Abstract
Inflammation generates various changes in body iron homeostasis, including iron sequestration in the reticuloendothelial system with ensuing hypoferremia and anemia of chronic disease. Increased iron accumulation is caused by hepcidin-mediated down-regulation of the iron export protein ferroportin and higher iron uptake. However, enhanced iron acquisition by macrophages cannot be accounted for by the previously reported transferrin receptor (TfR1) down-regulation in macrophages exposed to lipopolysaccharide (LPS)/interferon γ (IFNγ) because it impairs a major iron uptake mechanism. Because TfR1 is up-regulated by the hypoxia-inducible factor (HIF-1), we investigated the effect of inflammatory and anti-inflammatory signals on HIF-1-mediated TfR1 gene expression. Exposure of mouse macrophages (RAW 264.7 and J774A.1 cells or peritoneal macrophages) to LPS/IFNγ up-regulated NF-κB, which in turn rapidly and transiently activated HIF-1-dependent TfR1 expression and iron uptake. Activation of an anti-inflammatory pathway by pre-exposure to the adenosine A2A receptor agonist CGS21680 prevented the inducing effect of LPS/IFNγ on HIF-1 and TfR1 expression by inhibiting NF-κB activity, whereas treatment with CGS21680 alone increased HIF-1-mediated TfR1 expression by means of an NF-κB-independent signaling pathway. In conclusion, an interplay of the HIF-1 and NF-κB pathways controls TfR1 transcription in inflammation. The consequent changes in TfR1 expression may be involved in modulating iron retention in inflammatory macrophages, thus possibly contributing to the development of hypoferremia in the early phases preceding the down-regulation of macrophage ferroportin by hepcidin.
Inflammatory states are associated with changes in body iron homeostasis (1). The main systemic response is a rapid fall in plasma iron concentration accompanied by iron sequestration in the reticuloendothelial system. By restricting iron availability for erythroid progenitor cells, prolonged hypoferremia may limit hemoglobin synthesis and cause inflammation-related anemia (2, 3). Increased iron retention within inflammatory macrophages, which is favored by the induction of the iron storage protein ferritin (4–8) and regarded as a host attempt to withhold iron from the invading pathogens (1, 9, 10), may be due to increased iron uptake and decreased iron export (9). Characterization of the interaction between the acute phase protein hepcidin and the iron exporter ferroportin has shed light on the molecular mechanisms underlying the blockade of macrophage iron release (2, 3, 11). The cytokine-triggered increase in circulating hepcidin causes the internalization and degradation of ferroportin (12), the major iron exporter, thus blocking iron release from macrophages (13). However, although it has been shown that direct exposure to a bolus of hepcidin rapidly lowers serum iron (14) and that peak urinary hepcidin levels in LPS-treated subjects precede the development of hypoferremia (15), the rapid onset of hypoferremia in LPS-treated mice (16–19) suggests that factors other than hepcidin-dependent ferroportin down-regulation (e.g. iron uptake) may be important for iron sequestration within reticuloendothelial cells during the very early phase of the inflammatory response.
The pathways of iron acquisition by macrophages are less clear, as are the changes induced by inflammatory stimuli. This is particularly true in the case of the role of changes in the internalization of transferrin-bound iron through the transferrin receptor (TfR1)2 during the development of reticuloendothelial iron sequestration under inflammatory conditions. A number of studies have shown that exposure to inflammatory stimuli for 10–24 h down-regulates TfR1 expression (5, 6, 20–23) and, because this impairs a major iron uptake mechanism (21, 23), it cannot account for the increased accumulation of iron in macrophages. This inhibition of TfR1 expression is post-transcriptionally controlled by means of the well characterized interaction between iron regulatory proteins (IRPs) and the iron-responsive elements in the untranslated regions of iron-related mRNAs (24–27), as nitric oxide (NO)-dependent IRP2 down-regulation decreases TfR1 mRNA levels and increases ferritin synthesis (5–7, 22).
However, TfR1 expression is also regulated at the transcriptional level. We and others have previously shown that TfR1 expression is up-regulated by the hypoxia-inducible factor (HIF1) (28–30), which is typically activated under hypoxic conditions but can also be turned on by a number of non-hypoxic stimuli, including inflammatory signals such as NO and LPS (for review, see Dery et al. (31)).
On the basis of these considerations, we investigated the effect of LPS/IFNγ on the HIF-1-mediated activation of TfR1 gene expression in macrophages. In an attempt to clarify the effect of the interplay between inflammatory and anti-inflammatory modulators on HIF-1-mediated TfR1 expression, we also considered the effect of the anti-inflammatory molecule adenosine, as we have recently shown that adenosine A2A receptor-mediated signaling induces HIF-1 and activates HIF-1 target genes in macrophages (32). The results of the present study show that, by inducing NF-κB, inflammatory signals activate HIF-1-dependent and IRP-independent TfR1 expression and the uptake of transferrin-bound iron. They also show that exposure to the adenosine A2A receptor agonist CGS21680 alone increases HIF-1-mediated TfR1 expression, whereas pre-treatment with CGS21680 prevents the inducing effect of LPS/IFNγ on HIF-1 and TfR1 expression by inhibiting NF-κB activity.
EXPERIMENTAL PROCEDURES
Cell Cultures and Treatments—The J774A.1 and RAW 264.7 murine macrophage cell lines were obtained from the European Collection of Cell Cultures and cultured in endotoxin-free E-MEM or RPMI 1640 medium, respectively (Sigma), containing 10% fetal bovine serum, 2 mm glutamine, 100 units/ml penicillin, and 0.1 ng/ml streptomycin at 37 °C in 5% CO2. Proteose peptone-elicited peritoneal macrophages were harvested from 8-week-old pathogen-free female CD1 mice (Charles River Italia, Calco, Italy) housed, fed, and handled in compliance with the prescriptions for the care and use of laboratory animals. The macrophages were purified by means of adherence to plastic tissue culture clusters (Corning-Costar Italia, Milan, Italy) for 2 h at 37°C in 5% CO2 as previously described (33). Near-confluent J774A.1 or RAW 264.7 cells and peritoneal macrophages were exposed to 1 μg/ml LPS and 100 units/ml IFNγ or various concentrations (5–50 μm) of the A2A adenosine receptor agonist CGS21680 (Sigma) or 100 μm adenosine (Sigma) for different times. When appropriate, the cells were treated with 10 μm Bay 11-7082 (Sigma) or 100 nm chetomin (Alexis, Italy) or 100 μm desferrioxamine (DFO, Sigma).
Immunoblot Analyses—To detect TfR1 and β-actin, cytosolic extracts were prepared as described (34). HIF-1α, NF-κB p65, and TFIID were determined in nuclear extracts prepared according to Tacchini et al. (35). Aliquots of cytosolic or nuclear extracts containing equal amounts of proteins (as assessed using the Bio-Rad protein assay kit) were electrophoresed and electroblotted onto Hybond ECL membranes (Amersham Biosciences). After assessing transfer by means of Ponceau S staining, the membranes were incubated with TfR1 antibody (Zymed Laboratories Inc., San Francisco, CA) diluted 1:1000, HIF-1α antibody (H1α67, Novus Biologicals, Littleton, CO) diluted 1:1000, TFIID antibody (Santa Cruz Biotechnology, Santa Cruz, CA) diluted 1:500, NF-κB p65 antibody (Santa Cruz Biotechnology) diluted 1:500, and β-actin antibody (Sigma) diluted 1:20,000. After incubation with the appropriate secondary antibody, the antigens were detected using an immunodetection kit (ECL Plus, Amersham Biosciences) and quantitated by means of densitometry with the values being calculated after normalization to the amount of TFIID or β-actin.
Uptake of 55Fe-labeled Transferrin—To evaluate the incorporation of 55Fe-labeled transferrin (Tf), the cells were exposed to 1 μm 55Fe-labeled Tf during the last 2 h of the various treatments. Human apoTf (Sigma) was incubated with 55Fe-iron citrate, prepared by mixing 55FeCl3 (PerkinElmer Life Sciences) with citric acid in a 1:2 molar ratio under previously described conditions (36). At the end of the incubation, the medium was removed, and the cells were washed and homogenized in the lysis buffer used for the immunoblot analysis. Aliquots of the lysates were taken to measure the amount of cellular 55Fe by means of liquid scintillation counting using Ultima Gold (Packard Instrument Co.) and determine protein content.
Electrophoretic Mobility Shift Assay—Aliquots of the nuclear extracts were incubated with [γ-32P]ATP (Amersham Bioscience)-labeled oligonucleotides (Primm, Milan, Italy) corresponding to the sequence from nucleotide position –93 to nucleotide position –75 relative to the transcription start site in the human TfR1 gene encompassing the binding sites for HIF-1 (5′-AGCGTACGTGCCTCAGGA-3′) and oct-1 (5′-TGCGAATGCAAATCACTAGAA-3′), electrophoresed, and autoradiographed (35). The specificity of the assay was demonstrated by the fact that the signals disappeared after the addition of a 50-fold excess of specific (but not nonspecific) oligonucleotides. The quantitative determinations were made by means of direct nuclear counting using an InstantImager (Packard Instrument Co.), with the values being calculated after normalization to the activity of oct-1.
Plasmid Constructs—The pGL3PGK6TKp vector (a kind gift of P. J. Ratcliffe, Oxford, UK) contains a hypoxia-responsive element (HRE) multimer (37). The wild-type (–455 TfR1) construct containing the TfR1 promoter was generated from vectors kindly provided by Dr. L. Kuhn, as previously described (28). For the mutated (-455 mTfR1) plasmid, a site-directed mutation of the HRE sequence 5′-TACGTGC-3′ in the –455 TfR1, was introduced by designing one pair of complementary oligonucleotides including the desired mutation site to replace the bases TACGT with AATTC. The pNF-κB-Luc reporter vector designed for monitoring the activation of NF-κB was purchased from BD Biosciences. The expression vector pcDNA3ARNTdelta_b (ΔARNT) (obtained from M. Schwarz, Tübingen, Germany) codes for the dominant negative mutant form of the HIF-1 ARNT subunit (35). The RSVIkBαMMS supersuppressor of NF-κB (38) (ssNF-κB) was kindly provided by N. D. Perkins.
Transient Transfection Assay—Subconfluent RAW 264.7 cells maintained in complete medium were transiently transfected (using TransIT™ LT1, Mirus, Bologna, Italy) in 24-well multiwell plates with a 50:1 mixture of the various constructs and the pRL-TK reporter vector containing Renilla luciferase, which was used to normalize transfection efficiency. When appropriate, the cells were co-transfected with dominant negative expression vectors (see above). After 24 h, the cells were collected, washed, and lysed using the reporter lysis buffer (Promega, Milan, Italy), and luciferase activities were measured in a Promega luminometer using the Dual Luciferase Reporter Assay System (Promega) (37). The empty vectors showed practically undetectable luciferase activity. All of the transfection experiments were carried out on duplicate plates and were repeated at least three times (n = 6).
Short Hairpin RNA Knockdown—Short hairpin RNA (shRNA) constructs against Mus musculus HIF-1α (catalog number TR517255) were purchased from Origene, Technologies, Inc. (Rockville, MD). The targeted sequences were: CTGTTCACCAAAGTTGAATCAGAGGATA (#1), CTTCTGTTATGAGGCTCACCATCAGTTA (#2), TCAAGAAACGACCACTGCTAAGGCATCA (#3), TTACCTTCATCGGAAACTCCAAAGCCACT (#4). RAW 264.7 macrophages maintained in complete medium were plated onto 6-well culture dishes (400.000 cells/well) or onto T75 flasks (2.5 × 106 cells/flask) for TfR1 and HIF-1α immunoblot analysis, respectively. After 24 h the medium was changed, and cells were transfected with a mixture of the four plasmids (250 ng each) containing the shRNA or with the empty pRS vector using the transfection method described above. 48 h later the medium was changed, and the cells were treated for 4 h with LPS/IFNγ. The cytosolic and nuclear extracts were then prepared as described above.
RNA-protein Gel Retardation Assay—The cell lysates were prepared as described (34), and equal amounts of the supernatant proteins were incubated with a probe transcribed from the pSPT-fer plasmid containing the iron-responsive elements of the human ferritin H chain (39) using T7 RNA polymerase in the presence of [α-32P]UTP (Amersham Biosciences) and then treated with RNase T1 and heparin (34). After separation on non-denaturing polyacrylamide gels, the RNA-protein complexes were visualized by means of autoradiography and quantitated by means of direct nuclear counting using an InstantImager (Packard Instrument Co.).
Northern Blot Analysis—Total cell RNA was isolated, and equal amounts were electrophoresed under denaturing conditions (35). To confirm that each lane contained equal amounts of RNA, the rRNA content in each lane was estimated in the ethidium bromide-stained gels by means of laser densitometry. The RNA was transferred to Hybond-N filters (Amersham Biosciences) and hybridized with 32P-labeled mouse TfR1 cDNA. The quantitative determinations were made by means of direct nuclear counting using an InstantImager (Packard Instrument Co.), with the values calculated after normalization to the amount of ribosomal RNA.
Statistical Analysis—The data are expressed as the mean values ± S.D. and were compared using analysis of variance; p values of <0.05 were considered significant.
RESULTS
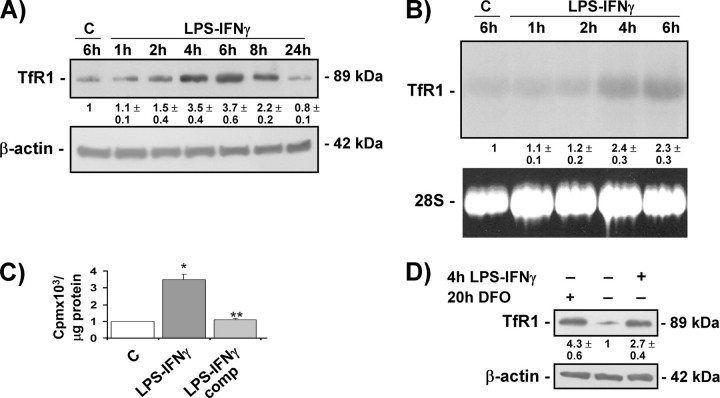
LPS-IFNγ Up-regulates TfR1 Expression and Transferrin-bound Iron Uptake—To study the effect of inflammatory stimuli on TfR1, mouse RAW 264.7 macrophages were treated with the concentrations of LPS-IFNγ normally used to stimulate macrophages (which have been previously demonstrated to affect iron metabolism) (5, 6, 21), and TfR1 expression was evaluated at protein and mRNA levels. Cell viability and proliferation rates were not affected by any of the treatments (results not shown). Immunoblot analysis showed that TfR1 protein levels increased in a time-dependent manner up to 6–8 h and then decreased to below those of untreated cells at 24 h (Fig. 1A). Northern blot analysis (Fig. 1B) showed that the increase in TfR1 protein levels observed in RAW 264.7 cells exposed to LPS-IFNγ for 4–6 h was accompanied by an increase in steady-state TfR1 mRNA levels. Similar results were found in another macrophage cell line (J774A.1) (results not shown). To determine whether the higher TfR1 expression also enhanced iron uptake, RAW 264.7 cells were exposed to 55Fe-labeled transferrin and the total cell content of radioactive iron was determined by means of liquid scintillation counting. The 55Fe content was more than three times higher in the LPS-IFNγ-treated RAW 264.7 cells, thus indicating that the higher TfR1 levels are functionally involved in favoring greater iron availability (Fig. 1C). Competition by a 100-fold excess of unlabeled transferrin effectively blocked iron incorporation. To investigate whether the TfR1 induction observed in the macrophage cell lines was also detectable in primary mononuclear cells, we analyzed extracts from mouse peritoneal macrophages treated with LPS-IFNγ for 4 h; exposure to LPS-IFNγ strongly increased TfR1 content with an effect similar to that obtained with the iron chelator DFO (Fig. 1D).
FIGURE 1.
LPS-IFNγ activates TfR1 expression and transferrin-bound iron uptake. A, immunoblot analysis of cytosolic extracts from RAW 264.7 cells untreated (C) or treated with LPS-IFNγ for different times using the antibody against TfR1. The blots were reprobed with the antibody against β-actin as a loading control. B, Northern blot analysis of total RNA extracted from RAW 264.7 cells untreated (C) or treated with LPS-IFNγ for different times and hybridized with 32P-labeled TfR1 cDNA. 28 S RNA was used as a control for RNA loading. C, 55 Fe-labeled transferrin uptake (expressed as cpm of radioactive iron/μg of protein) in RAW 264.7 cells untreated (C) or treated with LPS-IFNγ for 4 h in the absence or presence of a 100-fold excess of unlabeled transferrin (comp). The values are mean ± S.D. *, p < 0.001 versus untreated controls; **, p < 0.001 versus cells without competition. D, immunoblot analysis of cytosolic extracts from mouse peritoneal macrophages untreated or treated with LPS-IFNγ for 4 h or DFO for 20 h using the antibody against TfR1; β-actin was used as a loading control. All of the results are representative of at least three independent experiments. The values indicate the -fold difference ±S.D. in relation to the untreated controls.
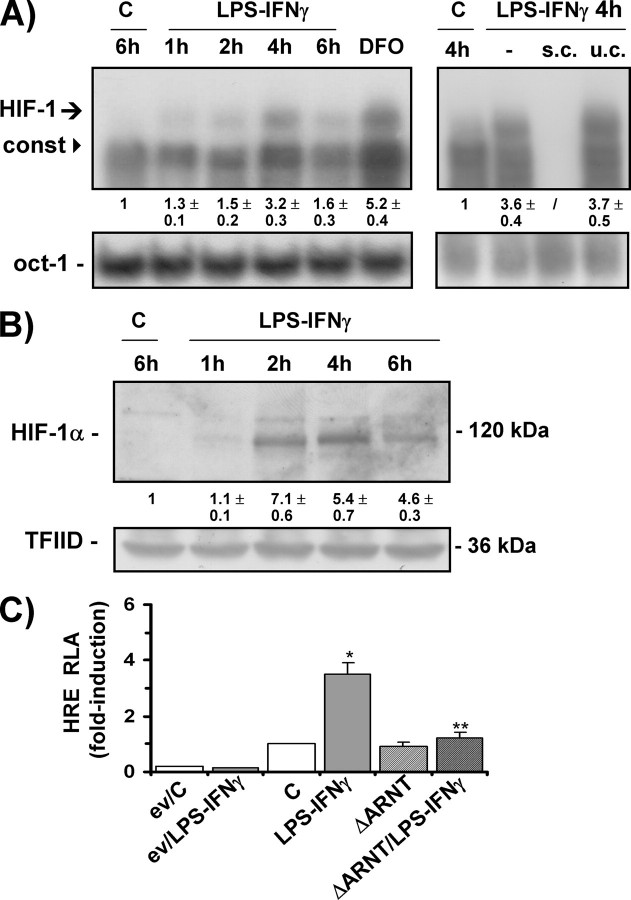
Increased TfR1 Expression Is Mediated by HIF-1—Given the previous demonstration that TfR1 is an HIF-1-inducible gene (28–30) and that HIF-1 is also activated by typical inflammatory stimuli such as LPS (40, 41), we investigated whether HIF-1 was involved in the TfR1 induction response to LPS-IFNγ. RAW 264.7 cells were exposed to LPS-IFNγ, and the DNA binding activity of HIF-1 was evaluated by electrophoretic mobility shift assay using a probe spanning the HRE present in the TfR1 promoter (28, 30). The treatment increased HIF-1 activity in a time-dependent manner, and activation was already detectable at 1–2 h; binding peaked after 4 h and then declined (Fig. 2A). Similar binding activity was found in extracts of cells exposed to the iron chelator DFO, a well known inducer of HIF-1 (42). Competition experiments using both specific and unspecific unlabeled oligonucleotides demonstrated the specificity of the interaction between the DNA probe and the induced nuclear factors. The complex that migrates faster is a so-called constitutive factor that is closely related or identical to the transcription factors ATF-1 and CREB-1 and has been shown to be induced by hypoxia, iron chelation, and adenosine (32, 43–45). Immunoblot analysis of the nuclear extracts showed that HIF-1 protein levels were also greatly increased in the cells exposed to LPS-IFNγ, with a time response reflecting that shown by DNA binding activity (Fig. 2B). Similar HIF-1 activation was found in LPS-IFNγ-treated J774A.1 macrophages (results not shown). We then used transactivation capacity experiments to verify whether the HIF-1 induced by LPS-IFNγ was transcriptionally active. In RAW 264.7 cells transiently transfected with a luciferase reporter gene controlled by a DNA fragment containing multiple consensus HREs, which has previously been shown to drive HIF-1-dependent transcription in response to hypoxia and hypoxiamimics (32, 37), the expression of the reporter gene increased more than 3-fold in response to LPS-IFNγ (Fig. 2C). Further indications of the involvement of HIF-1 in the LPS-IFNγ-dependent activation of luciferase activity were obtained by means of experiments in which the transactivating capacity of HIF-1 was almost completely abolished by the co-transfection of a plasmid expressing a dominant negative of the β subunit of the HIF-1 heterodimer (ΔARNT), which retains the capacity to form a heterodimer but cannot bind DNA (35) (Fig. 2C).
FIGURE 2.
LPS-IFNγ induces HIF-1 expression and transactivation capacity. A, electrophoretic mobility shift assay analysis of the HIF-1 DNA binding activity of nuclear extracts from RAW 264.7 cells untreated (C) or treated with LPS-IFNγ for different times or DFO for 20 h; s.c. and u.c. are the specific and unspecific competition, respectively, of the LPS-IFNγ/4-h sample. The arrow indicates the inducible HIF-1 complex, whereas the arrowhead indicates the constitutive (const) complex. The binding activity of the constitutively expressed transcription factor oct-1 was used to assess equal loading. B, immunoblot analysis of the nuclear extracts from RAW 264.7 cells untreated (C) or treated with LPS-IFNγ for different times using anti-HIF-1α antibody. The blots were reprobed using the antibody against TFIID as a loading control. C, relative luciferase activity (RLA) in RAW 264.7 cells untreated (C) or exposed to LPS/IFNγ. The cells were transiently transfected with the empty pGL3 vector (ev) or a construct in which luciferase was controlled by an HRE multimer; when appropriate, the cells were also co-transfected with an expression vector coding for a dominant negative mutant of the constitutive HIF-1 β subunit (ΔARNT). The cells were co-transfected using a control vector containing the Renilla luciferase gene. Luciferase activity was determined after 24 h, corrected for transfection efficiency on the basis of Renilla luciferase activity, and normalized to the activity recorded in untreated cells (arbitrarily defined as 1). *, p < 0.001 versus untreated controls; **, p < 0.001 versus cells treated with LPS/IFNγ. All of the results are representative of at least three independent experiments. The values indicate the -fold difference ±S.D. in relation to the untreated controls.
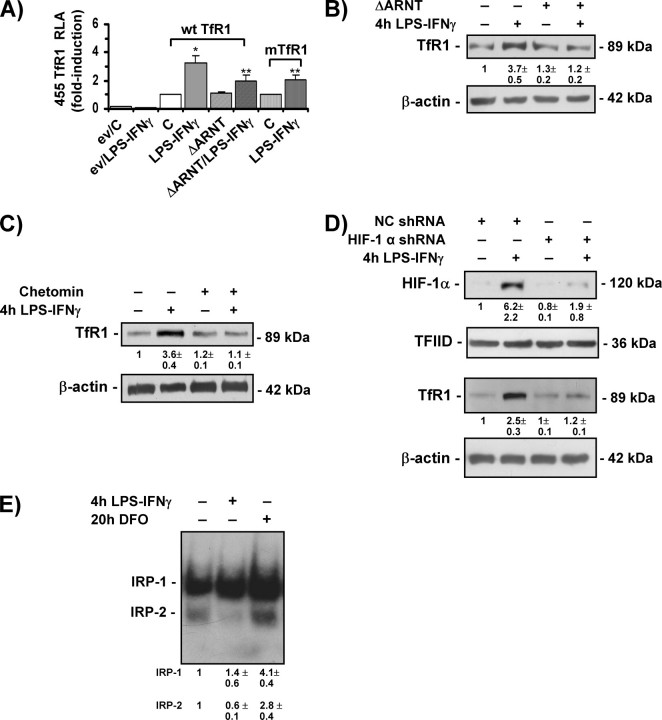
Having demonstrated that HIF-1 activity is induced by LPS-IFNγ in RAW 264.7 cells, we investigated the role of HIF-1-mediated transcriptional regulation in the induction of TfR1 expression. To this end, RAW 264.7 cells were transfected with a luciferase reporter gene under the control of a 455-bp fragment of the human TfR1 promoter (wild type or mutated in the HRE) (Fig. 3A); it has been previously demonstrated that this fragment is sufficient for the efficient transcriptional induction of a reporter gene in response to hypoxia and that the mutation is effective in suppressing the responsiveness to hypoxia and iron chelation (28, 30). We found that the wild-type construct was transcribed 3-fold more efficiently in RAW 264.7 cells treated with LPS-IFNγ than in control cells. Importantly, both the HRE mutation and co-transfection of the dominant negative ΔARNT significantly reduced the differences in transcription, thus indicating that HIF-1-mediated transcriptional activation is involved in TfR1 expression in LPS-IFNγ-treated RAW 264.7 cells (Fig. 3A). A further indication of the role of HIF-1 was provided by experiments showing that the LPS-IFNγ-mediated increase in TfR1 protein levels was abolished in RAW 264.7 cells transfected with the dominant negative ΔARNT (Fig. 3B). Similarly, the presence of chetomin, which prevents the formation of the transcriptionally competent HIF-1 complex (46), completely inhibited the increase in TfR1 protein levels triggered by exposure to LPS-IFNγ (Fig. 3C). To further verify the role of HIF-1 in the activation of TfR1 in response to LPS-IFNγ, we used shRNA technology to specifically knock down HIF-1α (Fig. 3D). Transfection of RAW 264.7 cells with a set of four expression vectors coding shRNAs against HIF-1α resulted in a 70% reduction in the inducible levels of HIF-1 as compared with cells transfected with a negative control plasmid. The down-regulation of HIF-1 prevented almost completely the increase in TfR1 triggered by exposure to LPS-IFNγ. The lack of effect on the expression of TFIID and β-actin indicates that silencing was specific.
FIGURE 3.
Increased TfR1 transcription in LPS-IFNγ-treated cells is mediated by HIF-1. A, RLA in RAW 264.7 cells untreated (C) or exposed to LPS/IFNγ. The cells were transiently transfected with the empty pGL2 basic vector (ev) or a construct in which luciferase was controlled by a 455-bp fragment of the wild-type (wtTfR1) or mutated (mTfR1) TfR1 promoter. The cells were co-transfected with the Renilla luciferase and, when appropriate, dominant negative ΔARNT vectors as described in Fig. 2C. Luciferase activity was determined after 24 h and calculated as described in Fig. 2C.*, p < 0.001 versus untreated controls; **, p < 0.001 versus cells treated with LPS/IFNγ. B, immunoblot analysis of cytosolic extracts from RAW 264.7 cells untreated or treated with LPS-IFNγ for 4 h in the presence or absence of the dominant negative ΔARNT using the antibody against TfR1. The blots were reprobed with the antibody against β-actin as a loading control. C, immunoblot analysis of cytosolic extracts from RAW 264.7 cells untreated or treated with LPS-IFNγ for 4 h in the presence or absence of the HIF-1 inhibitor chetomin (added 2 h before treatment) using the antibody against TfR1. The blots were reprobed with the antibody against β-actin as a loading control. D, immunoblot analysis of nuclear or cytosolic extracts from RAW 264.7 cells untreated or treated with LPS-IFNγ for 4 h in the presence of either the negative control shRNA (NC) or shRNA targeting HIF-1α. Nuclear extracts were probed using the antibody against HIF-1α and TFIID as a loading control; cytosolic extracts were incubated with the antibody against TfR1 and reprobed with the antibody against β-actin as a loading control. E, RNA band-shift analysis of IRP activity in RAW 264.7 cells untreated or treated with LPS/IFNγ for 4 h or DFO for 20 h. The cytosolic extracts were incubated with an excess of a 32P-labeled iron-responsive element probe and separated on non-denaturing polyacrylamide gels. All of the results are representative of at least three independent experiments. The values indicate the -fold difference ±S.D. in relation to the untreated controls.
To investigate the possibility that LPS-IFNγ leads to TfR1 and HIF-1 activation by reducing the intracellular iron pool, we used an RNA band shift assay to measure the activity of the iron regulatory proteins IRP1 and IRP2, key regulators of iron homeostasis that are activated by decreased intracellular iron availability and in turn up-regulate TfR1 expression (24, 25). In line with previous findings (5), Fig. 3D shows that the RNA binding activity of IRP1 slightly and not significantly increased, and that of IRP2 decreased in RAW 264.7 cells treated with LPS-IFNγ for 4 h but, as expected, IRP2 was remarkably activated in cells deprived of iron by exposure to the iron chelator DFO (24, 25). IRP binding to the 3′-untranslated region of TfR1 mRNA protects the transcript from degradation, thus increasing TfR1 expression (25); therefore, diminished levels of IRP2 are not expected to contribute to the up-regulation of TfR1 observed at early times of exposure to inflammatory stimuli. This finding indicates that the iron-dependent post-transcriptional control exerted by the iron-responsive element/IRPs pathway is not involved in the up-regulation of TfR1 expression observed soon after LPS-IFNγ stimulation.
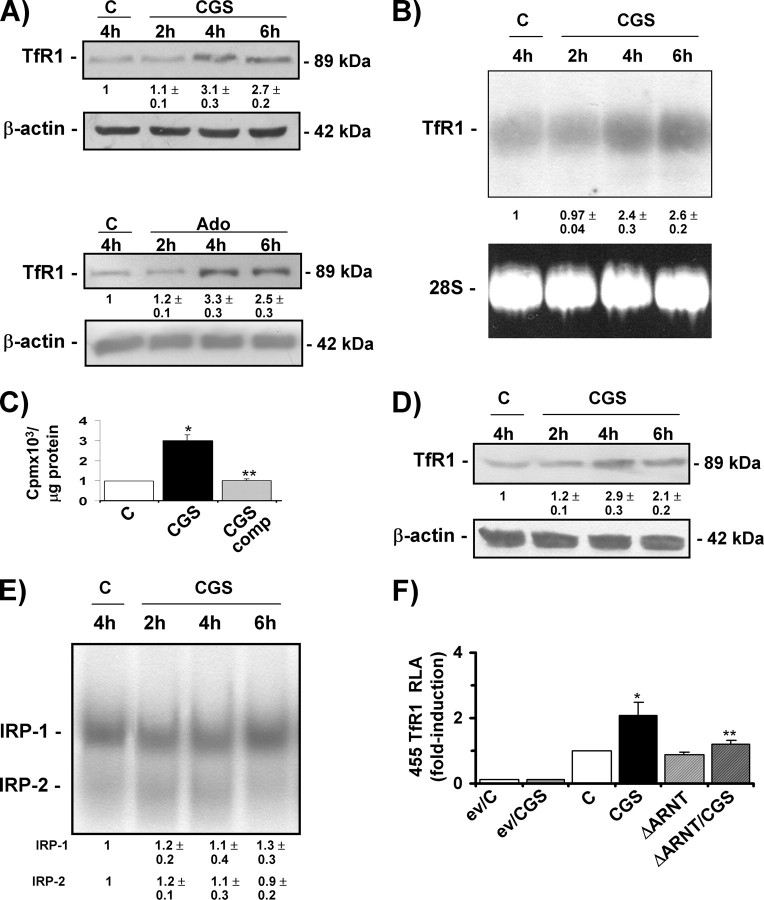
Adenosine A2A Receptor Activation Increases HIF-1-mediated TfR1 Expression and Transferrin-bound Iron Uptake—Under normoxic conditions, the HIF-1 pathway can be stimulated by both inflammatory mediators (40, 41, 47) and anti-inflammatory effectors, as we have recently found that by activating the A2A receptor, the tissue-protecting and anti-inflammatory molecule adenosine can induce HIF-1 and activate some HIF-1 target genes in J774A.1 and peritoneal macrophages (32). We, therefore, next investigated the role of adenosine pathway signaling in HIF-1-mediated TfR1 expression. RAW 264.7 cells, which express adenosine receptors (48) and are a widely used model for investigating the anti-inflammatory effects of adenosine, were incubated in the presence of 50 μm CGS21680, a selective receptor agonist of the A2A receptor that has been shown to activate HIF-1 (32). Time-course experiments showed that TfR1 protein (Fig. 4A) and mRNA (Fig. 4B) levels increased after 4–6 h of treatment and then decreased to control levels (data not shown). Exposure to adenosine also increased TfR1 protein levels (Fig. 4A). We also found that the uptake of transferrin-bound iron was stimulated because 55Fe content was about three times higher in CGS21680-treated RAW 264.7 cells (Fig. 4C). These results are in line with the findings of several studies in which high micromolar concentrations of adenosine analogues were used to elicit functional responses in the RAW 264.7 line (45, 49–51), but a similar increase in TfR1 protein levels was found in J774A.1 cells treated with a 10-fold lower concentration of CGS21680 (Fig. 4D).
FIGURE 4.
The adenosine A2A receptor agonist CGS21680 induces HIF-1-mediated TfR1 expression and transferrin-bound iron uptake. A, immunoblot analysis of cytosolic extracts from RAW 264.7 cells untreated (C) and treated with 50 μm CGS21680 (CGS) or 100 μm adenosine (Ado) for different times using the antibody against TfR1. The blots were reprobed with the antibody against β-actin as a loading control. B, Northern blot analysis of total RNA extracted from RAW 264.7 cells untreated (C) or treated with 50 μm CGS21680 for different times and hybridized with 32P-labeled TfR1 cDNA. 28 S RNA was used as a control for RNA loading. C, 55 Fe-labeled transferrin uptake (expressed as cpm of radioactive iron/μg of protein) in RAW 264.7 cells untreated (C) or treated with 50 μm CGS21680 for 4 h in the presence or absence of a 100-fold excess of unlabeled transferrin (comp). The values are the mean ± S.D. *, p < 0.001 versus untreated controls; **, p < 0.001 versus cells without competition. D, immunoblot analysis of cytosolic extracts from J774A.1 cells untreated (C) or treated with 5 μm CGS21680 for different times using the antibody against TfR1. The blots were reprobed with the antibody against β-actin as a loading control. E, RNA band-shift analysis of IRP activity in RAW 264.7 cells untreated or treated with 50 μm CGS21680 for different times. The experimental procedures were as described for Fig. 3D. F, RLA in RAW 264.7 cells untreated (C) or exposed to 50 μm CGS21680. The cells were transiently transfected with the empty pGL2 basic vector (ev) or a construct in which luciferase was controlled by a 455-bp fragment of the TfR1 promoter. The cells were co-transfected with the Renilla luciferase and, when appropriate, dominant negative ΔARNT vectors as described in Fig. 2C. Luciferase activity was determined after 24 h and calculated as described in Fig. 2C.*, p < 0.001 versus untreated controls; **, p < 0.001 versus cells treated with 50 μm CGS21680. All of the results are representative of at least three independent experiments. The values indicate the -fold difference ±S.D. in relation to the untreated controls.
To investigate the molecular mechanisms underlying these effects, we first assessed IRPs activity (Fig. 4E) and found that the RNA binding activity of both IRP1 and IRP2 remained unaltered in RAW 264.7 cells treated with CGS21680 for 2–6 h, i.e. when TfR1 up-regulation occurs. Further evidence that CGS21680 activates TfR1 at the transcriptional level in an IRP-independent manner was provided by transfection experiments showing that it doubled the luciferase activity driven by the TfR1 promoter (Fig. 4F); furthermore, the effect of CGS21680 was almost completely prevented when the cells were co-transfected with the dominant negative ΔARNT (Fig. 4F).
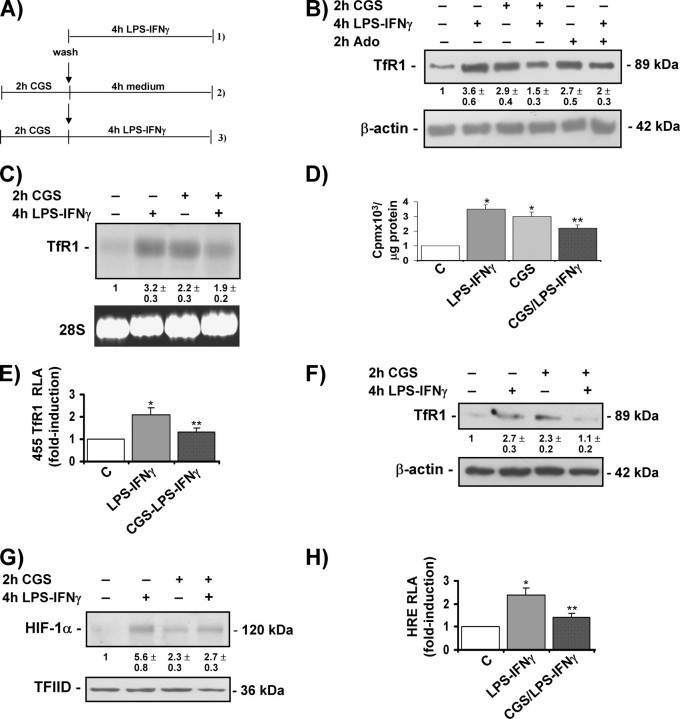
A2A Receptor Activation Inhibits LPS-IFNγ-induced TfR1 Expression and HIF-1 Activity—Given the observation that both LPS-IFNγ and the A2A receptor agonist (i.e. immunomodulators with inflammatory or anti-inflammatory properties) activate TfR1 expression through the HIF-1 pathway, additional experiments were performed to investigate TfR1 expression in cells treated with CGS21680 before the addition of LPS-IFNγ (Fig. 5A), an experimental model that is usually used to demonstrate the anti-inflammatory action of adenosine and adenosine analogues (52–54). The LPS-IFNγ-stimulated increase in the levels of TfR1 protein (Fig. 5B) and mRNA (Fig. 5C) was attenuated when the cultures were pretreated with CGS21680, which when present alone was nevertheless capable of inducing TfR1 expression. A similar effect on TfR1 protein levels was exerted by pretreatment with adenosine (Fig. 5B). Comparable results were obtained in J774A.1 cells pretreated with 5 μm CGS21680 (results not shown). Similarly, the uptake of transferrin-bound iron was reduced when the cells were pre-incubated with CGS21680 before the addition of LPS-IFNγ (Fig. 5D). The transfection experiments shown in Fig. 5E indicated that the effect of A2A receptor stimulation is mediated at the transcriptional level because the LPS-IFNγ-triggered increase in luciferase activity under the control of the TfR1 promoter was reduced by pre-exposure to CGS21680. The preventive effects of CGS21680 detected in RAW 264.7 cells were also found in freshly isolated primary macrophages, as TfR1 protein induction by LPS-IFNγ was impaired in peritoneal macrophages pretreated with CGS21680 (Fig. 5F).
FIGURE 5.
Activation of the A2A receptor inhibits LPS-IFNγ-induced TfR1 expression and HIF-1 activity. A, experimental design. RAW 264.7 cells were treated with 1) LPS-IFNγ for 4 h and 2) 50 μm CGS21680 for 2 h and then washed and incubated in fresh medium for a further 4 h and 3) 50 μm CGS21680 for 2 h and then washed and incubated in fresh medium containing LPS-IFNγ for a further 4 h. The untreated cells were incubated in fresh medium for 6 h. B, immunoblot analysis of cytosolic extracts from RAW 264.7 cells untreated and treated with CGS21680 as described in A or pretreated with 100 μm adenosine (Ado) using the antibody against TfR1. The blots were reprobed with the antibody against β-actin as a loading control. C, Northern blot analysis of total RNA extracted from RAW 264.7 cells untreated or treated as described in A and hybridized with 32P-labeled TfR1 cDNA. 28 S RNA was used as a control for RNA loading. D, 55 Fe-labeled transferrin uptake (expressed as cpm of radioactive iron/μg of protein) in RAW 264.7 cells untreated (C) or treated as described in A. *, p < 0.001 versus untreated controls; **, p < 0.001 versus cells treated with LPS/IFNγ. E, RLA in RAW 264.7 cells untreated (C) or treated as described in A. The cells were transiently transfected with a construct in which luciferase was controlled by a 455-bp fragment of the TfR1 promoter and co-transfected with the Renilla luciferase vector. Luciferase activity was determined after 24 h and calculated as described in Fig. 2C.*, p < 0.001 versus untreated controls; **, p < 0.001 versus cells treated with LPS/IFNγ. F, immunoblot analysis of cytosolic extracts from mouse peritoneal macrophages untreated or treated as described in A using the antibody against TfR1. The blots were reprobed with the antibody against β-actin as a loading control. G, immunoblot analysis of nuclear extracts from RAW 264.7 cells untreated or treated as described in A using the antibody against HIF-1α. The blots were reprobed with the antibody against TFIID as a loading control. H, RLA in RAW 264.7 cells untreated (C) or treated as described in A. The cells were transiently transfected with a construct in which luciferase was controlled by an HRE multimer and co-transfected with the Renilla luciferase vector. Luciferase activity was determined after 24 h and calculatedas described in Fig. 2C.*, p < 0.001 versus untreated controls; **, p < 0.001 versus cells treated with LPS/IFNγ. All of the results are representative of at least three independent experiments. The values indicate the -fold difference ±S.D. in relation to the untreated control.
To determine the molecular mechanisms underlying the effects of CGS21680 pretreatment on TfR1 transcription, we analyzed HIF-1 expression and transactivation capacity. As shown in Fig. 5G, the LPS-IFNγ-activated increase in HIF-1 protein levels was blunted when RAW 264.7 cells were pre-treated with CGS21680, and the same occurred in CGS21680-pre-treated J774A.1 cells (results not shown). Furthermore, partial inhibition of the induction of luciferase activity under the control of the multiple consensus HREs was observed in RAW 264.7 cells pre-treated with CGS21680 before stimulation with LPS-IFNγ (Fig. 5H).
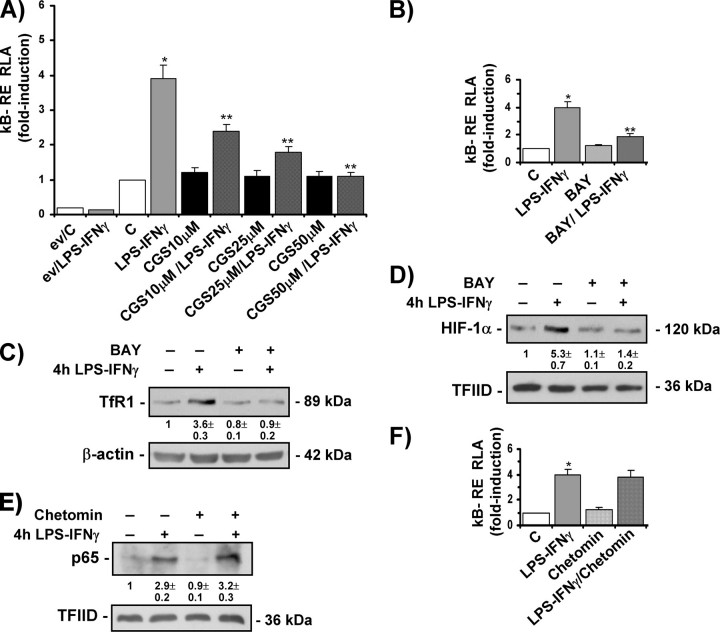
Activation of the A2A Receptor Prevents LPS-IFNγ-induced and HIF-1-mediated TfR1 Expression by Inhibiting NF-κB Activity—Because previous studies have indicated that the stimulation of adenosine receptors has anti-inflammatory effects by suppressing NF-κB activation (55, 56) and it has also been recently reported that LPS enhances HIF-1 transcription in human monocytic cell lines at normoxia by inducing the binding of NF-κB to the HIF-1 promoter (41), we tested whether the transactivation capacity of NF-κB was induced by LPS-IFNγ in RAW 264.7 macrophages and the effects of CGS21680 pretreatment. As expected, LPS-IFNγ greatly increased luciferase activity in cells transfected with a construct bearing multiple NF-κB responsive elements, whereas treatment with CGS21680 alone had no significant effects (Fig. 6A); on the other hand, pretreatment with increasing concentrations of the A2A receptor agonist progressively reduced the LPS-IFNγ-dependent activation of luciferase activity. To test whether NF-κB was involved in LPS-IFNγ-dependent HIF-1 induction and consequent TfR1 activation, we first determined whether the direct inhibition of NF-κB activity (independently of the CGS21680-mediated pathway) modified the response of TfR1 to LPS-IFNγ stimulation. When RAW 264.7 cells were stimulated with LPS-IFNγ in the presence of the NF-κB-specific inhibitor BAY117082, which suppressed the inducing effect of LPS-IFNγ on NF-κB transcriptional activity (Fig. 6B), the induction of TfR1 protein levels was completely prevented (Fig. 6C). Similar inhibition was observed in cells transfected with the RSVIkBαMMS vector, a dominant negative supersuppressor of NF-κB (ssNF-κB) (data not shown). The determination of HIF-1 levels in nuclear extracts demonstrated that the presence of BAY117082 also prevented LPS-IFNγ-activated HIF-1 induction (Fig. 6D). Exposure to BAY117082 alone at this concentration (10 μm) did not affect cell viability (results not shown) or TfR1 or HIF-1 levels (Figs. 6, C and D).
FIGURE 6.
Activation of the A2A receptor prevents LPS-IFNγ-induced and HIF-1-mediated TfR1 expression by inhibiting NF-κB activity. A, RLA in RAW 264.7 cells untreated (C) or treated as described in Fig. 5A. The cells were transiently transfected with the empty pTAL vector (ev) or a construct in which luciferase was controlled by a NF-κB multimer and co-transfected with the Renilla luciferase vector. Luciferase activity was determined after 24 h and calculated as described in Fig. 2C.*, p < 0.001 versus untreated controls; **, p < 0.001 versus cells treated with LPS/IFNγ. B, RLA in RAW 264.7 cells untreated (C) or treated with LPS-IFNγ for 4 h in the presence or absence of the NF-κB inhibitor Bay 11-7082 (BAY) (added 2 h before treatment). The cells were transiently transfected with a construct in which luciferase was controlled by a NF-κB multimer and co-transfected with the Renilla luciferase vector. Luciferase activity was determined as described above. *, p < 0.001 versus untreated controls; **, p < 0.001 versus cells treated with LPS/IFNγ. C, immunoblot analysis of cytosolic extracts from RAW 264.7 cells untreated or treated with LPS-IFNγ for 4 h in the presence or absence of BAY (added 2 h before treatment) using the antibody against TfR1. The blots were reprobed with the antibody against β-actin as a loading control. D, immunoblot analysis of nuclear extracts from RAW 264.7 cells untreated or treated as in panel C using the antibody against HIF-1α. The blots were reprobed with the antibody against TFIID as a loading control. E, immunoblot analysis of nuclear extracts from RAW 264.7 cells untreated or treated with LPS-IFNγ for 4 h in the presence or absence of chetomin (added 2 h before treatment) using the antibody against p65. The blots were reprobed with the antibody against TFIID as a loading control. F, RLA in RAW 264.7 cells untreated (C) or treated with LPS-IFNγ for 4 h in the presence or absence of chetomin (added 2 h before treatment). The cells were transiently transfected with a construct in which luciferase was controlled by a NF-κB multimer and co-transfected with the Renilla luciferase vector. Luciferase activity was determined as described above. *, p < 0.001 versus untreated control. All of the results are representative of at least three independent experiments. The values indicate the -fold difference ±S.D. in relation to the untreated control.
Taken together, these results suggested that HIF-1 is downstream of NF-κB in the response pathway to inflammatory stimuli leading to increased TfR1 expression, and evidence supporting the idea that NF-κB is upstream of HIF-1 was provided by experiments in which we measured NF-κB p65 subunit levels and luciferase activity driven by multiple NF-κB responsive elements in RAW 264.7 cells treated with chetomin. The lack of HIF-1 transcriptional activity caused by chetomin, which inhibited the increase in TfR1 protein levels triggered by exposure to LPS-IFNγ (see Fig. 3C), did not prevent the increase in NF-κB p65 subunit nuclear levels (Fig. 6E) or NF-κB transactivation activity (Fig. 6F) in response to LPS-IFNγ challenge.
DISCUSSION
Macrophages take up iron by means of three major pathways; this is, the acquisition of heme iron as a result of erythrophagocytosis or CD163-mediated hemoglobin-haptoglobin scavenging, the transmembrane transport of iron by DMT1, and the internalization of transferrin-bound iron through TfR1 (1, 9). The roles played by these three pathways in the increased iron storage and reduced circulating iron levels associated with inflammatory conditions are not fully understood. Enhanced non-transferrin-bound iron acquisition through the up-regulation of DMT1 has been described in cytokine-stimulated human monocytic cell lines (23). Similarly, the increased phagocytosis of red blood cells reported in tumor necrosis factor-α-stimulated macrophages could increase the acquisition of heme iron (57). On the other hand, although the up-regulation of TfR1 expression has been reported in fibroblasts exposed to tumor necrosis factor-α and interleukin 1α (58), a number of studies of murine and human reticuloendothelial cells (5, 6, 20–23) have shown that TfR1 expression is post-transcriptionally down-modulated by exposure to inflammatory stimuli for 10–24 h, an effect that would impair an important iron uptake mechanism.
In this study we found reduced TfR1 protein levels in RAW 264.7 cells treated for 24 h with LPS-IFNγ, which is in line with previous findings in murine macrophages (5, 6, 21, 22) but also showed that a transcriptionally regulated induction of TfR1 expression occurs early after treatment. In addition to a number of experiments and assays showing that TfR1 expression is under the transcriptional control of well defined transcription factors during the inflammatory response (see below), RNA band-shift analysis demonstrated that the IRPs that post-transcriptionally control TfR1 expression (24, 25) are not up-regulated by LPS-IFNγ (Fig. 3D and Refs. 5 and 6), thus suggesting that TfR1 mRNA stabilization is not involved.
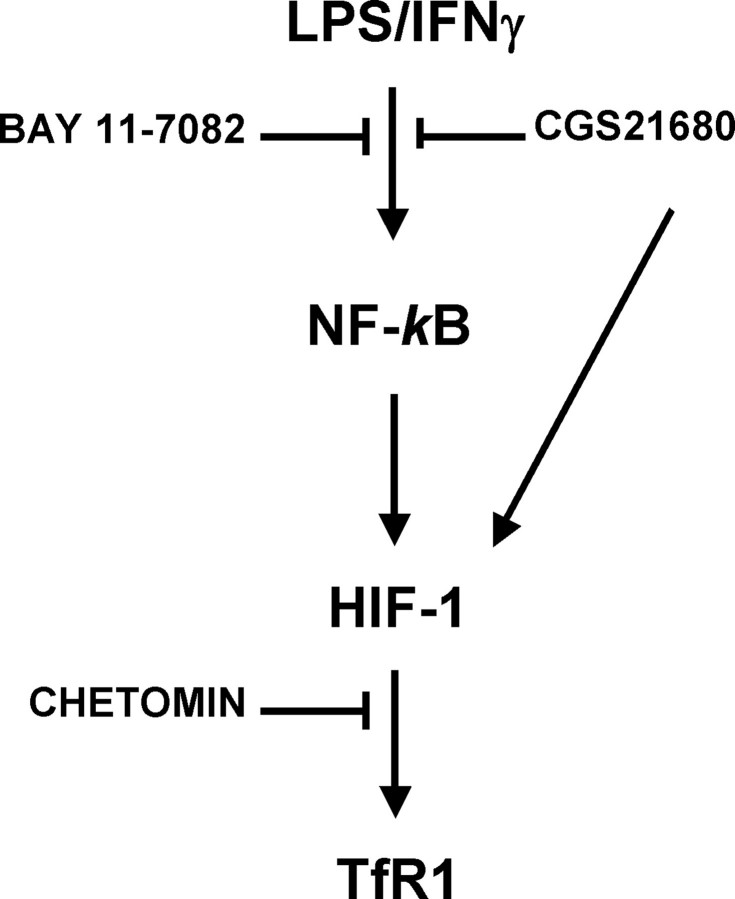
The molecular mechanisms of TfR1 transcriptional regulation under inflammatory conditions are complex and involve at least two pathways (Fig. 7). The induction of TfR1 expression in LPS-IFNγ-treated macrophage cells is regulated by a signaling pathway that successively involves NF-κB activation, HIF-1 induction, and TfR1 transcription. NF-κB activation, which is presumably induced through the TLR4-dependent signaling cascade (59) and possibly involves a transient and early increase in the low molecular weight iron pool (60), occurs very early (30 min after an LPS-IFNγ challenge, results not shown) and leads to a prompt increase in HIF-1 activity. On the basis of previous findings in other experimental models, this HIF-1 activity could be due to the increased transcription (32, 35, 41, 47, 61) and/or translation of HIF-1 mRNA (32, 35, 62), although we cannot exclude the possibility that other mechanisms, such as the stabilization of HIF-1 protein or the modulation of HIF-1 transcriptional activity by LPS-generated reactive oxygen species that has been recently described (63), may be involved.
FIGURE 7.
Proposed model of TfR1 transcriptional modulation by inflammatory and anti-inflammatory signals in macrophages. The induction of TfR1 expression in LPS-IFNγ-treated macrophage cells is regulated by a signaling pathway that successively involves NF-κB activation, HIF-1 induction, and TfR1 transcription. The order of these events is indicated by the fact that HIF-1 activation is prevented by the NF-κB inhibitor Bay 11-7082, whereas chetomin, which inhibits HIF-1 transcriptional activity, is unable to block NF-κB induction. Adenosine plays a dual role; on the one hand, adenosine receptor activation by the A2A receptor agonist CGS21680 may inhibit NF-κB activation by LPS/IFNγ and the cascade of events described above, thus possibly blunting the iron sequestration response typical of the inflammatory status. On the other hand, it may enhance TfR1 expression by activating HIF-1 directly through an A2A receptor-activated and NF-κB-independent signaling cascade (32) under situations of hypoxia, etc.
Higher nuclear levels of HIF-1 enhance TfR1 transcription by binding to the previously characterized HRE (28–30), as indicated by our electrophoretic mobility shift assay and reporter gene experiments using wild-type and mutated TfR1 promoter regions. The role of HIF-1 in TfR1 up-regulation was further demonstrated by experiments in which the amount or activity of HIF-1 was diminished using three different approaches, including HIF-1 silencing (see Figs. 2 and 3). Despite the strong evidence for a major role of HIF-1 in TfR1 induction, we cannot presently rule out a possible minor involvement of other transcription factors, which is suggested by the incomplete inhibition observed in transfection experiments with the mutated HRE and by the slight increase in CREB-1 binding (see constitutive complex in Fig. 2A).
It has been reported that HIF-1-dependent NF-κB activity is important for cell survival in hypoxic neutrophils (64), that NF-κB activity is induced as a result of the accumulation of HIF-1 expression in renal carcinoma cells (65), and that a signaling pathway from HIF-1 to NF-κB is involved in epithelial inflammation (66), but NF-κB seems to be upstream of HIF-1 in macrophages challenged with LPS-IFNγ. Evidence that these transcription factors are activated in the order described in Fig. 7 is provided by (a) the specific lack of TfR1 induction in cells unable to activate a HIF-1-dependent response (see Fig. 3), (b) the reduced or absent increase in both HIF-1 and TfR1 when NF-κB activity is prevented by a chemical inhibitor or the expression of a dominant negative (see Figs. 6, C and D), and (c) the unaffected induction of NF-κB in the presence of HIF-1 inactivation (see Figs. 6, E and F). Furthermore, a bioinformatic search for transcription factor binding sites revealed the absence of NF-κB consensus elements in the TfR1 promoter region used in our study (data not shown), thus suggesting that TfR1 is not a direct transcriptional target of NF-κB. In line with our findings, recent studies have shown that reactive oxygen species up-regulate HIF-1 transcription by activating NF-κB (61) and that HIF-1 is induced in LPS-treated macrophages in an NF-κB-dependent manner (47), although NF-κB-independent activation of HIF-1 was recently described in human THP-1 cells exposed to high concentrations of LPS (67).
Our results, therefore, show that NF-κB, the master regulator of inflammatory pathways, not only induces ferritin synthesis (for review, see Ref. 8) but also the expression of TfR1, the other major player of cellular iron homeostasis. These two proteins, which are inversely modulated by iron availability through the IRP regulatory network (24, 25), are therefore unidirectionally activated at transcriptional level by NF-κB-mediated inflammatory signals. In addition to being relevant to the regulation of systemic iron homeostasis in inflammation (see below), this finding offers new insights into the molecular mechanisms controlling intracellular iron metabolism.
Our findings also show that HIF-1 activation and the consequent TfR1 transcriptional up-regulation may be triggered by the activation of adenosine A2A receptor-dependent pathways (see Fig. 4), thus indicating the existence of another independent pathway of TfR1 induction (Fig. 7). Given that TfR1 is an HIF-1 target gene (28–30), this was not unexpected as we have recently demonstrated that A2A receptor-mediated activation leads to HIF-1 activation in normoxic macrophages (32). However, we also found that pretreatment with the same concentration of CGS21680 can block the activation of the NF-κB/HIF-1 axis that leads to increased TfR1 expression (Fig. 7), and therefore, the activity of HIF-1 (and, hence, TfR1 levels) in cultured macrophages exposed to inflammatory and anti-inflammatory stimuli seems to depend on the particular experimental context. HIF-1 induction, which according to previous findings (32) is mediated by the protein kinase C and phosphatidylinositol 3-kinase pathways and occurs in the absence of NF-κB activation (see Fig. 6), was observed when the anti-inflammatory signal of the A2A receptor agonist was present alone (Fig. 4), whereas down-modulation occurred when CGS21680 was administered before the inflammatory stimulus (see Fig. 5) because of its capacity for preventing NF-κB induction (Figs. 6A and Refs. 54 and 55). Moreover, synergistic up-regulation due to increased NF-κB-dependent HIF-1-transcription by LPS and the stabilization of HIF-1 mRNA by means of A2A receptor binding has been found in macrophages co-stimulated with adenosine and LPS administered together (47).3
Although the preventive effect of CGS21680 was also evident in primary macrophages (see Fig. 5F), our study was mainly done on macrophage cell lines, and thus, the present results do not allow us to draw any conclusions concerning the possible role of adenosine in HIF-1-mediated TfR1 expression in vivo, but we can envisage a dual role; 1) adenosine receptor activation inhibits NF-κB induction by LPS and the cascade of events described above and, thus, may represent a mechanism to keep the iron sequestration response typical of the inflammatory state under control, in line with the anti-inflammatory role of adenosine (53) and 2) in the absence of LPS, extracellular adenosine, which accumulates in areas with impaired blood supply and low oxygen tension (54, 68), enhances TfR1 expression by activating HIF-1 directly (32). This may occur under the non-infective inflammatory conditions associated with hypoxia, ischemia/reperfusion, trauma, etc. (54), in which HIF-1 is required for induction of protective genes and activation of salvaging pathways (32, 53). In these settings TfR1 up-regulation may represent an “unavoidable” response with presently undefined implications for iron homeostasis. Indeed, whereas the hypoxia-induced increase in TfR1 expression in erythroid cells and in rapidly dividing cells may provide iron for hemoglobin synthesis and for cell multiplication (3, 11), the physiological significance of TfR1 up-regulation in other cell types is unclear (28, 69).
Importantly, our findings also demonstrate that increased TfR1 protein expression is functionally involved in modulating iron accumulation in macrophages under inflammatory conditions because the RAW 264.7 cells showed a greater uptake of transferrin-bound iron when stimulated with LPS-IFNγ (see Fig. 1). As regards the mechanisms underlying iron sequestration within inflammatory phagocytes, hepcidin (which down-regulates iron efflux from macrophages through the internalization and degradation of ferroportin (12) and is responsive to inflammatory signals and cytokines) (2, 3) certainly plays a key role, but the sequestration of iron in the reticuloendothelial system is a dynamic process involving various mechanisms and pathways. Although an increase in urinary hepcidin levels in LPS-treated humans precedes the development of hypoferremia (15) and direct hepcidin administration rapidly lowers serum iron levels (14), this does not preclude the involvement of other mechanisms as well. A number of reports indicating a rapid onset of hypoferremia in animals treated with LPS (16–19) suggest that ferroportin down-regulation in reticuloendothelial cells is not the only factor responsible for initial hypoferremia. Our results support the concept that an increase in the transferrin-mediated uptake of blood iron may represent an early and transient mechanism that cooperates with reduced iron efflux from macrophages and diminished intestinal iron absorption (19) in causing initial hypoferremia. This increase in transferrin-bound iron uptake is relatively short-lived because HIF-1 activation is transient (see Fig. 2), and iron accumulation, which is also secondary to increased DMT1-mediated iron entry (23), may cooperate with high NO production (5, 6, 21, 22) to down-modulate IRP2 (and, hence, TfR1) at later times of exposure to inflammatory stimuli (5, 6, 20–23). However, at these times, hepcidin-dependent (2, 3, 70, 71) and hepcidin-independent (72) ferroportin down-modulation is probably sufficient to maintain iron sequestration in macrophages.
Taken together, our data reveal novel mechanisms underlying the transcriptional control of TfR1 expression that are regulated by an interplay of inflammatory and anti-inflammatory mediators. The consequent changes in TfR1 expression, which are functionally involved in modulating iron retention in macrophages under inflammatory conditions, may be relevant for the ensuing hypoferremia.
This study was supported by grants from Ministero dell'Università e della Ricerca Scientifica MIUR-PRIN (to G. C.), MIUR-FISR (to G. C.), and FIRST (to G. C., L. T., and S. R.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: TfR, transferrin (Tf) receptor; IRP, iron regulatory protein; LPS, lipopolysaccharide; IFN, interferon; HIF, hypoxia-inducible factor; HRE, hypoxia-responsive element; shRNA, short hairpin RNA; DFO, desferrioxamine; RLA, relative luciferase activity.
L. Tacchini, E. Gammella, and G. Cairo, unpublished observations.
References
- 1.Weiss, G. (2005) Best. Pract. Res. Clin. Haematol. 18 183–201 [DOI] [PubMed] [Google Scholar]
- 2.Ganz, T. (2003) Blood 102 783–788 [DOI] [PubMed] [Google Scholar]
- 3.Andrews, N. C., and Schmidt, P. J. (2007) Annu. Rev. Physiol. 69 69–85 [DOI] [PubMed] [Google Scholar]
- 4.Konijn, A. M., Carmel, N., Levy, R., and Hershko, C. (1981) Br. J. Haematol. 49 361–370 [DOI] [PubMed] [Google Scholar]
- 5.Recalcati, S., Taramelli, D., Conte, D., and Cairo, G. (1998) Blood 91 1059–1066 [PubMed] [Google Scholar]
- 6.Kim, S., and Ponka, P. (2000) J. Biol. Chem. 275 6220–6226 [DOI] [PubMed] [Google Scholar]
- 7.Kim, S., and Ponka, P. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 12214–12219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Torti, F. M., and Torti, S. V. (2002) Blood 99 3505–3516 [DOI] [PubMed] [Google Scholar]
- 9.Knutson, M., and Wessling-Resnick, M. (2003) Crit. Rev. Biochem. Mol. Biol. 38 61–88 [DOI] [PubMed] [Google Scholar]
- 10.Cairo, G., Bernuzzi, F., and Recalcati, S. (2006) Genes Nutrition 1 25–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Domenico, I., McVey Ward, D., and Kaplan, J. (2008) Nat. Rev. Mol. Cell Biol. 9 72–81 [DOI] [PubMed] [Google Scholar]
- 12.Nemeth, E., Tuttle, M. S., Powelson, J., Vaughn, M. B., Donovan, A., Ward, D. M., Ganz, T., and Kaplan, J. (2004) Science 306 2090–2093 [DOI] [PubMed] [Google Scholar]
- 13.Delaby, C., Pilard, N., Goncalves, A. S., Beaumont, C., and Canonne-Hergaux, F. (2005) Blood 106 3979–3984 [DOI] [PubMed] [Google Scholar]
- 14.Rivera, S., Nemeth, E., Gabayan, V., Lopez, M. A., Farshidi, D., and Ganz, T. (2005) Blood 106 2196–2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kemna, E., Pickkers, P., Nemeth, E., van der Hoeven, H., and Swinkels, D. (2005) Blood 106 1864–1866 [DOI] [PubMed] [Google Scholar]
- 16.Gutteberg, T. J., Rokke, O., Andersen, O., and Jorgensen, T. (1989) Scand. J. Infect. Dis. 21 709–715 [DOI] [PubMed] [Google Scholar]
- 17.Bertini, R., Bianchi, M., Erroi, A., Villa, P., and Ghezzi, P. (1989) J. Leukocyte Biol. 46 254–262 [DOI] [PubMed] [Google Scholar]
- 18.Yang, F., Liu, X. B., Quinones, M., Melby, P. C., Ghio, A., and Haile, D. J. (2002) J. Biol. Chem. 277 39786–39791 [DOI] [PubMed] [Google Scholar]
- 19.Laftah, A. H., Sharma, N., Brookes, M. J., McKie, A. T., Simpson, R. J., Iqbal, T. H., and Tselepis, C. (2006) Biochem. J. 397 61–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Byrd, T. F., and Horwitz, M. A. (1993) J. Clin. Investig. 91 969–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mulero, V., and Brock, J. H. (1999) Blood 94 2383–2389 [PubMed] [Google Scholar]
- 22.Kim, S., and Ponka, P. (1999) J. Biol. Chem. 274 33035–33042 [DOI] [PubMed] [Google Scholar]
- 23.Ludwiczek, S., Aigner, E., Theurl, I., and Weiss, G. (2003) Blood 101 4148–4154 [DOI] [PubMed] [Google Scholar]
- 24.Hentze, M. W., and Kuhn, L. C. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 8175–8182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cairo, G., and Pietrangelo, A. (2000) Biochem. J. 352 241–250 [PMC free article] [PubMed] [Google Scholar]
- 26.Cairo, G., Recalcati, S., Pietrangelo, A., and Minotti, G. (2002) Free Radic. Biol. Med. 32 1237–1243 [DOI] [PubMed] [Google Scholar]
- 27.Theil, E. C., and Leipuviene, R. (2007) Cell. Mol. Life Sci. 64 2945–2955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tacchini, L., Bianchi, L., Bernelli-Zazzera, A., and Cairo, G. (1999) J. Biol. Chem. 274 24142–24146 [DOI] [PubMed] [Google Scholar]
- 29.Lok, C. N., and Ponka, P. (1999) J. Biol. Chem. 274 24147–24152 [DOI] [PubMed] [Google Scholar]
- 30.Bianchi, L., Tacchini, L., and Cairo, G. (1999) Nucleic Acids Res. 27 4223–4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dery, M. A., Michaud, M. D., and Richard, D. E. (2005) Int. J. Biochem. Cell Biol. 37 535–540 [DOI] [PubMed] [Google Scholar]
- 32.De Ponti, C., Carini, R., Alchera, E., Nitti, M. P., Locati, M., Albano, E., Cairo, G., and Tacchini, L. (2007) J. Leukocyte Biol. 82 392–402 [DOI] [PubMed] [Google Scholar]
- 33.Taramelli, D., Recalcati, S., Basilico, N., Olliaro, P., and Cairo, G. (2000) Lab. Investig. 80 1781–1788 [DOI] [PubMed] [Google Scholar]
- 34.Cairo, G., Tacchini, L., Pogliaghi, G., Anzon, E., Tomasi, A., and Bernelli-Zazzera, A. (1995) J. Biol. Chem. 270 700–703 [DOI] [PubMed] [Google Scholar]
- 35.Tacchini, L., De Ponti, C., Matteucci, E., Follis, R., and Desiderio, M. A. (2004) Carcinogenesis 25 2089–2100 [DOI] [PubMed] [Google Scholar]
- 36.Alberghini, A., Recalcati, S., Tacchini, L., Santambrogio, P., Campanella, A., and Cairo, G. (2005) J. Biol. Chem. 280 30120–30128 [DOI] [PubMed] [Google Scholar]
- 37.Tacchini, L., Matteucci, E., De Ponti, C., and Desiderio, M. A. (2003) Exp. Cell Res. 290 391–401 [DOI] [PubMed] [Google Scholar]
- 38.Perkins, N. D. (2000) Trends Biochem. Sci. 25 434–440 [DOI] [PubMed] [Google Scholar]
- 39.Mullner, E. W., Neupert, B., and Kuhn, L. C. (1989) Cell 58 373–382 [DOI] [PubMed] [Google Scholar]
- 40.Blouin, C. C., Page, E. L., Soucy, G. M., and Richard, D. E. (2004) Blood 103 1124–1130 [DOI] [PubMed] [Google Scholar]
- 41.Frede, S., Stockmann, C., Freitag, P., and Fandrey, J. (2006) Biochem. J. 396 517–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang, G. L., and Semenza, G. L. (1993) Blood 82 3610–3615 [PubMed] [Google Scholar]
- 43.Kvietikova, I., Wenger, R. H., Marti, H. H., and Gassmann, M. (1995) Nucleic Acids Res. 23 4542–4550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Agani, F., and Semenza, G. L. (1998) Mol. Pharmacol. 54 749–754 [DOI] [PubMed] [Google Scholar]
- 45.Nemeth, Z. H., Leibovich, S. J., Deitch, E. A., Sperlagh, B., Virag, L., Vizi, E. S., Szabo, C., and Hasko, G. (2003) Biochem. Biophys. Res. Commun. 312 883–888 [DOI] [PubMed] [Google Scholar]
- 46.Kung, A. L., Zabludoff, S. D., France, D. S., Freedman, S. J., Tanner, E. A., Vieira, A., Cornell-Kennon, S., Lee, J., Wang, B., Wang, J., Memmert, K., Naegeli, H. U., Petersen, F., Eck, M. J., Bair, K. W., Wood, A. W., and Livingston, D. M. (2004) Cancer Cell 6 33–43 [DOI] [PubMed] [Google Scholar]
- 47.Ramanathan, M., Pinhal-Enfield, G., Hao, I., and Leibovich, S. J. (2007) Mol. Biol. Cell 18 14–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nemeth, Z. H., Lutz, C. S., Csoka, B., Deitch, E. A., Leibovich, S. J., Gause, W. C., Tone, M., Pacher, P., Vizi, E. S., and Hasko, G. (2005) J. Immunol. 175 8260–8270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hasko, G., Szabo, C., Nemeth, Z. H., Kvetan, V., Pastores, S. M., and Vizi, E. S. (1996) J. Immunol. 157 4634–4640 [PubMed] [Google Scholar]
- 50.Min, H. W., Moochhala, S., and Eng, K. H. (2000) Life Sci. 66 1781–1793 [DOI] [PubMed] [Google Scholar]
- 51.Martin, L., Pingle, S. C., Hallam, D. M., Rybak, L. P., and Ramkumar, V. (2006) J. Pharmacol. Exp. Ther. 316 71–78 [DOI] [PubMed] [Google Scholar]
- 52.Hasko, G., and Cronstein, B. N. (2004) Trends Immunol. 25 33–39 [DOI] [PubMed] [Google Scholar]
- 53.Sitkovsky, M. V., Lukashev, D., Apasov, S., Kojima, H., Koshiba, M., Caldwell, C., Ohta, A., and Thiel, M. (2004) Annu. Rev. Immunol. 22 657–682 [DOI] [PubMed] [Google Scholar]
- 54.Linden, J. (2005) Mol. Pharmacol. 67 1385–1387 [DOI] [PubMed] [Google Scholar]
- 55.Majumdar, S., and Aggarwal, B. B. (2003) Oncogene 22 1206–1218 [DOI] [PubMed] [Google Scholar]
- 56.Khoury, J., Ibla, J. C., Neish, A. S., and Colgan, S. P. (2007) J. Clin. Investig. 117 703–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moldawer, L. L., Marano, M. A., Wei, H., Fong, Y., Silen, M. L., Kuo, G., Manogue, K. R., Vlassara, H., Cohen, H., Cerami, A., and Lowry, S. F. (1989) FASEB J. 3 1637–1643 [DOI] [PubMed] [Google Scholar]
- 58.Tsuji, Y., Miller, L. L., Miller, S. C., Torti, S. V., and Torti, F. M. (1991) J. Biol. Chem. 266 7257–7261 [PubMed] [Google Scholar]
- 59.O'Neill, L. A., and Bowie, A. G. (2007) Nat. Rev. Immunol. 7 353–364 [DOI] [PubMed] [Google Scholar]
- 60.Xiong, S., She, H., Takeuchi, H., Han, B., Engelhardt, J. F., Barton, C. H., Zandi, E., Giulivi, C., and Tsukamoto, H. (2003) J. Biol. Chem. 278 17646–17654 [DOI] [PubMed] [Google Scholar]
- 61.Bonello, S., Zahringer, C., BelAiba, R. S., Djordjevic, T., Hess, J., Michiels, C., Kietzmann, T., and Gorlach, A. (2007) Arterioscler. Thromb. Vasc. Biol. 27 755–761 [DOI] [PubMed] [Google Scholar]
- 62.Zhou, J., Callapina, M., Goodall, G. J., and Brune, B. (2004) Cancer Res. 64 9041–9048 [DOI] [PubMed] [Google Scholar]
- 63.Mi, Z., Rapisarda, A., Taylor, L., Brooks, A., Creighton-Gutteridge, M., Melillo, G., and Varesio, L. (2008) Cell Cycle 7 232–241 [DOI] [PubMed] [Google Scholar]
- 64.Walmsley, S. R., Print, C., Farahi, N., Peyssonnaux, C., Johnson, R. S., Cramer, T., Sobolewski, A., Condliffe, A. M., Cowburn, A. S., Johnson, N., and Chilvers, E. R. (2005) J. Exp. Med. 201 105–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.An, J., and Rettig, M. B. (2005) Mol. Cell. Biol. 25 7546–7556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Scortegagna, M., Cataisson, C., Martin, R. J., Hicklin, D. J., Schreiber, R. D., Yuspa, S. H., and Arbeit, J. M. (2008) Blood 111 3343–3354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nishi, K., Oda, T., Takabuchi, S., Oda, S., Fukuda, K., Adachi, T., Semenza, G. L., Shingu, K., and Hirota, K. (2008) Antioxid. Redox Signal. 10 983–995 [DOI] [PubMed] [Google Scholar]
- 68.Sitkovsky, M., and Lukashev, D. (2005) Nat. Rev. Immunol. 5 712–721 [DOI] [PubMed] [Google Scholar]
- 69.Tacchini, L., Fusar Poli, D., Bernelli-Zazzera, A., and Cairo, G. (2002) Hepatology 36 103–111 [DOI] [PubMed] [Google Scholar]
- 70.Peyssonnaux, C., Zinkernagel, A. S., Datta, V., Lauth, X., Johnson, R. S., and Nizet, V. (2006) Blood 107 3727–3732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Theurl, I., Theurl, M., Seifert, M., Mair, S., Nairz, M., Rumpold, H., Zoller, H., Bellmann-Weiler, R., Niederegger, H., Talasz, H., and Weiss, G. (2008) Blood 111 2392–2399 [DOI] [PubMed] [Google Scholar]
- 72.Liu, X. B., Nguyen, N. B., Marquess, K. D., Yang, F., and Haile, D. J. (2005) Blood Cells Mol. Dis. 35 47–56 [DOI] [PubMed] [Google Scholar]