Abstract
The Gram-positive bacterium Geobacillus stearothermophilus NRS 2004/3a possesses a cell wall containing an oblique surface layer (S-layer) composed of glycoprotein subunits. O-Glycans with the structure [→2)-α-l-Rhap-(1→3)-β-l-Rhap-(1→2)-α-l-Rhap-(1→]n = 13-18, a2-O-methyl group capping the terminal repeating unit at the nonreducing end and a →2)-α-l-Rhap-[(1→3)-α-l-Rhap]n = 1-2(1→3)- adaptor are linked via a β-d-Galp residue to distinct sites of the S-layer protein SgsE. S-layer glycan biosynthesis is encoded by a polycistronic slg (surface layer glycosylation) gene cluster. Four assigned glycosyltransferases named WsaC-WsaF, were investigated by a combined biochemical and NMR approach, starting from synthetic octyl-linked saccharide precursors. We demonstrate that three of the enzymes are rhamnosyltransferases that are responsible for the transfer of l-rhamnose from a dTDP-β-l-Rha precursor to the nascent S-layer glycan, catalyzing the formation of the α1,3- (WsaC and WsaD) and β1,2-linkages (WsaF) present in the adaptor saccharide and in the repeating units of the mature S-layer glycan, respectively. These enzymes work in concert with a multifunctional methylrhamnosyltransferase (WsaE). The N-terminal portion of WsaE is responsible for the S-adenosylmethionine-dependent methylation reaction of the terminal α1,3-linked l-rhamnose residue, and the central and C-terminal portions are involved in the transfer of l-rhamnose from dTDP-β-l-rhamnose to the adaptor saccharide to form the α1,2- and α1,3-linkages during S-layer glycan chain elongation, with the methylation and the glycosylation reactions occurring independently. Characterization of these enzymes thus reveals the complete molecular basis for S-layer glycan biosynthesis.
Glycosylation is the most common posttranslational modification of proteins (1-3), playing important roles in all living organisms (4-7). Although glycosylation has popularly been considered to be restricted to eukaryotes, prokaryotic glycosylation is an emerging field that has opened new avenues in basic and applied research. Among the best investigated prokaryotic glycoproteins are surface layer (S-layer)6 glycoproteins of Bacillaceae (11, 12). Due to the ability of native and recombinant S-layer (glyco)proteins to self-assemble into two-dimensional arrays in solution and on various supports (e.g. silica, polymers, liposomes, lipid films, or membranous structures), they are considered promising candidates for in vivo and in vitro glycan display approaches with nanometer scale periodicity (13-15). Understanding the basic principles of prokaryotic protein glycosylation is a prerequisite for engineering of tailor-made (“functional”) glycan motifs on S-layer proteins for applications in microbiology, nanobiotechnology, and vaccinology (8-10).
The S-layer glycoprotein of the Gram-positive organism Geobacillus stearothermophilus NRS 2004/3a serves as a model system for elucidating the basic principles of prokaryotic protein glycosylation. The S-layer glycan with the structure, 2-OMe-α-l-Rhap-(1→3)-β-l-Rhap-(1→2)-α-l-Rhap-(1→[2)-α-l-Rhap-(1→3)-β-l-Rhap-(1→2)-α-l-Rhap-(1→] n = 13-18 2)-α-l-Rhap-(1→[3)-α-l-Rhap]n = 1-2-(1→3)-β-d-Galp-(1→ is O-glycosidically linked to threonine 590, threonine 620, and serine 794 of the S-layer protein SgsE (16, 17).
Based on sequence comparisons, we hypothesize that the genetic information for the enzymes involved in S-layer glycan biosynthesis of G. stearothermophilus NRS 2004/3a is organized in a polycistronic slg (S-layer glycosylation) gene cluster (GenBank™ accession number AF328862) (18). Besides four previously uncharacterized, putative glycosyltransferases, which are the focus of the present study, the slg gene cluster encodes enzymes catalyzing the biosynthesis of dTDP-β-l-rhamnose, ABC transporter components, an initiating glycosyltransferase (WsaP), and an oligosaccharyltransferase (WsaB). Additionally, UDP-Gal that is required for S-layer glycan formation is derived from the general metabolism of the bacterium (19). Generally, slg gene clusters show high homology to gene clusters involved in O-polysaccharide (O-PS) biosynthesis of Gram-negative bacteria (12, 20). The presence of a homopolymeric S-layer glycan chain (poly-l-rhamnan) in G. stearothermophilus NRS 2004/3a and the occurrence of the wzm/wzt genes encoding ABC transporter components in the slg gene cluster indicate that the biosynthesis of this S-layer glycan may follow a route similar to the ABC transporter-dependent pathway of O-PS chains (21). On the other hand, with the functional characterization of the initiation enzyme WsaP (a WbaP homologue) of S-layer glycan biosynthesis as a Gal-1-phosphate:lipid carrier transferase, it is evident that also a biosynthesis module from the ABC transporter-independent O-PS biosynthesis route is utilized (22). More precisely, in G. stearothermophilus NRS 2004/3a, WsaP replaces the GlcNAc-1-phosphate:undecaprenyl phosphate (und-P) transferase activity of WecA, which is the characteristic initiation reaction of ABC transporter dependent pathways of O-PS biosynthesis (23).
In this report, we demonstrate the function of the four assigned glycosyltransferases of the slg gene cluster of G. stearothermophilus NRS 2004/3a, named WsaC-WsaF, which should be responsible for the consecutive transfer of rhamnose residues from a nucleotide diphosphate-activated precursor to the growing S-layer glycan chain while catalyzing the formation of the α1,2-, α1,3-, and β1,2-linkages. In this context, we identify WsaE as a novel multifunctional enzyme, catalyzing not only the consecutive transfer of l-rhamnose residues from dTDP-β-l-rhamnose to the adaptor saccharide of the S-layer glycan chain but also the S-adenosylmethionine (SAM)-dependent methylation reaction of the terminal α-l-rhamnose residue of the glycan chain. It is shown that the glycosylation reaction and the methylation reaction occur independently. Further we show that the four glycosyltransferases working in concert are sufficient for S-layer poly-l-rhamnan biosynthesis in G. stearothermophilus NRS 2004/3a.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions—Bacterial strains and plasmids are listed in Table S1. G. stearothermophilus NRS 2004/3a was grown in S-VIII medium at 55 °C (16) and Escherichia coli was grown in Luria-Bertani broth at 37 °C. Growth media were supplemented with kanamycin (50 μg/ml), when appropriate.
Sequence Analysis—Protein sequences were analyzed using the BLASTP on-line sequence homology analysis tools (National Center for Biotechnology Information, Bethesda, MD). Putative transmembrane helices were identified with the TMHMM2.0 program (Center for Biological Sequence Analysis, Lyngby, Denmark). Sequence alignments were carried out with the ClustalW program (available on the World Wide Web).
Construction of Plasmids—Various plasmids for enzyme overexpression in E. coli were constructed (Tables S1 and S2). To obtain proteins with an N-terminal His6 tag, the entire coding sequences of WsaC, WsaD, WsaE, WsaF, RmlB, RmlC, and RmlD and/or truncated versions thereof were amplified by PCR with the primer pairs pET-WsaC_for/pET-WsaC_rev (WsaC), pET-WsaC_for/pET-WsaC_I_rev (WsaC_I), pET-WsaD_for/pET-WsaD_rev (WsaD), pET-WsaD_for/pET-WsaD_I_rev (WsaD_I), pET-WsaE_for/pET-WsaE_rev (WsaE), pET-WsaE_for/pET-WsaE_M_rev (WsaE_M), pET-WsaE_for/pET-WsaE_N_rev (WsaE_N), pET-WsaE_B_for/pET-WsaE_ rev (WsaE_B), pET-WsaE_C_for/pET-WsaE_rev (WsaE_C), pET-WsaE_B_for/pET-WsaE_A_rev (WsaE_A), pET-WsaF_ for/pET-WsaF_rev (WsaF), RmlB_for/RmlB_rev (RmlB), RmlC_for/RmlC_rev (RmlC), and RmlD_for/RmlD_rev (RmlD), respectively (Table S2). Amplification products were digested with NdeI/XhoI (WsaC, WsaD, RmlC, and RmlD), NheI/SstI (WsaF), or NheI/XhoI (WsaE and RmlB) and inserted into the dephosphorylated expression vector pET28a (Novagen, Madison, WI), which was linearized with the same restriction enzymes. The resulting plasmids were named pNGB220 (pET28a-WsaC) and pNGB221 (pET28a-WsaC_I), pNGB230 (pET28a-WsaD) and pNGB231 (pET28a-WsaD_I), pNGB240 (pET28a-WsaE), pNGB241 (pET28a-WsaE_M), pNGB242 (pET28a-WsaE_ N), pNGB243 (pET28a-WsaE_B), pNGB244 (pET28a-WsaE_C), pNGB245 (pET28a-WsaE_A), pNGB250 (pET28a-WsaF), pNGB261 (pET28a-RmlB), pNGB262 (pET28a-RmlC), and pNGB263 (pET28a-RmlD) and were transformed into the E. coli expression host BL21 Star (DE3).
Protein Overexpression—Expression of recombinant proteins in E. coli BL21 Star (DE3) was initiated by the addition of 1 mm isopropyl-β-d-thiogalactopyranoside to A600 ∼ 0.8 cultures, and cultivation was continued for an additional 4 h. Expression of recombinant proteins was monitored by SDS-PAGE (24). Protein bands were visualized with Coomassie Blue R-250 staining reagent. Semidry blotting of proteins to a polyvinylidene difluoride membrane (Bio-Rad) was performed as described previously (22). Development of the blot with anti-His tag monoclonal antibody (Novagen) was done according to the manufacturer's instructions.
Purification of Recombinant Proteins—Biomass from 400 ml of E. coli BL21Star (DE3) expression cultures was harvested by centrifugation (15 min, 4,500 × g, 4 °C). The pellet was washed with 40 ml of cold saline and resuspended in 10 ml of cold buffer A (50 mm ammonium/acetate, pH 7.5, 1 mm EDTA). Cells were lysed by ultrasonication (Branson sonifier 450; Branson, Danbury, CT; duty cycle 60%, output 6), applying eight cycles of 10 pulses with 1-min breaks, each. Unlysed cells were removed by centrifugation (4,500 × g, 15 min, 4 °C), and the membrane fraction was separated from the cell-free lysate by ultracentrifugation (200,000 × g, 60 min, 4 °C), followed by a washing step with buffer A. Soluble enzymes (WsaE, WsaF, RmlB, RmlC, and RmlD) were directly purified on 1-ml HisTrap™ HP prepacked columns (GE Healthcare), using an Amersham Biosciences FPLC™ system (flow rate 1 ml/min). The column was equilibrated in His A buffer (20 mm sodium phosphate, 0.5 m NaCl, 20 mm imidazole, pH 7.4). Proteins were eluted using a step gradient up to 0.5 m imidazole in His A buffer. Isolated membranes harboring WsaC or WsaD were extracted on ice for 3 h to overnight with 2% CHAPS or LDAO in His A buffer (Sigma). After centrifugation at 20,800 × g for 1 h, the supernatants of WsaC and WsaD were purified by HisTrap™ HP chromatography as described above. The addition of 0.2% CHAPS or 0.1 mm LDAO to all buffers was necessary to keep the proteins in solution. Fractions were analyzed by SDS-PAGE, and the protein content of the pools was determined using the Bio-Rad Bradford reagent.
Biosynthesis of dTDP-β-l-Rhamnose—dTDP-β-l-rhamnose was synthesized from dTDP-glucose using purified RmlB, RmlC, and RmlD enzymes as described previously (25).
Chemical Synthesis of Octyl Saccharide Substrates—For the functional enzyme assays, the following synthetic octyl-linked saccharides were used (Fig. S1): β-d-Gal-(1→O)-octyl (Mr 292.37) (I), α-l-Rha-(1→3)-β-d-Gal-(1→O)-octyl (Mr = 438.51) (II), and α-l-Rha-(1→3)-α-l-Rha-(1→3)-β-d-Gal-(1→O)-octyl (Mr = 584.65) (III). The chemical synthesis of (I-III) was based on commonly used monosaccharide building blocks (26-29).
Rhamnosyltransferase Assay with Octyl Saccharides—The synthetic substrates were dissolved in 2-propanol (I and III) or methanol (II) at a concentration of 20 nmol/μl (0.29 mg/50 μl (I), 0.44 mg/50 μl (II), and 0.28 mg/25 μl (III)). Reaction products of rhamnosyltransferases used as substrates in subsequent assays were dissolved in water. For the assay, either lysed cells (WsaC and WsaD) or cell-free extract and purified protein (WsaE and WsaF), respectively, were used. The reaction mixture contained 1 μl (20 nmol) of substrate, 2 μl (40 nmol) of dTDP-β-l-Rha, the relevant transferases, and 10 μl of 100 mm MnCl2 in a final volume of 100 μl of buffer A (50 mm ammonium acetate, pH 7.5, 1 mm EDTA). Incubation was performed at 37 °C for 1 h, and the reaction was stopped by the addition of 1.2 ml of chloroform/methanol (3:2) followed by extraction for 20 min. After removal of insoluble material, the supernatant was dried under a stream of nitrogen. Samples were resuspended in 10 μl of chloroform/methanol (3:2) and analyzed by TLC. Octyl-linked products were separated on silica Gel 60 aluminum TLC plates (20 × 20 cm; thickness, 0.25 mm; Merck) using the solvent system chloroform/methanol/water (65:25:4). Carbohydrates were detected with thymol reagent (30).
Methyltransferase Activity Assay—This assay was based on the rhamnosyltransferase activity assay. Different forms of WsaE (WsaE, WsaE_M and WsaE_N) were mixed with dTDP-β-l-Rha, octyl substrates (α-l-Rha-(1→3)-α-l-Rha-(1→3)-β-d-Gal-(1→O)-octyl (III), α-l-Rha-(1→2)-α-l-Rha-(1→3)-α-l-Rha-(1→3)-β-d-Gal-(1→O)-octyl (VII), or α-l-Rha-(1→3)-β-l-Rha-(1→2)-α-l-Rha-(1→3)-β-d-Gal-(1→O)-octyl (IX)), and MnCl2, and 1 μl of labeled S-[methyl-3H]adenosylmethionine (10 Ci/mmol; PerkinElmer Life Sciences) was added to a final volume of 100 μl. Incubation was performed at 37 °C for 1 h, and the reaction products were separated by TLC. For autoradiography, the TLC plates were exposed to an Eastman Kodak Co. BioMax MS film (Sigma) for 2 days at -70 °C.
ESI-QTOF MS—For MS analysis of the rhamnosyltransferase reaction products, the assays were scaled up 10-fold. When purified enzyme was used (WsaE and WsaF), the reaction products were directly applied to preparative TLC. When WsaC was used, octyl-linked reaction products were extracted and hydrolyzed under alkaline conditions to destroy phospholipids. Subsequently, the products were dissolved in chloroform/methanol (2:1), mixed with 8 parts of ethanol and 1 part of 1 m NaOH (final concentration 0.1 m NaOH, pH 12), and incubated for 30 min at 37 °C (31). After the addition of ethyl acetate, alkalistable products were extracted with chloroform/methanol, as described for the enzyme assay, and analyzed by TLC. For removal of remaining substrates, the samples were applied to preparative TLC. One of the lanes was stained with thymol, and the products were scratched off of the unstained plate at the corresponding height and extracted with 1 ml of chloroform/methanol (3:2), chloroform/methanol (1:2), methanol, and methanol/water (1:1), respectively. After removal of the silica material by centrifugation, the supernatant was dried under a stream of nitrogen and analyzed by ESI-QTOF MS.
For MS analysis of the products from the methyltransferase reaction, the assay was scaled up 20-fold using HisTrap-purified methyltransferase and unlabeled SAM (Sigma). After completion of the reaction, the protein was removed using Microcon® centrifugal filter units (Mr cut-off 10,000; Millipore). The sample was desalted using RP-18 Sep-Pak cartridges (Waters, Milford, MA). After activating the cartridge with 50% acetonitrile, it was equilibrated with MilliQ water, and the sample dissolved in 100 μl of MilliQ water was applied. After washing with 3 ml of MilliQ water, the product was eluted with 1 ml of 25, 50, 75, and 100% acetonitrile, respectively. Fractions of interest were combined and dried under a stream of nitrogen.
Positive mode mass spectrometry was performed on a Waters Micromass Q-TOF Ultima Global apparatus (Waters Micromass, Manchester, UK). Samples were subjected to offline infusion ESI-QTOF MS after dilution in 75% methanol containing 0.1% formic acid to a final concentration of about 50 pmol/μl. Spectra acquisition was performed using 2-3 kV capillary and 100 V cone voltage. Desolvation gas flow was set at 450 liters/h and cone gas at 50 liters/h. Samples were injected at a flow rate of 3-5 μl/min. The instrument was controlled by MassLynx 4.0 software (Waters Micromass).
Sample Preparation for NMR Studies—All large scale reaction mixtures contained 10 mm MnCl2 and were incubated overnight. Lysed cells containing WsaC (8.5 ml) were incubated with 5 mg (8.5 μmol) of α-l-Rha-(1→3)-α-l-Rha-(1→3)-β-d-Gal-(1→O)-octyl (III) and 15.5 μmol of dTDP-β-l-Rha. Purified WsaF (6 ml) was either reacted with 3 mg (6.8 μmol) of α-l-Rha-(1→3)-β-d-Gal-(1→O)-octyl (II) and 10 μmol of dTDP-β-l-Rha, or WsaF (4.3 ml) was reacted with 0.7 mg of the α-l-Rha-(1→2)-α-l-Rha-(1→3)-α-l-Rha-(1→3)-β-d-Gal-(1→O)-octyl (VII) reaction product of WsaE and 2 μmol of dTDP-β-l-Rha. For investigation of the α1,2-rhamnosyltransferase activity of WsaE, 8.3 ml of cell-free supernatant of E. coli expressing WsaE was incubated with 5 mg (8.5 μmol) of α-l-Rha-(1→3)-α-l-Rha-(1→3)-β-d-Gal-(1→O)-octyl (III) dissolved in 200 μl of methanol together with 510 μl of dTDP-β-l-Rha (10.2 μmol). For identification of α1,3-rhamnosyltransferase activity, 4.3 ml of WsaE-containing lysate were reacted with 0.5 mg of the β-l-Rha-(1→2)-α-l-Rha-(1→3)-β-d-Gal-(1→O)-octyl (IV) reaction product of WsaF and 1.5 μmol of dTDP-β-l-Rha. Membranes and aggregates were removed by ultracentrifugation at 200,000 × g for 45 min, and the pellet was reextracted with methanol. This fraction was combined with the supernatant and centrifuged using Microcon® centrifugal filter units (Mr cut-off 10,000) to remove the proteins. The filtrate was dried and analyzed by TLC, as described above. Dried reaction mixtures were resuspended in 0.5 ml of 20% methanol and applied to a Sephadex G10 column (1.5 × 117 cm) with 20% methanol as eluent. Elution was monitored by spotting 1 μl of each fraction on a TLC plate followed by staining with thymol reagent. Fractions positive for carbohydrate were analyzed by TLC, and the fractions containing the products were pooled, dried in a SpeedVac vacuum concentrator, and desalted using RP-18 Sep-Pak Cartridges (Waters) with a discontinuous gradient of acetonitrile (25, 50, 75, and 100% acetonitrile) for elution of the reaction products.
NMR Spectroscopy—All NMR spectra were recorded at 297 K on a Bruker DPX 400 instrument at 400.13 MHz for proton and 100.62 MHz for carbon using a 5-mm broad band probe with z-gradients. The sample contained freeze-dried material dissolved in 600 μl of D2O in a 5-mm NMR tube. The ppm scales were calibrated to 2,2-dimethyl-2-silapentane-5-sulfonic acid for proton (0.00 ppm) and external 1,4-dioxane for carbon (67.40 ppm). The NMR spectra were recorded using Bruker standard homonuclear pulse programs such as COSY and mlevph. For heteronuclear experiments, Bruker standard pulse programs were used as heteronuclear single quantum coherence spectroscopy for one-bond proton-carbon correlations and heteronuclear multiple bond correlation spectroscopy for multiple-bond proton-carbon correlations. All homonuclear and heteronuclear spectra were recorded with 1K × 1K data points and zero-filled in both dimensions.
Rhamnosyltransferase Assay with E. coli Membranes Harboring WsaP—E. coli C43 (DE3) cells harboring either pET28_WsaP, coding for the Gal-1-phosphate:lipid carrier transferase WsaP of G. stearothermophilus NRS 2004/3a (22), or pET28a_WsaD were lysed separately. For the assay, 88 μl of each or both lysates were mixed with 100 mm MnCl2 (final concentration 10 mm), 84 pmol (25 nCi) of radiolabeled UDP-d-[14C]galactose (GE Healthcare), and, optionally, 20 nmol of dTDP-β-l-Rha. The reaction was performed for 20 h at 37 °C and terminated by the addition of 1.25 ml of chloroform/methanol (3:2) (32). After shaking for 20 min, the samples were centrifuged, and the organic phase was transferred to a new tube. The organic phase was washed with 150 μl of 40 mm MgCl2. The upper phase obtained after centrifugation was removed, and the lower organic phase was washed with 400 μl of pure solvent upper phase chloroform/methanol/water/1 m MgCl2 (18:294:282:1). The organic phase containing the lipid-linked reaction products was dried under a stream of nitrogen, resuspended in 10 μl of chloroform/methanol (3:2), and applied to TLC and autoradiography as described above.
RESULTS
Sequence Analysis of the Transferases WsaC, WsaD, WsaE, and WsaF—Considering that the S-layer glycan of G. stearothermophilus NRS 2004/3a contains α- and β-linked l-rhamnose residues and the putative donor required for rhamnosyltransferases would be dTDP-β-l-rhamnose, a data base search for retaining and inverting glycosyltransferases was performed. Furthermore, the presence of a methylated terminal rhamnose residue let us hypothesize that a SAM-dependent methyltransferase may be present in the slg gene cluster of this organism. The results of the various BLAST searches revealed the presence of GT-2 and GT-4 transferases in the slg gene cluster. During our previous annotations of this gene cluster, we named the glycosyltransferase-encoding genes wsaC, wsaD, wsaE, and wsaF (12). The predicted primary structures of the encoded WsaC, WsaD, and WsaF proteins reveal proteins of the range of 300-400 amino acids, which is an expected size for a glycosyltransferase catalyzing a single reaction; these sequences also contain the DXD signature motif, which is commonly found in both inverting (i.e. inverting the stereochemistry of the donor) and retaining (i.e. retaining the stereochemistry of the donor) glycosyltransferases.
WsaC (protein accession number AAR99605) is assigned to the inverting transferases of the GT-2 family (33). An aspartate residue, Asp39, in the model protein SpsA (34), potentially corresponding to Asp40-Asp41 in WsaC, may coordinate N-3 of the uracil base, providing specificity for U and T as the sugar donor. In the so-called DXDD motif (DQDD; aa 96-99), the middle Asp residue binds the hydroxyl groups on the ribose moiety, whereas the third Asp residue binds a divalent metal ion. An additional Asp residue at position 191 is thought to function as a catalytic base activating the acceptor for nucleophilic attack at C-1 by deprotonation (35). As the substrate for WsaC is dTDP-β-l-Rha, an inverting transferase would be an α-rhamnosyltransferase. In the case of WsaD (AAR99614), there is homology to the inverting α-rhamnosyltransferases of Shigella flexneri, specifically matching the N-terminal (V/I)X(V/I)XDX2S signature motif (36). Members of GT-2 family have a highly conserved N-terminal domain and a less conserved C terminus, due to the differences in substrate specificity (Fig. S2). The topology model of both the WsaC and the WsaD protein predicts a single, transmembrane-spanning domain at the C-terminal region. The predicted cytoplasmic WsaF (AAR99609) protein contains EX7E motifs typically found in retaining glycosyltransferases of the GT-4 family (37). Since WsaF is putatively a retaining glycosyltransferase, we hypothesize that it would be responsible for the formation of the β1,2-linkage in the repeating unit trisaccharide of the G. stearothermophilus NRS 2004/3a S-layer protein glycan.
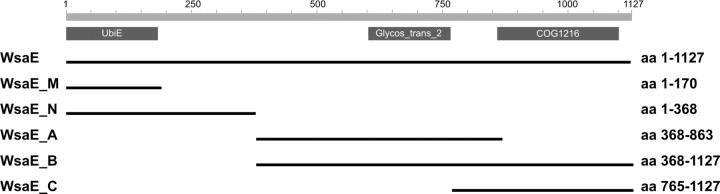
Finally, the wsaE gene encodes a predicted cytoplasmic protein of 1,127 amino acids (AAR99608); data base analysis of its sequence suggested that it may contain three functional domains (Fig. 1). The N-terminal portion of WsaE (aa 70-150) revealed homology to SAM-dependent methyltransferases (Fig. S3), whereas the central and C-terminal portions contain a glycosyltransferase domain, each belonging to the inverting GT-2 family (aa 604-765 and 863-1,043). The first glycosyltransferase domain contains a conserved DD motif and the DXDD motif. The second glycosyltransferase domain contains ED motifs and a DXE motif (Fig. S4).
FIGURE 1.
Predicted topology of the WsaE protein of G. stearothermophilus NRS 2004/3a as a basis for designing different forms of WsaE to assess functional domains of the enzyme. WsaE, aa 1-1,127, full size; WsaE_M, aa 1-170, including the C terminus with the UbiE motive; WsaE_N, aa 1-368, including the C terminus with the UbiE motive and additional 200 aa; WsaE_A, aa 368-863, including the first rhamnosyltransferase domain; WsaE_B, aa 368-1127, including both rhamnosyltransferase domains; WsaE_C, aa 765-1127, including the second rhamnosyltransferase domain.
Expression, Localization, and Purification of the Transferases WsaC, WsaD, WsaE, and WsaF—N-terminally His6-tagged forms of WsaC, WsaD, and WsaE and truncated versions thereof as well as WsaF were produced in E. coli BL21 Star (DE3). According to the Western immunoblot evidence (Fig. S5), both WsaC and WsaD were smaller than expected, corresponding to proteins of ∼37 and 30 kDa, respectively (Fig. S5A, lanes 2 and 4), as opposed to the calculated molecular masses of 40 kDa (WsaC) and 35.6 kDa (WsaD). A similar migration behavior has already been reported for the O-antigen biosynthesis enzyme WbbE from Salmonella enterica, for which also one transmembrane domain is predicted (38). The influence of the protein transmembrane domain resulting in too low molecular mass estimates was confirmed through analysis of C-terminally truncated forms of WsaC (WsaC_I; Fig. S5A, lane 3) and WsaD (WsaD_I; Fig. S5A, lane 5), both of which were devoid of the transmembrane region and could be detected at the calculated size on the Western immunoblot. WsaF (Fig. S5A, lane 7) as well as WsaE and its truncated forms (for details about the truncations, see Fig. 1 and Fig. S5B, lanes 2-6) and WsaF migrated at the correct size. This supports the topology models predicting WsaC and WsaD to be transmembrane proteins and WsaE and WsaF to be soluble proteins.
The cytoplasmic proteins (WsaC_I, WsaD_I, WsaE, and WsaF) could be directly purified by nickel affinity chromatography, whereas full-size WsaC and WsaD were shown to be accumulated in the membrane fraction obtained from lysed cells of E. coli BL21 Star (DE3) expression cultures (data not shown). After numerous attempts with different detergents (Triton X-100, octyl glucopyranoside, CHAPS, and LDAO) at various concentrations, WsaC and WsaD could be purified by using 2% CHAPS or LDAO for membrane extraction and the addition of detergent to all buffers used for nickel affinity chromatography. Detergent was necessary to keep the purified enzymes in solution.
Substrate Specificity of the Rhamnosyltransferases WsaC, WsaD, WsaF, and WsaE—Due to our hypothesis that the four predicted proteins WsaC-F would possess rhamnosyltransferase activity, we tested the activity of the recombinant forms of these proteins with dTDP-β-l-Rha (as donor) and various octyl-linked saccharides (as acceptors). The following conclusions regarding substrate specificity of the individual enzymes have to be interpreted in light of the in vitro system under study and of the lack of kinetic characterization of the enzyme reactions due to limited amounts of enzymes and substrates. Enzyme preparations (either cell-free lysates for WsaC and WsaD or supernatants after ultracentrifugation in the case of WsaE and WsaF) or FPLC-purified enzymes were assayed for their ability to transfer dTDP-β-l-Rha to the synthetic octyl-linked saccharides β-d-Gal-(1→O)-octyl (I), α-l-Rha-(1→3)-β-d-Gal-(1→O)-octyl (II), and α-l-Rha-(1→3)-α-l-Rha-(1→3)-β-d-Gal-(1→O)-octyl (III). According to the TLC evidence using chloroform/methanol/water (65:25:4) as solvent system, none of the enzymes used β-d-Gal-(1→O)-octyl (I) as substrate (Fig. S6, lanes 1, 12, 24, and 28). This result suggested another mechanism for the generation of α-l-Rha-(1→3)-β-d-Gal-(1→O)-octyl, which is discussed below.
WsaF was able to transfer one Rha residue to the α-l-Rha-(1→3)-β-d-Gal-(1→O)-octyl substrate (II), resulting in a new product (IV) with a slightly higher Rf value (Rf ∼ 0.40) than that of α-l-Rha-(1→3)-α-l-Rha-(1→3)-β-d-Gal-(1→O)-octyl (III) (Rf ∼ 0.37) (Fig. 2, compare lanes 2 and 3, and Fig. S6, lanes 28-30), indicating that the newly formed linkage is not α1,3. At this stage of experimentation, this finding would be consistent with the formation of either an α1,2- or a β1,2-linkage, according to the S-layer glycan structure of G. stearothermophilus NRS 2004/3a (16).
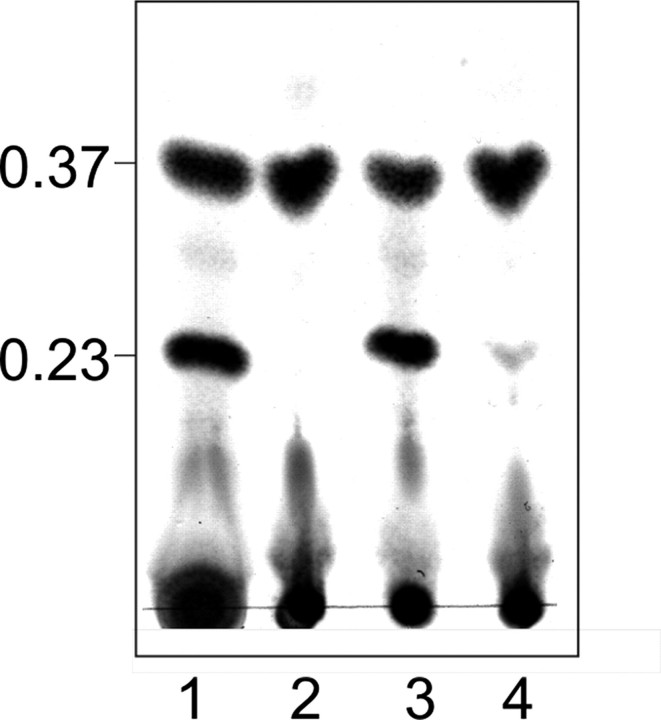
FIGURE 2.

TLC pattern of purified products from WsaC and WsaF activity assays using octyl-linked oligosaccharides. Products were separated on silica TLC plates with chloroform/methanol/water (65:25:4) as solvent. Lane 1, substrate (II) α-l-Rha-(1→3)-β-d-Gal-(1→O)-octyl; lane 2, product of WsaF (IV); lane 3, substrate (III)α-l-Rha-(1→3)-α-l-Rha-(1→3)-β-d-Gal-(1→O)-octyl; lane 4, products of WsaC (V and VI). Staining was performed with thymol reagent.
WsaC generated a new product (V) when incubated with α-l-Rha-(1→3)-α-l-Rha-(1→3)-β-d-Gal-(1→O)-octyl (III) and dTDP-β-l-Rha (Fig. S6, lanes 12-14). The new product (V) had a lower Rf value (Rf ∼ 0.25) than the substrate (Fig. 2, lane 4). When incubated overnight, a second, minor product (VI) with an even lower Rf value of ∼0.15 was synthesized in lower yield (Fig. 2, lane 4). Interestingly, the truncated form WsaC_I did not react (Fig. S6, lanes 20-22). Apparently, as is also the case for WsaP, the transmembrane domain is essential for anchoring of the enzyme in the membrane to maintain its activity (22).
WsaE was shown to react with α-l-Rha-(1→3)-α-l-Rha-(1→3)-β-d-Gal-(1→O)-octyl (III), yielding one product (VII) migrating with an Rf value of ∼0.23 (Fig. 3, lane 1, and Fig. S6, lanes 24-26). To identify the enzymatic activity of the two rhamnosyltransferase domains, the three truncated forms WsaE_A (N-terminal domain), WsaE_B (both domains), and WsaE_C (C-terminal domain) were assayed. WsaE_B showed with substrate (III) activity comparable with that of full-length WsaE (Fig. 3, lane 3); product formation with WsaE_C was less pronounced (Fig. 3, lane 4), whereas WsaE_A did not react at all (Fig. 3, lane 2). These results indicate that both glycosyltransferase domains are necessary for full activity of the enzyme, with WsaE_C being the critical domain for formation of the α1,2-linkage. Obviously, the N-terminal methyltransferase domain is not crucial for the transfer of rhamnose from dTDP-β-l-Rha, since WsaE_B, lacking the methyltransferase domain, is fully active. Since none of the synthetic substrates contained a β1,2-linked rhamnose residue, which would be the substrate for the α1,3-rhamnosyltransferase required to form the repeats of the S-layer glycan, the reaction product of WsaF, which is β-l-Rha-(1→2)-α-l-Rha-(1→3)-β-d-Gal-(1→O)-octyl (IV), was used as a substrate for WsaC and WsaE. Unexpectedly, no product was formed by WsaC, whereas two products were formed by WsaE according to the TLC evidence (data not shown; see NMR data below).
FIGURE 3.
TLC pattern of rhamnosyltransferase activity assays of the different truncated forms of WsaE using α-l-Rha-(1→3)-α-l-Rha-(1→3)-β-d-Gal-(1→O)-octyl (III) as substrate. The products were separated on silica TLC plates with chloroform/methanol/water 65:25:4 as solvent; the representative part of the TLC plate is shown. Lane 1, WsaE; lane 2, WsaE_A; lane 3, WsaE_B; lane 4, WsaE_C. Staining was performed with thymol.
WsaD did not react with any of the substrates (Fig. S6, lanes 1-3). None of the transferases gave a new product when incubated with dTDP-β-l-Rha alone or with the donor substrates alone (Fig. S6, lanes 4-7, 15-18, 27, and 31) as negative controls.
WsaE and WsaF retained their activity after purification by affinity chromatography. In contrast, according to an in vitro assay of purified WsaC containing LDAO or CHAPS, the enzyme activity of these preparations was lost, suggesting that a lipid environment is necessary for enzymatic activity of WsaC or that the enzyme loses critical folding properties during purification. This was confirmed by the inactivity of WsaC_I lacking the transmembrane domain (see above). Concerning usage of CHAPS, conflicting data exist in the literature, ranging from promotion (39) to loss of enzyme activity (40).
Analysis of the Rhamnosyltransferase Products by ESI-QTOF MS—To further support the composition of the different octyl-linked reaction products, purified products obtained from large scale in vitro assays were analyzed by ESI-QTOF MS. The masses obtained for the different products were in accordance with the glycan chain length predicted from the migration of the samples on TLC plates (Table S3). To confirm the identity of the sugar residues, the product ions were fragmented by MS2. For instance, the MS2 spectrum of the singly charged ion at m/z 899.40 corresponds to the product (VI) of the in vitro assay of WsaC (Fig. S7). The singly charged ions at m/z 753.34, 607.29, 461.23, and 315.18 correspond to the subsequent loss of four rhamnoses.
Analysis of the Rhamnosyltransferase Products by NMR—For the characterization of the linkage specificity of WsaC, WsaE and WsaF, respectively, the newly synthesized octyl-linked saccharides were analyzed by NMR spectroscopy. To obtain sufficient amounts of material, the large scale reaction mixtures were applied to chromatography on Sephadex G-10 with 20% methanol as solvent. Although separation of remaining substrate and products was possible for all enzymes, the two products (V and VI) of WsaC could not be separated. The yields of individual reactions given below do not reflect absolute reaction yields, because some of the enzyme products were sacrificed in order to achieve purity, and due to limited amounts of substrates the reaction conditions were not optimized. For comparison with the enzyme products, NMR data of the substrate octyl trisaccharide (III) were fully assigned and compared favorably with those of the corresponding glycosyl units in the S-layer core glycan (41) (Table 1).
TABLE 1.
400.13 MHz 1H and 100.62 MHz 13C NMR chemical shift data (in ppm) and J coupling constants of the rhamnosyl oligosaccharides (III, V, VII, and IV)
Chemical shift data were measured at 297 K for solutions in D2O. 13C NMR data were obtained from a proton-decoupled one-dimensional spectrum for compounds III, IV, and V and from HSQC spectra of compound VII.
|
Carbohydrate signals |
Octyl |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 1 | 2 | CH3 | |
| Compound III | |||||||||
| A →3)-β-d-Galp-(1 → | |||||||||
| 1H | 4.42 | 3.61 | 3.68 | 4.00 | 3.70 | 3.77/3.73 | 3.89/3.67 | 1.62 | 0.85 |
| JH,H (Hz) | (7.8) | (9.9) | 81.36 | (3.1, 1.0) | 75.83 | 61.63 | 71.46 | 29.51 | 14.19 |
| 13C | 103.32 | 70.89a | 69.23 | ||||||
| B →3)-α-l-Rhap-(1 → | |||||||||
| 1H | 5.00 | 4.15 | 3.89 | 3.54 | 3.80 | 1.28 | |||
| JH,H (Hz) | (1.8) | (3.1) | (9.7) | (9.7) | 70.12 | (6.2) | |||
| 13C | 103.05 | 70.67 | 79.15 | 72.08 | 17.46b | ||||
| C α-l-Rhap-(1 → | |||||||||
| 1H | 5.04 | 4.06 | 3.84 | 3.45 | 3.80 | 1.29 | |||
| JH,H (Hz) | (1.7) | (3.3) | (9.7) | (9.6) | 69.93 | (6.3) | |||
|
13C
|
103.22
|
70.96a |
70.96a |
72.81
|
17.38b |
||||
| Compound V | |||||||||
| A →3)-β-d-Galp-(1 → | |||||||||
| 1H | 4.43 | 3.61 | 3.68 | 4.00 | 3.69 | 3.78-3.72 | 3.94/3.68 | 1.65-1.58 | 0.85 |
| JH,H (Hz) | (7.6) | (9.6) | 81.38 | (3.2) | 75.84 | 61.63 | 71.47 | 29.57 | 14.19 |
| 13C | 103.32 | 70.89a | 69.24 | ||||||
| B →3)-α-l-Rhap-(1 → | |||||||||
| 1H | 5.02b | 4.14c | 3.90 | 3.56d | 3.88-3.82 | 1.29c | |||
| JH,H (Hz) | (2.0) | (3.6) | (9.6) | (9.6) | 70.12 | (6.2) | |||
| 13C | 103.05f | 70.76a | 79.17 | 72.08 | 17.44 | ||||
| C →3)-α-l-Rhap-(1 → | |||||||||
| 1H | 5.01b | 4.17c | 3.90 | 3.54d | 3.92-3.88 | 1.28e | |||
| JH,H (Hz) | (2.0) | (3.2) | (9.6) | (9.6) | 70.12 | 17.44g | |||
| 13C | 103.26f | 70.96a | 79.17 | 72.08 | |||||
| D α-l-Rhap-(1 → | |||||||||
| 1H | 5.04 | 4.07 | 3.84 | 3.46 | NDh | 1.29e | |||
| JH,H (Hz) | (1.6) | (3.2) | (9.6) | (9.6) | 69.93 | 17.37g | |||
|
13C
|
100.82
|
70.96a |
70.65a |
72.79
|
|||||
| Compound VII | |||||||||
| A →3)-β-d-Galp-(1 → | |||||||||
| 1H | 4.40 | 3.55 | 3.66 | 3.97 | ∼3.66 | ∼3.70 | 3.83/3.64 | 1.59 | 0.82 |
| JH,H (Hz) | (7.6) | (9.6) | 81.5 | (2.9, 1.0) | 75.9 | 61.7 | 71.5 | 29.5 | 14.8 |
| 13C | 103.4 | 70.7 | 69.3 | ||||||
| B →3)-α-l-Rhap-(1 → | |||||||||
| 1H | 4.97 | 4.10 | 3.76 | 3.52 | 3.80-3.89 | 1.26c | |||
| JH,H (Hz) | (1.6) | (3.3) | (10.1) | (10.1) | ND | (6.0) | |||
| 13C | 103.1 | 70.7 | 79.1 | 72.5 | 17.4 | ||||
| C →2)-α-l-Rhap-(1 → | |||||||||
| 1H | 5.17 | 4.03 | 3.91 | 3.45 | 3.80-3.89 | 1.25c | |||
| JH,H (Hz) | (1.6) | (3.2) | (9.8) | (9.9) | ND | (6.1) | |||
| 13C | 101.7 | 78.5 | 70.0 | 73.0 | 17.4 | ||||
| D α-l-Rhap-(1 → | |||||||||
| 1H | 4.92 | 4.03 | 3.86 | 3.40 | 3.63 | 1.22 | |||
| JH,H (Hz) | (1.6) | (3.2) | (9.6) | (9.6) | 70.6 | (6.4) | |||
|
13C
|
103.1
|
71.0
|
71.0
|
73.0
|
17.4
|
||||
| Compound IV | |||||||||
| A →3)-β-d-Galp-(1 → | |||||||||
| 1H | 4.43 | 3.59 | 3.68 | 4.01 | 3.70 | 3.75/3.65 | 3.92/3.67 | 1.59 | 0.82 |
| JH,H (Hz) | (7.8) | (9.9) | 81.45 | (3.0, 1.0) | 75.74 | 61.51 | 71.37b | 29.41 | 14.11 |
| 13C | 103.20 | 70.86a | 69.12 | ||||||
| JC,H (Hz) | (163.8) | ||||||||
| B →2)-α-l-Rhap-(1 → | |||||||||
| 1H | 5.14 | 4.25 | 3.84 | 3.48 | 3.84 | 1.28 | |||
| JH,H (Hz) | (1.5) | (3.5) | (9.8) | (9.7) | 70.04c | (6.3) | |||
| 13C | 100.74 | 79.14 | 70.16c | 73.01d | 17.28 | ||||
| JC,H (Hz) | (172.5) | ||||||||
| C β-l-Rhap-(1 → | |||||||||
| 1H | 4.70 | 4.01 | 3.58 | 3.38 | 3.38 | 1.31 | |||
| JH,H (Hz) | (1.5) | (3.3) | 71.61 | (9.5) | 72.91d | (5.7) | |||
| 13C | 99.13 | 70.69a | 72.61d | 17.28 | |||||
| JC,H (Hz) | (160.5) | ||||||||
Assignments are interchangeable.
Assignments are interchangeable.
Assignments are interchangeable.
ND, not determined.
For WsaC, the conversion of 5 mg of α-l-Rha-(1→3)-α-l-Rha-(1→3)-β-d-Gal-(1→O)-octyl (III) yielded 2.9 mg of purified product. Within the detection limit of the NMR spectrometer, only signals of product V were observed in the NMR spectra (Fig. 4a). Product V was shown to be a tetrasaccharide fully amenable for NMR analysis. The 1H NMR spectrum recorded at 400 MHz revealed three closely spaced signals in the anomeric region attributable to α-rhamnopyranosyl units. Heteronuclear single quantum coherence spectroscopy and COSY experiments allowed a complete assignment of the spin-spin correlations and provided evidence for the substitution at carbon-3 due to the glycosylation shifts observed for units B and C, respectively, whereas the 13C NMR shifts for residue D were consistent with a terminal rhamnose moiety (Table 1 and Fig. 5). Moreover, due to the similar substitution pattern of both units B and C, the intensities of the C-2, C-3, C-4, and C-5 13C NMR signals corresponded to two atoms, respectively (data not shown). Thus, NMR analysis of the major WsaC product (V) clearly demonstrated that WsaC is an α1,3-rhamnosyltransferase, which is able to transfer at least one rhamnose residue to an α1,3-linked rhamnose. Considering the results from TLC and ESI-QTOF MS analysis of WsaC products (Fig. S7), it is very likely that WsaC is also capable of transferring one more α1,3-linked rhamnose to product V, yielding compound VI, which implies that this enzyme is responsible for the core variability present in the S-layer glycan (17).
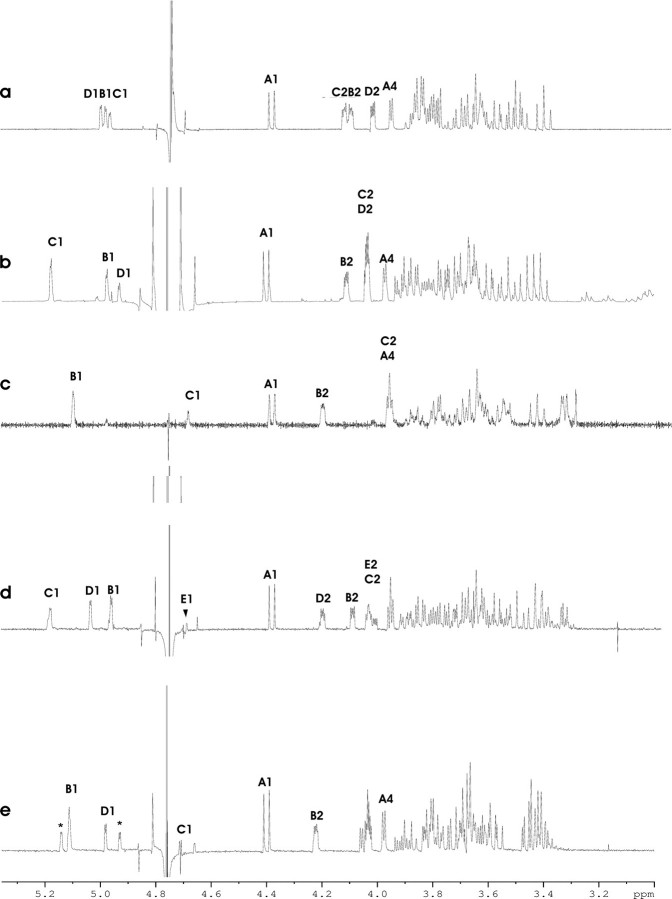
FIGURE 4.
400 MHz 1H NMR spectra of the anomeric and bulk region of the products V (a), VII (b), IV (c), VIII (d), and IX (e), measured in D2O at 297 K. The Arabic numerals refer to protons in sugar residues denoted by letters, as shown in Fig. 5. *, the anomeric proton signals of the terminal disaccharide part in the tentative pentasaccharide component (X).
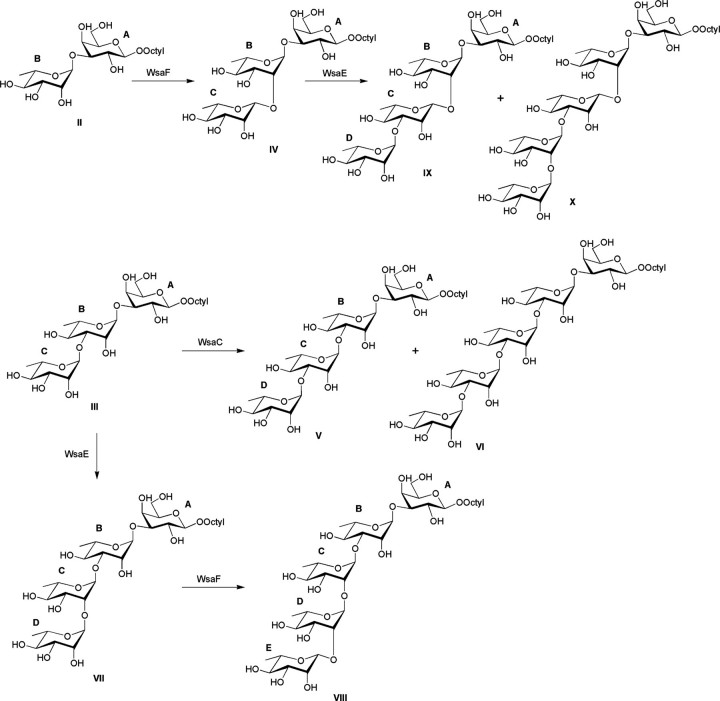
FIGURE 5.
Reaction scheme for the conversion of the synthetic substrates II and III to products IV, V, and VII and subsequently to VI, VIII, IX, and X, involving the enzymes WsaC, WsaE, and WsaF of G. stearothermophilus NRS 2004/3a. For the assignment of glycoses A, B, C, D, and E, see Table 1.
Incubation of WsaE with 5 mg of starting material (III) and dTDP-β-l-Rha yielded 0.96 mg of purified reaction product (VII). Analysis of the spin-spin connectivities by homo- and heteronuclear correlation experiments allowed the identification of unit B as a 3-substituted α-rhamnosyl moiety based on the downfield-shifted 13C NMR signal of C-3 (Table 1 and Fig. 4b), whereas the anomeric proton of unit C was connected to a C-2 carbon atom displaying a downfield shift to 78.5 ppm. The remaining anomeric proton of residue D could be assigned to a terminal α-rhamnosyl unit (Fig. 5). Since both H-2 protons of units C and D had identical chemical shifts, the assignment of H-1 of unit C was inferred from the upfield-shifted signal of the corresponding anomeric carbon signal in unit C at 101.7 ppm (Table 1). The NMR analysis thus clearly established the structure of the tetrasaccharide octyl glycoside (VII) and thereby identified WsaE as an enzyme with α1,2-rhamnosyltransferase activity.
WsaF converted 3 mg of α-l-Rha-(1→3)-β-d-Gal-(1→O)-octyl (II) to 0.59 mg of new product (IV). The 1H NMR spectrum of the product indicated a new, upfield-shifted signal hidden under the solvent water peak (Fig. 4c). Measurement of the heteronuclear JC-1,H-1 coupling constants clearly proved that residue C had the β-anomeric configuration and confirmed the α-configuration of unit B (Table 1 and Fig. 5). In addition, the downfield-shifted 13C NMR signal of carbon-2 in unit B proved C-2 as the linkage site. Thus, WsaF is a retaining glycosyltransferase responsible for the β1,2-linkage in the polyrhamnan. However, α-l-Rha-(1→3)-β-d-Gal-(1→O)-octyl (II) is not the natural substrate for the β1,2-rhamnosyltransferase in the biosynthesis of the glycan chain. It would need an α1,2-linked rhamnose substrate according to the glycan structure of G. stearothermophilus NRS 2004/3a (16). Since WsaF could only use α-l-Rha-(1→3)-β-d-Gal-(1→O)-octyl (II) and not α-l-Rha-(1→3)-α-l-Rha-(1→3)-β-d-Gal-(1→O)-octyl (III) (Fig. S6, lanes 29 and 30), it does not generally recognize α1,3-linked rhamnoses. An explanation for the recognition could be that the combination of α1,3-linked rhamnose with galactose and octyl can bind to the catalytic domain. Due to the unavailability of structural data on WsaF or a homologous protein, this explanation remains speculative.
The natural substrate for WsaF is the α-l-Rha-(1→2)-α-l-Rha-(1→3)-α-l-Rha-(1→3)-β-d-Gal-(1→O)-octyl product (VII) of WsaE (see above). After NMR analysis, this product (0.7 mg) was used for an in vitro activity assay of WsaF. The new product (VIII) (0.31 mg) was analyzed by TLC (data not shown), purified, and applied to NMR analysis, which confirmed that WsaF is indeed a β1,2-rhamnosyltransferase. Although a full assignment of the product mixture could not be accomplished, the spectral features of the distal disaccharide part structure in VIII were similar to that of compound IV (Fig. 4d). In particular, upfield-shifted anomeric 1H and 13C NMR signals of the terminal rhamnose were consistent with the β-anomeric configuration, whereas a downfield-shifted C-2 signal (77.8 ppm) was observed for unit D (data not shown).
To analyze the second reaction product of WsaE, a large scale reaction with 0.5 mg of β-l-Rha-(1→2)-α-l-Rha-(1→3)-β-d-Gal-(1→O)-octyl (IV; see above) yielded 0.25 mg of a purified product mixture. The 1H NMR spectrum of the mixture revealed the presence of additional anomeric protons indicative of a mixture containing a tetrasaccharide (IX) and a pentasaccharide (X) (Fig. 4e). Although a full assignment of the tetrasaccharide (IX) could not be accomplished, the 13C NMR signal of carbon-3 of the β-rhamnosyl residue C was observed at 81.1 ppm, indicative of α-substitution at position 3. The anomeric 1H/13C signals of the terminal α-linked rhamnose unit D were observed at 5.01/101.61 ppm, respectively (Fig. 5). Two minor additional anomeric proton and carbon signals were observed with chemical shifts (5.16/100.88 ppm and 4.95/102.65 ppm, respectively) similar to the terminal disaccharide part of compound VII (Fig. 4b). These data thus suggest a further chain elongation of unit D by a rhamnose unit in α1,2-linkage. Thus, NMR analysis provided evidence that WsaE is obviously a multifunctional enzyme, which is also responsible for the transfer of an α1,3-linked rhamnose residue to the β1,2-linked rhamnose and, most probably, the subsequent addition of an α1,2-linked rhamnose.
Methyltransferase Activity of WsaE—To determine if the putative methyltransferase domain of WsaE is responsible for the methylation of the non-reducing end of the terminal repeating unit of the glycan chain of G. stearothermophilus NRS 2004/3a, a cell free lysate of E. coli BL21 Star (DE3) expressing WsaE was incubated with [3H]SAM and α-l-Rha-(1→3)-α-l-Rha-(1→3)-β-d-Gal-(1→O)-octyl (III), either with or without addition of dTDP-β-l-Rha. According to the TLC evidence WsaE was able to methylate α-l-Rha-(1→3)-α-l-Rha-(1→3)-β-d-Gal-(1→O)-octyl (III) (Fig. 6, lanes 1, 2 and 11, 12). Only this product with an Rf value of 0.47 was formed, even if dTDP-β-l-Rha was added and one additional α1,2-linked rhamnose residue was linked to the substrate (compare with Fig. 3, lane 1), indicating that WsaE cannot methylate α1,2-linked rhamnose. This result was verified by MS analysis of a scaled up reaction mixture containing unlabeled SAM and dTDP-β-l-Rha (Table 2). Peaks corresponding to [M + H]+,[M + NH4]+, and [M + Na]+ of unmethylated substrate l-Rha-(1→3)-α-l-Rha-(1→3)-β-d-Gal-(1→O)-octyl (III) and product α-l-Rha-(1→2)-α-l-Rha-(1→3)-α-l-Rha-(1→3)-β-d-Gal-(1→O)-octyl (VII) as well as methylated substrate (III) could be observed, whereas no peak corresponded to the expected mass of methylated product (VII), showing that this reaction product of WsaE, which was proven by NMR to contain a terminal α1,2-rhamnose, could not be methylated.
FIGURE 6.
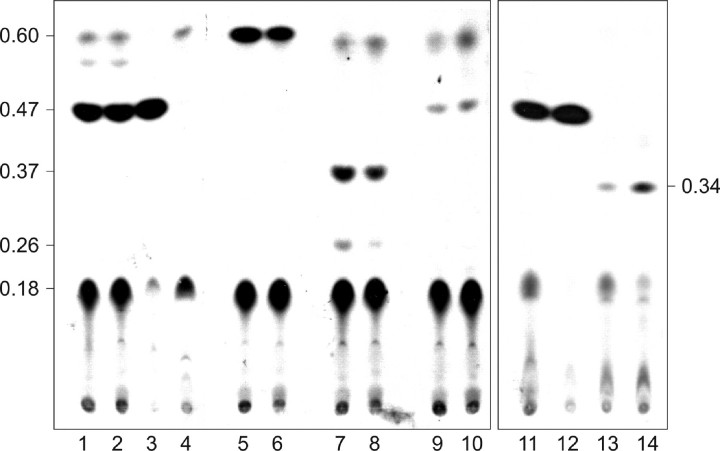
Autoradiogram of methyltransferase activity assays of WsaE and the different truncated versions of WsaE, using α-l-Rha-(1→3)-α-l-Rha-(1→3)-β-d-Gal-(1→O)-octyl (III) (lanes 1-4, 11, and 12), α-l-Rha-(1→3)-β-d-Gal-(1→O)-octyl (II) (lanes 5 and 6), the reaction products of WsaC α-l-Rha-(1→3)-α-l-Rha-(1→3)-α-l-Rha-(1→3)-β-d-Gal-(1→O)-octyl (V and VI) (lanes 7 and 8), α-l-Rha-(1→2)-α-l-Rha-(1→3)-α-l-Rha-(1→3)-β-d-Gal-(1→O)-octyl (VII) (lanes 9 and 10), or α-l-Rha-(1→3)-β-l-Rha-(1→2)-α-l-Rha-(1→3)-β-d-Gal-(1→O)-octyl (IX) (lanes 13 and 14) as substrate. In lanes 1, 6, 8, 10, 12, and 14, dTDP-β-l-Rha was added; all lanes except for lanes 3 and 4 contain WsaE. Lane 3, WsaE_N; lane 4, WsaE_M. The products were separated on silica TLC plates with chloroform/methanol/water (65:25:4) as solvent.
TABLE 2.
ESI-QTOF MS analysis of octyl-linked products of an in vitro methylrhamnosyltransferase activity assay of WsaE with substrate III
|
Assignment |
[M + H]+ |
[M + NH4]+ |
[M + Na]+ |
|||
|---|---|---|---|---|---|---|
| Experimental | Theoretical | Experimental | Theoretical | Experimental | Theoretical | |
| Rha-Rha-Gal-octyl (III) | 585.35 | 585.31 | 602.33 | 607.36 | 607.29 | |
| Methyl-Rha-Rha-Gal-octyl | 599.36 | 599.33 | 616.39 | 616.35 | 621.34 | 621.31 |
| Rha-Rha-Rha-Gal-octyl (VII) | 731.40 | 731.37 | 748.44 | 748.39 | 753.39 | 753.35 |
| Methyl-Rha-Rha-Rha-Gal-octyl | 745.39 | 762.41 | 767.37 | |||
To confirm that the N terminus of WsaE is responsible for the methyltransferase activity of the enzyme, the two truncated forms WsaE_M (including the UbiE motive) and WsaE_N (including the UbiE motive and additional 200 aa) were incubated with [3H]SAM. Compared with full-size WsaE, WsaE_N (Fig. 6, lane 3) showed an even higher activity, whereas the shorter form WsaE_M was inactive (Fig. 6, lane 4). These results confirmed that the N terminus is sufficient for methyltransferase activity. To further establish that WsaE methylates only α1,3-linked rhamnose residues, independent of the glycan chain length, α-l-Rha-(1→3)-β-d-Gal-(1→O)-octyl (II); the reaction products of WsaC, which are α-l-Rha-(1→3)-α-l-Rha-(1→3)-α-l-Rha-(1→3)-β-d-Gal-(1→O)-octyl (V) and (VI); and both reaction products of the rhamnosyltransferase domains of WsaE, which are α-l-Rha-(1→2)-α-l-Rha-(1→3)-α-l-Rha-(1→3)-β-d-Gal-(1→O)-octyl (VII) and α-l-Rha-(1→3)-β-l-Rha-(1→2)-α-l-Rha-(1→3)-β-d-Gal-(1→O)-octyl (IX), were used as substrates for WsaE. All reactions were done with and without dTDP-β-l-Rha but did not show any difference (compare neighboring lanes in Fig. 6). WsaE methylated all α1,3-linked rhamnoses, resulting in products with an Rf value of 0.60 for substrate (II) (Fig. 6, lanes 5 and 6), with Rf values of 0.37 and 0.26 for the products (V) and (VI) of WsaC (Fig. 6, lanes 7 and 8) and with an Rf value of 0.34 for the α1,3-linked product (IX) of WsaE (Fig. 6, lanes 13 and 14). Note that all Rf values of methylated products are increased as compared with the Rf values of the unmethylated substrates (compare with Figs. 2 and 4). WsaE was not able to methylate the α1,2-linked rhamnose of (VII) (Fig. 6, lanes 9 and 10). The faint band at the same Rf value (Rf ∼ 0.47) as the reaction product of WsaE with α-l-Rha-(1→3)-α-l-Rha-(1→3)-β-d-Gal-(1→O)-octyl (III) most likely corresponds to methylation of residual substrate (III) being present as minor impurity in the tetrasaccharide sample α-l-Rha-(1→2)-α-l-Rha-(1→3)-α-l-Rha-(1→3)-β-d-Gal-(1→O)-octyl (VII). These results confirm the methylation of the α1,3-linked rhamnose of the terminal repeating unit of the glycan chain of G. stearothermophilus NRS 2004/3a (16) and furthermore show that WsaE indeed possesses a relaxed specificity for the glycan chain length.
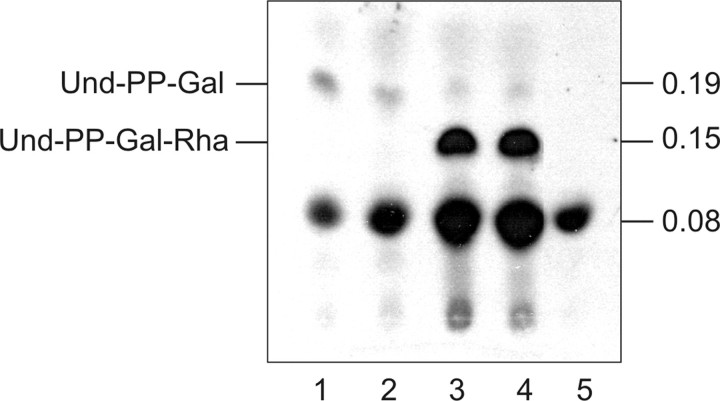
Substrate Specificity of WsaD—Considering the overall composition of the S-layer glycan of G. stearothermophilus NRS 2004/3a, the specificities that could be attributed to the transferases WsaC, WsaE, and WsaF (this study), and the lack of activity of WsaD in the aforementioned experiments, it was hypothesized that WsaD should correspond to the rhamnosyltransferase, which transfers the first rhamnose residue from dTDP-β-l-Rha to the lipid-linked galactose in the course of S-layer glycan biosynthesis. Consequently, WsaD should be able to recognize the reaction product of WsaP, which is lipid-PP-linked galactose, as substrate. For this experiment, equal amounts of lysed E. coli C43 (DE3) cells expressing either of the enzymes were mixed with 14C-labeled UDP-Gal and unlabeled dTDP-β-l-Rha and incubated overnight. As negative controls, each enzyme was incubated alone. The reaction products were analyzed by TLC and autoradiography (Fig. 7, lanes 1-5). In all reaction mixtures, a spot at an Rf value of 0.08 was visible. However, the reaction product with an Rf value of 0.19 was formed only in the presence of WsaP (Fig. 7, lanes 1-4), indicating that it corresponds to und-PP-Gal as described before (22). The formation of this product was not dependent on the addition of dTDP-β-l-Rha (Fig. 7, compare lanes 1 and 2). A new product with a lower Rf value (Rf ∼ 0.15) than the reaction product of WsaP (Rf ∼ 0.19) was synthesized when WsaD was added to the reaction mixture (Fig. 7, lanes 3 and 4). Interestingly, this product was also formed without the addition of dTDP-β-l-Rha (Fig. 7, compare lanes 3 and 4). Apparently, dTDP-β-l-Rha present in E. coli C43 (DE3) cells can be used for the reaction (42). The negative control WsaD alone did not give any new product (Fig. 7, lane 5). These findings indicate that WsaD is responsible for the transfer of the first rhamnose to the lipid-linked und-PP-Gal intermediate.
FIGURE 7.
TLC autoradiogram showing lipid carrier-linked intermediates synthesized in an in vitro assay using membranes isolated from E. coli C43 (DE3) harboring WsaP and/or WsaD and 14C-labeled-UDP-Gal. Lane 1, WsaP; lane 2, WsaP with dTDP-Rha; lane 3, WsaP and WsaD; lane 4, WsaP and WsaD with dTDP-Rha; lane 5, WsaD with dTDP-Rha. Products were separated on silica plates by TLC using the solvent system chloroform/methanol/water (65:25:4). The representative part of the TLC plate is shown.
DISCUSSION
In our study to determine the molecular basis for S-layer glycoprotein glycan biosynthesis in G. stearothermophilus NRS 2004/3a, we used recombinant enzymes as well as chemically synthesized substrates, linked to a hydrophobic octyl spacer, representing partial structural elements of the mature S-layer glycan. In G. stearothermophilus NRS 2004/3a, the glycan chain is linked to the protein via β-galactose (Gal) (16). Thus, β-Gal-octyl (I), was tested as a first substrate. It was unexpected to find that β-Gal-octyl (I) was not recognized by any of the transferases encoded by the slg gene cluster of the organism. A possible explanation might be that for the activity of the first rhamnosyltransferase, a pyrophosphate linkage of the galactose to the lipid carrier as is present in the native substrate would be essential. Another explanation for the lack of reactivity with β-Gal-octyl (I) might be that actually an α-linked Gal residue would be needed as a substrate for the first rhamnosyltransferases to act on, in the case that the possible oligosaccharyltransferase is inverting the linkage type during the transfer of the glycan onto the S-layer protein. Recently, the pgl (protein N-glycosylation) gene cluster of Campylobacter jejuni was described (43), where for the in vitro characterization of the glycosyltransferase PglA, synthetic und-PP-α-Bac was used as a substrate (44, 45). Although the glycan chain is linked via β-bacillosamine to Asn of the protein (46), the α-substrate is recognized by PglA, indicating that the glycan chain is α-linked to und-P and that the oligosaccharyltransferase PglB (47) is inverting the anomeric configuration of the linkage. Although nothing is known about the reaction mechanism of the respective oligosaccharyltransferase WsaB of G. stearothermophilus NRS 2004/3a, it might well be that protein glycosylation generally follows a common mechanism, involving the inversion of the stereochemistry at the reducing end of the glycan chain during its transfer from the lipid carrier onto the protein.
There have been previous models for LPS biosynthesis in Gram-negative bacteria (21) but none to date for a cell wall glycoconjugate of Gram-positive bacteria. Based on our data on a Gram-positive bacterium, we can propose a stepwise biosynthesis of the S-layer glycan of G. stearothermophilus NRS 2004/3a. The presence of a predicted ABC-2-type transporter and the absence of a putative polymerase in the slg gene cluster of G. stearothermophilus NRS 2004/3a (19) indicate that the S-layer glycan chains are most probably synthesized in a process comparable with the ABC transporter-dependent pathway of LPS O-PS biosynthesis (21). In our system, the previously characterized WsaP protein, which is a homologue of WbaP, serves as the initiation enzyme instead of a WecA, which usually acts as the initiation enzyme in that pathway (21, 22, 48). With the knowledge of the substrate specificities and the linkage types formed by the action of the different rhamnosyltransferases (this study), we can propose the following biosynthesis model for the S-layer glycan assembly (Fig. 8). WsaP is implicated in the first step of S-layer glycan synthesis, whereby galactose is transferred from its nucleotide-activated form (UDP-Gal) to a membrane-associated lipid carrier at the cytoplasmic face of the plasma membrane (22). In the next step, the first adaptor rhamnose is α1,3-linked from dTDP-β-l-rhamnose to the lipid-bound galactose by the action of WsaD, followed by the transfer of one or two additional α1,3-linked rhamnoses to the first α1,3-linked rhamnose by the action of WsaC to complete the adaptor. The different numbers of α1,3-linked rhamnoses transferred by WsaC would be reflected by the core variability of the S-layer glycan (17). Adaptor formation has also been described for other homopolymeric glycan chains, such as E. coli O8, O9a, and O9 antigen (49) or Klebsiella pneumoniae O3 and O5, where the adaptor is synthesized by the action of the mannosyltransferases WbdC and WbdB or homologues thereof, to transfer the initial mannosyl residue to the priming und-PP-GlcNAc, followed by the addition of another α1,3-linked mannosyl residue. In G. stearothermophilus NRS 2004/3a, chain extension and formation of repeating unit-like structures presumably continues in the cytoplasm by the processive action of the two rhamnosyltransferases WsaE and WsaF, whereby WsaE is a multifunctional enzyme forming the α1,2- and the α1,3-linkage, and WsaF is a β1,2-rhamnosyltransferase. We can imagine a scenario in which an enzyme complex is formed that exerts multiple reaction cycles, with each transferase recognizing the reaction product of the foregoing enzyme. First, WsaE recognizes the α1,3-linked rhamnose product from the adaptor formed by WsaC and adds an α1,2-linked rhamnose to the saccharide intermediate, which is subsequently recognized by WsaF, which β1,2-links the next following rhamnose. The β1,2-linked rhamnose would then serve as acceptor for the transfer of rhamnose in α1,3-linkage and, subsequently, of another rhamnose in α1,2-linkage, both catalyzed by WsaE, with the latter transfer reaction being already the first step of the next cycle.
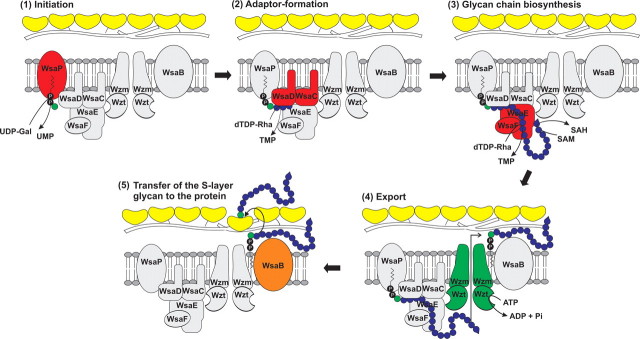
FIGURE 8.
Proposed model of S-layer glycoprotein glycan biosynthesis in G. stearothermophilus NRS 2004/3a. 1, initiation; transfer of a Gal residue from UDP-α-d-Gal to a lipid carrier catalyzed by WsaP. 2, adaptor formation; α1,3-linkage of a rhamnose residue from dTDP-β-l-Rha to the primer by the action of WsaD, followed by the transfer of one or two additional α1,3-linked rhamnoses by the action of WsaC. 3, glycan chain biosynthesis; formation of repeating unit-like structures by action of the rhamnosyltransferases WsaE and WsaF, whereby WsaE is forming the α1,2- and the α1,3-linkages, and WsaF is forming the β1,2-linkage. Chain growth is terminated by 2-O-methylation of the terminal repeating unit, catalyzed by the O-methyltransferase domain of WsaE. 4, export; the Wzt component of the Wzm/Wzt ABC transporter system is predicted to be responsible for binding of the 2-O-methylated glycan chain and its subsequent export through the membrane. 5, transfer of the S-layer glycan to the protein. The final transfer of the completed S-layer glycan to the S-layer protein would be catalyzed by the oligosaccharyltransferase WsaB.
The variation in the final chain length as shown by analysis of the native glycan (17) is potentially governed by methylation of the terminal rhamnose residue; specifically, 2-O-methylation directly blocks the hydroxyl group that would be the acceptor for any subsequent rhamnosyltransferase reaction (16). This reaction is catalyzed by an O-methyltransferase, which is located at the N-terminal protein portion of WsaE. Nothing is known, so far, about how the methylation event and S-layer glycan chain length termination are coordinated and regulated in G. stearothermophilus NRS 2004/3a. The in vitro assays of this study have shown that WsaE seems to methylate terminal α1,3-linked rhamnoses, even of very short glycan chains. Since G. stearothermophilus NRS 2004/3a is not amenable to transformation, no knock-out mutants of this WsaE domain could be designed so far to prove the influence of WsaE on the glycan chain length in vivo and to show effects on the organism. Nonreducing terminal modifications, such as methylation, have been identified on a number of glycans exported by ABC transporters in Gram-positive as well as in Gram-negative organisms; on the S-layer glycans of Thermoanaerobacter thermohydrosulfuricus L111-69 (41), Aneurinibacillus thermoaerophilus L420-91T (50), and G. tepidamans GS5-97T (51); on the O9a and O8 polysaccharides of E. coli (52, 53); and several other glycans (54). The gene clusters responsible for the biosynthesis of these polysaccharides encode a Wzt homologue with an extended C-terminal domain (12, 55), which was described to be responsible for the binding of the modified glycan chain and its subsequent export through the membrane in the biosynthesis of O8 and O9a O-PS (54).
The involvement of multifunctional enzymes in the biosynthesis of polysaccharides is not unusual. In E. coli O8 and O9a polysaccharides, for instance, chain extension is performed by a multidomain serotype-specific mannosyltransferase WbdA (56). In contrast to WsaE of G. stearothermophilus NRS 2004/3a, methylation or phosphomethylation of the nonreducing end occurs in separate steps by the action of WdbD. Although it is reported that methylation plays a critical role in chain length determination and that overexpression of WdbD reduces O-PS chain length in serotypes O8 and O9a (57), it is unclear how WdbD activity is regulated.
The described enzymatic reactions give a complete picture of how the structure of the S-layer glycan of G. stearothermophilus NRS 2004/3a, consisting of trisaccharide repeats with the structure →[2)-α-l-Rhap-(1→3)-β-l-Rhap-(1→2)-α-l-Rhap-(1→]n, with the terminal rhamnose residue at the nonreducing end modified at the hydroxyl group of carbon-2 by O-methylation, a short adaptor saccharide of α1,3-linked l-Rhap residues, and a β-d-galactose residue serving as linker to threonine 590, threonine 620, and serine 794 of the S-layer polypeptide backbone (16, 17) (Fig. 9), might be assembled. The completed methylated glycan chain would then be recognized and transported across the membrane by a process involving an ABC transporter (55) and, eventually, be transferred to the S-layer protein by an oligosaccharyltransferase WsaB. A comparable transferase function was described recently for PglL and PilO, which are involved in protein O-glycosylation in Neisseria meningitidis and Pseudomonas aeruginosa, respectively (58), and for PglB involved in N-glycosylation in C. jejuni (47, 59, 60).
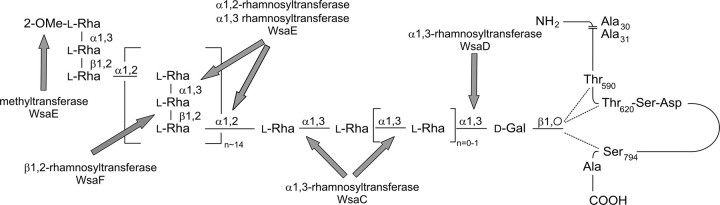
FIGURE 9.
S-layer glycoprotein glycan of G. stearothermophilus NRS 2004/3a (16). The sites of action of the different rhamnosyltransferases and the methyltransferase are indicated with arrows. Note that the α1,2- and α1,3-rhamnosyltransferase activities of WsaE and the β1,2-rhamnosyltransferase activity of WsaF also apply for the terminal repeating unit; for simplification, this is not indicated in the figure.
Considering that several other slg gene clusters contain an ABC transporter (19) and numerous S-layer glycan chains are modified at the nonreducing end (16, 41, 50, 51), the biosynthesis model proposed for G. stearothermophilus NRS 2004/3a might be more generally valid for S-layer protein glycosylation. In future studies, it will be interesting to show how these enzymes interact and work in concert in S-layer glycan biosynthesis of G. stearothermophilus NRS 2004/3a and how the modality of the glycan chain length is regulated. The understanding of S-layer glycan biosynthesis and the characterization of the involved enzymes are important steps toward the goal of engineering functional S-layer neo-glycoproteins (11, 12, 61) for applications in the fields of nanobiotechnology, biomimetics, and vaccine and drug design.
Supplementary Material
This work was supported by Austrian Science Fund Projects P19047-B12 (to C. S.) and P18013-B10 (to P. M.), Hochschuljubilaümsstiftung der Stadt Wien Project H-1809/2006 (to C. S.), ETH Zürich, and the Swiss National Science Foundation (to P. H. S.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1-S3 and Figs. S1-S7.
Footnotes
The abbreviations used are: S-layer, bacterial cell surface layer; ABC, ATP-binding cassette; CHAPS, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate; COSY, correlation spectroscopy; dTDP-β-l-Rha, dTDP-β-l-rhamnose; ESI-QTOF, electrospray ionization quadrupole time-of-fight; MS, mass spectrometry; Galp, galactopyranose; Gal, galactose; LDAO, lauryldimethylamine N-oxide; LPS, lipopolysaccharide; O-PS, O-polysaccharide; MS2, MS/MS; Rhap, rhamnopyranose; SAM, S-adenosylmethionine; und-P, undecaprenyl phosphate; und-PP, undecaprenylpyrophosphate.
References
- 1.Helenius, A., and Aebi, M. (2004) Annu. Rev. Biochem. 73 1019-1049 [DOI] [PubMed] [Google Scholar]
- 2.Szymanski, C. M., and Wren, B. W. (2005) Nat. Rev. Microbiol. 3 225-237 [DOI] [PubMed] [Google Scholar]
- 3.Eichler, J., and Adams, M. W. (2005) Microbiol. Mol. Biol. Rev. 69 393-425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rudd, P. M., Elliott, T., Cresswell, P., Wilson, I. A., and Dwek, R. A. (2001) Science 291 2370-2376 [DOI] [PubMed] [Google Scholar]
- 5.Sharon, N., and Lis, H. (2004) Glycobiology 14 53R-62R [DOI] [PubMed] [Google Scholar]
- 6.Ohtsubo, K., and Marth, J. D. (2006) Cell 126 855-867 [DOI] [PubMed] [Google Scholar]
- 7.Seeberger, P. H., and Werz, D. B. (2007) Nature 446 1046-1051 [DOI] [PubMed] [Google Scholar]
- 8.Elliott, S., Lorenzini, T., Asher, S., Aoki, K., Brankow, D., Buck, L., Busse, L., Chang, D., Fuller, J., Grant, J., Hernday, N., Hokum, M., Hu, S., Knudten, A., Levin, N., Komorowski, R., Martin, F., Navarro, R., Osslund, T., Rogers, G., Rogers, N., Trail, G., and Egrie, J. (2003) Nat. Biotechnol. 21 414-421 [DOI] [PubMed] [Google Scholar]
- 9.Stevens, J., Blixt, O., Tumpey, T. M., Taubenberger, J. K., Paulson, J. C., and Wilson, I. A. (2006) Science 312 404-410 [DOI] [PubMed] [Google Scholar]
- 10.Schäffer, C., Novotny, R., Küpcü, S., Zayni, S., Scheberl, A., Friedmann, J., Sleytr, U. B., and Messner, P. (2007) Small 3 1549-1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schäffer, C., and Messner, P. (2004) Glycobiology 14 31R-42R [DOI] [PubMed] [Google Scholar]
- 12.Messner, P., Steiner, K., Zarschler, K., and Schäffer, C. (2008) Carbohydr. Res. 343 1934-1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schuster, B., Pum, D., Sára, M., and Sleytr, U. B. (2006) Mini Rev. Med. Chem. 6 909-920 [DOI] [PubMed] [Google Scholar]
- 14.Sleytr, U. B., Egelseer, E. M., Ilk, N., Pum, D., and Schuster, B. (2007) FEBS J. 274 323-334 [DOI] [PubMed] [Google Scholar]
- 15.Sleytr, U. B., Huber, C., Ilk, N., Pum, D., Schuster, B., and Egelseer, E. M. (2007) FEMS Microbiol. Lett. 267 131-144 [DOI] [PubMed] [Google Scholar]
- 16.Schäffer, C., Wugeditsch, T., Kählig, H., Scheberl, A., Zayni, S., and Messner, P. (2002) J. Biol. Chem. 277 6230-6239 [DOI] [PubMed] [Google Scholar]
- 17.Steiner, K., Pohlentz, G., Dreisewerd, K., Berkenkamp, S., Messner, P., Peter-Katalinić, J., and Schäffer, C. (2006) J. Bacteriol. 188 7914-7921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Novotny, R., Schäffer, C., Strauss, J., and Messner, P. (2004) Microbiology 150 953-965 [DOI] [PubMed] [Google Scholar]
- 19.Novotny, R., Pfoestl, A., Messner, P., and Schäffer, C. (2004) Glycoconj. J. 20 435-447 [DOI] [PubMed] [Google Scholar]
- 20.Power, P. M., and Jennings, M. P. (2003) FEMS Microbiol. Lett. 218 211-222 [DOI] [PubMed] [Google Scholar]
- 21.Raetz, C. R. H., and Whitfield, C. (2002) Annu. Rev. Biochem. 71 635-700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steiner, K., Novotny, R., Patel, K., Vinogradov, E., Whitfield, C., Valvano, M. A., Messner, P., and Schäffer, C. (2007) J. Bacteriol. 189 2590-2598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valvano, M. A. (2003) Front. Biosci. 8 S452-S471 [DOI] [PubMed] [Google Scholar]
- 24.Laemmli, U. K. (1970) Nature 227 680-685 [DOI] [PubMed] [Google Scholar]
- 25.Graninger, M., Kneidinger, B., Bruno, K., Scheberl, A., and Messner, P. (2002) Appl. Environ. Microbiol. 68 3708-3715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fürstner, A., and Müller, T. (1999) J. Am. Chem. Soc. 121 7814-7821 [Google Scholar]
- 27.Love, K. R., and Seeberger, P. H. (2004) Angew. Chem. Int. Ed. 43 602-605 [DOI] [PubMed] [Google Scholar]
- 28.Werz, D. B., and Seeberger, P. H. (2005) Angew. Chem. Int. Ed. 44 6315-6318 [DOI] [PubMed] [Google Scholar]
- 29.Werz, D. B., Ranzinger, R., Herget, S., Adibekian, A., von der Lieth, C.-W., and Seeberger, P. H. (2007) ACS Chem. Biol. 2 685-691 [DOI] [PubMed] [Google Scholar]
- 30.Adachi, S. (1965) J. Chromatogr. 17 295-299 [DOI] [PubMed] [Google Scholar]
- 31.Wolucka, B. A., and de Hoffmann, E. (1998) Glycobiology 8 955-962 [DOI] [PubMed] [Google Scholar]
- 32.Folch, J., Lees, M., and Sloane-Stanley, G. H. (1957) J. Biol. Chem. 226 497-509 [PubMed] [Google Scholar]
- 33.Saxena, I. M., Brown, R. M., Jr., Fevre, M., Geremia, R. A., and Henrissat, B. (1995) J. Bacteriol. 177 1419-1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Charnock, S. J., and Davies, G. J. (1999) Biochemistry 38 6380-6385 [DOI] [PubMed] [Google Scholar]
- 35.Tarbouriech, N., Charnock, S. J., and Davies, G. J. (2001) J. Mol. Biol. 314 655-661 [DOI] [PubMed] [Google Scholar]
- 36.Morona, R., Macpherson, D. F., Van Den, B. L., Carlin, N. I., and Manning, P. A. (1995) Mol. Microbiol. 18 209-223 [DOI] [PubMed] [Google Scholar]
- 37.Geremia, R. A., Petroni, E. A., Ielpi, L., and Henrissat, B. (1996) Biochem. J. 318 133-138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keenleyside, W. J., Clarke, A. J., and Whitfield, C. (2001) J. Bacteriol. 183 77-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rush, J. S., Rick, P. D., and Waechter, C. J. (1997) Glycobiology 7 315-322 [DOI] [PubMed] [Google Scholar]
- 40.Riley, J. G., Menggad, M., Montoya-Peleaz, P. J., Szarek, W. A., Marolda, C. L., Valvano, M. A., Schutzbach, J. S., and Brockhausen, I. (2005) Glycobiology 15 605-613 [DOI] [PubMed] [Google Scholar]
- 41.Bock, K., Schuster-Kolbe, J., Altman, E., Allmaier, G., Stahl, B., Christian, R., Sleytr, U. B., and Messner, P. (1994) J. Biol. Chem. 269 7137-7144 [PubMed] [Google Scholar]
- 42.Wang, Q., Fang, X., Bai, B., Liang, X., Shuler, P. J., Goddard, W. A., III, and Tang, Y. (2007) Biotechnol. Bioeng. 98 842-853 [DOI] [PubMed] [Google Scholar]
- 43.Szymanski, C. M., Logan, S. M., Linton, D., and Wren, B. W. (2003) Trends Microbiol. 11 233-238 [DOI] [PubMed] [Google Scholar]
- 44.Glover, K. J., Weerapana, E., and Imperiali, B. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 14255-14259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weerapana, E., Glover, K. J., Chen, M. M., and Imperiali, B. (2005) J. Am. Chem. Soc. 127 13766-13767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Young, N. M., Brisson, J. R., Kelly, J., Watson, D. C., Tessier, L., Lanthier, P. H., Jarrell, H. C., Cadotte, N., St Michael, F., Aberg, E., and Szymanski, C. M. (2002) J. Biol. Chem. 277 42530-42539 [DOI] [PubMed] [Google Scholar]
- 47.Wacker, M., Feldman, M. F., Callewaert, N., Kowarik, M., Clarke, B. R., Pohl, N. L., Hernandez, M., Vines, E. D., Valvano, M. A., Whitfield, C., and Aebi, M. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 7088-7093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Whitfield, C. (2006) Annu. Rev. Biochem. 75 39-68 [DOI] [PubMed] [Google Scholar]
- 49.Kido, N., Torgov, V. I., Sugiyama, T., Uchiya, K., Sugihara, H., Komatsu, T., Kato, N., and Jann, K. (1995) J. Bacteriol. 177 2178-2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schäffer, C., Müller, N., Christian, R., Graninger, M., Wugeditsch, T., Scheberl, A., and Messner, P. (1999) Glycobiology 9 407-414 [DOI] [PubMed] [Google Scholar]
- 51.Kählig, H., Kolarich, D., Zayni, S., Scheberl, A., Kosma, P., Schäffer, C., and Messner, P. (2005) J. Biol. Chem. 280 20292-20299 [DOI] [PubMed] [Google Scholar]
- 52.Sugiyama, T., Kido, N., Kato, Y., Koide, N., Yoshida, T., and Yokochi, T. (1998) J. Bacteriol. 180 2775-2778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vinogradov, E., Frirdich, E., MacLean, L. L., Perry, M. B., Petersen, B. O., Duus, J. O., and Whitfield, C. (2002) J. Biol. Chem. 277 25070-25081 [DOI] [PubMed] [Google Scholar]
- 54.Cuthbertson, L., Kimber, M. S., and Whitfield, C. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 19529-19534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cuthbertson, L., Powers, J., and Whitfield, C. (2005) J. Biol. Chem. 280 30310-30319 [DOI] [PubMed] [Google Scholar]
- 56.Kido, N., Sugiyama, T., Yokochi, T., Kobayashi, H., and Okawa, Y. (1998) Mol. Microbiol. 27 1213-1221 [DOI] [PubMed] [Google Scholar]
- 57.Clarke, B. R., Cuthbertson, L., and Whitfield, C. (2004) J. Biol. Chem. 279 35709-35718 [DOI] [PubMed] [Google Scholar]
- 58.Faridmoayer, A., Fentabil, M. A., Mills, D. C., Klassen, J. S., and Feldman, M. F. (2007) J. Bacteriol. 189 8088-8098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wacker, M., Linton, D., Hitchen, P. G., Nita-Lazar, M., Haslam, S. M., North, S. J., Panico, M., Morris, H. R., Dell, A., Wren, B. W., and Aebi, M. (2002) Science 298 1790-1793 [DOI] [PubMed] [Google Scholar]
- 60.Kowarik, M., Numao, S., Feldman, M. F., Schulz, B. L., Callewaert, N., Kiermaier, E., Catrein, I., and Aebi, M. (2006) Science 314 1148-1150 [DOI] [PubMed] [Google Scholar]
- 61.Steiner, K., Hanreich, A., Kainz, B., Hitchen, P. G., Dell, A., Messner, P., and Schäffer, C. (2007) Small, in press [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.