Abstract
Bone morphogenetic proteins (BMPs) are members of the transforming growth factor-β superfamily of growth factors and are used clinically to induce new bone formation. The purpose of this study was to evaluate receptor utilization by BMP-2, BMP-4, BMP-6, and BMP-7 in primary human mesenchymal stem cells (hMSC), a physiologically relevant cell type that probably mediates the in vivo effects of BMPs. RNA interference-mediated gene knockdown revealed that osteoinductive BMP activities in hMSC are elicited through the type I receptors ACVR1A and BMPR1A and the type II receptors ACVR2A and BMPR2. BMPR1B and ACVR2B were expressed at low levels and were not found to play a significant role in signaling by any of the BMPs evaluated in this study. Type II receptor utilization differed significantly between BMP-2/4 and BMP-6/7. A greater reliance on BMPR2 was observed for BMP-2/4 relative to BMP-6/7, whereas ACVR2A was more critical to signaling by BMP-6/7 than BMP-2/4. Significant differences were also observed for the type I receptors. Although BMP-2/4 used predominantly BMPR1A for signaling, ACVR1A was the preferred type I receptor for BMP-6/7. Signaling by both BMP-2/4 and BMP-6/7 was mediated by homodimers of ACVR1A or BMPR1A. A portion of BMP-2/4 signaling also required concurrent BMPR1A and ACVR1A expression, suggesting that BMP-2/4 signal in part through ACVR1A/BMPR1A heterodimers. The capacity of ACVR1A and BMPR1A to form homodimers and heterodimers was confirmed by bioluminescence resonance energy transfer analyses. These results suggest different mechanisms for BMP-2/4- and BMP-6/7-induced osteoblastic differentiation in primary hMSC.
Bone morphogenetic proteins (BMPs)2 are members of the transforming growth factor-β superfamily of growth factors. BMPs are key regulators of cellular growth and differentiation and regulate tissue formation in both developing and mature organisms. To date, ∼20 unique BMP ligands have been identified and categorized into numerous subclasses based on amino acid sequence similarity (1, 2). BMP-7 (osteogenic protein-1) and BMP-2 are well studied members of this family of growth factors and are now being used clinically to induce new bone formation in spine fusions and long bone nonunion fractures (3, 4). BMP-2 and BMP-7 belong to two closely related BMP subclasses, namely the BMP-2/4 subclass and the BMP-5/6/7 subclass (1). The capacity of ligands from both BMP subclasses to induce osteoblastic differentiation has been rigorously demonstrated (5). However, a thorough understanding of the mechanism through which distinct BMP ligands affect target cells is lacking. Such information is central to realizing the potential of individual BMPs as therapeutic agents and for the rational targeting of a specific BMP to the appropriate clinical indication.
BMP activities are mediated by tetramers of serine/threonine kinase receptors, consisting of two type I and two type II receptors. Three type I receptors (BMPR1A (ALK-3), BMPR1B (ALK-6), and ACVR1A (ALK-2)) and three type II receptors (BMPR2, ACVR2A, and ACVR2B) have been identified (6). Receptor co-patching studies have revealed the presence of both preformed and BMP-induced type I/type II receptor oligomers (7). Binding of BMP ligands to receptor complexes leads to phosphorylation of the type I receptors by constitutively active type II receptors (8). BMP-activated type I receptors phosphorylate intracellular signaling proteins, including the receptor-regulated Smads, Smad-1, -5, and -8 (9), which form heteromeric complexes with the common mediator Smad, Smad-4. Activated Smad complexes then translocate to the nucleus and act as transcription factors to induce the expression of BMP-responsive genes. Other BMP signaling pathways have also been identified and shown to mediate the osteoinductive signals of BMPs. These include the Smad-independent p38 mitogen-activated protein kinase pathway (7, 10) and the phosphatidylinositol 3-kinase/AKT pathway (11).
Several studies have demonstrated that BMP ligands discriminate among individual type I and type II receptors. In COS-7 cells co-transfected with different type I receptor cDNAs and the C. elegans Type II receptor Daf-4, BMP-7 bound more efficiently to ACVR1A and BMPR1A than to BMPR1B, whereas BMP-4 bound only to BMPR1A and BMPR1B (12). In contrast, when co-transfected with ACVR2A or ACVR2B, BMP-7 bound BMPR1B and ACVR1A more efficiently than BMPR1A (13). The association of BMP-7 with ACVR1A in the presence of ACVR2A or ACVR2B was also observed in P19 embryonic carcinoma cells (14). In a similar study, differences in type II receptor affinities were observed between BMP-7 and BMP-4, with BMPR2 binding BMP-7 more effectively than BMP-4 (15).
Mesenchymal stem cells (MSC) are a multipotent cell type that can differentiate down the osteoblastic, chondrogenic, myogenic, or adipogenic lineages. Primary human bone marrow-derived mesenchymal stem cell (hMSC) differentiation is an important model for BMP bioactivity, since it is likely that this cell type contributes to healing and bone formation following the clinical administration of BMPs. The present study was designed to evaluate BMP receptor utilization by osteoinductive BMPs, including BMP-2, BMP-4, BMP-6, and BMP-7, during the osteoblastic differentiation of primary hMSC. A model was developed to systematically knock down all type I or type II BMP receptors, alone or in combination, to elucidate receptor utilization by each ligand. The results obtained reveal significant differences in type I and type II receptor usage among the four BMPs evaluated. Interestingly, this work also suggests distinct type I receptor dimerization patterns within receptor complexes utilized by BMP-2/4 and BMP-5/6/7 subclass members. The model employed in these studies could be broadly applied to better understand BMP signaling potentials in other clinically relevant cell and tissue types.
EXPERIMENTAL PROCEDURES
Cell Culture and Culture Media—Primary hMSC and hMSC culture media, including mesenchymal stem cell growth medium (MSCGM) and osteogenic differentiation medium (ODM), were purchased from Lonza (Walkersville, MD). Cells were expanded in vitro and used for experimentation within five passages of the initial thaw. HEK-293 cells were purchased from ATCC (Manassas, VA) and cultured in minimal essential medium (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum, 1.5 g/liter NaHCO3, 2 mm l-glutamine, 1 mm sodium pyruvate and penicillin/streptomycin.
BMP Treatment—Recombinant BMP-2, BMP-6, and BMP-7 were produced in Chinese hamster ovary cells and are available at Stryker Biotech (16). BMP-4 was obtained from R&D Systems (Minneapolis, MN). ODM was prepared according to the manufacturer's instructions using the provided supplements of ascorbic acid and β-glycerophosphate but excluding the dexamethasone. Unless stated otherwise, the concentration of fetal bovine serum in ODM was ∼10%. BMPs were diluted in ODM to the indicated concentrations.
Alizarin Red Staining—hMSC were seeded into 48-well dishes at 1.0 × 104 cells/well in MSCGM. Twenty-four hours later, MSCGM was replaced with ODM alone or ODM containing the indicated concentration of BMP. Media changes were performed every 3–4 days. Alizarin red staining was performed on day 17 using an osteogenesis quantitation kit (Chemicon International, Temecula, CA) according to the manufacturer's instructions.
Quantification of Gene Expression—RNA was isolated using the TurboCapture 96 mRNA Kit (Qiagen, Valencia, CA) according to the manufacturer's protocol. Reverse transcription was performed using 40 units of Moloney murine leukemia virus reverse transcriptase (Promega, Madison, WI) in a buffer containing 20 mm Tris-HCl, 50 mm KCl, 5 mm MgCl2, 500 μm each dNTP (Invitrogen, Carlsbad, CA), and 5 ng/μl random primers (Promega, Madison, WI). Reverse transcription was carried out at 23 °C for 10 min and 42 °C for 50 min followed by a 5-min inactivation step at 85 °C. All reagents and instrumentation for gene expression analysis were obtained from Applied Biosystems (Foster City, CA). Quantitative PCR was carried out using a 7900HT fast real time PCR system and predesigned TaqMan gene expression assays according to the manufacturer's specifications. Reference numbers for assays used in this study are as follows: GAPDH (Hs99999905_m1), cyclophilin (Hs99999904_ m1), BMPR1A (Hs00831730_s1), BMPR1B (Hs00176144_m1), BMPR2 (Hs00176148_m1), ACVR1A (Hs00153836_m1), ACVR2A (Hs00155658_m1), ACVR2B (Hs00609603_m1), ID-1 (Hs00357821_g1), NOGGIN (Hs00271352_s1), PTHR1 (Hs00174895_m1), IBSP (bone sialoprotein) (Hs00173720_m1), and DLX-5 (Hs00193291_m1).
The analysis of osteoblast marker gene expression and siRNA-mediated receptor knockdown was performed using the standard curve method of relative quantification, according to the procedure recommended by Applied Biosystems. The analysis of BMP receptor expression in hMSC and tissue cDNAs was performed using the absolute standard curve method. Briefly, DNA plasmids containing the human sequences of each BMP receptor, GAPDH or cyclophilin, were used as templates in PCRs to amplify target DNA for standard curve preparation. TrueClone cDNAs encoding BMPR1A (accession number NM_004329.2), BMPR1B (accession number NM_001203.1), BMPR2 (accession number NM_ 001204.5), GAPDH (accession number NM_002046.3), and Cyclophilin A (accession number NM_021130.3) were obtained from OriGene (Rockville, MD). cDNAs for ACVR1A (accession number NM_001105), ACVR2A (accession number NM_001616.3), and ACVR2B (accession number NM_ 001106.3) were obtained from GenScript (Piscataway, NJ). DNA primers (IDT, Coralville, IA) were designed to flank the relevant TaqMan amplicon. Primer sequences are shown in supplemental Table 1. 10 ng of each DNA were exposed to 25 cycles of PCR according to the following thermal profile: denaturation at 95 °C for 20 s, annealing at 55 °C for 10 s, and extension at 70 °C for 15 s. PCR products were gel-purified using a Qiaquick gel extraction kit (Qiagen, Valencia, CA) and quantified by spectrophotometry. Standard curves were run using serial 1:10 dilutions of target DNA from 3 × 106 to 30 copies. The number of expressed molecules of each target gene in experimental samples was quantified against the appropriate standard curve and normalized to an arbitrary copy number (1000) of either GAPDH or cyclophilin from the same sample.
Transient Gene Knockdown—Stealth RNAi DuoPaks (Invitrogen) containing two unique, prevalidated siRNA sequences per gene, were used to target the type I BMP receptors ACVR1A, BMPR1A, and BMPR1B and the type II receptors BMPR2, ACVR2A, and ACVR2B. Two Stealth RNAi negative controls (LO and Medium GC content) were utilized as controls to confirm the specificity of each targeted knockdown. Phenotypic results for ACVR1A and ACVR2A were additionally confirmed with a third pre-designed siRNA (Dharmacon, Lafayette, CO), using a chemistry-matched negative control from the same vendor.
hMSC were transfected with siRNA using a Nucleofector II (Amaxa Biosystems, Gaithersburg, MD) and employing the manufacturer's hMSC kit. A total of 6 μg of siRNA was delivered to 5 × 105 hMSC, and transfected cells were cultured for 48 h in MSCGM to allow down-regulation of gene targets. Cells were then stimulated with 100 ng/ml of either BMP-2, -4, -6, or -7 for 24 h in MSCGM with 0.2% FBS or 96 h in ODM with 5% fetal bovine serum. BMP receptor and osteoblastic marker gene expression was measured by qPCR. To quantify the phenotypic effect of receptor knockdown, ID-1, IBSP, NOGGIN, and DLX-5 qPCR data were expressed as the percentage of inhibition of BMP-mediated induction of each gene in the targeted siRNA treatments relative to the control siRNA treatments, according to Equation 1,
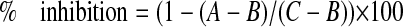 |
(Eq. 1) |
where A represents the quantity of ID-1 mRNA expression following receptor knockdown and BMP treatment, B represents the base line quantity of ID-1 mRNA expression without BMP treatment, and C represents the quantity of ID-1 mRNA expression following control transfection and BMP treatment.
Experiments were performed on three separate occasions using hMSC from multiple donors. The average treatment values from the three replicate experiments were analyzed by two-sample t-tests and two-way analysis of variance using a Bonferroni adjustment for multiple comparisons.
Generation of Renilla Luciferase (Rluc) and Green Fluorescent Protein (GFP)2 Fusion Expression Constructs—TrueClone cDNA encoding FGFR1 (accession number NM_023106.1) was obtained from OriGene (Rockville, MD). cDNAs for ACVR1A and BMPR1A were obtained as described above. Using these constructs as template, the open reading frame corresponding to each gene was amplified by PCR using DNA primers (IDT, Coralville, IA) containing an appropriate restriction site for subsequent cloning into pRluc-N1 and pGFP2-N1 fusion protein expression vectors (PerkinElmer Life Sciences). Primer sequences are shown in supplemental Table 2.
PCR products were digested with the appropriate restriction enzymes, gel-purified, and ligated in frame into pRluc-N1 and pGFP2-N1. In brief, pRluc-N1 and pGFP2-N1 were restriction-digested and dephosphorylated using Antarctic phosphatase (New England Biolabs, Ipswich, MA). PCR products were cloned at the amino terminus of Rluc or GFP2, such that the expressed proteins were tagged with Rluc or GFP2 at the carboxyl terminus. Untagged expression constructs for each receptor were then created by incorporating a stop codon at the end of the receptor coding sequence. Recombinant constructs were transformed into One Shot TOP10 E. coli (Invitrogen) and plated on agar containing kanamycin (pRluc vectors) or zeocin (pGFP2 vectors). Plasmid DNA was prepared using the EndoFree Plasmid Maxi kit (Qiagen, Valencia, CA). All recombinant constructs were verified by sequencing the full-length open reading frames.
Bioluminescence Resonance Energy Transfer (BRET)2 Assays—Unless stated otherwise, all reagents, materials, and instrumentation used in the BRET2 assay were purchased from PerkinElmer Life Sciences. HEK-293 cells were seeded into 6-well dishes at 6 × 105 cells/well. The following day, cells were co-transfected with a total of 2 μg of DNA consisting of the indicated amount of recombinant receptor fusion DNA constructs together with pcDNA as a filler. Transfections were carried out using 6 μl of FuGENE HD (Roche Applied Science). After 48 h, cells were detached using phosphate-buffered saline glucose (Invitrogen), centrifuged, and resuspended in phosphate-buffered saline with glucose. Cells were then distributed in parallel to 96-well white or black CulturPlates for analysis of luminescence and fluorescence emissions, respectively. BRET2 assays were performed using a VICTOR Light luminescence counter. Briefly, DeepBlueC substrate was added by autoinjection to a final concentration of 5 μm, and luminescence was read immediately at 515/30 nm and 410/80 nm. The BRET2 ratio was calculated as the difference of emission at 515 nm/410 nm between the co-transfected Rluc and GFP2 fusion proteins and the Rluc fusion protein alone (17). Results were expressed in milli-BRET2 units, where 1 milli-BRET2 unit corresponds to the BRET2 ratio values multiplied by 1000.
For saturation assays, cells were transfected with 10 ng of the donor Rluc receptor fusion construct and increasing quantities (from 50 to 1600 ng) of the acceptor GFP2-receptor fusion construct. GFP2 expression was quantified using a SpectraMax M5 multimode reader (Molecular Devices, Sunnyvale, CA). Data were analyzed by nonlinear regression, and BRET2 ratio values were plotted against the ratio of GFP2 DNA to Rluc DNA. For competition assays, cells were transfected with 10 ng of the donor Rluc-receptor fusion construct, 400 ng of the acceptor GFP2-receptor fusion construct, and 0, 800, or 1600 ng of the competitor receptor construct. Competitor receptors consisted of untagged BMP receptors or FGFR1. For competition assays, treatment averages were compared with the control using two-sample t tests and one-way analysis of variance, applying Dunnett's method for multiple comparisons.
RESULTS
BMP-2, BMP-4, BMP-6, and BMP-7 Induce Osteoblastic Differentiation in Primary hMSC—The ability of BMP-2, BMP-4, BMP-6, and BMP-7 to drive the osteoblastic differentiation of primary hMSC was compared at both the molecular and cellular level. The expression of six osteoblast marker genes was evaluated by qPCR at either 24 or 72 h, following treatment with the four osteoinductive BMPs (Fig. 1A). Expression of the genes ID-1, DLX-5, and NOGGIN was up-regulated to equivalent levels over the controls by all four BMPs within 24 h of treatment. Likewise, the expression of AP (alkaline phosphatase), PTHR1 (parathyroid hormone receptor 1), and IBSP was increased after 3 days of treatment by all four ligands. The observed gene regulation is in keeping with the reported expression of osteoblast-associated genes during in vitro osteoblastic differentiation (18–22) and suggests that all four BMPs induce a similar gene expression response in hMSC.
FIGURE 1.

BMP-2, BMP-4, BMP-6, and BMP-7 induce osteoblastic differentiation in primary hMSC. A, primary hMSC were seeded in 48-well dishes at 1.5 × 104 cells/well and cultured in MSCGM (No Trt), ODM alone, or ODM supplemented with 400 ng/ml BMP-2, BMP-4, BMP-6, or BMP-7. Cells were lysed after 24 h of treatment to assess ID-1, DLX-5, or NOGGIN gene expression (left), and after 3 days to assess AP, PTHR1, and IBSP gene expression (right). Levels of gene expression were quantified by RT-qPCR (RQ) with normalization to GAPDH. Values shown represent the mean ± S.D. of triplicate measurements and are expressed relative to treatment with ODM alone. B, primary hMSC were seeded in 48-well dishes at 1.0 × 104 cells/well and treated with ODM alone or ODM supplemented with 100, 200, or 500 ng/ml BMP-2, BMP-4, BMP-6, or BMP-7 for 17 days. Cells were stained with Alizarin red to detect calcium deposition.
We next evaluated the capacity of the four BMPs to induce matrix mineralization. Calcium deposits were detected in all BMP treatments by Alizarin red staining (Fig. 1B) but not in hMSC cultured in ODM alone. The observed mineralization was BMP dose-dependent. Collectively, these results demonstrate that all four BMPs induce robust osteoblastic differentiation of primary hMSC and, further, that the quality and magnitude of this differentiation is similar among all four ligands. A systematic investigation of receptor utilization in hMSC was next undertaken to determine whether the four BMPs were exerting their osteoinductive activities via the same or different cellular receptors.
Characterization of BMP Receptor Expression Profiles in Primary hMSC and Human Tissue cDNAs— We first assessed the expression of six known BMP receptors in primary hMSC from three separate donors. The copy number of expressed mRNA encoding each receptor was quantified and normalized to GAPDH. Although some variability was observed among donors, the relative expression levels of the six receptors were comparable (Fig. 2A). ACVR1A and BMPR2 were the most abundantly expressed type I and type II receptors, respectively, with normalized expression levels ranging from ∼29 to 67 copies for ACVR1A and ∼24 to 44 copies for BMPR2. BMPR1A and ACVR2A were expressed at intermediate levels, with normalized levels ranging from ∼15 to 20 copies for BMPR1A and ∼7 to 10 copies for ACVR2A. Expression of BMPR1B and ACVR2B mRNA was consistently the lowest of the six BMP receptors, with levels ranging from 1 to 2 copies for BMPR1B and <1 copy for ACVR2B.
FIGURE 2.

Characterization of BMP receptor expression profiles in primary hMSC and human tissue cDNAs. A–C, expression of BMP receptors and endogenous control mRNA was measured by RT-qPCR using a standard curve prepared with known quantities of the appropriate target DNA. Values shown represent the mean ± S.D. of triplicate measurements. A, receptor expression levels in untreated hMSC from three donors. Values are expressed as the number of copies of target mRNA per 1000 copies of GAPDH. B and C, expression of type I (B) and type II (C) BMP receptors in 10 human tissue cDNAs. Values are expressed as the number of copies of target mRNA per 1000 copies of cyclophilin.
We then evaluated the BMP receptor expression profile in 10 human tissue cDNA preparations. For each tissue cDNA, the mRNA copy number for each receptor was quantified and normalized to cyclophilin. ACVR1A was the most abundantly expressed type I receptor in the majority of tissues, including brain, heart, kidney, liver, lung, and ovary (Fig. 2B). The normalized ACVR1A copy number ranged from <1 in testis to 38 in skeletal muscle. BMPR1A tended to be more abundantly expressed than BMPR1B, with copy numbers ranging from <1 to 41 and from <1 to 11, respectively, across tissue types. Tissues with notable patterns of type I receptor expression include skeletal muscle, with highly abundant ACVR1A and BMPR1A expression, as well as prostate and spinal cord, which both demonstrated roughly equal expression of all three type I receptors.
Either ACVR2B or BMPR2 was the most abundantly expressed type II receptor in all tissues, with normalized mRNA copy numbers ranging from ∼5 in lung to 82 in skeletal muscle for ACVR2B and from 4 in liver to 201 in lung for BMPR2 (Fig. 2C). The expression level of BMPR2 in lung was the highest among the tissues tested. Skeletal muscle was notable for an unusual receptor expression pattern, with a high level of expression of both BMPR2 and ACVR2B. ACVR2A exhibited a low level of expression in most tissues, with copy numbers ranging from 3 to 21 copies. These data reveal significant variability among human tissues with regard to overall expression levels of each receptor as well as the relative BMP receptor expression levels within each tissue. This diversity of receptor expression patterns could provide some insight into the broad array of bioactivities reported for BMP ligands.
hMSC BMP Receptors Are Specifically and Potently Inhibited by Nucleoporation of Targeted siRNA—siRNAs targeting the six BMP receptors were delivered to hMSC by nucleoporation to down-regulate receptor expression prior to BMP treatment. Receptors were knocked down both individually and in all possible combinations of type II (Fig. 3A) or type I (Fig. 3B) receptors. Receptor knockdown was confirmed by qPCR at the time of phenotypic analysis, typically 72 h postnucleofection. Target knockdown for ACVR1A, BMPR1A, ACVR2A, and BMPR2 ranged from ∼70 to 99% in all experiments relative to control nucleoporations. Knockdown for BMPR1B and ACVR2B ranged from 65 to 83% and from 30 to 77%, respectively, in one experiment. Knockdown overall for both of these low abundance receptors was probably higher, since mRNA expression following targeted nucleoporation fell below quantifiable levels in the remaining two of three experiments. In most instances, nontargeted receptors were not affected by the inhibition of other receptors. However, BMPR1B mRNA appeared to be consistently up-regulated following the knockdown of the other type I receptors (Fig. 3B). It is possible that the observed up-regulation reflects a positive feedback response representing compensation for the loss of other type I receptors.
FIGURE 3.
BMP type I and type II receptors are potently inhibited by nucleoporation of targeted siRNA. A and B, primary hMSC were nucleoporated with a total of 6 μg of siRNA targeting each of the six receptors individually or as all possible combinations of type II (A) or type I (B) receptors or control siRNA. Target mRNA for each of the receptors was measured by RT-qPCR (RQ) 3 days after nucleoporation, at the time of phenotypic analysis. Values are shown relative to the quantity of receptor expression in control nucleoporations. Control values were set to 1 by dividing all nucleoporation replicates in the qPCR analysis of each receptor by the mean value of the control treatment. Values represent the mean ± S.D. of triplicate measurements of ODM-treated cells from one representative experiment. +, the targeted siRNA was included in the nucleoporation treatment; -, the targeted siRNA was omitted from the nucleoporation.
BMP-2/4 and BMP-6/7 Differentially Utilize BMP Receptors to Stimulate Osteoblastic Differentiation of hMSC—The effect of knocking down type I or type II receptors on signaling by BMP-2, BMP-4, BMP-6, or BMP-7 was initially assessed using ID-1 mRNA expression as a primary end point marker (Fig. 4). The concurrent knockdown of the type II receptors ACVR2A, ACVR2B, and BMPR2 completely blocked the capacity of all four BMPs (p < 0.001) to induce ID-1 (Fig. 4A). Similarly, the simultaneous nucleoporation of siRNA targeting ACVR1A, BMPR1A, and BMPR1B inhibited ID-1 induction by 98–100% (p < 0.001) following treatment with all four BMPs (Fig. 4B). These results indicate that no other endogenous type I or type II receptors are able to mediate BMP signaling in primary hMSC in the absence of the receptors targeted in this study. Interestingly, the phenotypic effects on signaling by BMP-2 and BMP-4 were strikingly similar within each of the receptor knockdown treatments. Likewise, the phenotypic effects of receptor knockdown were similar for BMP-6 and BMP-7. Collectively, these data suggest that although all four BMPs utilize the same limited set of BMP receptors, the degree of reliance on each individual receptor differs between BMP-2/4 and BMP-6/7.
FIGURE 4.

Receptor down-regulation in hMSC differentially inhibits ID-1 induction by the BMP-2/4 and BMP-6/7 subfamilies. A and B, primary hMSC were nucleoporated with a total of 6 μg of siRNA targeting each of the six BMP receptors individually or as all possible combinations of type II (A) or type I (B) receptors or control siRNA. 48 h after nucleoporation, cells were treated with MSCGM alone or MSCGM containing 100 ng/ml BMP-2, BMP-4, BMP-6, or BMP-7 for 24 h, and expression of ID-1 mRNA was quantified by RT-qPCR and normalized to GAPDH. Data are expressed as the percentage inhibition of ID-1 induction for each receptor knockdown treatment relative to the control transfection as described under “Experimental Procedures.” Values shown represent the mean ± S.D. of three replicate experiments performed on different days using hMSC from multiple donors. Knockdown treatments that were found to be significantly different from the control transfections are indicated with an asterisk, with p values provided under “Results.” Within receptor knockdown treatments, significant differences between the BMP-2/4 and BMP-6/7 subclasses are indicated by brackets, with p values provided under “Results.”
Pronounced differences in type II receptor utilization were observed (Fig. 4A). Knockdown of ACVR2A alone significantly inhibited signaling by BMP-6 and BMP-7 by ∼27% (p = 0.009) and 36% (p = 0.002), respectively, but did not significantly affect signaling by BMP-2 and BMP-4. The difference between BMP-2/4 and BMP-6/7 was statistically significant (p < 0.001), indicating a greater reliance on ACVR2A for signaling by BMP-6/7 versus BMP-2/4. In contrast, knockdown of BMPR2 resulted in an approximately 78% (p < 0.001) and 84% (p < 0.001) inhibition of ID-1 induction following treatment with BMP-2 and BMP-4, respectively, versus 53% (p < 0.001) and 62% (p < 0.001) following treatment with BMP-6 and BMP7, respectively. The difference between BMP-2/4 and BMP-6/7 was statistically significant (p < 0.001), indicating a greater reliance on BMPR2 for signaling by BMP-2/4 versus BMP-6/7. No significant effect of ACVR2B knockdown was observed for any of the BMPs investigated. Phenotypic data for BMPR2 and ACVR2B were confirmed using two unique siRNA sequences, and phenotypic data for ACVR2A were confirmed using three unique siRNA sequences (data not shown). Taken together, these data indicate that ACVR2A and BMPR2 are the primary type II receptors mediating the signaling of all four BMPs in hMSC. Interestingly, the complete loss of signaling for all four BMPs, observed when ACVR2A and BMPR2 were simultaneously knocked down, represented a greater than additive effect of knocking down ACVR2A and BMPR2 individually. These data suggest that BMPR2 and ACVR2A have the capacity to partially compensate for one another during BMP signaling.
The relative contribution of each type I receptor to signaling also differed considerably between BMP-2/4 and BMP-6/7 (Fig. 4B). Down-regulation of ACVR1A alone resulted in partial inhibition of signaling by all four BMPs. However, the magnitude of this inhibition was significantly different (p = 0.002) between the two subclasses. When ACVR1A was knocked down, an approximately 59–60% inhibition of signaling for BMP-2 (p < 0.001) and BMP-4 (p < 0.001) was observed relative to the control transfection versus 76–77% for BMP-6 (p < 0.001) and BMP-7 (p < 0.001), indicating a greater reliance on ACVR1A for signaling by BMP-6/7 compared with BMP-2/4. In contrast, knocking down BMPR1A alone resulted in a marked inhibition of signaling by BMP-2 (87%, p < 0.001) and BMP-4 (90%, p < 0.001) but had only a marginal effect on ID-1 induction following treatment with BMP-6 (28%, p < 0.005) or BMP-7 (18%, p = 0.552). The effect for BMP-7 did not reach statistical significance. These data demonstrate that, in human bone marrow MSC, BMPR1A is utilized principally by BMP-2/4 and only minimally by BMP-6/7. No significant effect of BMPR1B knockdown was observed for any of the BMPs. The effect of BMPR1A and BMPR1B knockdown on BMP-induced ID-1 expression was confirmed using two unique siRNA sequences, and phenotypic read-outs following ACVR1A knockdown were confirmed using three unique siRNA sequences (data not shown). Results were also confirmed 4 days after BMP treatment using the later stage osteoblastic marker genes IBSP, NOGGIN, and DLX-5 as phenotypic readouts (Fig. 5).
FIGURE 5.

Type I receptor knockdown phenotypes persist into later stages of osteoblast differentiation and affect multiple downstream genes. Primary hMSC were nucleoporated with a total of 6 μg of siRNA targeting ACVR1A and BMPR1A alone or in combination or control siRNA. 48 h after nucleoporation, cells were treated with ODM alone or ODM supplemented with 100 ng/ml BMP-2, BMP-4, BMP-6, or BMP-7. Expression of ID-1, IBSP, NOGGIN, and DLX-5 mRNA after 4 days of treatment was quantified by RT-qPCR and normalized to GAPDH. Data are expressed as the percentage inhibition of target gene induction for each receptor knockdown treatment relative to the control transfection as described under “Experimental Procedures.” Values shown represent the mean ± S.D. of triplicate measurements from one representative experiment. +, the targeted siRNA was included in the nucleoporation treatment; -, the targeted siRNA was omitted from the nucleoporation.
BMP-6 and BMP-7 Utilize Homodimers of Type I BMP Receptors, whereas BMP-2 and BMP-4 Utilize Homodimers and Heterodimers of Type I BMP Receptors—For BMP-6 and BMP-7, the simultaneous knockdown of BMPR1A/BMPR1B resulted in 24–28% inhibition of signaling, suggesting that the remaining 72–76% of signaling might be mediated by homodimers of ACVR1A, the only type I receptor expressed under these conditions (Fig. 4B). This was confirmed by the fact that ACVR1A knockdown caused a 76–77% reduction in ID-1 induction following treatment with BMP-6 or BMP-7. Similarly, the simultaneous knockdown of ACVR1A/BMPR1B resulted in a 75–82% reduction of signaling for BMP-6 and BMP-7, suggesting that the remaining 18–25% of signaling is mediated by homodimers of BMPR1A, the only type I receptor expressed in this treatment. This was confirmed by the fact that BMPR1A knockdown resulted in a 18–28% reduction in ID-1 induction after signaling by BMP-6 and BMP-7. These data indicate that BMP-6 and BMP-7 utilize predominantly type I receptor homodimers for signaling in hMSC, with ACVR1A homodimers mediating the majority of signaling (∼76%) and BMPR1A homodimers mediating ∼18–25% (Fig. 6A).
FIGURE 6.
Proposed model of type I receptor utilization by the BMP-2/4 and BMP-6/7 subfamilies in primary hMSC. A, BMP-6- and BMP-7-induced osteoblastic differentiation in hMSC is mediated by homodimers of ACVR1A (72–76%) and homodimers of BMPR1A (18–25%). B, BMP-2- and BMP-4-induced osteoblastic differentiation in hMSC is mediated by homodimers of ACVR1A (13–14%), homodimers of BMPR1A (36–40%), and heterodimers of BMPR1A/ACVR1A (46–51%).
In contrast, homodimers of ACVR1A and BMPR1A do not appear to be sufficient to mediate the full signaling potential of BMP-2 and BMP-4. For BMP-2 and BMP-4, the simultaneous knockdown of BMPR1A/BMPR1B resulted in 86–87% inhibition of signaling, suggesting that the remaining 13–14% is mediated by ACVR1A homodimers (Fig. 4B). Interestingly, knocking down ACVR1A alone resulted in a 59–60% inhibition of signaling, a significantly higher percentage than what was found to be mediated by ACVR1A homodimers. Likewise, the simultaneous knockdown of ACVR1A/BMPR1B resulted in 60–64% inhibition of BMP-2/4 signaling, suggesting that the remaining 36–40% of signaling is mediated by BMPR1A homodimers. Knocking down BMPR1A alone resulted in an 87–90% inhibition of signaling for BMP-2 and BMP-4, a much greater inhibitory effect than would be predicted for the loss of BMPR1A homodimers. These data suggest that only a portion of BMP-2 and BMP-4 signaling can be mediated by homodimers of ACVR1A (13–14%) and BMPR1A (36–40%). The remaining portion of the BMP-2 and BMP-4 signaling potential requires that both ACVR1A and BMPR1A be expressed concurrently by hMSC. We hypothesized therefore that a percentage of BMP-2 and BMP-4 signaling in hMSC might require a heterodimer of ACVR1A and BMPR1A (Fig. 6B).
BMPR1A and ACVR1A Interact to Form Type I Receptor Homodimers and Heterodimers—In order to test this hypothesis, we employed the BRET2 technology to determine whether ACVR1A and BMPR1A heterodimerize in live cells. A BRET2 signal is produced when an Rluc fusion protein comes into close proximity, generally less than 100 Å, with a GFP2 fusion protein and an energy transfer from luciferase to GFP2 takes place (17, 23). BRET technologies have been applied in recent years to effectively characterize the homo- and hetero-oligomerization of TβRII variants (24) and β-arrestins (25), as well as interactions between TβRII with αvβ3 integrins (26).
Full-length ACVR1A, BMPR1A, or FGFR1 were expressed in HEK-293 cells as fusion proteins with either Rluc (ACVR1A-Rluc, BMPR1A-Rluc, FGFR1-Rluc) or GFP2 (ACVR1A-GFP2, BMPR1A-GFP2, FGFR1-GFP2). Expression from all Rluc and GFP2 constructs was confirmed by assessing luciferase activity or fluorescence, respectively. In addition, receptor-GFP2 fusion proteins were localized to the plasma membrane using fluorescence microscopy (data not shown).
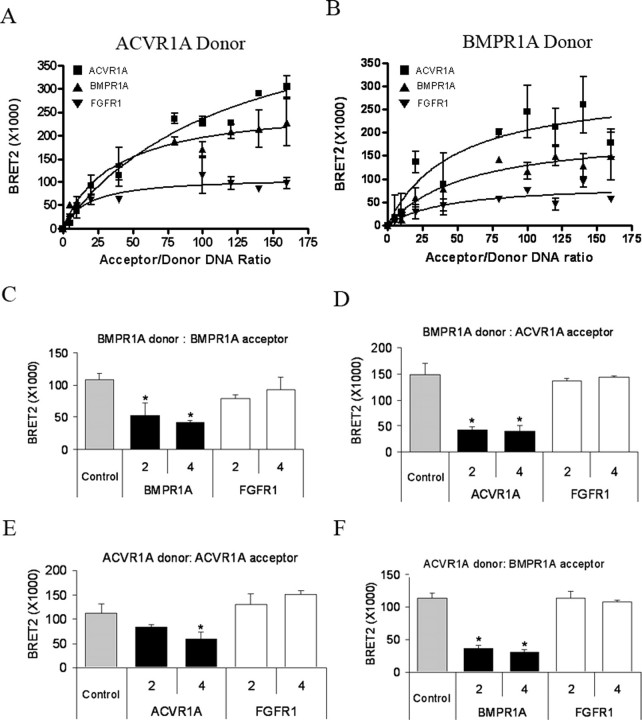
Type I receptor interactions were initially evaluated in BRET2 saturation assays using ACVR1A-Rluc as donor (Fig. 7A). A maximal BRET2 signal was obtained by co-transfection of ACVR1A-Rluc and ACVR1A-GFP2. A robust BRET2 signal was also observed following co-transfection of ACVR1A-Rluc with BMPR1A-GFP2. These data demonstrate that ACVR1A forms homodimers with other ACVR1A monomers and heterodimers with BMPR1A. No significant association of ACVR1A-Rluc with the negative control FGFR1-GFP2 was observed. The greater magnitude of the BRET2 signal obtained for the ACVR1A homodimer, relative to the ACVR1A/BMPR1A heterodimer, suggests that either ACVR1A homodimers are formed with greater frequency than ACVR1A/BMPR1A heterodimers or that homodimerization of ACVR1A-Rluc/ACVR1A-GFP2 brings the Rluc and GFP2 tags into closer proximity than does the heterodimerization of ACVR1A-Rluc/BMPR1A-GFP2, allowing for a more efficient energy transfer from Rluc to GFP2.
FIGURE 7.
Constitutive homo- and heterodimerization of BMPR1A and ACVR1A is detected using BRET2 technology. A–F, constitutive interactions between BMPR1A and ACVR1A monomers were evaluated using BRET2. HEK-293 cells were plated in 6-well dishes at 6 × 105 cells/well. The following day, cells were co-transfected with 2 μg of DNA, consisting of the indicated amount of recombinant receptor fusion DNA constructs together with pcDNA as filler. Luminescence, fluorescence, and BRET2 values were determined as described under “Experimental Procedures.” A and B, saturation curves were generated by co-transfecting a fixed amount (10 ng) of ACVR1A-Rluc donor (A) or BMPR1A-Rluc (B) with increasing quantities (from 50 to 1600 ng) of acceptor, either ACVR1A-GFP2, BMPR1A-GFP2, or FGFR1-GFP2. Data were analyzed by nonlinear regression, and BRET2 ratio values were plotted against the ratio of GFP2 to Rluc DNA. Values shown represent the mean ± S.D. of triplicate measurements from one representative experiment. C–F, competition assays were performed by co-transfecting either 10 ng of BMPR1A-Rluc donor with 400 ng of BMPR1A-GFP2 acceptor (C), 10 ng of BMPR1A-Rluc donor with 400 ng of ACVR1A-GFP2 acceptor (D), 10 ng of ACVR1A-Rluc donor with 400 ng of ACVR1A-GFP2 acceptor (E), or 10 ng of ACVR1A-Rluc donor with 400 ng of BMPR1A-GFP2 acceptor (F), witha0-(Control) or 2- or 4-fold excess of relevant unlabeled receptor or FGFR1 as competitor. Values shown represent the mean BRET2 signal ± S.D. of triplicate measurements from one representative experiment. BRET2 values that were statistically different from the control are indicated with an asterisk; p < 0.05.
Saturation studies were repeated in the reverse configuration, using BMPR1A-Rluc as donor (Fig. 7B). In this series of experiments, co-transfection of ACVR1A-GFP2 as acceptor resulted in the strongest BRET2 signal, followed by that of BMPR1A-GFP2. These data demonstrate that BMPR1A forms homodimers and heterodimers with ACVR1A. No significant association of BMPR1A-Rluc with the control FGFR1-GFP2 was observed. The relative strength of the BRET2 signals obtained in this study might indicate that BMPR1A/ACVR1A heterodimers form with greater frequency than BMPR1A homodimers. A similar finding was observed for the murine transcript variant TβRIIB, which associates with TβRII more readily than with other TβRIIB monomers (24).
BRET2 competition assays were then performed to further verify the specificity of type I homodimer and heterodimer formation. Co-transfection of BMPR1A-Rluc/BMPR1A-GFP2 with a 2- or 4-fold excess of unlabeled BMPR1A (Fig. 7C) and co-transfection of BMPR1A-Rluc/ACVR1A-GFP2 with an excess of unlabeled ACVR1A (Fig. 7D) both led to significant reductions in the observed BRET2 signal. Co-transfection with unlabeled FGFR1 competitor had no effect on the BRET2 signal in either case. Similarly, co-transfection of ACVR1A-Rluc/ACVR1A-GFP2 (Fig. 7E) or ACVR1A-Rluc/BMPR1A-GFP2 (Fig. 7F) with an excess of unlabeled ACVR1A or BMPR1A, respectively, markedly reduced the observed BRET2 signal, whereas transfection with the unlabeled FGFR1 had no effect. These data demonstrate specificity in the formation of ACVR1A and BMPR1A homodimers and also in the formation of type I heterodimers composed of BMPR1A and ACVR1A.
DISCUSSION
The objective of this study was to evaluate BMP receptor utilization during the initiation of osteoblastic differentiation in primary hMSC. RNAi-mediated receptor knockdown revealed that the osteoinductive activities of BMPs in primary hMSC are elicited through the type I receptors ACVR1A and BMPR1A and the type II receptors ACVR2A and BMPR2. BMPR1B and ACVR2B did not contribute significantly to BMP-mediated osteoblastic differentiation in this system, possibly due to the low level of expression of these receptors.
The relative importance of the BMP receptors might be shifted in other precursor cell types, should a different representation of BMP receptors be present. Indeed, in murine adipose-derived adult stromal cells, BMPR1B is expressed at levels equivalent to those of BMPR1A and BMPR2 and becomes significantly up-regulated during osteogenic differentiation (27). Progression down the osteoblast lineage is highly dependent on BMPR1B in this cell type as well as in murine osteoblast precursor cells (28). Likewise, BMP-2-induced osteogenic activity is enhanced following growth factor-mediated up-regulation of BMPR1B in primary human bone cells (29). These studies collectively underscore the complexity and flexibility of receptor utilization in BMP-induced osteoblastic differentiation in diverse cellular environments.
Although all four BMPs evaluated in this study utilized the same set of receptors, the degree of reliance on each individual receptor varied between members of the two BMP subclasses. With regard to the type I receptors, BMPR1A was found to be more critical to signaling by BMP-2/4, whereas ACVR1A was more crucial for BMP-6/7 signaling. For the type II receptors, all four BMPs utilized primarily BMPR2 to mediate osteoinductive signaling in hMSC. The loss of BMPR2, however, led to a greater reduction in signaling for BMP-2/4 than for BMP-6/7, whereas down-regulation of ACVR2A inhibited signaling by BMP-6/7 but not BMP-2/4. Overall, these data are in accordance with previously published work (12, 14, 30, 31).
Data from the simultaneous type I receptor knockdown treatments suggested differences in oligomerization patterns within receptor complexes utilized by BMP-2/4 or BMP-6/7. Although both BMP-6/7 and BMP-2/4 appear to signal through homodimers of ACVR1A and BMPR1A, BMP-2 and BMP-4 might also require a heterodimer of BMPR1A/ACVR1A to achieve their full signaling potential. BMPR1A and ACVR1A homodimer and heterodimer formation was demonstrated in live mammalian cells using BRET2. Similar associations of BMP type I receptors were previously reported by Gilboa et al. (32), who used co-immunoprecipitation studies to detect homodimers of BMPR1A/BMPR1A and BMPR1B/BMPR1B as well as heterodimers of BMPR1A/BMPR1B at the membrane surface of COS7 cells following overexpression of the receptor monomers. These data indicate that a range of receptor complexes might be present concurrently in BMP-responsive cells to meet the diverse signaling needs of the various BMP family members.
It is interesting to speculate that BMP signaling through a type I heterodimer might have functional consequences distinct from those of signaling exclusively through receptor homodimers. This appears to be the case in Drosophila embryonic development, where heterodimers of the BMP-like ligands Dpp and Scw signal through a receptor tetramer consisting of a homodimer of the type II receptor Punt and one monomer each of the type I receptors Sax and Tkv. Activation of the type I receptor heterodimer produces a synergistic response that triggers expression of a distinct set of genes from those stimulated by signaling through the type I homodimer (33). Should such a scenario also be true in mammalian cells, it would provide an additional mechanism through which BMP signaling might be translated into the multitude of diverse phenotypes observed in BMP-responsive cells and tissues.
In the present study, all four BMPs were found to be equally osteoinductive in primary hMSC, despite differences between members of the BMP-2/4 and BMP-5/6/7 subclasses with respect to receptor preferences and potential utilization of type I heterodimers. In contrast, significant differences in the bioactivities of BMPs have been demonstrated in a number of other physiological systems. The kidney is one organ in which such differences are widely recognized. In chronic kidney disease, BMP-2 appears to induce osteoblastic differentiation of vascular smooth muscle cells (34, 35), leading to vascular calcification, whereas BMP-7 demonstrates opposing effects (36). In addition, during renal development, low doses of BMP-7 (0.25 nm) have been found to stimulate branching morphogenesis, whereas higher doses of BMP-7 or BMP-2 at any tested dose were inhibitory (36). In the adult kidney, BMP-7, but not BMP-4 or BMP-6, was found to counteract transforming growth factor-β1-mediated epithelial-to-mesenchymal transition and reverse the fibrosis and tissue damage associated with chronic renal injury (37, 38). A similar remedial effect of BMP-7 on fibrosis has been documented in the heart (39), liver (40), and lung (41).
One potential determinant of whether BMP activities parallel each other or diverge from each other in a given tissue might be the tissue-specific pattern of receptor expression. We quantified receptor expression in 10 human tissues to compare the representation of BMP receptors. Qualitative differences in the relative expression level of receptors within each tissue were observed as well as significant quantitative differences in the expression of each individual receptor among the 10 tissues. It is highly plausible that substantial differences in receptor expression among human tissues, as documented here, might explain why BMP ligands have similar activities in some physiological systems and unique activities in others.
In summary, the data reported in this study indicate that BMP-2, BMP-4, BMP-6, and BMP-7 all induce osteoblastic differentiation of primary hMSC but display distinct preferences in the BMP receptors utilized to mediate an activation signal. Furthermore, BMP-2/4 might require a heterodimer of type I receptors, whereas BMP-6/7 signal exclusively through type I homodimers. Future studies will be aimed at elucidating the downstream effects of these differences in receptor utilization and in further clarifying the mechanism through which the different BMP ligands transmit their signaling messages in clinically relevant systems.
Supplementary Material
Acknowledgments
We thank Dr. Kening Song for preparing the receptor constructs used in the BRET2 assays and Steve Creech of Statistically Significant Consulting for statistical analysis of the RNAi knockdown data.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1 and 2.
Footnotes
The abbreviations used are: BMP, bone morphogenetic protein; MSC, mesenchymal stem cell(s); hMSC, human MSC; MSCGM, mesenchymal stem cell growth medium; ODM, osteogenic differentiation medium; RNAi, RNA interference; siRNA, small interfering RNA; RT, reverse transcription; qPCR, quantitative PCR; Rluc, Renilla luciferase; GFP2, green fluorescent protein; BRET2, bioluminescence resonance energy transfer; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
References
- 1.Wordinger, R. J., and Clark, A. F. (2007) Exp. Biol. Med. 232 979-992 [DOI] [PubMed] [Google Scholar]
- 2.Kawabata, M., Imamura, T., and Miyazono, K. (1998) Cytokine Growth Factor Rev. 9 49-61 [DOI] [PubMed] [Google Scholar]
- 3.Gautschi, O. P., Frey, S. P., and Zellweger, R. (2007) ANZ J. Surg. 77 626-631 [DOI] [PubMed] [Google Scholar]
- 4.Garrison, K. R., Donell, S., Ryder, J., Shemilt, I., Mugford, M., Harvey, I., and Song, F. (2007) Health Technol. Assess. 11 1-168 [DOI] [PubMed] [Google Scholar]
- 5.Canalis, E., Economides, A. N., and Gazzerro, E. (2003) Endocr. Rev. 24 218-235 [DOI] [PubMed] [Google Scholar]
- 6.Goumans, M. J., and Mummery, C. (2000) Int. J. Dev. Biol. 44 253-265 [PubMed] [Google Scholar]
- 7.Nohe, A., Hassel, S., Ehrlich, M., Neubauer, F., Sebald, W., Henis, Y. I., and Knaus, P. (2002) J. Biol. Chem. 277 5330-5338 [DOI] [PubMed] [Google Scholar]
- 8.Wrana, J. L., Attisano, L., Wieser, R., Ventura, F., and Massague, J. (1994) Nature 370 341-347 [DOI] [PubMed] [Google Scholar]
- 9.Miyazawa, K., Shinozaki, M., Hara, T., Furuya, T., and Miyazono, K. (2002) Genes Cells 7 1191-1204 [DOI] [PubMed] [Google Scholar]
- 10.Guicheux, J., Lemonnier, J., Ghayor, C., Suzuki, A., Palmer, G., and Caverzasio, J. (2003) J Bone Miner. Res. 18 2060-2068 [DOI] [PubMed] [Google Scholar]
- 11.Osyczka, A. M., and Leboy, P. S. (2005) Endocrinology 146 3428-3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.ten Dijke, P., Yamashita, H., Sampath, T. K., Reddi, A. H., Estevez, M., Riddle, D. L., Ichijo, H., Heldin, C. H., and Miyazono, K. (1994) J. Biol. Chem. 269 16985-16988 [PubMed] [Google Scholar]
- 13.Yamashita, H., ten Dijke, P., Huylebroeck, D., Sampath, T. K., Andries, M., Smith, J. C., Heldin, C. H., and Miyazono, K. (1995) J. Cell Biol. 130 217-226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Macias-Silva, M., Hoodless, P. A., Tang, S. J., Buchwald, M., and Wrana, J. L. (1998) J. Biol. Chem. 273 25628-25636 [DOI] [PubMed] [Google Scholar]
- 15.Rosenzweig, B. L., Imamura, T., Okadome, T., Cox, G. N., Yamashita, H., ten Dijke, P., Heldin, C. H., and Miyazono, K. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 7632-7636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sampath, T. K., Maliakal, J. C., Hauschka, P. V., Jones, W. K., Sasak, H., Tucker, R. F., White, K. H., Coughlin, J. E., Tucker, M. M., and Pang, R. H. (1992) J. Biol. Chem. 267 20352-20362 [PubMed] [Google Scholar]
- 17.Pfleger, K. D., Seeber, R. M., and Eidne, K. A. (2006) Nat. Protoc. 1 337-345 [DOI] [PubMed] [Google Scholar]
- 18.Friedman, M. S., Long, M. W., and Hankenson, K. D. (2006) J. Cell. Biochem. 98 538-554 [DOI] [PubMed] [Google Scholar]
- 19.Lee, M. H., Kim, Y. J., Kim, H. J., Park, H. D., Kang, A. R., Kyung, H. M., Sung, J. H., Wozney, J. M., and Ryoo, H. M. (2003) J. Biol. Chem. 278 34387-34394 [DOI] [PubMed] [Google Scholar]
- 20.Yeh, L. C., Tsai, A. D., and Lee, J. C. (2002) J. Cell. Biochem. 87 292-304 [DOI] [PubMed] [Google Scholar]
- 21.Gazzerro, E., Gangji, V., and Canalis, E. (1998) J. Clin. Invest. 102 2106-2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qi, H., Aguiar, D. J., Williams, S. M., La Pean, A., Pan, W., and Verfaillie, C. M. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 3305-3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu, Y., Piston, D. W., and Johnson, C. H. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 151-156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krishnaveni, M. S., Hansen, J. L., Seeger, W., Morty, R. E., Sheikh, S. P., and Eickelberg, O. (2006) Biochem. Biophys. Res. Commun. 351 651-657 [DOI] [PubMed] [Google Scholar]
- 25.Storez, H., Scott, M. G., Issafras, H., Burtey, A., Benmerah, A., Muntaner, O., Piolot, T., Tramier, M., Coppey-Moisan, M., Bouvier, M., Labbe-Jullie, C., and Marullo, S. (2005) J. Biol. Chem. 280 40210-40215 [DOI] [PubMed] [Google Scholar]
- 26.Scaffidi, A. K., Petrovic, N., Moodley, Y. P., Fogel-Petrovic, M., Kroeger, K. M., Seeber, R. M., Eidne, K. A., Thompson, P. J., and Knight, D. A. (2004) J. Biol. Chem. 279 37726-37733 [DOI] [PubMed] [Google Scholar]
- 27.Wan, D. C., Shi, Y. Y., Nacamuli, R. P., Quarto, N., Lyons, K. M., and Longaker, M. T. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 12335-12340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen, D., Ji, X., Harris, M. A., Feng, J. Q., Karsenty, G., Celeste, A. J., Rosen, V., Mundy, G. R., and Harris, S. E. (1998) J. Cell Biol. 142 295-305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singhatanadgit, W., Salih, V., and Olsen, I. (2006) J. Cell. Physiol. 209 912-922 [DOI] [PubMed] [Google Scholar]
- 30.Liu, F., Ventura, F., Doody, J., and Massague, J. (1995) Mol. Cell. Biol. 15 3479-3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aoki, H., Fujii, M., Imamura, T., Yagi, K., Takehara, K., Kato, M., and Miyazono, K. (2001) J. Cell Sci. 114 1483-1489 [DOI] [PubMed] [Google Scholar]
- 32.Gilboa, L., Nohe, A., Geissendorfer, T., Sebald, W., Henis, Y. I., and Knaus, P. (2000) Mol. Biol. Cell 11 1023-1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Connor, M. B., Umulis, D., Othmer, H. G., and Blair, S. S. (2006) Development 133 183-193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li, X., Yang, H. Y., and Giachelli, C. M. (2008) Atherosclerosis, in press [DOI] [PMC free article] [PubMed]
- 35.Hruska, K. A., Mathew, S., and Saab, G. (2005) Circ. Res. 97 105-114 [DOI] [PubMed] [Google Scholar]
- 36.Piscione, T. D., Yager, T. D., Gupta, I. R., Grinfeld, B., Pei, Y., Attisano, L., Wrana, J. L., and Rosenblum, N. D. (1997) Am. J. Physiol. 273 F961-F975 [DOI] [PubMed] [Google Scholar]
- 37.Zeisberg, M., Hanai, J., Sugimoto, H., Mammoto, T., Charytan, D., Strutz, F., and Kalluri, R. (2003) Nat. Med. 9 964-968 [DOI] [PubMed] [Google Scholar]
- 38.Zeisberg, M., Shah, A. A., and Kalluri, R. (2005) J. Biol. Chem. 280 8094-8100 [DOI] [PubMed] [Google Scholar]
- 39.Zeisberg, E. M., Tarnavski, O., Zeisberg, M., Dorfman, A. L., McMullen, J. R., Gustafsson, E., Chandraker, A., Yuan, X., Pu, W. T., Roberts, A. B., Neilson, E. G., Sayegh, M. H., Izumo, S., and Kalluri, R. (2007) Nat. Med. 13 952-961 [DOI] [PubMed] [Google Scholar]
- 40.Zeisberg, M., Yang, C., Martino, M., Duncan, M., Rieder, F., Tanjore, H., and Kalluri, R. (2007) J. Biol. Chem. 282 23337-23347 [DOI] [PubMed] [Google Scholar]
- 41.Izumi, N., Mizuguchi, S., Inagaki, Y., Saika, S., Kawada, N., Nakajima, Y., Inoue, K., Suehiro, S., Friedman, S. L., and Ikeda, K. (2006) Am. J. Physiol. 290 L120-L126 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.