Abstract
Plant isoprenoids are derived from two biosynthetic pathways, the cytoplasmic mevalonate (MVA) and the plastidial methylerythritol phosphate (MEP) pathway. In this study their respective contributions toward formation of dolichols in Coluria geoides hairy root culture were estimated using in vivo labeling with 13C-labeled glucose as a general precursor. NMR and mass spectrometry showed that both the MVA and MEP pathways were the sources of isopentenyl diphosphate incorporated into polyisoprenoid chains. The involvement of the MEP pathway was found to be substantial at the initiation stage of dolichol chain synthesis, but it was virtually nil at the terminal steps; statistically, 6–8 isoprene units within the dolichol molecule (i.e. 40–50% of the total) were derived from the MEP pathway. These results were further verified by incorporation of [5-2H]mevalonate or [5,5-2H2]deoxyxylulose into dolichols as well as by the observed decreased accumulation of dolichols upon treatment with mevinolin or fosmidomycin, selective inhibitors of either pathway. The presented data indicate that the synthesis of dolichols in C. geoides roots involves a continuous exchange of intermediates between the MVA and MEP pathways. According to our model, oligoprenyl diphosphate chains of a length not exceeding 13 isoprene units are synthesized in plastids from isopentenyl diphosphate derived from both the MEP and MVA pathways, and then are completed in the cytoplasm with several units derived solely from the MVA pathway. This study also illustrates an innovative application of mass spectrometry for qualitative and quantitative evaluation of the contribution of individual metabolic pathways to the biosynthesis of natural products.
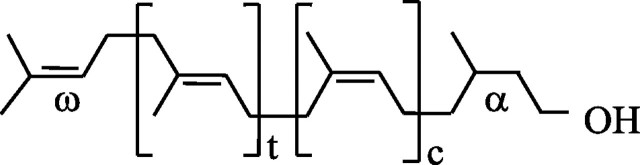
Polyisoprenoid alcohols together with sterols and quinone side chains constitute three main branches of terpene products originating from farnesyl diphosphate (FPP)4 (1). These linear five-carbon unit polymers are divided into two groups, i.e. polyprenols and dolichols, according to the hydrogenation status of the α-terminal isoprene unit (dolichol structure is shown in Fig. 1). In cells, polyprenols and dolichols are always found as mixtures of prenologues, and data collected so far show polyprenols to be typical for bacteria and plants, whereas dolichols are generally attributed to animals and yeast (2). Nevertheless, it should be remembered that dolichols are the predominant form in some plant organs like roots (3). Data on the occurrence and functions of polyisoprenoids are summarized in recently published reviews (4, 5). The formation of the polyisoprenoid chain, starting from the ω-end of the molecule (Fig. 1), proceeds in a biphasic manner with farnesyl-diphosphate synthase responsible for the synthesis of the all-trans-FPP (three isoprene units of ω-t2 structure, t stands for trans-isoprene unit), and its further elongation by cis-prenyltransferase. The latter enzyme, cloned from several prokaryotic and eukaryotic organisms (see Refs. 6, 7 and references therein), including Arabidopsis thaliana (8, 9) and Hevea brasiliensis (10), utilizes isopentenyl diphosphate (IPP) for elongation of FPP up to the desired chain length, thus producing a family of polyprenyl diphosphates (n isoprene units of ω-t2-cn-3 structure, c stands for cis-isoprene unit), which are subsequently converted to polyprenols or dolichols according to the “tissue-specific requirements” by a still unknown mechanism.
FIGURE 1.
Structure of dolichols. t and c indicate the number of internal trans- and cis-isoprene units; α and ω indicate the terminal isoprene units.
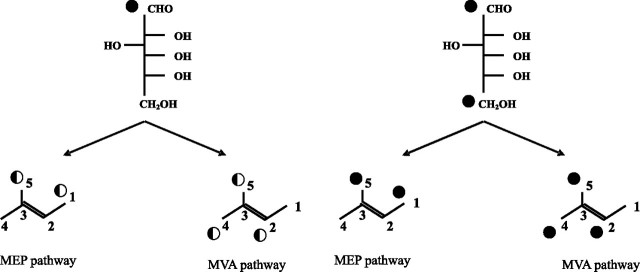
In plant cells two pathways are known to produce IPP utilized by numerous enzymes to finally give more than 50,000 different isoprenoid structures, the mevalonate pathway (MVA) and the mevalonate-independent methylerythritol phosphate pathway (MEP) (for reviews, see Refs. 11–13). Both pathways are compartmentalized as follows: the MVA in the cytoplasm to provide sterols, the many sesquiterpenes, and the prenyl chains of ubiquinones, and the MEP one in the plastids giving hemi-, mono-, and diterpenes, carotenoids, and the side chain of plastoquinone. Although both pathways are thought to operate independently under normal conditions, interactions between them have been reported repeatedly. As a result of the exchange of intermediates derived from both pathways, “mixed origin” isoprenoids are described (reviewed in Ref. 12). More recently, the potential for the cross-talk between the MEP and MVA pathways has been directly proven by genetic modifications of pathway-specific enzymes and by application of pathway-specific inhibitors, namely mevinolin (14), also referred to as lovastatin, a highly specific inhibitor of 3-hydroxy-3-methylglutaryl-coenzyme A reductase in the MVA pathway, and fosmidomycin (15), a specific inhibitor of 1-deoxy-d-xylulose-5-phosphate reductoisomerase in the MEP pathway. Both inhibitors have recently been used to perturb biosynthetic flux in hairy roots (16, 17). The involvement of both pathways leading to the formation of a studied isoprenoid compound is most often estimated by the application of specifically labeled [13C]glucose. A pathway-specific pattern of 13C label within the isoprenoid residues (Fig. 2) allows their origins via the MEP or MVA route to be discerned.
FIGURE 2.
MVA and MEP pathway-specific labeling pattern of isoprene units resulting from glucose catabolism via glycolysis (according to Ref, 27) shown separately for [1-13C]glucose and [1,6-13C2]glucose. Half-black circle indicates 13C abundance in isoprene units, which is 50% of the initial one in [1-13C]glucose (see “Results”). Numbering of carbon atoms is indicated.
Using [3H]mevalonate, it has been shown that in plants mainly cis-polyisoprenoid alcohols are synthesized via the MVA pathway (18, 19), similarly to mammalian dolichols (20, 21), although the possibility of an input from the alternative MEP pathway has not been addressed. Localization of polyprenols to plastids (22–24) and accompanying dolichols (a small fraction of total polyisoprenoid pool) to microsomes (25) might be the indication of their MEP and MVA origin, respectively. On the other hand, the MEP pathway has been found to contribute significantly to the synthesis of solanesol, an all-trans-pren-9 (26), whereas the solanesyl-like side chains of ubiquinone turned out to be synthesized from IPP derived from the MVA pathway (27).
A hairy root culture of Coluria geoides was established for eugenol production (28); and because it also produces dolichols, it could be a useful model for studies of the early steps of dolichol biosynthesis in plants. The recently developed HPLC/electrospray ionization-mass spectrometry methods (3) should greatly facilitate precise investigation of their structure. Despite many efforts, the biosynthetic origin of polyisoprenoid alcohols in plants remains unclear. Therefore, we have decided to analyze which pathway is the source of IPP built into dolichol.
Here we report that both pathways are involved in the biosynthesis of dolichols in hairy roots of C. geoides. The ω-terminal isoprene unit and several subsequent ones are synthesized with an involvement of both the MEP and MVA pathways, in contrast to the very last α-terminal and a few preceding units, where contribution of the MEP pathway is negligible. According to our MS data, on average 6–8 isoprene units per dolichol molecule (ranging from 14 to 18 isoprene units) are formed in the MEP-dependent manner. A model is discussed, suggesting spatial regulation of dolichol synthesis and a unidirectional influx of IPP into plastids.
EXPERIMENTAL PROCEDURES
Chemicals and Isotope-labeled Compounds—[1-13C]Glucose (99%), [1,6-13C2]glucose (99 and 97%), and [U-13C6]glucose (99%), together with C62H6 (2H 99.5%) were obtained from Cambridge Isotope Laboratories (Andover, MA); the inhibitors mevinolin and fosmidomycin were obtained from Merck and Molecular Probes (Eugene, OR), respectively. (RS)-[5-2H]Mevalonate was prepared according to Ref. 29 using NaB[2H]4 (Cambridge Isotope Laboratories, Andover, MA). 1-Deoxy-[5,5-2H2]d-xylulose was synthesized as described earlier (30). All dolichol and polyprenol standards were from the Collection of Polyprenols (Institute of Biochemistry and Biophysics, Polish Academy of Sciences, Warsaw, Poland). Chromatographic materials were from Merck and HPLC solvents from Baker B.V. (Deventer, Holland).
Plant Material, Labeling Experiments, and Analysis of [13C]Polyisoprenoids—Hairy roots of C. geoides were cultivated in liquid medium (modified B5 medium containing 0.75% glucose) for 21 days (logarithmic growth stage) in darkness at 22 °C on a rotary shaker at 105 rpm (3). All the biosynthetic precursors were added to the medium from the beginning of culture growth, and labeling was continued for 3 weeks. For NMR measurements the medium contained a mixture of natural abundance glucose (1.1% 13C) and [1-13C]glucose (99% 13C) at a 9:1 ratio (w/w). The maximal possible 13C abundance in the products was thus 5.4% (see “Results”). For mass spectrometry either [13C]glucose alone or, when indicated, native glucose supplemented with [5-2H]mevalonate (3.9 mg/flask) or 1-deoxy-[5,5-2H2]xylulose (4.6 mg/flask) was used. For inhibitor studies regular medium was supplemented either with mevinolin (30 μm) or fosmidomycin (100 μm) for the last 3 days of the culture, and no symptoms of toxicity were observed during these treatments. Two independent feeding experiments were carried out, each performed in duplicate. Non-saponifiable lipids obtained from dried hairy roots were purified chromatographically as described earlier (3). For NMR analysis the dolichol-enriched fraction eluted from the silica gel column was further subjected to flash chromatography on RP-18 Lichroprep gel suspended in methanol. Pure dolichol mixture was eluted with acetone.
NMR Analysis—1H and 13C NMR spectra of metabolically 13C-labeled dolichols (7 mm) were obtained with a Varian INOVA 400 MHz (Palo Alto, CA) spectrometer at 25 °C in C62H6. Two-dimensional {1H,13C} gradient heteronuclear single quantum correlation experiments (31–33) were performed in proton-decoupled mode with a carbon spectral width of 25 kHz and 256 increments. One-dimensional 13C experiment was performed with 64 K data points and a spectral width of 25 kHz in one-dimensional proton-decoupled mode. The measurement was carefully calibrated to achieve integrals for quantitative analysis. For CH2 and CH3 carbons of dolichol, T1 was measured yielding values between 1.1 and 2.7 s. The pulse sequence was optimized according to the literature (34, 35); delay of 5.0 s, pulse width of 90°, and acquisition time of 0.2 s were applied avoiding nuclear Overhauser effects and presaturation. One hundred and twenty thousand scans (168 h) were collected yielding S/N = 130. Spectra were calibrated against the chemical shift of benzene in 13C spectra (128.0 ppm). Assignment of all 13C NMR signals (Table 2, for sequential numbering of carbon atoms, and see also supplemental Table 1 and supplemental Fig. 1) was done by combined use of two-dimensional {1H,13C} gradient heteronuclear single quantum correlation spectra and literature data (36). Four residual signals were assigned to the traces of the organic solvent used for dolichol isolation, n-hexane (δ(C-α H3), 14.4 ppm; δ(C-β H2), 23.1 ppm, and δ(C-γ H2), 32.3 ppm), and to traces of “grease” accumulated during sample preparation (δ(CH2), 30.2 ppm) as confirmed with 1H,13C correlation spectra, whereas two low intensity signals (δ 77.6 and 29.5 ppm) remained unassigned.
TABLE 2.
Isotopic abundances in dolichol 13C NMR spectrum Isotopic abundance of [1-13C]glucose used was 10.9% and thus maximal possible abundance was 5.4%, see “Results.” Numbering of carbon atom positions is given in Fig. 2. Average isotopic abundance of C-3 signals was assumed to correspond to the natural 13C abundance (∼1.1%) and was further used as a standard for calibration. α and ω stand for OH- and hydrocarbon-terminal isoprene residues, respectively (see Fig. 1 and supplemental Fig. 1 and Table 1). Values in bold highlight 13C labeling.
| Position in isoprene unit | Carbon atom type and localization in dolichol chaina | Chemical shift | Total 13C abundance | No. of carbon atoms in Dol-16 | 13C abundance per carbon atom | Metabolic pathway involved |
|---|---|---|---|---|---|---|
| ppm | % | % | ||||
| C-1 | CH2 ω | 27.2 | 2.0 | 1 | 2.0 | MEP |
| CH2trans ω-1, ω-2 | 27.1 | 4.1 | 2 | 2.1 | MEP | |
| CH2cis | 26.9 | 13.3 | 12 | 1.1 | Not labeled (MVA?) | |
|
CH2 α
|
60.8
|
1.5
|
1
|
1.5
|
Not labeled (MVA?) | |
|
Total
|
20.8
|
16
|
1.3 ± 0.4
|
|||
| C-2 | CH ω | 124.9 | 2.2 | 1 | 2.2 | MVA |
| CH trans ω-1, ω-2 | 124.7 | 4.8 | 2 | 2.4 | MVA | |
| 124.8 | ||||||
| CH cis | 125.6 | 33.8 | 11 | 3.1 | MVA | |
| CH cis α +1 | 126.2 | 3.4 | 1 | 3.4 | MVA | |
|
CH2 α
|
40.3
|
4.0
|
1
|
4.0
|
MVA | |
|
Total
|
48.3
|
16
|
3.0 ± 0.4
|
|||
| C-3 | C ω | 131.1 | NDb | 1*b | NDb | Natural abundance |
| C trans ω-1 | 135.4 | 11.1 | 1 | 0.9 | ||
| C cis | 135.3 | 11 | ||||
| C trans ω-2 | 134.8 | 1.1 | 1 | 1.1 | ||
| C cis α+1 | 135.0 | 1.5 | 1 | 1.5 | ||
|
CH α
|
29.5
|
2.4c |
1
|
2.4c |
||
|
Total
|
16.1
|
15
|
1.1 ± 0.4
|
|||
| C-4 | CH3 ω trans | 17.7 | 2.3 | 1 | 2.3 | MVA |
| CH2trans ω-1, ω-2 | 40.2 | 5.9 | 2 | 3.0 | MVA | |
| CH2cis ω-3 | 32.4 | 3.0c | 1 | 3.0c | MVA | |
| CH2cis | 32.6 | 33.2 | 11 | 3.0 | MVA | |
|
CH2 α
|
37.9
|
3.0
|
1
|
3.0
|
MVA | |
|
Total
|
47.4
|
16
|
3.0 ± 0.2
|
|||
| C-5 | CH3 ω cis | 25.8 | 4.1 | 1 | 4.1 | MVA + MEP |
| CH3trans ω-1, ω-2 | 16.1 | 5.8 | 2 | 2.9 | MVA + MEP | |
| CH3cis | 23.7 | 43.4 | 12 | 3.6 | MVA + MEP | |
|
CH3 α
|
19.7
|
3.1
|
1
|
3.1
|
MVA + MEP | |
| Total | 56.4 | 16 | 3.5 ± 0.3 |
Sequential numbers of carbon atoms in Dol-16 chain are given in supplemental Fig. 1 and Table 1
ND indicates not determined due to too low a signal
Approximate integration value resulting from overlapping signals is shown
HPLC/ESI-MS analysis was performed as described for native dolichols (3) with the following modifications. Briefly, when indicated potassium acetate dissolved in solvent B was introduced post-column by a syringe pump (flow rate 5 μl/min) through a T union into the LC flow before entering the mass spectrometer instead of the sodium salt. For statistical calculations, all the m/z data were normalized for [M + Na]+ pseudomolecular ions. Because in all measurements only singly charged dolichol pseudomolecular ions were observed, the values of m/z were considered as molecular masses (in daltons) of pseudomolecular ions (adducts with sodium or potassium cations, as indicated).
Analysis of MS Spectra, Modeling of the Theoretical Envelope of MS Spectra—The theoretical distribution of isoptomers, Ψ(M), so-called “theoretical MS spectra,” is mathematically given by a binomial distribution describing the probability of k successes (i.e. number of 13C atoms replacing 12C ones) in a hypothetical experiment of N = n × z tries, i.e. synthesis of Dol-n consisting of n isoprene units, each unit containing “z” carbon atoms which, theoretically, might be 13C-enriched (see the Table 1) with the unitary probability of success, p, according to Equation 1,
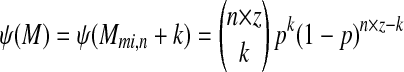 |
(Eq. 1) |
where Mmi,n is the mass of monoisotopic [12C]Dol-n; a binomial coefficient is given by Equation 2,
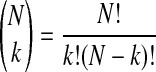 |
(Eq. 2) |
and p is the probability of labeling of the specified carbon atom, which depends on the feeding conditions (summarized in Table 1). For the statistical analysis, the isotopomer distributions calculated for [1-13C]glucose and [1,6-13C2]glucose experiments were additionally corrected for the natural 1.1% 13C abundance at “nonenriched” positions (cf. Fig. 2).
TABLE 1.
Probability of 13C labeling of specified carbon atoms within an isoprene unit synthesized via the MEP or the MVA pathway For isoprene units obtained from [1-13C]- and [1,6-13C2]glucose feeding, two types of carbon atoms, labeled with different probabilities (p), are expected, unlike for native and [U-13C6]glucose-derived ones, where all carbon atoms are labeled with equal probability. Numbering of carbon atom positions is given in Fig. 2.
|
Feeding conditions |
Position in isoprene unit |
Isoprene unit |
|||
|---|---|---|---|---|---|
|
MEP-derived |
MVA-derived |
||||
| p probability of 13C labeling | No. of carbon atoms labeled with indicated p | p probability of 13C labeling | No. of carbon atoms labeled with indicated p | ||
| Native glucose | C-1 | 0.01 | |||
| C-2 | all 5 atoms labeled with p = 0.01 | ||||
| C-3 | |||||
| C-4 | |||||
| C-5 | |||||
| [1-13C]Glucose | C-1 | 0.49 | 2 atoms labeled with p = 0.49 | 0.01 | 3 atoms labeled with p = 0.49 |
| C-2 | 0.01 | 3 atoms labeled with p = 0.01 | 0.49 | 2 atoms labeled with p = 0.01 | |
| C-3 | 0.01 | 0.01 | |||
| C-4 | 0.01 | 0.49 | |||
| C-5 | 0.49 | 0.49 | |||
| [1,6-13C2]Glucose | C-1 | 0.98 | 2 atoms labeled with p = 0.98 | 0.01 | 3 atoms labeled with p = 0.98 |
| C-2 | 0.01 | 3 atoms labeled with p = 0.01 | 0.98 | 2 atoms labeled with p = 0.01 | |
| C-3 | 0.01 | 0.01 | |||
| C-4 | 0.01 | 0.98 | |||
| C-5 | 0.98 | 0.98 | |||
| [U-13C6]Glucose | C-1 | 0.99 | |||
| C-2 | all 5 atoms labeled with p = 0.99 | ||||
| C-3 | |||||
| C-4 | |||||
| C-5 | |||||
Analysis of MS Spectra, Estimation of the Average Molecular Mass of Dolichol—For each dolichol Dol-n, the average molecular mass was estimated as the location of the center of a Gaussian distribution fitted to the pattern of 10–20 highest intensity signals recorded experimentally. The values were estimated with the standard deviation 0.1–0.3 Da.
Meta-analysis of MS Spectra, Linear Regression Analysis— The values of the estimated average molecular masses of dolichols were used to analyze the trends of the increase of dolichol molecular mass using two alternative linear regression procedures.
In the first linear regression procedure, the average molecular masses of dolichols, M(n,i), were analyzed as a function of the number of the isoprene units in the dolichol molecule, n, according to Equation 3,
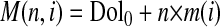 |
(Eq. 3) |
Thus, the average molecular mass of a dolichol molecule, consisting of n isoprene units M(n,i), is the sum of the masses of all the isoprenoid units, n × m(i), and the intercept, called Dol0, which represents the mass difference obtained by subtraction of the mass of n repeating C5H8 units from the total mass of the Dol-n pseudomolecular ion [MDol-n + Na]+ (see the chemical formula below Table 3). It is also worth stressing that Dol0 contains no carbon atoms and consequently its mass cannot be dependent on feeding conditions. These calculations were performed separately for all the feeding conditions, enumerated by index i, and resulted in two sets of parameters, i.e. slopes m(i), which correspond to the average molecular mass of the isoprenoid unit specific for a given feeding experiment, and intercepts, Dol0. This approach permitted a qualitative estimation of the involvement of the metabolic pathways in the synthesis of individual isoprene units.
TABLE 3.
Expected and experimental values of molecular masses and molecular mass enrichment of isoprene unit
|
Glucose |
Isoprene unit (C5H8) (Da) |
Dol0
(Da)a |
||||||
|---|---|---|---|---|---|---|---|---|
|
Molecular mass |
Molecular mass enrichment |
|||||||
|
Expected |
Experimental |
Expected |
Experimental |
Expected |
Experimental |
|||
| MVA | MEP | MVA | MEP | |||||
| Nonlabeled | 68.08 | 68.08 ± 0.05 | 0 | 0 | 43.04 | 43.1 | ||
| [1-13C]Glucose | 69.57 | 69.06 | 69.32 ± 0.09 | 1.49 | 0.98 | 1.24 ± 0.09 | 41.4 | |
| [1,6-13C2]Glucose | 71.07 | 70.06 | 71.17 ± 0.21 | 2.99 | 1.98 | 3.09 ± 0.21 | 33.7 | |
| [U-13C6]Glucose | 73.03 | 72.87 ± 0.42 | 4.95 | 4.79 ± 0.42 | 45.5 | |||
Dol0 represents the mass difference obtained by subtraction of the mass of n repeating C5H8 units from the total mass of the Dol-n pseudomolecular ion [MDol-n + Na]+ and corresponds to the sum of masses of atoms highlighted in bold: [H(C5H8)n-1-CH2-CH(CH3)-CHH-CHH-OH · Na]+
In the second linear regression procedure, the average molecular masses of dolichols were analyzed as a function of the theoretical isotopic enrichments, εMVA and εMEP, expected for the MVA and MEP pathways, respectively, according Equation 4; see Table 3 for the theoretical ε values.
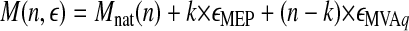 |
(Eq. 4) |
According to Equation 4, the average molecular mass of a Dol-n molecule is the sum of the following three components: the molecular mass of native dolichol, Mnat(n), the mass increment from k units derived via the MEP pathway, k × εMEP, and the mass increment from (n – k) units derived from the MVA pathway, (n - k) × εMVA, and (εMEP = ⅔εMVA).
All the numerical analyses, including linear regression and fitting to the Gaussian distribution via conjugated gradient method (37), were performed with the aid of GnuPlot 4.0 software (©2004 Thomas Williams, Colin Kelley, available on line).
RESULTS
13C NMR Analysis of Polyisoprenoid Alcohols Labeled with [1-13C]Glucose—Previous analysis of polyisoprenoid composition showed that hairy roots of C. geoides contained a family of dolichols (Dol-13 to Dol-29, with Dol-16 dominating) accompanied by traces of polyprenols of similar chain lengths (3). Because ∼60% of dolichols accumulated in Coluria roots were in the form of esters with carboxylic acids (data not shown), alkaline hydrolysis was always performed prior to further analysis. Root cultures were grown for no longer than 3 weeks to keep glucose concentration in the medium sufficiently high to ensure that it was the main carbon and energy source.
It is well established in the literature that after labeling with [1-13C]glucose, carbon atoms derived from C-3 of IPP and DMAPP (Fig. 2 and supplemental Fig. 1) should not be labeled either via the MVA or the MEP pathway. Indeed, analysis of the NMR spectrum of [13C]-labeled dolichol (Table 2) confirmed that their isotope abundances were the lowest of all the carbon atoms. Thus, the signals in 13C-labeled dolichol (Dol) spectrum assigned to the IPP/DMAPP C-3 atoms were considered as reference signals of the natural 13C isotope abundance, and the mean value of their integration calculated per a single carbon atom was considered as the natural abundance of ∼1.1% and used for the subsequent normalization of the abundances of other signals.
High 13C abundances in 13C-labeled dolichol spectrum were recorded for C-2 and C-4 atoms of IPP and DMAPP; the calculated average abundance for C-2 and C-4 atoms was 3.0 ± 0.4 and 3.0 ± 0.2%, respectively. Somewhat higher values were observed for C-5 atoms of IPP and DMAPP (average value 3.5 ± 0.3%). In contrast, the isotopic abundance of C-1 of IPP and DMAPP was considerably lower (average 1.3 ± 0.4%) than those of C-2, -4, and -5. Notable differences between subgroups of the C-1 type atoms were recorded however. The isotopic abundance of the C-1 atoms located in the trans-isoprene units (∼2%) was notably higher than that of C-1 type atoms of the cis-isoprene units (∼1%), and in fact the latter one was at the level of the natural abundance. Nonetheless, it should be remembered that at the applied experimental conditions (10.9% isotopic 13C abundance in the feeding medium), the maximal expected 13C abundance in the products equals 5.4% because of glycolysis. In glycolysis, [1-13C]glucose gives [3-13C]glyceraldehyde 3-phosphate and [3-13C]pyruvate, two precursors of the MEP pathway, and subsequently formed [2-13C]acetyl-CoA, the direct precursor of the MVA pathway. Because of the isomerization of the triose phosphates, dihydroxyacetone phosphate and glyceraldehyde 3-phosphate are interconverted, and C-1 and C-6 of glucose are metabolically equivalent. Consequently, the 13C abundance in the final product is ∼50% that used in the supplied glucose, and the probability of labeling of each potentially labeled carbon atom is ∼0.5. It should be noticed, however, that the expected 13C abundance (5.4%) was never observed for any of the labeled positions, indicating possible activity of the pentose phosphate pathway that resulted in a partial loss of 13C from C-1 of glucose, formation of 13CO2, and consequently isotopic dilution. Nevertheless, the 13CO2 resulting from [13C]glucose catabolism is not efficiently recycled in nonphotosynthetic tissues like roots, which in turn precludes scrambling in the labeling pattern in fact not observed in our labeling experiments (see “Discussion”).
Conclusions Arising from 13C NMR Spectrum of Dolichol— The observed labeling pattern was consistent with a dual pathway origin of dolichols; however, the intensity of labeling of selected carbon atoms was not constant throughout the length of the dolichol molecule. The lower isotopic abundances found for C-1, -2, and -4 of IPP and DMAPP than that of C-5 are well explained by their different labeling pattern (Fig. 2). Although C-5 is labeled via either pathway, C-1 will become labeled only when synthesized via the MEP one, whereas C-2 and -4 only via MVA. Consequently, if a dolichol molecule contains isoprene units derived from both pathways, its C-1, C-2, and C-4 positions will be labeled to a lesser extent than the C-5 positions of IPP/DMAPP because of isotopic dilution.
Thanks to the high resolution of the NMR spectra of dolichols, signals of carbon atoms could be assigned unambiguously to all the positions C-1 to C-5 of IPP/DMAPP and also could be differentiated between those deriving from the trans and the cis units (i.e. those near the ω terminus of the dolichol molecule and those near the α terminus, respectively; see Fig. 1) (Table 2 and supplemental Table 1). Keeping these data in mind, the results of the analysis of the 13C NMR spectrum suggested that the ω-terminal part of the dolichol molecule contains both MEP- and MVA-derived isoprene units, in contrast to the α-terminal part exclusively derived from the MVA pathway (see supplemental Fig. 2). Simultaneously, the characteristic profile of this spectrum, especially no labeling of C-3 atoms of IPP, indicated that virtually no 13C scrambling occurred during the labeling experiments. Because only approximate estimation of the relative contributions of both pathways to dolichol biosynthesis may be based on 13C NMR analysis, 13C-labeled dolichols were analyzed in parallel by mass spectrometry to further evaluate the relative involvement of both pathways to their biosynthesis.
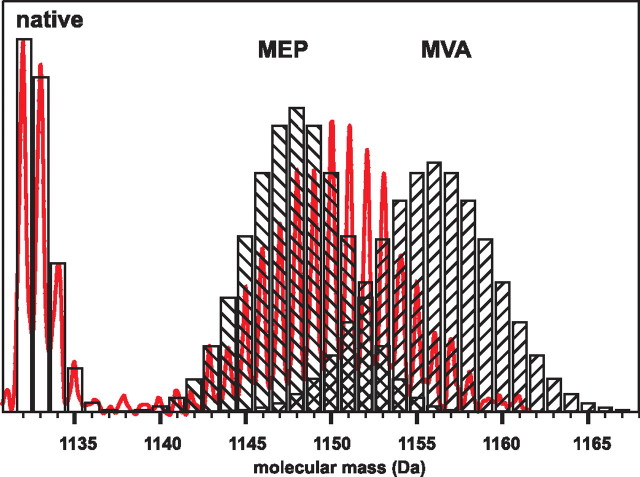
MS Analysis of Polyisoprenoid Alcohols Labeled by [U-13C6]Glucose—As expected, the m/z values for pseudomolecular ions of dolichols obtained from roots grown on [U-13C6]glucose as the sole carbon source (99% isotope abundance) were increased compared with native dolichols and equaled the maximal m/z predicted for uniformly labeled [U-13C5n]Dol-n (supplemental Fig. 3A). In addition to the signals of the labeled dolichols, signals of lower intensity of native Dol-n, originating from the inoculum, were recorded in the same spectrum. A similar effect has recently been reported for sitosterol in Catharanthus roseus cells where 30% of this lipid originated from the inoculum (38). This observation indicated that native and [13C]dolichol of the same chain length were not separated in the HPLC conditions applied. The profile of the spectrum indicated efficient formation of uniformly labeled dolichol proving that the pool of nonlabeled natural abundance intermediates in cells of the root inoculum was negligible. Additionally, a higher number of isotopic signals of “hypo-labeled” [13C]dolichol isotopomers (e.g. seven signals for Dol-16) were observed than for native dolichol (four signals resulting from the natural, ∼1.1%, abundance of 13C), which (Fig. 3B) might reflect slight discrepancies in the labeling of individual carbon atoms in the precursor [U-13C6]glucose. In fact, the experimental mass distribution perfectly agrees with the envelope of the theoretical MS spectrum, calculated according to Equation 1 under the assumption of the average 13C labeling probability, p, of 0.95, which is close to the expected value of 0.99.
FIGURE 3.
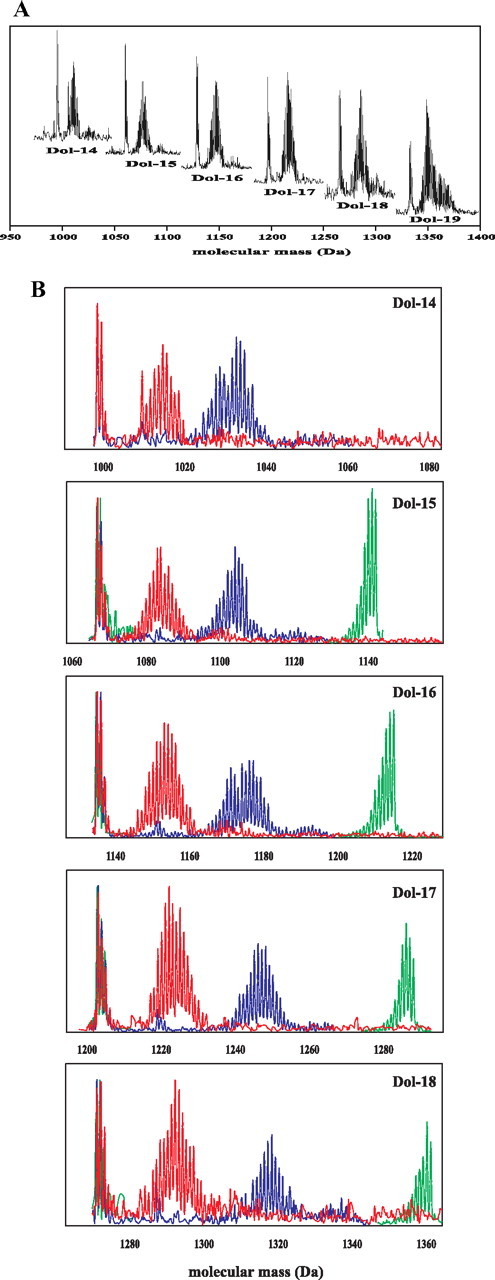
Profiles of dolichol isotopomer signals obtained from in vivo labeling with glucose. A, overlaid HPLC/ESI-MS spectra of Dol-n (-14 up to -19) obtained from [1-13C]glucose. B, spectra acquired after all three feeding experiments: red, [1-13C]glucose; blue, [1,6-13C2]glucose, and green, [U-13C6]glucose. In all samples, the group of low mass signals represents residual population of native dolichols remaining from root inoculum.
MS Analysis of Polyisoprenoid Alcohols Labeled by [1-13C]Glucose—The pseudomolecular ions of dolichols obtained after [1-13C]glucose feeding formed a relatively complex “family” of signals, e.g. for [13C]Dol-16 they centered around m/z 1151.4 Da (supplemental Fig. 3B) and were accompanied by signals corresponding to native Dol-16 with a maximum at 1132.3 Da. Spectra of several homologous dolichols (Dol-14 to Dol-19) were recorded (Fig. 3A); however, the latter one, because of its low intensity, was not further used for quantitative analysis. The observed shift of isotopomer distribution toward higher molecular mass indicated 13C enrichment of the dolichol molecules. The complex distribution pattern of the signals, which mirrors the stochastic dispersion of the probability of formation of differentially labeled [13C]dolichol isotopomers, is understandable bearing in mind that the probability of labeling of each potentially labeled carbon atom is ∼0.5, as the result of stochastic mixing of triose pools during glycolysis (see “Results,” “13C NMR Analysis of Polyisoprenoid Alcohols Labeled with [1-13C]Glucose”). For each IPP molecule derived from the MEP pathway (C-1 and C-5 labeled, Fig. 2), the number of combinations of 13C enrichment patterns is 22, and the number of differently labeled IPP isotopomers is 3, with the 13C distribution profile [13C2]:[13C1]:[13C0] in a 1:2:1 ratio. In the case of the MVA pathway (C-2-, -4-, and -5-labeled), the number of combinations is 23, whereas the number of IPP isotopomers is 4, and the corresponding profile is [13C3]:[13C2]: [13C1]:[13C0] in a 1:3:3:1 ratio. The condensation of the differently labeled IPP molecules leading to the polyisoprenoid backbone is also further probabilistic, resulting in a complex mixture of differentially labeled dolichol isotopomers producing a complex pattern of m/z signals in MS analysis, as presented in Fig. 3.
MS Analysis of Polyisoprenoid Alcohols Labeled by [1,6-13C2]Glucose—To simplify the dolichol mass distribution patterns, [1,6-13C2]glucose was used for feeding. As expected, the m/z values were higher than those obtained with [1-13C]glucose for all dolichols studied (Fig. 3B and supplemental Fig. 3C). Unexpectedly, however, the MS spectrum was still rather complex, which might result from the interference of two phenomena. First, 13C abundance in the doubly labeled glucose molecule was different at C-1 and C-6 (99 versus 97%, respectively). Second, the 13CO2 released by the oxidative pentose phosphate pathway might have been reincorporated via the activity of the malic enzyme, yielding a small but noticeable redistribution of 13C within the intermediates resulting in higher-than-expected formation of hypo- and hyper-labeled 13C-labeled dolichol. Additionally, the effect of stochastic mixing of the pools of [13C2]IPP and [13C3]IPP, respectively, derived from the MEP and MVA pathways and incorporated into the polyisoprenoid chains, should also be considered.
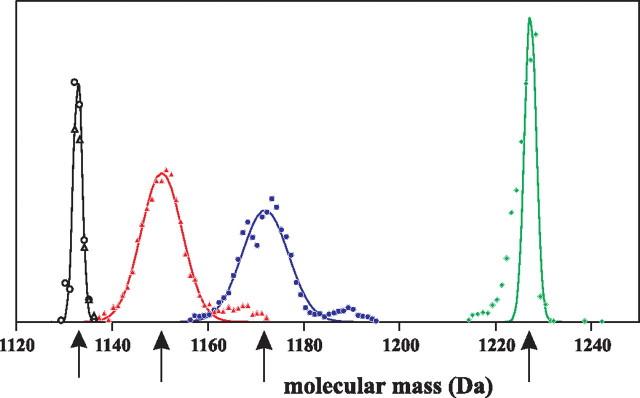
Global Analysis of Mass Spectrometry Data—The complex mass spectra recorded for the dolichols provoked us to generate “theoretical spectra,” i.e. the theoretical distributions of [13C]dolichol isotopomers expected for dolichol synthesized exclusively via either the MEP or the MVA pathway. For this calculation, glycolysis was assumed as the only source of the intermediates for IPP formation. Representative theoretical spectra calculated separately for Dol-16 synthesized either via the MVA or the MEP pathways with [1-13C]glucose feeding are shown in Fig. 4. The experimentally recorded Dol-16 spectrum was located between the two theoretical distributions, indicating a mixed biosynthetic origin of this dolichol. Similar correlation was obtained for [1,6-13C2]glucose labeling (not shown). This observation indicates that neither the MVA nor the MEP pathway was the sole source of IPP utilized for the formation of the dolichol molecule, which is in accordance with the NMR data described above. On the other hand, the complexity of the experimental MS spectra prompted us to apply numerical methods for their analysis.
FIGURE 4.
Comparison of experimental and predicted MS spectra of Dol-16. Overlay of experimentally recorded MS spectrum of Dol-16 obtained from [1-13C]glucose feeding (red curve, cf. Fig. 3B) with two model theoretical spectra (hatched bars) calculated for Dol-16 synthesized exclusively by either the MEP or the MVA pathway. Location of the experimental data between the two theoretical distributions indicates mixed biosynthetic origin of dolichol. The signals of native dolichol (Fig. 3B) are overlaid with the theoretical spectrum (open bars) calculated for native Dol-16.
Estimation of Average Molecular Masses of [13C]Dolichols— Because each dolichol species always occurs as a mixture of isotopomers, a knowledge of its average molecular mass was required for all the calculations described below. This estimation was done for the theoretical spectra generated above for each dolichol (Dol-14–18) separately for each labeling conditions (supplemental Table 2). Thus for native or uniformly labeled glucose, for which the expected average molecular mass of Dol-n is the same regardless of the pathway used for its biosynthesis, 10 values of the expected average molecular masses of Dol-14–18, 5 for either of these conditions, were obtained. For selectively labeled glucose, the theoretical average molecular masses expected for dolichols (Dol-14–18) obtained exclusively either from the MEP or the MVA pathway gave two values for each dolichol, thus in total 10 values for [1-13C]glucose and similarly 10 values for [1,6-13C2]glucose feeding were obtained (supplemental Table 2). The experimental spectra were treated in a manner analogous to that applied to the theoretical ones described above. The average experimental molecular masses were estimated separately for each dolichol labeled at all four labeling conditions (feeding by [1-13C]-, [1,6-13C2]-, [U-13C6]-, and native glucose). Namely, for Dol-15–18, four mixtures of isotopomers each and three mixtures for Dol-14 were used, as shown in Fig. 3B; for technical reasons the spectrum of uniformly labeled Dol-14 obtained after [U-13C6]glucose feeding could not be recorded. A representative determination of the four values of average experimental molecular masses of Dol-16 (three labeling conditions overlaid) is shown in Fig. 5. As the final result a set of 19 experimental values of molecular masses of dolichols was obtained (supplemental Table 2). The location of the experimental values of the Dol-n average molecular masses between the two theoretical ones (Fig. 4) indicates that each Dol-n molecule was of a mixed biosynthetic origin, via both the MEP and MVA pathways. The obtained experimental average molecular masses of dolichols were further analyzed to estimate the involvement of the two pathways in dolichol biosynthesis. It is worthwhile to emphasize that the value of the average experimental molecular mass of Dol-n permits to unambiguously determine the number of isoprene units derived from the MEP and the MVA pathway, respectively (for explanation see supplemental text).
FIGURE 5.
Estimation of average molecular mass of dolichol. Gaussian distribution was fitted to experimentally recorded signals of isotopomers with highest intensity, as presented for representative Dol-16. Overlaid spectra obtained for Dol-16 isolated from roots grown on [1-13C]glucose (filled triangles), [1,6-13C2]glucose (filled circles), and [U-13C6]glucose (filled diamonds). Empty triangles and circles represent residual native dolichols from the inoculum of [1-13C]glucose and [1,6-13C2]glucose. Solid lines represent Gaussian profiles for Dol-16 obtained after feeding by [1-13C]glucose (red lines), [1,6-13C2]glucose (blue lines), [U-13C6]glucose (green lines), and native dolichols (black lines). Arrows indicate the estimated average masses. Each point represents the intensity of a single Dol-16 isotopomer signal in the mass spectrum.
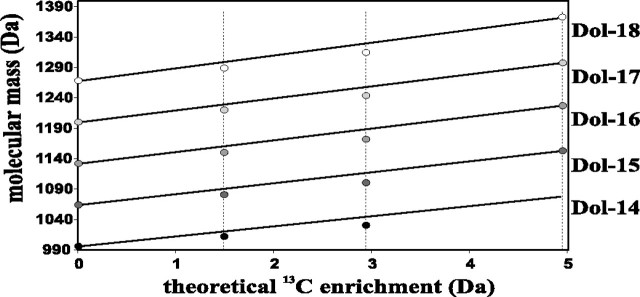
Qualitative Analysis of Mass Spectrometry Data—Using the experimental average molecular masses of 13C-labeled dolichols (supplemental Table 2), it was possible to estimate the average increase of the molecular mass between sequential dolichols, which corresponds to the average mass of a single isoprene unit. The mass of the average isoprene unit, m(i), will of course be dependent on the feeding condition. The values of the average molecular masses of dolichols (supplemental Table 2) were analyzed according to the linear regression method. The dolichol molecular mass is thus described by Equation 3. Individual trends estimated for each labeling experiment are presented in supplemental Fig. 4. The linear regression analysis leads to the estimation of four average masses of isoprene units and four values of Dol0, summarized in Table 3. The theoretically expected enrichment of the average molecular mass of a single isoprene unit was calculated based on the labeling pattern of the isoprene unit for [1-13C]glucose and [1,6-13C2]glucose (Fig. 2). The experimental values of the enrichment of the mass of the isoprene unit (1.24 ± 0.09 and 3.09 ± 0.21 Da for the singly and doubly labeled glucose, respectively) were in better agreement with the theoretical values predicted for the MVA pathway (correspondingly 1.49 and 2.99 Da) than with those for the MEP pathway (0.99 and 1.49 Da, respectively). It should be stressed here that the average molecular mass of a single isoprene unit calculated above concerns isoprene units located in the proximity of the α terminus of the Dol-n molecule because all the Dol-n species are synthesized by elongation of precursors from a common pool of polyprenyl diphosphates. Thus, Dol-n is formed directly from Dol-(n-1) via addition of a single isoprene unit to the α terminus and consequently the estimated increments concern the α-terminal units.
On the other hand, the average molecular masses estimated experimentally for the entire Dol-n molecules (see above) were lower than those calculated for the MVA-derived dolichols, indicating that some part of these molecules must have been synthesized via the MEP pathway, which produces “lighter” isoprenoid units. It should be remembered that isoprenoid units derived from feeding by either [1-13C]glucose or [1,6-13C2]glucose might be either “light,” originating from the MEP pathway and so containing up to two 13C atoms per unit, or “heavy,” originating from the MVA pathway thus containing up to three 13C atoms per unit. Too low a mass of the experimentally estimated average molecular mass of Dol-n is also mirrored by the lower-than-expected experimental values of Dol0, presented in Table 3 (further discussed in comments to supplemental Fig. 4). By elimination, because we know from the earlier considerations that the α-terminal isoprene units were derived from the MVA pathway, the MEP-derived ones could only have been used for the synthesis of the ω-proximal portion of the dolichol molecules. It should be also pointed out that good agreement of the values of the observed increase of the molecular mass between sequential dolichols with theoretical calculations clearly indicates that there was no sizeable loss of 13C atoms along the biosynthetic pathway from mono- or doubly labeled glucose toward dolichol, which in turn indicates negligible 13C scrambling.
Quantitative Analysis of Mass Spectrometry Data—We proceeded to estimate the number of the two types of isoprene units in the dolichol molecules. For the isoprene unit synthesized via the MVA pathway, the expected isotopic enrichment, εMVA, should be 1.49 and 2.94 Da for [1-13C]glucose and [1,6-13C2]glucose, respectively, with 0.05- and 4.95-Da enrichment for native and uniformly labeled glucose (Table 3). For a dolichol molecule, Dol-n, synthesized entirely via the MVA pathway, its average molecular mass, M(n) after [1-13C]glucose feeding should equal Mnat(n) + n × 1.49, because it is the sum of the molecular mass of native Dol-n and the total enrichment of all the isoprene units. Similarly it should be Mnat(n) + n × 2.94 for [1,6-13C2]glucose feeding and Mnat(n) + n × 4.95 for feeding with uniformly labeled glucose. A graphic presentation of this linear increase of molecular mass plotted as a function of the enrichment expected for the MVA pathway is shown separately for each dolichol in Fig. 6 (solid lines). The experimental average molecular masses of dolichols were lower than those calculated for the MVA labeling, as discussed above. To estimate the number of the lighter (k) and heavier (n - k) isoprene units per Dol-n molecule, Equation 4 giving the average molecular mass of each dolichol species obtained with all three types of [13C]glucose and native glucose feeding was used. Equations describing the individual trends estimated for each dolichol are presented in the supplemental text. Because εMEP = ⅔εMVA (see Table 3 for the theoretical ε values), the slope value for the linear regression (Equation 4) against εMEP (n - ⅓ × k) permitted estimation of k, the average number of isoprene units derived from the MEP pathway (Table 4). According to these calculations, six to eight (lighter) isoprene units per dolichol molecule are derived from the MEP pathway.
FIGURE 6.
The effect of 13C labeling on the increase of dolichol molecular mass. Markers represent average molecular masses of individual dolichols; Dol-14, Dol-15, Dol-16, Dol-17, and Dol-18 relate to the theoretical enrichment in the MVA pathway indicated by vertical dotted lines, 1.49, 2.99, and 4.95 Da for mono-, di-, and uniformly labeled glucose, respectively (see Table 3). Solid lines follow the expected values calculated for masses of dolichols synthesized exclusively via the MVA pathway. The observed mass deficit of 7–15 Da found for mono- and di-labeled glucose experiments indicates substantial participation of the alternative MEP pathway in dolichol synthesis.
TABLE 4.
Number of isoprenoid units in a dolichol molecule biosynthesized via the MVA and the MEP pathway Estimations were done separately for each Dol-n according to Equation 4.
|
Dol-n |
No. of isoprene units |
||
|---|---|---|---|
| n | MVA-derived (n-k) | MEP-derived k | |
| Dol-14 | 14 | 7.3 ± 1.8 | 6.7 ± 1.8 |
| Dol-15 | 15 | 7.1 ± 1.8 | 7.9 ± 1.8 |
| Dol-16 | 16 | 8.6 ± 2.4 | 7.4 ± 2.4 |
| Dol-17 | 17 | 10.5 ± 2.7 | 6.5 ± 2.7 |
| Dol-18 | 18 | 11.4 ± 3.3 | 6.6 ± 3.3 |
In summary, the described meta-analysis of mass spectrometry data revealed that in dolichols longer than Dol-14 successive isoprene units used for elongation of the molecule (i.e. a single α-terminal unit for Dol-15; two units. α and α + 1 for Dol-16; three α-terminal ones for Dol-17; four for Dol-18, see supplemental Fig. 1) are derived exclusively from the MVA pathway. The 13C NMR spectroscopy data indicated that also the α-terminal unit of Dol-14 is of the MVA origin, similarly as in the longer dolichols. Other units, incorporated into the growing Dol molecule at earlier steps, may be derived from either pathway; on average between six and eight such units come from the MEP pathway and the balance (i.e. seven to five for the 13-unit oligoprenyl precursor) is from MVA. A model summarizing the involvement of the MVA and MEP pathways in the biosynthesis of a dolichol molecule is presented in supplemental Fig. 2. This conclusion on the involvement of both pathways in dolichol biosynthesis was verified by application of pathway-specific precursors and inhibitors.
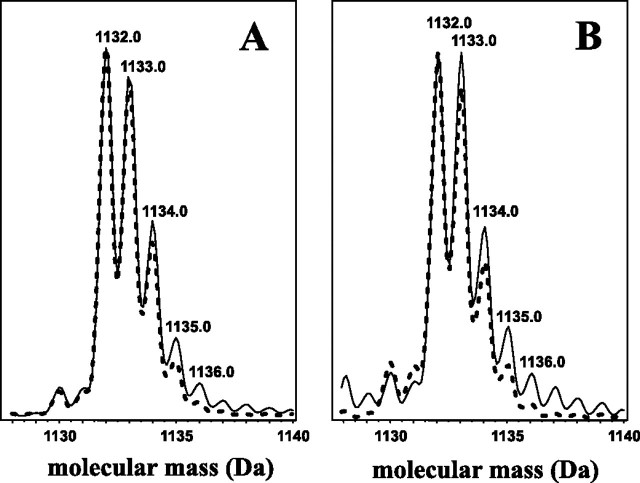
Labeling of Polyisoprenoids with Pathway-specific Precursors— Supplementation of the feeding medium with deuterated compounds (i.e. either with [5,5-2H2]deoxyxylulose (DX) or [5-2H]mevalonate, precursors of the MEP and the MVA pathway, respectively) resulted in a slight increase (∼10–15%) of the relative intensity of respective signals. For [5,5-2H2]DX, the intensities of [M + 2]+ as well as the following [M + 3]+ and [M + 4]+ isotopic signals were increased (Fig. 7A), whereas for [5-2H]mevalonate all but the monoisotopic signals were enhanced (Fig. 7B) for each of the dolichols analyzed. In contrast to mevalonate, feeding experiments with DX indicated incorporation of an even number of deuterium atoms per dolichol molecule, which is in agreement with the known maintenance of the two hydrogen atoms linked to C-5 of DX at the C-1 position (Fig. 2) of the isoprene unit (39, 40). The observed enhancement of the odd ion [M + 3] resulted from the incorporation of two deuterium atoms into a dolichol isotopomer containing one 13C atom per molecule. The low incorporation of deuterium was because of the high dilution of the deuterium-labeled precursors because they constituted only 0.06% (by mass) of glucose in the feeding medium. Effective incorporation of tritium from [3H]mevalonate into both polyisoprenoid alcohols and sterols was also noted (supplemental Fig. 5). The highest labeling was found for 21-day-old cultures.
FIGURE 7.
Incorporation of deuterium-labeled pathway-specific precursors into dolichols. Overlaid HPLC/ESI-MS spectra of representative Dol-16 of the deuterium-enriched (solid line) and native (dotted line) samples obtained from feeding with [5,5-2H2]deoxyxylulose (A) and[5-2H]mevalonate (B).
Effect of Pathway-specific Inhibitors on Isoprenoid Accumulation—Mevinolin (30 μm), a specific inhibitor of 3-hydroxy-3-methylglutaryl-CoA reductase of the MVA pathway, efficiently inhibited accumulation of both polyisoprenoid alcohols and sterols in the oldest culture, resulting in a remarkable decrease of their content (by 85 and 82% for polyisoprenoids and sterols, respectively, see Table 5). A higher concentration of mevinolin (60 μm) gave similar results (not shown); however, surprisingly, de novo synthesis of both lipids from [3H]mevalonate was also inhibited suggesting a pleiotropic effect of this drug on isoprenoid metabolism. On the one hand this might be an indication of the inhibition of other enzymes of the pathway besides 3-hydroxy-3-methylglutaryl-coenzyme A reductase, as was earlier found for sesquiterpene cyclase (41) or in mammalian systems for geranylgeranyl diphosphate synthase (42). On the other hand, possible toxic effects of mevinolin should be also considered (43). Fosmidomycin, a specific inhibitor of 1-deoxy-d-xylulose 5-phosphate reductoisomerase decreased the accumulation of both lipids in the youngest culture (by 73 and 78% for dolichols and sterols, respectively), whereas an increased content of both lipids was found for 2- and 3-week-old cultures. The variable effects of fosmidomycin most probably mirror metabolic shifts between both pathways upon stress caused by the inhibitor.
TABLE 5.
Effect of mevinolin and fosmidomycin on accumulation of polyisoprenoid alcohols and sterols Values represent mean ± S.D. of three experiments performed in triplicate. d.w. indicates dry weight.
|
Age of culture |
Polyisoprenoid alcohols |
Sterols |
||||
|---|---|---|---|---|---|---|
| No inhibitor | Mevinolin | Fosmidomycin | No inhibitor | Mevinolin | Fosmidomycin | |
| days | μg/g d.w. | % control | μg/g d.w. | % control | ||
| 7 | 12.0 ± 2.1 | 133.3 ± 16.5 | 26.7 ± 7.5 | 2522 ± 100 | 76.2 ± 5.1 | 22.2 ± 4.5 |
| 14 | 23.9 ± 3.1 | 20.9 ± 4.6 | 150.0 ± 16.1 | 2769 ± 152 | 42.3 ± 4.4 | 111.0 ± 11.1 |
| 21 | 27.9 ± 2.9 | 15.4 ± 9.6 | 225.2 ± 27.5 | 3271 ± 220 | 18.0 ± 3.5 | 133.1 ± 12.3 |
DISCUSSION
In this study the biosynthetic origin of plant dolichols was investigated. We analyzed the contribution of the two alternative pathways known to produce IPP in plant cells, the MEP and the MVA pathway, to dolichol biosynthesis. Dolichol synthesis was studied by in vivo labeling with a general precursor (glucose), pathway-specific precursors (MVA or DX), and by inhibition of the metabolic flow with pathway-specific inhibitors. Our conclusion indicating a “mosaic” structure of plant dolichols is based on the following observations. First, isoprenoid units located at and near the ω-end of the dolichol molecules could be synthesized via either pathway as shown by the high intensities of the corresponding signals in the 13C NMR spectrum. Second, isoprenoid units localized at the proximity of the α terminus were exclusively of the MVA origin as shown by the numerical analysis of the 13C-enriched masses of pseudomolecular ions and in parallel by NMR. Third, the calculated average number of lighter (i.e. derived from MEP) isoprenoid units per dolichol molecule was between 6 and 8 as based on the MS spectra, which was also supported by the 13C NMR data. The efficient incorporation of 13C atoms from uniformly labeled glucose into dolichols confirmed the applicability of Coluria roots as a model for biosynthetic studies. Dolichols were labeled almost exclusively by products derived from the glycolytic pathway as confirmed by the NMR spectrum (synchronized labeling of C-2 and C-4 carbon atoms, with proportionally higher labeling of C-5 atoms of IPP, see Table 2) and MS spectra (increase of the dolichol molecular mass, see Table 3). All these observations exclude any substantial scrambling of 13C during the course of labeling. Yet the general conclusions drawn from such a model should take its limits into consideration. In vitro propagated organs might differ from their physiological equivalents. Heterotrophic growth of the tissue stops the influx of intermediates derived from photosynthesis as well as the light-driven regulation of metabolism. Summarizing the validity of the meta-analysis of MS data applied here, it should be pointed out that by knowing the value of the enrichment of the molecular mass of a compound of interest and the number of isoprene units constituting its molecule one can estimate the relative input of both the MEP and MVA pathways to its biosynthesis (for details see supplemental text).
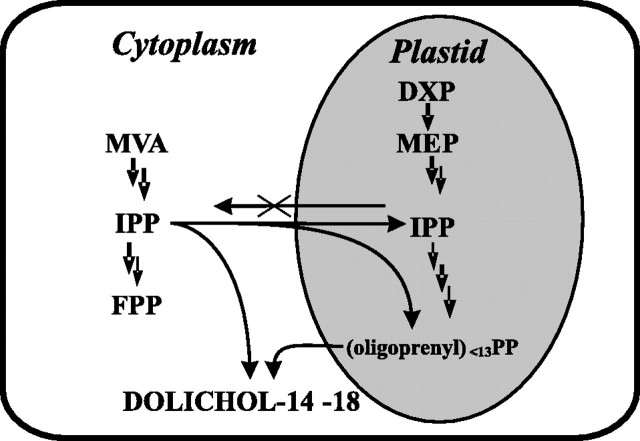
Finally, the proposed mechanism of dolichol biosynthesis is as follows. A mixture of IPPs derived from the MVA and MEP pathways is used in plastids for the initiation of the process and sequential condensations, and thus-formed oligoprenyl diphosphates (surely shorter than 14 isoprene units, see supplemental Fig. 2) are exported to the cytoplasm where they are finally elongated and terminated with IPP of exclusively MVA origin. Such a mechanism, besides compartmentalization of the sequential steps, also requires a unidirectional import of IPP from the cytoplasm to plastids. A spatial model of the organization of dolichol biosynthesis is presented in Fig. 8. Such a model assuming stochastic mixing of IPP molecules within the plastid compartment explains well the broad signal distribution pattern in the MS spectra obtained for [1,6-13C2]glucose feeding (Fig. 3B), which results from randomized insertion of the doubly (MEP-derived) and triply (MVA-derived) labeled IPP in the growing oligoprenyl chain. A further argument supporting the dual-pathway biosynthetic origin of dolichol is given by the localization of the experimental distribution profile of [13C]dolichol isotopomers between the theoretical profiles (Fig. 4) calculated separately for each pathway. In line with the above model (Fig. 8) is a recent observation of an export of MEP-derived IPP and possibly also geranyl diphosphate from plastids to the cytoplasm during sesquiterpene biosynthesis in carrot roots (44).
FIGURE 8.
Compartmentalization of dolichol biosynthesis in root cells. Proposed cooperation of MEP and MVA pathways.
The dolichol biosynthetic scenario described above raises the intriguing question of the nature of at least two independent cis-prenyltransferases which should be involved. Their existence seems plausible in the light of the in silico predicted occurrence of a family of six genes encoding cis-prenyltransferase in the Arabidopsis genome (45). However, the mechanism of the reaction remains unclear because the cytoplasm/endoplasmic reticulum localized cis-prenyltransferase should accept medium chain length oligoprenyl diphosphate as a substrate, although it is generally believed that only all-trans-FPP can serve as a starter for the polyisoprenoid chain.
Another interesting but still unresolved aspect of the presented model is the plastidial source of farnesyl diphosphate, the substrate for the first putative cis-prenyltransferase. Two well characterized Arabidopsis FPP synthases were localized to the cytoplasm and mitochondria (46). Therefore, FPP in plastids could possibly be synthesized as a leaked side product formed either by geranyl or geranylgeranyl-diphosphate synthases because in Arabidopsis both these enzymes were found in this compartment (47, 48); however, plastidial localization of farnesyl-diphosphate synthase has also been postulated (49). Moreover, some studies on incorporation of 13C-labeled glucose showed that sesquiterpenes were derived exclusively from the MEP pathway, indirectly indicating that synthesis of farnesyl diphosphate is possible in plastids (50).
In addition to [13C]glucose labeling, stimulation of each pathway with specific precursors or perturbations of the metabolite flux by specific inhibitors of both the MVA and MEP pathways further supported their involvement in dolichol biosynthesis. The presented model proposing the spatial regulation of dolichol biosynthesis is based on an analysis of the dolichol labeling pattern obtained after a 3-week labeling period, at steady-state conditions for incorporation of the labeled precursor. This does not exclude possible temporal and/or physiologically regulated fluctuations of the respective contributions of the MEP and MVA pathways to the dolichol synthesis. Concomitant regulation of both pathways has already been documented in several systems for biosynthesis of several plastid isoprenoids such as carotenoids, chlorophyll, plastoquinone (43, 51), and cytoplasmic isoprenoids, e.g. gibberellins and sterols (43, 52).
Mechanisms of regulation of the metabolic cross-talk are extensively studied. Recently, a temporal model of the cross-talk showing up-regulation of the MVA pathway during seed germination in darkness followed by its repression upon illumination accompanied by up-regulation of the MEP pathway (53) was presented. Besides light (see excellent discussion in Ref. 13), the cross-talk between both pathways might also be regulated by the circadian clock (54) and environmental stress (17). There are complementary results showing the capability of plastids to import (52, 53, 55–58) as well as to export IPP (59, 60).
The complex mechanism of dolichol biosynthesis emerging from the data presented above suggests the existence of a constitutive transfer of oligoprenyl diphosphates from plastids to the cytoplasm with simultaneous efficient unidirectional transport of IPP from the cytoplasm toward plastids. With several especially terminal steps of this process remaining elusive, such a tentative model still awaits experimental confirmation.
Supplementary Material
Acknowledgments
We thank Professor Andrzej Ejchart (Institute of Biochemistry and Biophysics, Polish Academy of Sciences) and Professor Renata Bogatek-Leszczynska (Warsaw Agricultural University) for stimulating discussions and anonymous reviewers for their highly stimulating suggestions. We are thankful to Professor Jan Fronk (Warsaw University) for critical reading of the manuscript and valuable comments.
This work was supported by the State Committee for Scientific Research Grant PBZ-KBN-110/P04/19 and by European Community Grant LSHB-CT-2004-005151. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–5, Tables 1 and 2, and additional text.
Footnotes
The abbreviations used are: FPP, farnesyl diphosphate; MVA, mevalonate; MEP, methylerythritol phosphate; IPP, isopentenyl diphosphate; DMAPP, dimethyl allyl diphosphate; Dol-n, dolichol composed of n isoprenoid units; HPLC/ESI-MS, high performance liquid chromatography/electrospray ionization mass spectrometry; Dol, dolichol; DX, deoxyxylulose.
References
- 1.Bach, T. J. (1995) Lipids 30 191-202 [DOI] [PubMed] [Google Scholar]
- 2.Hemming, F. W. (1992) Biochem. Cell Biol. 70 377-381 [DOI] [PubMed] [Google Scholar]
- 3.Skorupinska-Tudek, K., Bienkowski, T., Olszowska, O., Furmanowa, M., Chojnacki, T., Danikiewicz, W., and Swiezewska, E. (2003) Lipids 38 981-990 [DOI] [PubMed] [Google Scholar]
- 4.Rezanka, T., and Votruba, J. (2001) J. Chromatogr. A 936 95-110 [DOI] [PubMed] [Google Scholar]
- 5.Swiezewska, E., and Danikiewicz, W. (2005) Prog. Lipid Res. 44 235-258 [DOI] [PubMed] [Google Scholar]
- 6.Liang, P.-H., Ko, T.-P., and Wang, A. H.-J. (2002) Eur. J. Biochem. 269 3339-3354 [DOI] [PubMed] [Google Scholar]
- 7.Kharel, Y., and Koyama, T. (2003) Nat. Prod. Rep. 20 111-118 [DOI] [PubMed] [Google Scholar]
- 8.Oh, S. K., Han, K. H., Ryu, S. B., and Kang, H. (2000) J. Biol. Chem. 275 18482-18488 [DOI] [PubMed] [Google Scholar]
- 9.Cunillera, N., Arro, M., Fores, O., Manzano, D., and Ferrer, A. (2000) FEBS Lett. 477 170-174 [DOI] [PubMed] [Google Scholar]
- 10.Asawatreratanakul, K., Zhang, Y. W., Wititsuwannakul, D., Wititsuwannakul, R., Takahashi, S., Rattanapittayaporn, A., and Koyama, T. (2003) Eur. J. Biochem. 270 4671-4680 [DOI] [PubMed] [Google Scholar]
- 11.Rohmer, M. (1999) Nat. Prod. Rep. 16 565-574 [DOI] [PubMed] [Google Scholar]
- 12.Eisenreich, W., Bacher, A., Arigoni, D., and Rohdisch, F. (2004) Cell. Mol. Life Sci. 61 1401-1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodriguez-Concepción, M. (2006) Phytochem. Rev. 5 1-15 [Google Scholar]
- 14.Alberts, A. W., Chen, J., Kuron, G., Hunt, V., Huff, J., Hoffman, C., Rothrock, J., Lopez, M., Joshua, H., Harris, E., Patchett, A., Monaghan, R., Currie, S., Stapley, E., Albers-Schonberg, G., Hensens, O., Hirshfield, J., Hoogsteen, K., Liesch, J., and Springer, J. (1980) Proc. Natl. Acad. Sci. U. S. A. 77 3957-3961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuzuyama, T., Shimizu, T., Takahashi, S., and Seto, H. (1998) Tetrahedron Lett. 39 7913-7916 [Google Scholar]
- 16.Yamazaki, Y., Kitajima, M., Arita, M., Takayama, H., Sudo, H., Yamazaki, M., Aimi, N., and Saito, K. (2004) Plant Physiol. 134 161-170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ge, X., and Wu, J. (2005) Plant Sci. 168 487-491 [Google Scholar]
- 18.Gough, D. P., and Hemming, F. W. (1970) Biochem. J. 117 309-317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swiezewska, E., Thelin, A., Dallner, G., Andersson, B., and Ernster, L. (1993) Biochem. Biophys. Res. Commun. 192 161-166 [DOI] [PubMed] [Google Scholar]
- 20.Adair, W. L., and Keller, R. K. (1982) J. Biol. Chem. 257 8990-9000 [PubMed] [Google Scholar]
- 21.Chojnacki, T., and Dallner, G. (1988) Biochem. J. 251 1-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hemming, F. W. (1983) Biochem. Soc. Trans. 11 497-504 [DOI] [PubMed] [Google Scholar]
- 23.Swiezewska, E., Dallner, G., Andersson, B., and Ernster, L. (1993) J. Biol. Chem. 268 1494-1499 [PubMed] [Google Scholar]
- 24.Kurisaki, A., Sagami, H., and Ogura, K. (1997) Phytochemistry 44 45-50 [Google Scholar]
- 25.Sakaihara, T., Honda, A., Tateyama, S., and Sagami, H. (2000) J. Biochem. (Tokyo) 128 1073-1078 [DOI] [PubMed] [Google Scholar]
- 26.Fukusaki, E., Takeno, S., Bamba, T., Okumoto, H., Katto, H., Kajiyama, S., and Kobayashi, A. (2004) Biosci. Biotechnol. Biochem. 68 1988-1990 [DOI] [PubMed] [Google Scholar]
- 27.Disch, A., Hemmerlin, A., Bach, T. J., and Rohmer, M. (1998) Biochem. J. 331 615-621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olszowska, O., Alferman, A. W., and Furmanowa, M. (1996) Plant Cell Tissue Organ Cult. 45 273-276 [Google Scholar]
- 29.Keller, R. K. (1986) J. Biol. Chem. 261 12053-12059 [PubMed] [Google Scholar]
- 30.Meyer, O., Hoeffler, J. F., Grosdemange-Billiard, C., and Rohmer, M. (2004) Tetrahedron 60 12153-12162 [Google Scholar]
- 31.Kay, L. E., Keifer, P., and Saarinen, T. (1992) J. Am. Chem. Soc. 114 10663-10665 [Google Scholar]
- 32.Palmer, A. G., III, Cavanagh, J., Wright, P. E., and Rance, M. (1991) J. Magn. Reson. 93 151-170 [Google Scholar]
- 33.Kontaxis, G., Stonehouse, J., Laue, E. D., and Keeler, J. (1994) J. Magn. Reson. 111 70-76 [Google Scholar]
- 34.Canet, D. (1976) J. Magn. Reson. 23 361-364 [Google Scholar]
- 35.Freeman, R., Hill, H. D. W., and Kaptein, R. (1972) J. Magn. Reson. 7 327-329 [Google Scholar]
- 36.Tanaka, Y., Sato, H., Kageyu, A., and Tomita, T. (1987) Biochem. J. 243 481-485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brandt, S. (1999) Data Analysis: Statistical and Computational Methods, 2nd Ed., pp. 423-427, Springer-Verlag Inc., Berlin
- 38.Schuhr, C. A., Radykewicz, T., Sagner, S., Latzel, C., Zenk, M. H., Arigoni, D., Bacher, A., Rohdich, F., and Eisenreich, W. (2003) Phytochem. Rev. 2 3-16 [Google Scholar]
- 39.Duvold, T., Cali, P., Bravo, J. M., and Rohmer, M. (1997) Tetrahedron Lett. 38 6181-6184 [Google Scholar]
- 40.Piel, J., Donath, J., Bandemer, K., and Boland, W. (1998) Angew. Chem. Int. Ed. 37 2478-2481 [DOI] [PubMed] [Google Scholar]
- 41.Vogeli, U., and Chappell, J. (1991) Arch. Biochem. Biophys. 288 157-162 [DOI] [PubMed] [Google Scholar]
- 42.Ericsson, J., Runquist, M., Thelin, A., Andersson, M., Chojnacki, T., and Dallner, G. (1993) J. Biol. Chem. 268 832-838 [PubMed] [Google Scholar]
- 43.Hemmerlin, A., Hoeffler, J.-F., Meyer, O., Trisch, D., Kagan, I. A., Grosdemange-Billiard, C., Rohmer, M., and Bach, T. (2003) J. Biol. Chem. 278 26666-26676 [DOI] [PubMed] [Google Scholar]
- 44.Hampel, D., Mosandl, A., and Wust, M. (2005) Phytochemistry 66 305-311 [DOI] [PubMed] [Google Scholar]
- 45.Lange, M., and Ghassemian, M. (2003) Plant Mol. Biol. 51 925-948 [DOI] [PubMed] [Google Scholar]
- 46.Cunillera, N., Boronat, A., and Ferrer, A. (1997) J. Biol. Chem. 272 15381-15388 [DOI] [PubMed] [Google Scholar]
- 47.Bouvier, F., Suire, C., d'Harlingue, A., Backhaus, R. A., and Camara, B. (2000) Plant J. 24 241-252 [DOI] [PubMed] [Google Scholar]
- 48.Okada, K., Saito, T., Nakagawa, T., Kawamukai, M., and Kamiya, Y. (2000) Plant Physiol. 122 1045-1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sanmiya, K., Ueno, O., Matsuoka, M., and Yamamoto, N. (1999) Plant Cell Physiol. 40 348-354 [DOI] [PubMed] [Google Scholar]
- 50.Barlow, A. J., Lorimer, S. D., Morgan, E. R., and Weavers, R. T. (2003) Phytochemistry 63 25-29 [DOI] [PubMed] [Google Scholar]
- 51.Laule, O., Furholz, A., Chang, H.-S., Zhu, T., Wang, X., Heifetz, P. B., Gruissem, W., and Lange, B. M. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 6866-6871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kasahara, H., Hanada, A., Kuzuyama, T., Takagi, M., Kamiya, Y., and Yamaguchi, S. (2002) J. Biol. Chem. 277 45188-45194 [DOI] [PubMed] [Google Scholar]
- 53.Rodriguez-Concepción, M., Fores, O., Martinez-Garcia, J. F., Gonzalez, V., Phillips, M. A., Ferrer, A., and Boronat, A. (2004) Plant Cell 16 144-156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dudareva, N., Andersson, S., Orlova, I., Gatto, N., Reichelt, M., Rhodes, D., Boland, W., and Gershenzon, J. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 933-938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kreuz, K., and Kleinig, H. (1984) Eur. J. Biochem. 141 531-535 [DOI] [PubMed] [Google Scholar]
- 56.Soler, E., Clastre, M., Bantignies, B., Marigo, G., and Ambid, C. (1993) Planta 191 324-329 [Google Scholar]
- 57.Milborrow, B. V. (2001) J. Exp. Bot. 52 1145-1164 [PubMed] [Google Scholar]
- 58.Nagata, N., Suzuki, M., Yoshida, S., and Muranaka, T. (2002) Planta 216 345-350 [DOI] [PubMed] [Google Scholar]
- 59.Bick, J. A., and Lange, B. M. (2003) Arch. Biochem. Biophys. 415 146-154 [DOI] [PubMed] [Google Scholar]
- 60.Ferro, M., Salvi, D., Rivière-Rolland, H., Vermat, T., Seigneurin-Berny, D., Grunwald, D., Garin, J., Joyard, J., and Rolland, N. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 11487-11492 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.