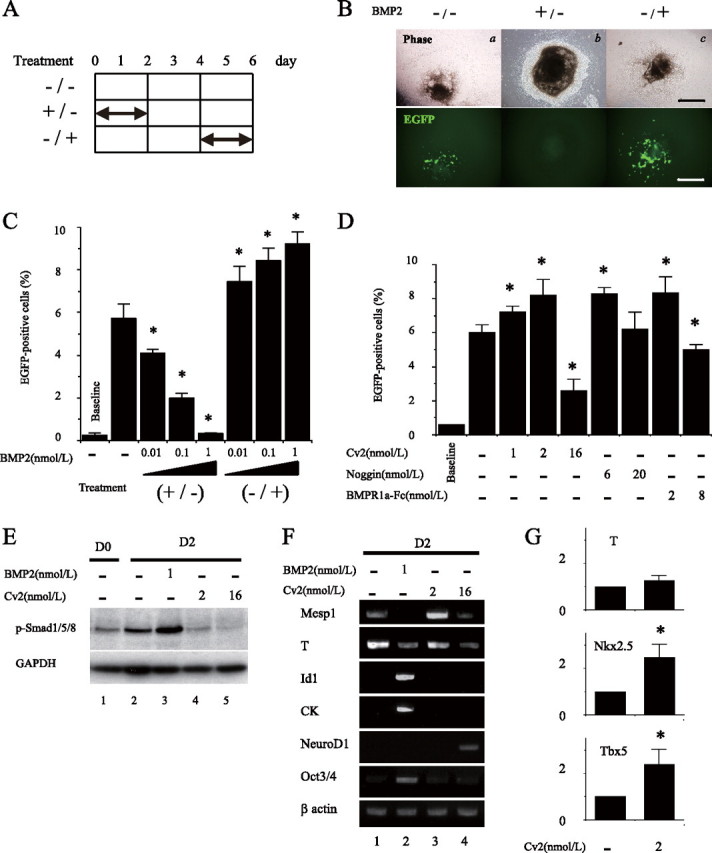
BMP2 plays distinct roles in early and late cardiomyocyte differentiation, and Cv2 regulates BMP signaling for cardiac lineage decision. A, experimental procedure of treatment. B, representative figures showing that effects of exposure to BMP on cardiomyocyte differentiation at day 6. Treatment with 1 nmol/liter BMP2 during the first 2 days (b) (+/-) resulted in no cardiac differentiation at day 6, whereas treatment without BMP2 (a) (-/-) generated cardiac cells. Contrarily, treatment with 1 nmol/liter BMP2 from day 5 to 6 (c) (-/+) increased the generation of EGFP-positive cells, suggesting enhanced cardiac differentiation. Scale bars equal 250 μm in panels. C and D, percentage of EGFP-positive cells assessed by FACS at day 7 (n = 3; *, p < 0.05 versus no treatment control). Baseline means no-treatment control at day 0. E, effects of either BMP2 or Cv2 on pSmad1/5/8 at day 2. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. F, effects of either BMP2 or Cv2 on gene expression at day 2. G, real-time PCR analysis of T (day 2), Nkx2.5 (day 6), and Tbx5 (day 6) after treatment with 2 nmol/liter Cv2 (n = 3; *, p < 0.05 versus no treatment control).
Progenitor cells destined to the cardiac lineage emerge from the primitive streak during gastrulation (31). Because there is a period that is sensitive to BMP concentrations in the early stage of cardiomyocyte differentiation of the P19 system, we hypothesized Cv2 modulation of BMP signaling in this period vulnerable to BMP activity. Therefore, with the focus on the time frame from day 1 to day 2 of early differentiation (Fig. 2B), we examined whether the suppression of BMP signals is required to generate the cardiac myocytes in further detail. For this purpose, we cultured differentiating P19 cells in the presence of Cv2, Noggin, or the BMPRIa-Fc protein during this period to evaluate cardiac differentiation by FACS. Increasing doses of the BMP antagonists led to successive peaks of cardiomyocyte differentiation at 2 nmol/liter Cv2, 6 nmol/liter Noggin, and 2 nmol/liter BMPRIa-Fc protein (Fig. 4D). In contrast, treatment of three BMP antagonists with higher doses than IC50 independently diminished the generation of cardiac cells. These results suggest that the dose of suppression of BMP signals with Cv2 as well as Noggin and BMPRIa-Fc at this early stage is critical in effective cardiac myocytes generation.
Cv2 Regulates BMP Signaling for Cell Lineage Determination—Analogous to the dorsoventral patterning of mesoderm by BMP signal gradients in Xenopus (14), we hypothesized that the manipulation of BMP levels either by BMP2 or Cv2 would reproduce the fate decision of cardiac mesoderm. To gain insights into the molecular mechanisms by which the modulation of BMP2 activity plays a potential lineage determinant at the early time point of differentiation, we detected and measured pSmad1/5/8 at day 2 of differentiation as an indicator of activated BMP signaling. Exposure to BMP2 (1 nmol/liter) markedly induced the pSmad1/5/8 (Fig. 4E, lane 3) compared with no-treatment control (Fig. 4E, lane 2). Conversely, treatment with both 2 and 16 nmol/liter Cv2 reduced pSmad1/5/8 to the base-line level at day 0 and to the level beneath the base-line level at day 0, respectively (Fig. 4E, lanes 4 and 5). RT-PCR analysis of day 2 aggregates demonstrated that exposure to BMP2 completely blocked Mesp1 expression and reduced T expression, whereas the Id1 and CK, high-dose responders of BMPs, were remarkably up-regulated (Fig. 4F, lane 2). In addition, expression of Oct3/4 sustained the high level relative to the others, suggesting that BMP-treated cells still sustain in the undifferentiated state. Conversely, treatment with Cv2 progressively down-regulated Oct3/4 expression and did not induce expression of Id1 and CK. Two nmol/liter Cv2 enhanced expression of T and Mesp1 (Fig. 4F, lane 3), whereas 16 nmol/liter Cv2 diminished expression of mesoderm markers and advanced expression of neuronal marker NeuroD1 (Fig. 4F, lane 4). When treated with 2 nmol/liter Cv2, real-time PCR analysis demonstrated the statistically significant increase in the mRNA levels of Nkx2.5 and Tbx5 (Fig. 4G). These results suggest the varying levels of BMP activity for priming differential genetic specification, including cardiac induction.
Loss of Cv2 Leads to Impaired Cardiomyocyte Differentiation—To further assess the involvement of endogenous Cv2 in cardiomyocyte differentiation, we performed loss-of-function experiments using short hairpin RNA specific for Cv2 (Fig. 5A). RNAi-mediated knockdown (KD) of Cv2 expression showed severely decreased cardiomyocyte differentiation at day 6 (Fig. 5, B and C). At day 2 of differentiation, increased pSmad1/5/8 and Id1 expression were observed in KD cells but not in control cells (Fig. 5, D and E), whereas there were no differences between both cell types in the phosphorylation of ERK and p38 (data not shown). In parallel with impaired cardiomyocyte differentiation, expression of cardiac mesoderm markers was reduced, but expression of visceral endoderm markers such as Sox7 and Hnf4 and pan-endoderm marker E-cadherin was induced in Cv2-KD cells (Fig. 5E). These results suggest that Cv2 KD preferentially gives rise to cells destined to the visceral endoderm through the up-regulation of BMP signals (32). These phenotypes in KD cells were reversed by the addition of recombinant Cv2 during the first 2 days of differentiation (Fig. 5C), and this was confirmed by mixing KD cells with control cells just before cardiac differentiation. Reduced cardiomyocyte differentiation in Cv2-KD cells was also reversed by co-culture with control cells KD (Fig. 5, F and G). These data suggest that Cv2-KD cells substituted loss of Cv2 for secreted Cv2 from control cells, and Cv2 itself can act in trans as an intercellular modulator in the extracellular space.
FIGURE 5.
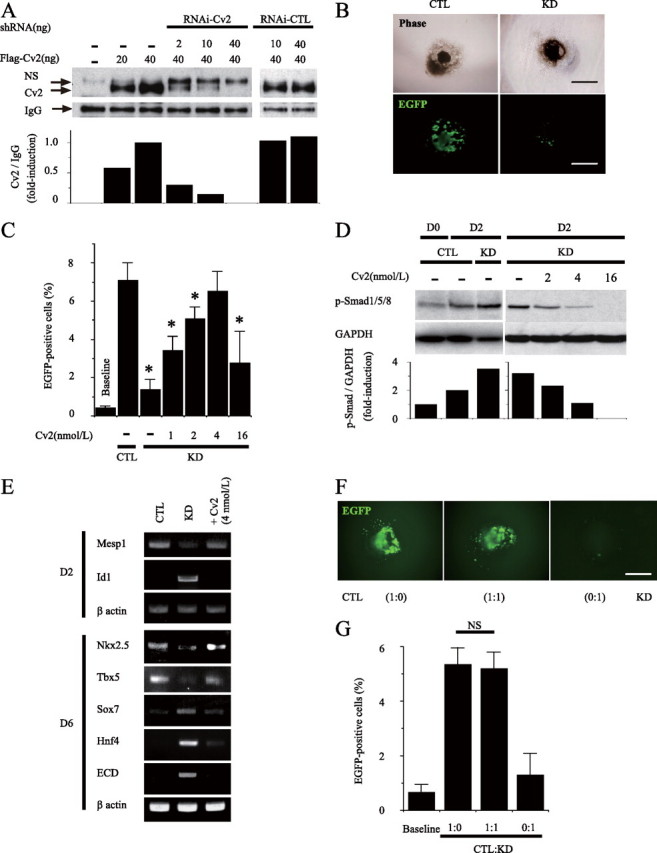
Loss of Cv2 leads to impaired cardiac differentiation that is rescued by addition of Cv2 proteins or co-culture with parental cells. A, representative western bolt shows Cv2 was successful knockdown of Cv2 by RNAi. Densitometry analysis shows significant reduction of secreted Cv2 proteins by RNAi in dose-dependent manner. shRNA, short hairpin RNA. CTL, control;NS, non-specific band. B, Cv2 RNAi clone showed impaired cardiac differentiation at day 6. Scale bars equal 250 μm in panels. C, treatment of KD cells with Cv2 during the first 2 days. Percentage of EGFP-positive cells assessed by FACS at day 7 (n = 3; *, p < 0.05 versus CTL). Baseline means no-treatment control at day 0. D, representative Western blot of pSmad1/5/8 at day 2. Densitometry analysis shows increased pSmad1/5/8 in KD cells more than control cells at day 2 of differentiation. Treatment of KD cells with Cv2 during the first 2 days inhibited pSmad1/5/8. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. E, gene expression analysis by RT-PCR at day 2 and day 6 ECD, E-cadherin. F, representative figures showing that effects of co-culture on cardiomyocyte differentiation at day 6. G, percentage of EGFP-positive cells assessed by FACS at day 7 (n = 3). NS, not significant.
DISCUSSION
There are still many questions about the molecular mechanisms of cardiac induction of mesodermal progenitor cells into the cardiac lineage despite emerging evidence that several cardiac specific transcription factors and growth factors act as key regulators (1-3). A clear understanding about how stem cells differentiate into cardiac cells is essential for the clinical applications of stem cells and would be connected with innovation in novel medical treatments.
Evidence suggests that signaling of BMPs is crucial for regulating cardiac induction, differentiation, and development (2-7). The functional significance of BMP2/4 appears to be unique on the basis of opposite effects on cardiogenesis in the developmental stage-dependent manner, the inhibitory effect at early stage, and the promotive effect at late stage (7). Although the promotive effect of BMP signaling has been studied extensively (2-6), how the inhibitory effect of BMP signaling is regulated remains unclear. In the present study we show that distinct roles of BMP signaling for cardiomyocyte differentiation; BMP2 blocks cardiomyocyte differentiation before up-regulation of cardiac mesoderm markers, whereas BMP2 enhances cardiomyocyte differentiation when cardiac mesoderm was specified. We also show that Cv2, known as a suppressor for BMP signaling, plays a key role in the specification of cardiac cell lineage by inhibiting the anti-cardiogenic effect of BMP signaling at an early stage of cardiomyocyte differentiation.
Cardiac cell lineage is determined in the early developmental stage (such as gastrulation) when endo-, meso-, and epidermal cells differentiate and dynamically move under complex signal networks (31, 33). During the gastrulation stage, BMPs, in concert with BMP-binding proteins, specify mesoderm subdivision by demarcating the spatial extent (13, 14). Our careful stepwise approaches recapitulating endogenous signals in the P19 cells revealed that the increase in BMP activity plays a repressive role for cardiomyocyte differentiation during the first 2 days of differentiation, at which specification of cardiac mesoderm occurs in embryos, evaluated by expression of T/brachyury (30) and Mesp1 (34). Together with findings that up-regulation of Cv2 transcription as well as Bmp transcription was detectable already at day 1 in the P19 system, these results suggest that the interaction between BMP and Cv2 plays a role in the decision of cardiac lineage fate.
We also found strong induction of Id1 expression and blockade of Mesp1 expression by administration of BMP2 during the first 2 days of cardiac differentiation, in concert with increased BMP activity. Because BMP signaling is known to suppress differentiation of stem cells and sustain self-renewal by Id1 (35), a negative regulator of basic helix-loop-helix transcription factors (36), this strong induction of Id1 expression might block cardiac induction by suppressing differentiation itself. Alternatively, with findings that transcription of the basic helix-loop-helix transcription factor Mesp1 but not T was specifically blocked, activation of BMP2 signaling at this timing might induce negative feedback repression of Mesp1 expression by unknown factors, resulting in the blockade of differentiation into cardiac mesoderm from mesendoderm but not into endoderm lineages. In this model the level of BMP activity may be a critical determinant of the effects on cardiomyocyte differentiation, and it is also possible that timing of its signal is critical. Conversely, complete ablation of BMP signal at an early stage of development leads cells to different specification from cardiac lineage; in BMP receptor 1 a null mouse embryos, the mesodermal formation is abolished at the onset of gastrulation, and no heart is formed (8). In our study, the optimized blockade of BMP activity with Cv2 (2 nmol/liter) enhanced cardiac myogenesis by promoting the specification of cardiac mesoderm but not by promoting the induction of undifferentiated mesoderm. Complete blockade of BMP activity by administration of Cv2 (16 nmol/liter) during the first 2 days of differentiation preferentially proceeded to neuronal cell lineage against cardiac cell lineage. These results support the emerging idea that early bursts of Cv2 acts to fine-tune the level of BMP activity to a dose that specifies commitment of cardiac cell fate rather than simply blocking BMPs.
The loss of Cv2 by RNAi inhibited cardiomyocyte differentiation and gave rise to cells destined to the endodermal lineage. In addition, we observed the up-regulation of Id1 followed by predominant expression of endoderm marker but not mesoderm marker. Coupled with findings that administration of Cv2 during the first 2 days of differentiation or co-culture with wild-type cells restored impaired cardiomyocyte differentiation of Cv2-KD cells, these data implicate the functional importance of Cv2 in cardiac fate decision at the early stage of cardiomyocyte differentiation. Cv2 likely inhibits an unnecessary increase in BMP activity in the state of precardiac mesoderm. Currently, the genes that are regulated by this unnecessary BMP activity and may inhibit cardiomyocyte differentiation remain unknown.
Our concept that Cv2 antagonism of anti-cardiogenic BMP signaling at early cardiomyocyte differentiation seems to be similar to the results of recent studies, which found that transient inhibition by noggin significantly promoted cardiogenesis from embryonic stem cells (37). In our study, although we failed to detect transcripts of noggin at early stages of differentiation in the P19 system, biochemical analysis showed that Cv2 as well as noggin acts as an antagonist, at least, suggesting that P19 cells might substitute Cv2 for noggin in this process. Nevertheless, both might have different roles from each other in embryogenesis. Previous studies demonstrated that in vivo pattern of their transcriptions is quite different; noggin transcripts restrict in the node at gastrulation in the mouse embryo (38, 39), and contrary to this, Cv2 transcripts detect in the precardiac mesoderm as well as the posterior primitive streak (16, this study). In addition, based on structural characteristics (16, 19), it is supposed that Cv2 might be a bifunctional modulator of BMP activity. Recent studies have reported that Cv2 can be proteolytically cleaved and that both cleaved and uncleaved Cv2 display similar affinities to BMPs (17). Together with our findings that Cv2 is expressed autologously in the population of cardiac cells and is proteolytically cleaved in concert with cardiac differentiation, these results suggest a spatial and temporal control of Cv2 cleavage might cause context-dependent switching of a BMP antagonist to a BMP agonist at different stages of cardiomyocyte differentiation. Additionally, the functions of the long C terminus module of CV2 are still unknown (16, 19). It is likely that these domains mediate the interaction with other unknown proteins leading to further modification of BMP signals for cardiomyocyte differentiation. Elucidation of their functions may help to explain the roles of Cv2 on the complex anti- and pro-cardiogenic BMP activities in early stage of cardiomyocyte differentiation.
In summary, our results provide the evidence for the unique requirement of Cv2 antagonism of BMP signals at the early stage of cardiomyocyte differentiation in P19 cells. In addition, our results demonstrate that Cv2 acts to fine-tune levels of BMP activity for decision of differential lineages. This study will allow for the innovation of future technologies to develop cardiac cells efficiently from stem cells.
Acknowledgments
We greatly appreciate the gift of the BMP-responsive luciferase reporter plasmid from T. Katagiri (Saitama Medical School). We thank Y. Nakano and M. Kuramoto for expert technical assistance. K. H. is grateful to Christopher C. Hill for critical editing of the manuscript.
This work was supported by grants from Japan Cardiovascular Research Foundation, grants-in-aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, and grants-in-aid from the Ministry of Health, Labor, and Welfare of Japan. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: BMP, bone morphogenetic protein; Cv2, Crossveinless-2; pSmad1/5/8, phosphorylated Smad1/5/8; FACS, fluorescent-assisted cell sorting; Id1, Inhibitor of DNA binding/differentiation 1; KD, knockdown; RNAi, RNA interference; αMHC, α-myosin heavy chain; EGFP, enhanced green fluorescent protein; CK, cytokeratin 18; KD, knockdown; RT, reverse transcription.
References
- 1.Srivastava, D., and Olson, E. (2000) Nature 407 221-226 [DOI] [PubMed] [Google Scholar]
- 2.Brand, T. (2003) Dev. Biol. 258 1-19 [DOI] [PubMed] [Google Scholar]
- 3.Buckingham, M., Meilhac, S., and Zaffran, S. (2005) Nat. Rev. Genet 6 826-835 [DOI] [PubMed] [Google Scholar]
- 4.Lough, J., and Sugi, Y. (2000) Dev. Dyn. 217 327-342 [DOI] [PubMed] [Google Scholar]
- 5.Schultheiss, T. M., Burch, J. B. E., and Lassar, A. B. (1997) Genes Dev. 11 451-462 [DOI] [PubMed] [Google Scholar]
- 6.Andree, B., Duprez, D., Vorbusch, B., Arnold, H., and Brand, T. (1998) Mech. Dev. 70 119-131 [DOI] [PubMed] [Google Scholar]
- 7.Ladd, A. N., Yatskievych, T. A., and Antin, P. B. (1998) Dev. Biol. 204 407-419 [DOI] [PubMed] [Google Scholar]
- 8.Mishina, Y., Suzuki, A., Ueno, N., and Behringer, R. R. (1995) Genes Dev. 9 3027-3037 [DOI] [PubMed] [Google Scholar]
- 9.Monzen, K., Hiroi, Y., Kudoh, S., Akazawa, H., Oka, T., Takimoto, E., Hayashi, D., Hosoda, T., Kawabata, M., Miyazono, K., Ishii, S., Yazaki, Y., Nagai, R., and Komuro, I. (2001) J. Cell Biol. 153 687-698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Wijk, B., Moorman, A. F. M., and van den Hoff, M. J. B. (2007) Cardiovasc. Res. 74 244-255 [DOI] [PubMed] [Google Scholar]
- 11.Zhang, H., and Bradley, A. (1996) Development 122 2977-2986 [DOI] [PubMed] [Google Scholar]
- 12.Hogan, B. L. (1996) Genes Dev. 10 1580-1594 [DOI] [PubMed] [Google Scholar]
- 13.De Robertis, E. M., and Kuroda, H. (2004) Annu. Rev. Cell Dev. Biol. 20 285-308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dosch, R., Gawantka, V., Delius, H., Blumenstock, C., and Niehrs, C. (1997) Development 124 2325-2334 [DOI] [PubMed] [Google Scholar]
- 15.Conley, C. A., Silburn, R., Singer, M. A., Ralston, A., Rohwer-Nutter, D., Olson, D. J., Gelbart, W., and Blair, S. S. (2000) Development 127 3947-3959 [DOI] [PubMed] [Google Scholar]
- 16.Coffinier, C., Ketpura, N., Tran, U., Geissert, D., and De Robertis, E. M. (2002) Mech. Dev. 119 179-184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rentzsch, F., Zhang, J., Kramer, C., Sebald, W., and Hammerschmidt, M. (2006) Development 133 801-811 [DOI] [PubMed] [Google Scholar]
- 18.Ikeya, M., Kawada, M., Kiyonari, H., Sasai, N., Nakao, K., Furuta, Y., and Sasai, Y. (2006) Development 133 4463-4473 [DOI] [PubMed] [Google Scholar]
- 19.Moser, M., Binder, O., Wu, Y., Aitsebaomo, J., Ren, R., Bode, C., Bautch, V. L., Conlon, F. L., and Patterson, C. (2003) Mol. Cell. Biol. 23 5664-5679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Binnerts, M. E., Wen, X., Cante-Barrett, K., Bright, J., Chen, H. T., Asundi, V., Sattari, P., Tang, T., Boyle, B., Funk, W., and Rupp, F. (2004) Biochem. Biophys. Res. Commun. 315 272-280 [DOI] [PubMed] [Google Scholar]
- 21.Coles, E., Christiansen, J., Economou, A., Bronner-Fraser, M., and Wilkinson, D. G. (2004) Development 131 5309-5317 [DOI] [PubMed] [Google Scholar]
- 22.Kamimura, M., Matsumoto, K., Koshiba-Takeuchi, K., and Ogura, T. (2004) Dev. Dyn. 230 434-445 [DOI] [PubMed] [Google Scholar]
- 23.Rudnicki, M. A., and McBurney, M. W. (1987) in Teratocarcinomas and Embryonic Stem Cells; A Practical Approach (Robertson, E. J., ed) pp. 19-49, IRL Press, Oxford
- 24.Takahashi, T., Lord, B., Schulze, C. P., Fryer, R. M., Sarang, S. S., Gullans, S. R., and Lee, R. T. (2003) Circulation 107 1912-1916 [DOI] [PubMed] [Google Scholar]
- 25.Katagiri, T., Imada, M., Yanai, T., Suda, T., Takahashi, N., and Kamijo, R. (2002) Genes Cells 7 949-960 [DOI] [PubMed] [Google Scholar]
- 26.Tada, S., Era, T., Furusawa, C., Sakurai, H., Nishikawa, S., Kinoshita, M., Nakao, K., Chiba, T., and Nishikawa, S. (2005) Development 132 4363-4374 [DOI] [PubMed] [Google Scholar]
- 27.Naito, A. T., Akazawa, H., Takano, H., Minamino, T., Nagai, T., Aburatani, H., and Komuro, I. (2005) Circ. Res. 97 144-151 [DOI] [PubMed] [Google Scholar]
- 28.Stary, M., Pasteiner, W., Summer, A., Hrdina, A., Eger, A., and Weitzer, G. (2005) Exp. Cell Res. 310 331-343 [DOI] [PubMed] [Google Scholar]
- 29.Angello, J. C., Kaestner, S., Welikson, R. E., Buskin, J. N., and Hauschka, S. D. (2006) Dev. Dyn. 235 2122-2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilkinson, D. G., Bhatt, S., and Herrmann, B. G. (1990) Nature 343 657-659 [DOI] [PubMed] [Google Scholar]
- 31.Parameswaran, M., and Tam, P. P. (1995) Dev. Genet. 17 16-28 [DOI] [PubMed] [Google Scholar]
- 32.Coucouvanis, E., and Martin, G. R. (1999) Development 126 535-546 [DOI] [PubMed] [Google Scholar]
- 33.Sater, A. K., and Jacobson, A. G. (1989) Development 105 821-830 [DOI] [PubMed] [Google Scholar]
- 34.Saga, Y., Hata, N., Kobayashi, S., Magnuson, T., Seldin, M. F., and Taketo, M. M. (1996) Development 122 2769-2778 [DOI] [PubMed] [Google Scholar]
- 35.Ying, Q. L., Nichols, J., Chambers, I., and Smith, A. (2003) Cell 115 281-292 [DOI] [PubMed] [Google Scholar]
- 36.Norton, J. D. (2000) J. Cell Sci. 113 3897-3905 [DOI] [PubMed] [Google Scholar]
- 37.Yuasa, S., Itabashi, Y., Koshimizu, U., Tanaka, T., Sugimura, K., Kinoshita, M., Hattori, F., Fukami, S., Shimazaki, T., Ogawa, S., Okano, H., and Fukuda, K. (2005) Nat. Biotechnol. 23 607-611 [DOI] [PubMed] [Google Scholar]
- 38.McMahon, J. A., Takada, S., Zimmerman, L. B., Fan, C. M., Harland, R. M., and McMahon, A. P. (1998) Genes Dev. 12 1438-1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miura, S., Davis, S., Klingensmith, J., and Mishina, Y. (2006) Development 133 3767-3775 [DOI] [PubMed] [Google Scholar]