Abstract
One of the processes that regulate intracellular levels of polyamines in mammalian cells is polyamine uptake. We have measured polyamine uptake in COS7 cells for putrescine, spermidine, and spermine, obtaining Km values of 4.5, 1.0, and 0.8 μm, respectively. Treatment of nonconfluent cells with cycloheximide stimulated polyamine uptake and prevented the inhibitory effect found in cells preloaded with polyamines, suggesting the existence of a feedback repression mechanism mediated by antizymes. Transient transfected cells with mutated antizyme forms of AZ1, AZ2, and AZ3, which do not require frameshifting, showed a total blockade of polyamine uptake. Transfection of COS7 cells with mouse or human AZIN2, a novel member of the antizyme inhibitor family, recently characterized by our group, markedly stimulated polyamine uptake and counteracted the action of any of the three antizymes in co-transfected cells. The stimulatory effect of AZIN2 on polyamine uptake was abrogated when the putative antizyme binding sequence, formed by residues 117–140 in AZIN2, was deleted. Real time reverse transcription-PCR analysis of antizyme inhibitor transcripts revealed that in brain and testes AZIN2 is more expressed than AZIN1, especially in the testes where the relative expression was about 25-fold higher. Collectively, our results clearly indicate that AZIN2 affects polyamine homeostasis not only by increasing ornithine decarboxylase activity but also by stimulating polyamine uptake, through negating the inhibitory effect of the antizymes. This finding may have physiological relevance, mostly in testes where AZ3 and AZIN2 are mainly expressed.
The polyamines putrescine, spermidine, and spermine are ubiquitous constituents of mammalian cells, which are essential for normal cell physiology and cell growth (1–3). These cationic molecules play multiple functions, including the regulation of nucleic acids and protein synthesis and the modulation of ion channels and receptors (4–6). In normal cells, the intracellular levels of polyamines are tightly controlled by biosynthesis, catabolism, uptake, and excretion (3, 7, 8). In rapidly proliferating cells, increased amounts of polyamines are obtained by activating both polyamine biosynthesis and uptake (9–11). Ornithine decarboxylase (ODC),4 a key enzyme in polyamine biosynthesis, is elevated in many kinds of malignancies, and the forced overexpression of ODC can transform mouse fibroblast cells (12). Growth factors, hormones, and polyamines themselves regulate ODC by mechanisms acting at transcriptional, translational, and post-translational levels (13). On the other hand, tumor cells exhibit enhanced polyamine transport activity in comparison with normal cells, and the pharmacological inhibition of polyamine biosynthesis leads to a compensatory increase in polyamine uptake activity (9, 14). Although polyamine transport systems have been described and characterized at the molecular level in bacteria and yeast (15), no mammalian polyamine carrier has yet been molecularly characterized. Different studies have revealed that mammalian cells operate in polyamine uptake systems that are energy-dependent, saturable, and carrier-mediated (14). Most recently, endocytic pathways have also been implicated in polyamine transport in mammalian cells (16).
A critical regulator of both polyamine biosynthesis and transport is the ODC antizyme (AZ) (7, 17). AZ protein is synthesized, when cellular polyamine levels increase, by stimulation of an unusual translational frameshift of the AZ messenger RNA (18). AZ induction decreases polyamine biosynthesis by inhibiting ODC and promoting the degradation of this enzyme by the 26 S proteasome (19). AZ also negatively regulates polyamine transport into the cells (20, 21), although the mechanism of antizyme inhibition of polyamine uptake is totally unknown. Three different antizymes, named AZ1, AZ2, and AZ3, have been characterized (17). AZ1 and AZ2 are widely expressed, whereas AZ3 expression is restricted to the testes (22, 23). Another factor, named antizyme inhibitor (AZIN), first discovered in the liver (24), participates in the regulation of polyamine metabolism in different mammalian tissues (25). Although the AZIN is highly homologous to ODC, it has no intrinsic ODC activity (26). However, this protein, which has a higher affinity for AZ than ODC, blocks the ability of the antizyme to both inhibit ODC activity and to promote ODC degradation (26), increasing accordingly polyamine biosynthesis. Experiments based on the forced induction of AZIN in cell cultures have also shown that AZIN overexpression resulted in elevation of polyamine uptake (27), demonstrating that this protein is a positive regulator of polyamine metabolism.
We recently showed that a new paralogue gene of ODC, named ODCp or ODC-like, that had been postulated to code for arginine decarboxylase (28) was devoid of ornithine or arginine decarboxylase activity but acted as an antizyme inhibitor; consequently we proposed to name it antizyme inhibitor 2 (AZIN2) (29). These results have later been corroborated for human ODCp by others (30). AZIN2/ODCp is mainly expressed in testes and brain both in mice and human (29, 31), and by both functional and co-immunoprecipitation experiments, we demonstrate that this protein is able to interact with the three antizyme isoforms as is also shown for the former antizyme inhibitor, the AZIN1 (32). As stated above, AZ3 is a testis-specific protein that is believed to participate in the maintenance of appropriate levels of polyamines during spermiogenesis (22). However, although it is known that both AZ1 and AZ2 inhibit ODC and decrease polyamine transport (20, 21, 33), the influence of AZ3 in polyamine uptake is mostly unknown. In this study, we have analyzed first the kinetic parameters of the polyamine uptake in COS7 cells, and second, by using transient transfection assays of COS7 cells with different AZIN2 and AZs constructs, we have studied the influence of AZIN2 and AZ3 on polyamine uptake. Our results indicate that AZ3 inhibits polyamine uptake and that AZIN2 acts as a positive regulator of polyamine transport. This action of AZIN2 is related with the abrogation of the negative effects of the three antizymes, which suggests that this protein may play a relevant role, at least in the regulation of testicular polyamine metabolism.
EXPERIMENTAL PROCEDURES
Materials—Moloney murine leukemia virus reverse transcriptase, GenElute mammalian total RNA Miniprep kit, anti-FLAG M2 monoclonal antibody peroxidase, protease inhibitor mixture (4-(2-aminoethyl) benzenesulfonyl fluoride, EDTA, bestatin, E-64, leupeptin, aprotinin), Igepal CA-360, nonradioactive polyamines and cycloheximide were purchased from Sigma. Pfu DNA polymerase was obtained from Biotools (Madrid, Spain). SYBR Green® PCR Master Mix was from Applied Biosystems (Warrington, UK). Restriction endonucleases EcoRI, XbaI, HindIII, and BamHI were from Fermentas Life Sciences (Vilnius, Lithuania). Lipofectamine 2000 transfection reagent and trypsin-EDTA were purchased from Invitrogen. QuikChange site-directed mutagenesis kit was from Stratagene (La Jolla, CA). [14C]Putrescine (specific activity 107 mCi/mmol), [14C]spermidine (specific activity 112 mCi/mmol), [14C]spermine (specific activity 113 mCi/mmol), ECL+ detection reagent, developing reagents and films were from Amersham Biosciences. Primers were purchased from Sigma Genosys. Scintillation solution Ecoscint-H was obtained from National Diagnostics (Atlanta, GA).
Cloning of AZ1, AZ2, AZ3, AZIN1, and AZIN2—The desired mouse genes were cloned into the expression vector pcDNA3 (Invitrogen) following standard procedures (34) and using the primers described elsewhere (29). Antizyme constructs with an appropriate deletion of one nucleotide in the frameshifting site, for full-length and functional expression, were obtained by the QuickChange site-directed mutagenesis kit. Human AZIN2 clone was obtained from the human expression plasmid library of Open Biosystems (Huntsville, AL). AZIN1 with the FLAG epitope fused to its C terminus was generated by inserting a double-stranded synthetic oligonucleotide encoding the epitope (in boldface) to the pcDNA3 containing the AZN1 sequence. This was designed for annealing with EcoRI and XbaI cohesive ends of the appropriate open vector. The sense oligomer had the sequence 5′-AATTCGACTATAAGGACGATGATGACAAGTGAT-3′ and the antisense oligomer the 5′-CTAGATCACTTGTCATCATCGTCCTTATAGTCG-3′ sequence.
Generation of AZIN2 Mutants—Several constructs of AZIN2 were generated using different strategies. The FLAG epitope was introduced to the N terminus of AZIN2 as described previously (29). AZIN2 with a streptavidin epitope (STrEP Tag) in its C terminus was obtained by subcloning the ORF of AZIN2 into the pEXPR-IBA 103 vector (Novagen) using the following primers: STag, forward, 5′-CTGTCTAGA (XbaI)ATGGCTGG CTATCTGAGTG-3′, and reverse, 5′-GCCGGATCC(BamHI)-CATGATGCTTGCTGGGGTG-3′). The green fluorescent protein sequence (GFP) was also added to the C terminus of AZIN2, in this case by subcloning in the vector pEGFP-N2 (Clontech), between the restriction sites EcoRI and BamHI, using as forward primer the original AZIN2 cloning primer and as reverse primer 5′-GGCGGATCC(BamHI)ACATGATGCTTGCTGGGGTG-3′. The following constructs were generated from the FLAG-tagged AZIN2: N-terminal deletions (Δ39N; Δ113N) and C-terminal deletions (Δ45C; Δ162C) were obtained by PCR of the appropriate fragment and subcloning into the pcDNA3 vector. For deletion of antizyme-binding site of AZIN2 (Δ117–140), two HindIII restriction sites flanking that region were introduced by PCR amplification of two fragments; the N-terminal fragment was digested with the restriction enzymes EcoRI and HindIII and the C-terminal fragment with HindIII and XbaI, and then a triple ligation was performed resulting in the deletion of the region between the introduced HindIII restriction sites. The primers used were as follows: Δ39N, 5′-AAGGAATTC(EcoRI)GCCTTCTTCGTGGCCGACCTG-3′; Δ113N, 5′-TATGAATTC(EcoRI)CCCTGTAAGCAAGTTGCACAG-3′; Δ45C, 5′-TCGTCTAGA(XbaI)TTACCAGGCTAGCCGGGACATG-3′; Δ162C, 5′-GGCTCTAGA(XbaI)TTAAACCTCCCTCTTGGCGACGATG-3′; Δ117, 5′-CTGAAGCTT(HindIII)TTGCTTACAGGGGTTGGCAC-3′; and Δ140, 5′-GAGAAGCTT(HindIII)AAGGTGGTCAAGAGCCACC-3′. In each case, one of the above primers and the forward or reverse cloning primers of the full AZIN2 ORF were used for obtaining the desired amplification fragment. The identity of the constructs was ascertained by DNA sequencing of the cloned inserts by means of an ABI PRISM 310 Genetic Analyzer (Applied Biosystems) at the “Servicio de Apoyo a las Ciencias Experimentales,” University of Murcia.
Cell Culture and Transient Transfection—The monkey kidney fibroblast-like COS7 cell line was obtained from the ATCC. Cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum, 100 units/ml penicillin, and 100 μg/ml streptomycin, at 37 °C under a 5% CO2-humidified atmosphere. Cells were grown to about 80% confluence. Transient transfections were carried out with Lipofectamine 2000 transfection reagent with 0.15 μg of pcDNA3 plasmid and 2 μl of Lipofectamine per well, in 24-well plates. After 6 h of incubation the transfection medium was removed, and fresh complete medium was added, and cells were cultured for 16 h after transfection. The cells were then used for Western blotting or for polyamine uptake assays. In co-transfection experiments, 0.3 μg of DNA per well were used, with the mixtures containing equimolecular amounts of each construct. The plasmid pcDNA3 without gene insertion was used as control. To assess transfection efficiency, a plasmid containing the GFP was transfected using the same conditions as above, and the percentage of cell expressing the GFP was determined by fluorescence microscopy.
Polyamine Uptake Assay—Cells were plated in 24-well plates and grown for 2 days to about 80% confluence. Then they were transfected or directly assessed for polyamine uptake. Cells were washed with DMEM (serum-free) to remove all traces of serum, and then 200 μl of fresh DMEM (serum-free) was added to the cells. The uptake assay was started by the addition of [14C]putrescine, [14C]spermidine, or [14C]spermine at a final concentration of 2 μm. Because the Km values calculated for putrescine was 4.5 μm, in some cases, 10 μm [14C]putrescine was also used as substrate. After incubation at 37 °C for different periods of time, the cells were washed three times with cold phosphate-buffered saline. Washed cell were lysed by incubation with trypsin for 30 min. Finally, 3 ml of the scintillation solution Ecoscint-H was added, and the radioactivity was measured in a Tri-Carb 2900 TR analyzer (PerkinElmer Life Sciences). In some treatments, unlabeled polyamines or cycloheximide was added to cells dissolved in DMEM (serum-free) at 100 μm and incubated for 90 min just before the radioactive uptake assay. The nonspecific accumulation of 14C-polyamines was measured by incubation of the cells at 4 °C, and the specific uptake was calculated by subtracting nonspecific from total accumulation. Each condition was assayed in triplicate, and in every experiment, 2 wells were used to determine the protein content by the method of Bradford (35) using bovine serum albumin as standard.
Western Blotting—Cells were solubilized in 50 mm Tris-HCl (pH 8), 1% Igepal, 1 mm EDTA, and protease inhibitor mixture and centrifuged at 12,000 × g for 20 min. Reducing SDS-PAGE was performed in 10% polyacrylamide gels. Gels were transferred to polyvinylidene difluoride membranes, blocked with 5% nonfat dry milk in phosphate-buffered saline, and incubated overnight at 4 °C with the anti-FLAG antibody peroxidase-labeled (1:5000). Immunoreactive bands were detected by using ECL+ detection reagent and commercial developing reagents and films. For loading controls, Erk2 was determined by means of polyclonal anti-Erk2 antibody (Santa Cruz Biotechnology Inc., Santa Cruz, CA).
RNA Extraction and Real Time RT-PCR—Total RNA was extracted from tissues with GenElute mammalian total RNA Miniprep kit following the manufacturer's instructions. Total RNA was reverse-transcribed using (dT)18 as primer and Moloney murine leukemia virus reverse transcriptase. Real time RT-PCR was carried out by the use of primers that amplified about 100-bp fragments, one of them hybridizing on the boundary between two exons to discard genomic amplifications. The PCR mixture contained 2.25 μl of 100 μm primers, 12.5 μl of SYBR Green® PCR master mix, 1 μl of template cDNA and RNase free water to 25 μl. The reaction was performed in a 7500 real time machine (Applied Biosystems) under the following cycling conditions: 1 cycle at 95 °C for 10 min and 40 cycles at 95 °C for 15 s and 60 °C for 1 min. Fluorescence data were collected from each cycle and were analyzed by means of 7500 SDS software (Applied Biosystems). The expression data of the genes were normalized to β-actin. Different concentrations of plasmids containing the ORF of ODC, AZIN1, and AZIN2 were used as templates to determine the efficiency of the amplification for each pair of primers.
The following primers were used: β-actin (forward, 5′-GATTACTGCTCTGGCTCCTAGCA-3′; reverse, 5′-GCTCAGGAGGAGCAATGATCTT-3′); AZIN1 (forward, 5′-CTTTCCACGAACCATCTGCT-3′; reverse, 5′-TTCCAGCATCTTGCATCTCA-3′); AZIN2 (forward, 5′-GCTTAGAGGGAGCCAAAGTG-3′; reverse, 5′-CTCAGCAAGGATGTCCACAC-3′); ODC (forward, 5′-ATGGGTTCCAGAGGCCAAA-3′; reverse, 5′-CTGCTTCATGAGTTGCCACATT-3′).
RESULTS
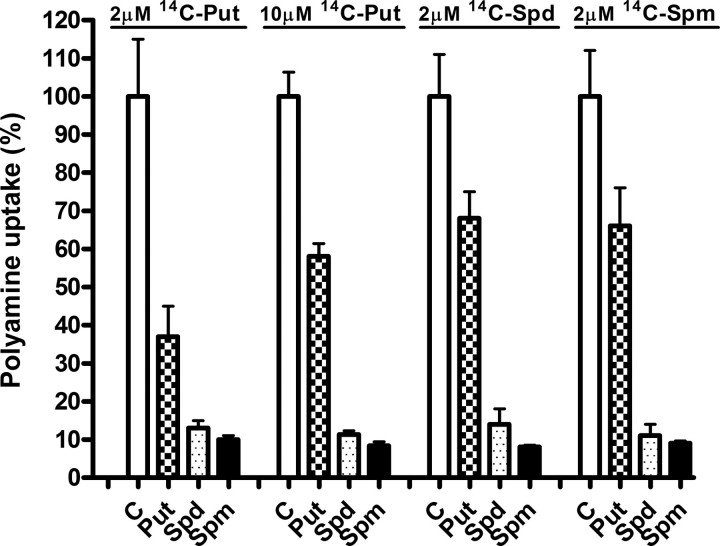
Polyamine Uptake by COS7 Cells—Because transfection assays were carried out in COS7 cells, we first studied some general properties of the polyamine transport system in this cell line. The uptake of 14C-labeled putrescine, -spermidine, and -spermine by COS7 cells at 37 °C and 2 μm concentration of each polyamine was linear over a 120-min course (data not shown). The apparent kinetic parameters Michaelis-Menten constant (Km) and maximal velocity (Vmax) of putrescine, spermidine, and spermine uptake by COS7 cells, determined by measuring the rate of uptake at different polyamine concentrations, are summarized in Table 1. The Km values were 4.5, 1.0, and 0.8 μm for putrescine, spermidine, and spermine, respectively. This indicates that the affinity of putrescine was lower than those of spermidine or spermine. However, Vmax for putrescine was about 2-fold higher than for the other polyamines. Fig. 1 shows that the uptake of each polyamine was markedly inhibited by excess of the other polyamines, which suggests the existence of a common carrier for putrescine, spermidine, and spermine in COS7 cells. In contrast, 20 μm agmatine did not inhibit the rate of polyamine uptake, excluding an unspecific blocking of the carrier by cationic amines (data not shown).
TABLE 1.
Kinetic parameters of polyamine uptake by COS7 cells
Polyamine uptake assays were carried out as described under “Experimental Procedures,” using 14C-labeled putrescine, spermidine, or spermine, in a range of concentrations from 0.1 to 20 μm. The apparent kinetic parameters Km and Vmax were determined by measuring the uptake rate of radioactive polyamines and Lineweaver-Burk analyses. Data are shown as mean ± S.E. of triplicate determinations.
| Polyamine | Km | Vmax |
|---|---|---|
| μm | pmol/h mg protein | |
| Putrescine | 4.50 ± 1.15 | 1248 ± 309 |
| Spermidine | 1.00 ± 0.07 | 733 ± 25 |
| Spermine | 0.81 ± 0.05 | 599 ± 16 |
FIGURE 1.
Influence of unlabeled polyamines in the assay medium on polyamine uptake by COS7 cells. Polyamine uptake assays were carried out as described under “Experimental Procedures,” using 2 μm of 14C-labeled putrescine (Put), spermidine (Spd), or spermine (Spm) in the absence or presence of 20 μm of unlabeled polyamines. Note that in the case of putrescine, 10 μm of 14C-labeled amine was also used. Data are shown as mean ± S.E. of triplicate determinations. C, control.
To examine the role of antizymes on polyamine uptake, protein synthesis in COS7 cells was inhibited by treatment with cycloheximide prior to uptake assays, because early studies had shown that cycloheximide treatment abrogated the feedback repression of polyamine uptake in different mammalian cell lines (36), and this repression was mediated by antizyme (20, 21). Fig. 2 shows that the rate of uptake of the three polyamines was rapidly increased after cycloheximide treatment, reaching a maximum (∼4-fold) around 4 h and decreasing slowly thereafter. This decrease could not be reversed by a second addition of cycloheximide 18 h after the first one. This suggests that the decay in the polyamine uptake observed in the second period of observation could be related with the turnover of the transport system rather than to antizyme recovery.
FIGURE 2.
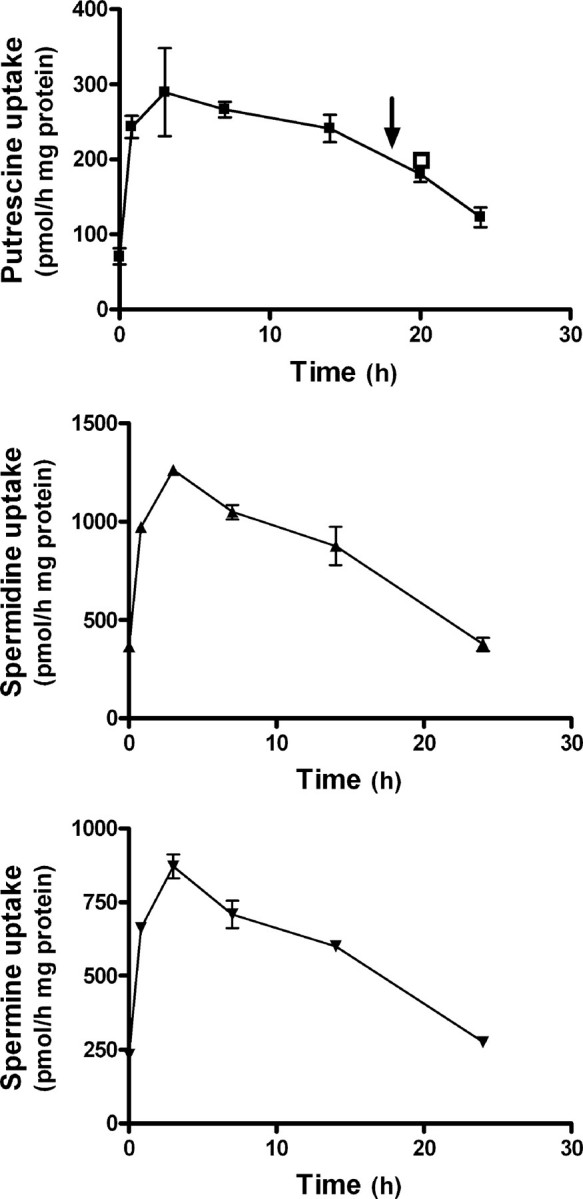
Influence of cycloheximide treatment on the rate of polyamine uptake by COS7 cells. Cells at 80% of confluency were treated with 100 μm cycloheximide (Chx), and the rate of polyamine uptake was measured at different times after cycloheximide addition, using 2 μm of labeled polyamine for a 30-min assay. In same cases, fresh cycloheximide was added 18 h after the first addition (see arrow), and polyamine uptake was measured 2 h later (open square). Data are shown as mean ± S.E. of triplicate determinations.
When COS7 cells were uploaded with polyamines by prior incubation with 100 μm of unlabeled putrescine, spermidine, or spermine for 90 min, the uptake of labeled putrescine was markedly reduced (Fig. 3A). This effect could be ascribed either to a direct trans-inhibitory effect on the polyamine transporter produced by the increase in intracellular polyamines or to the induction of antizymes in response to the rise in intracellular levels of polyamines. The fact that the inhibitory effect produced by the preincubation with polyamines was prevented by cycloheximide treatment (Fig. 3B) supports the latter possibility.
FIGURE 3.
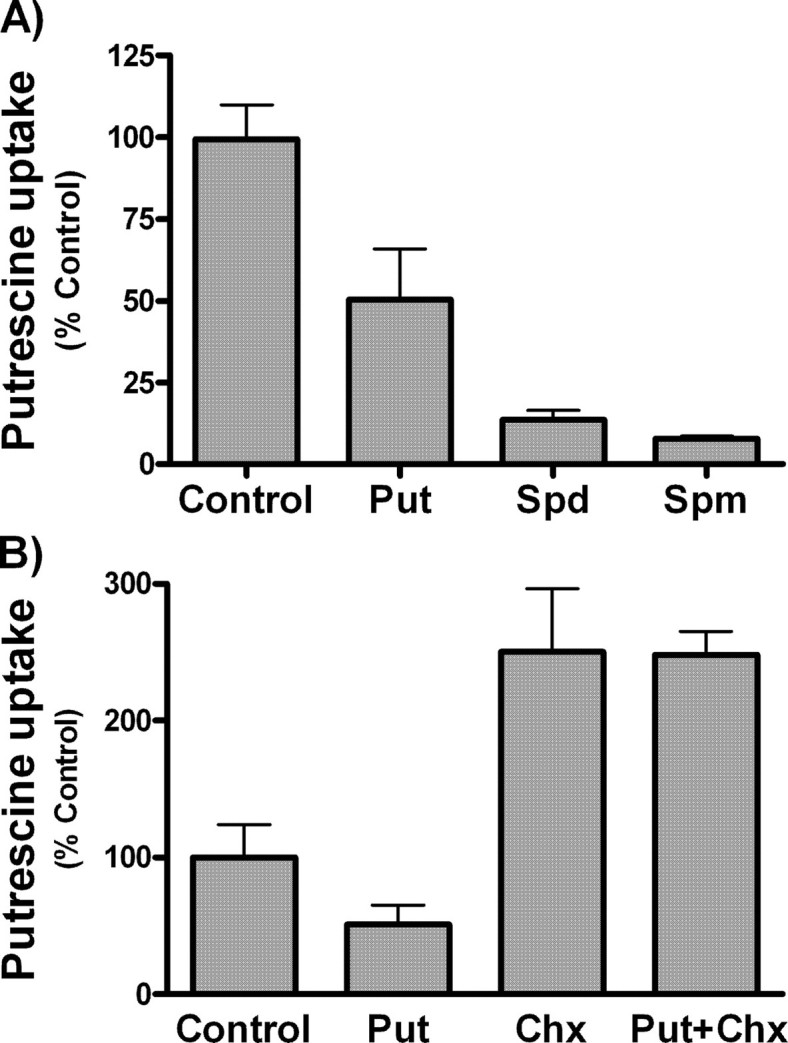
Influence of preincubation of COS7 cells with unlabeled polyamines in polyamine uptake. A, cells at 80% of confluency were incubated for 90 min with 100 μm of putrescine (Put), spermidine (Spd), or spermine (Spm). After washing twice with DMEM, polyamine uptake was carried out using 2 μm 14C-labeled putrescine for 30 min. B, cells were pretreated with 100 μm of unlabeled putrescine plus 100 μm of cycloheximide (Chx), and putrescine uptake was compared with their controls in the absence of cycloheximide. Data are shown as mean ± S.E. of triplicate determinations.
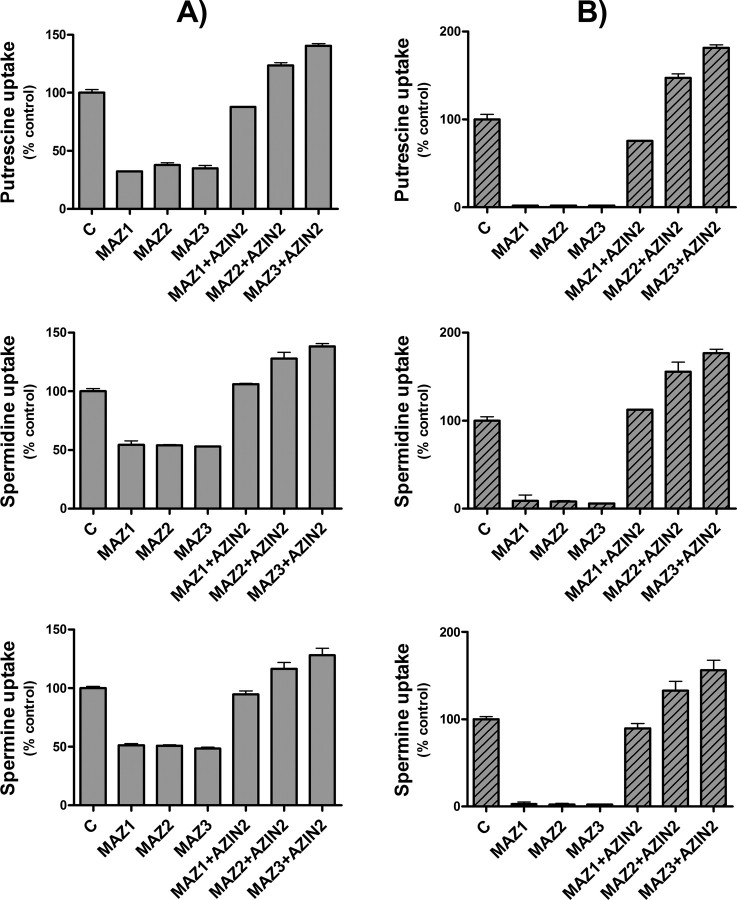
Influence of Single Transfection of COS7 Cells with Antizymes or Antizyme Inhibitors on Polyamine Uptake—To study the influence of the antizymes and antizyme inhibitors on polyamine uptake, COS7 cell were transiently transfected with plasmids containing the cDNA corresponding to mouse AZ1, AZ2, AZ3, AZIN1, and AZIN2, and human AZIN2 under the cytomegalovirus promoter. In the case of the antizymes, mutated versions lacking the stop codon of the ORF1 that synthesize antizymes without frameshift were used. These constructs were named as MAZ1, MAZ2, and MAZ3. Polyamine uptake was measured in the transfected cells using conditions similar to those used with nontransfected cells. In the cells transfected with the empty vector pcDNA3, no significant differences in the uptake of putrescine, spermidine, or spermine were noted, in comparison with nontransfected cells (data not shown). Fig. 4A shows that the uptake of putrescine, spermidine, and spermine was markedly reduced in the cells transfected with any of the three mutated antizymes. On the contrary, in the cells transfected with the murine AZIN2, polyamine uptake was significantly enhanced (∼2–3-fold). A similar effect was observed in transfections with human AZIN2 or mouse AZIN1. If one takes into consideration that the transfection efficiency of COS7 cells was around 50%, as estimated by measuring by laser confocal microscopy, the percentage of positively transfected cells expressing the GFP under the control of the cytomegalovirus promoter, the data of Fig. 4A can be corrected, subtracting the uptake corresponding to the nontransfected cells. The new plot shown in Fig. 4B indicates that the overexpression of any of the three antizymes in COS7 cells almost abolished polyamine uptake. On the other hand, the corrected values in the case of the antizyme inhibitors clearly indicate that these proteins markedly enhanced polyamine uptake, reaching levels similar to those obtained in nontransfected cells treated with cycloheximide. This result suggests that antizyme inhibitor overexpression totally blocked the endogenous levels of antizymes existing in COS7 cells. Similar results to those described above were obtained when other mammalian cell lines such as HEK293T or NIH 3T3 were used (results not shown).
FIGURE 4.
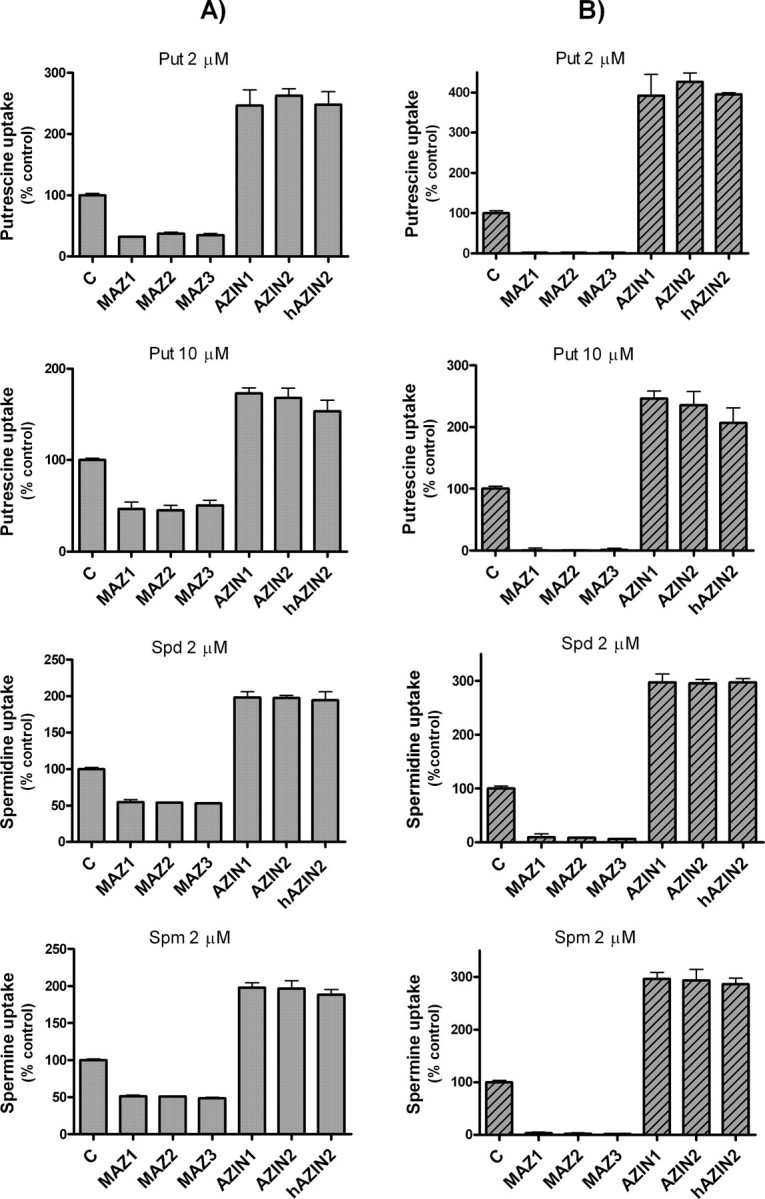
Polyamine uptake by COS7 cells transfected with antizymes or antizyme inhibitors. A, cells were transfected, as described under “Experimental Procedures,” with the expression plasmid pcDNA3 containing the ORF of mutated mouse antizymes without frameshift (MAZ1, MAZ2, and MAZ3) or the ORF of mouse AZIN1 and AZIN2 and human AZIN2 (hAZIN2) in each case. Control cells were transfected with the empty vector. Polyamine uptake assays were carried out using 2 μm 14C-labeled putrescine, spermidine, or spermine. Note that in the case of putrescine, 10 μm of 14C-labeled amine was also used. B, polyamine uptake plots using corrected data calculated by subtracting polyamine uptake corresponding to nontransfected cells, considering the transfection efficiency estimated by fluorescence microscopy of GFP protein overexpressed under the same transfection conditions. Data are shown as mean ± S.E. of triplicate determinations. C, control.
Polyamine Uptake in COS7 Cells Co-transfected with Antizymes and AZIN2—To confirm that AZIN2 is a positive regulator of the uptake of putrescine, spermidine, and spermine by negating the inhibitory effect of antizymes on polyamine transport, COS7 cells were co-transfected with equimolar mixtures of plasmids coding for AZIN2 and each one of the antizymes, and polyamine uptake was compared with cells transfected with an equivalent amount of antizymes. As expected, AZIN2 prevented the negative effect of antizymes on polyamine uptake (Fig. 5). However, in this case, the stimulation of polyamine uptake with respect to control cells was not as high as in the case of the single transfectants of AZIN2. Moreover, the capacity to counteract the action of the three antizymes appeared to be different, with lower effect on AZ1. The apparent lower effect of AZIN2 on polyamine uptake in the double transfectants (Fig. 5), in comparison with the single transfectants (Fig. 4), can be explained, taking into consideration that in the double transfectants the amount of each antizyme should be much higher than in the single transfectants, where there is no forced expression of the antizymes.
FIGURE 5.
Polyamine uptake by COS7 cells co-transfected with antizymes and AZIN2. A, cells were transfected with equimolecular amounts of each of the mutated antizymes (MAZ1, MAZ2 and MAZ3) and of the empty vector or AZIN2. Control cells were transfected with the empty vector. Polyamine uptake assays were carried out using 2 μm 14C-labeled putrescine, spermidine or spermine. B, polyamine uptake plots using corrected data subtracting polyamine uptake corresponding to nontransfected cells considering transfection efficiency, that was estimated by fluorescence microscopy of GFP protein overexpressed under the same transfection conditions. Data are shown as mean ± S.E. of triplicate determinations. C, control.
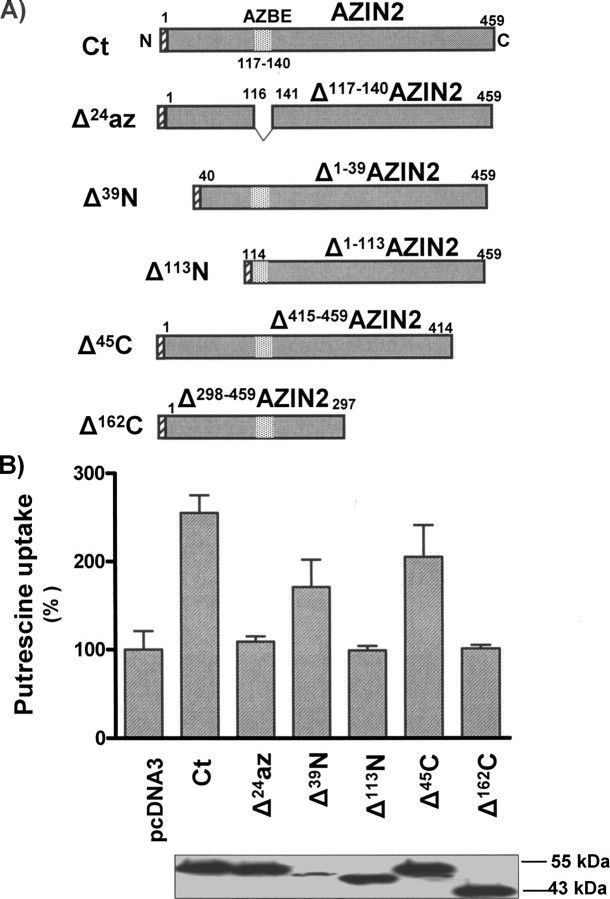
Influence of AZIN2 Modifications on Polyamine Uptake—To assess the possible implication of the different regions of AZIN2 protein on polyamine uptake, several constructs of AZIN2 were generated, including both additions and deletions (Figs. 6 and 7). Fusion proteins that conserved the whole AZIN2 sequence but carried different tags at C or N terminus exerted the same effect on polyamine uptake as AZIN2 (Fig. 6). In fact, the appended FLAG epitope at the N terminus of AZIN2 did not significantly change the antizyme inhibiting function of the protein. Similarly, both of the two chimeric proteins carrying the streptavidin-binding peptide or the GFP tagged at the C-terminal part of AZIN2 conserved the capacity to stimulate polyamine uptake. Besides, when cells were transiently transfected with AZIN1 and AZIN2 tagged with the FLAG epitope, both constructs produced a similar increase in polyamine uptake rate for equivalent levels of protein expression, as estimated by Western blot (see Fig. 6). This indicates that under our experimental conditions, both antizyme inhibitors appear to have a similar capacity for stimulating polyamine uptake. On the other hand, the mutant AZIN2 lacking the sequence corresponding to the putative antizyme binding domain (AZBE) of ODC (37) or AZIN1 (38, 39), lying between residues 117 and 140, was unable to increase polyamine uptake (Fig. 7). Truncated AZIN2 proteins with a lack of 39 or 45 residues in the N or C terminus, respectively, were still able to stimulate putrescine uptake. In the first case, the smaller effect could be the consequence of the lower level of expression. However, the removal of larger sequences in both ends abolished the capacity of AZIN2 to affect antizyme action, although they still contain the AZBE (Fig. 7).
FIGURE 6.
Influence of AZIN2 modifications on polyamine uptake by COS7 cells. A, scheme of the AZIN2 constructions. The FLAG epitope was added to the N terminus of AZIN2, and the streptavidin-binding peptide (SBP) or the green fluorescent protein (GFP) were fused to the C terminus of AZIN2 by the appropriate subcloning of AZIN2 in tagged plasmids (see “Experimental Procedures”). B, cells were transfected with the wild type AZIN2 or the different constructs generated. The comparison of protein levels of AZIN1-FLAG and AZIN2-FLAG, measured by Western blot using an anti-FLAG antibody, is shown above their respective columns. Loading controls were performed using anti Erk2 antibody (data not shown). Control cells (C) were transfected with the empty vector. Polyamine uptake assays were carried out using 2 μm 14C-labeled putrescine for 30 min. Data are shown as mean ± S.E. of triplicate determinations.
FIGURE 7.
Influence of AZIN2 deleted forms on polyamine uptake by COS7 cells. A, scheme of the AZIN2 deletions. N-terminal deletions (Δ39N; Δ113N), C-terminal deletions (Δ45C; Δ162C), and antizyme-binding site deletion of AZIN2 (Δ117–140) were obtained by PCR and subcloning into the pcDNA3 plasmid containing the FLAG epitope (see “Experimental Procedures”). B, cells were transfected with the wild type AZIN2 or the different constructs generated. Control cells were transfected with the empty vector. Polyamine uptake assays were carried out using 2 μm 14C-labeled putrescine for 30 min. Data are shown as mean ± S.E. of triplicate determinations. Protein levels of the different AZIN2 truncated forms were determined by Western blot using anti FLAG antibody. Loading controls were performed using anti Erk2 antibody (data not shown). Ct indicates wild type AZIN2.
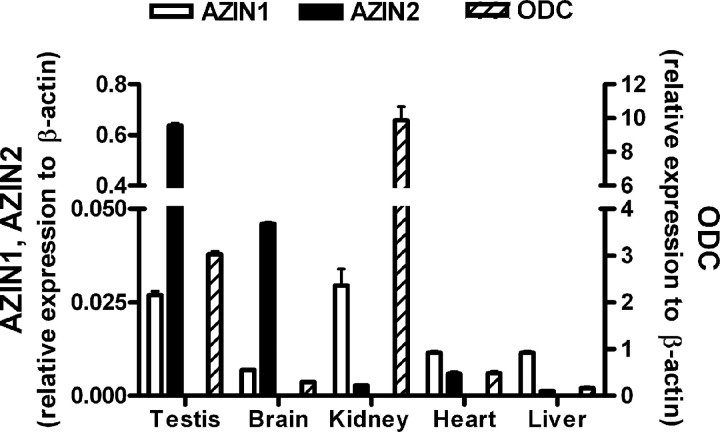
Quantification of the Expression Levels of Antizyme Inhibitors in Mouse Tissues—To compare the expression levels of the two antizyme inhibitors and ODC in different mouse tissues, total RNA was extracted from these tissues, and mRNA levels were quantified by using real time RT-PCR. Fig. 8 shows that in all tissues studied, the levels of ODC transcripts were higher than those of antizyme inhibitors, especially in the kidneys of males. AZIN2 was most expressed in the testes, followed by the brain. In these two tissues the expression of AZIN2 was about 23-fold higher than AZIN1 in the testes and 6-fold in the brain. However, in kidney, heart, and liver, AZIN1 was more expressed than AZIN2.
FIGURE 8.
Comparative gene expression of AZIN1, AZIN2, and ODC in different mouse tissues by real time RT-PCR. RNA from testes, brain, kidney, heart, and liver was isolated and analyzed by real time RT-PCR. Expression values of the genes were normalized with respect to β-actin and corrected using the efficiency of amplification of each gene calculated using known standards. Data are shown as mean ± S.E. of triplicate determinations.
DISCUSSION
We have previously described that mouse ODCp, a paralogue gene of ODC, is devoid of enzymatic activity but acts as a novel antizyme inhibitor, which we termed AZIN2 (29). The objective of this study was to determine whether AZIN2, by counteracting the action of the three antizymes, plays a role in regulating polyamine uptake in mammalian cells. For that purpose, after some trials, we selected COS7 cells first to characterize the polyamine uptake system and second to study the effect of the transfection of COS7 cells with different constructs of antizymes and antizyme inhibitors on the uptake of putrescine, spermidine, and spermine. Our data suggest that in COS7 cells there is a common transport system for the three polyamines, and after inhibiting protein synthesis by cycloheximide, the polyamine uptake was markedly stimulated. These results indicate that in nonconfluent growing COS7 cells, a labile protein, presumably antizyme, is negatively affecting polyamine uptake, as described for other mammalian cells (20, 21, 34).
Our results clearly indicate that the expression of AZIN2/ODCp in COS7 cells markedly increases the uptake of the three polyamines putrescine, spermidine, and spermine, counteracting the negative effect of the antizymes on polyamine uptake. These findings corroborate our previous claim that ODCp or ODC-like acts as an antizyme inhibitor, not only because it binds to the three antizymes and stimulates ODC activity (29) but also by negating the repressing effect of any of the three antizymes on polyamine uptake. Moreover, the fact that the deletion in AZIN2 of the sequence equivalent to the antizyme binding domain, existing in mouse ODC (37) and in AZIN1 (38, 39), abolished the stimulatory effect of AZIN2 on polyamine uptake also supports that this domain is critical for the binding of AZIN2 to the antizyme molecule. This result and previous findings on the binding of mouse ODC and AZIN1 to antizyme (7, 17, 19, 26) support the contention that ODC, and their homologous proteins AZIN1 and AZIN2, share the ability to bind to antizymes and that in this function the AZBE is a common and indispensable element. On the other hand, our results also provide evidence that in AZIN2, the sequences containing the ∼40 terminal residues in both N- and C-terminal ends are not essential for binding to antizymes.
After our study that mouse ODCp acted as an antizyme inhibitor devoid of both ornithine decarboxylase and arginine decarboxylase activities (29), it was published that the human orthologue of ODCp also inhibited AZ1-dependent degradation of ODC as the murine protein (30), confirming our previous results. Our present findings have also revealed that the human AZIN2 is able to stimulate polyamine uptake, apparently as efficiently as mouse AZIN2 or AZIN1. This suggests that the sophisticated network regulating polyamine metabolism mediated by antizymes and antizyme-binding proteins is conserved from mice to humans. In fact, ODC and the two antizyme inhibitors can bind to any of the three antizymes characterized so far. Among the antizyme family members, AZ1 and AZ2 are expressed in numerous mammalian tissues (40), whereas AZ3 expression is restricted to the testes (22). Both AZ1 and AZ2 inhibit ODC and polyamine uptake (20, 21, 33). AZ3 also possesses ODC inhibitory activity (22), but no data are available about its possible role as inhibitor of polyamine uptake. Our results on polyamine uptake by COS7 cells transfected with AZ3 clearly indicated that AZ3 may also negatively affect polyamine transport.
Because antizymes are able to decrease polyamine uptake, it was believed that antizyme inhibitor should prevent the negative effect of antizymes on polyamine transport. This prediction was confirmed by showing that the overexpression of AZIN1 in Chinese hamster ovary cells produced a moderate increase in the uptake of spermine and some polyamine analogues (41). Other experiments using NIH 3T3 fibroblasts transfected with AZIN1 have corroborated that overexpression of AZIN1 increases the uptake of spermidine (27). In contrast to these results, no increase in spermine uptake was observed in rat prostate carcinoma cells overexpressing AZIN1 (39). Our results on COS7 cells also support that AZIN1 increases polyamine uptake, at least as efficiently as AZIN2. Recently, Snapir et al. (42) found that AZIN2, stably transfected into NIH 3T3 cells, exerted a modest increase on the spermidine uptake rate (less than 20%), when compared with the effect of AZIN1 (about 100%), concluding that AZIN2 is less potent than AZIN1 in regulating polyamine uptake. The higher increase in polyamine uptake rate produced by both AZINs (about 300%) in our system in comparison with that observed in stably transfected cells could be related with a lower AZs/AZINs ratio. Nevertheless, in COS7 cells transiently co-transfected with AZ1 and AZIN1 or AZIN2, both antizyme inhibitors exerted a similar effect (result not shown). In this regard, both mouse and human AZIN1 and AZIN2 showed similar capacity in counteracting the effects of AZs on ODC activity (29, 30).
The physiological role of antizyme inhibitors is far from being understood. Different experiments have indicated that AZIN1 may be related with the control of cellular growth, not only by acting on antizymes but also through antizyme-independent effects (27). In fact, overexpression of AZIN1 increased the growth rate and induced the transformed phenotype in different types of cells (27, 39), whereas silencing its expression using specific small interfering RNA decreased cell proliferation (27, 39, 43). Moreover, AZIN1 is up-regulated in several human cancers (44) and is rapidly stimulated in cultured fibroblasts by serum and phorbol esters (45). AZIN1 is also up-regulated in capillary endothelial cells by fibroblast growth factor 2, a known angiogenic factor (46). Coincidentally, the human AZIN1 gene is located in a region of chromosome 8 that is amplified in some tumors (47, 48). In normal tissues, AZIN1 transcripts have been found in the liver, lung, kidney, and heart of rats (26), but little is known about its expression in mouse tissues. On the other hand, AZIN2 appears to be expressed exclusively in the brain and testes, both in mouse and humans (29, 31). However, there are no quantitative data comparing the expression of both genes in mammalian tissues. Our results of real time RT-PCR revealed that in the mouse the expression levels of ODC and antizyme inhibitors among the different tissues assayed varied significantly. ODC was the most expressed in all tissues analyzed. Interestingly, AZIN2 was much more expressed than AZIN1 in brain and testes. In kidney and heart the expression level of AZIN2 was lower than AZIN1, although the expression of the latter in these tissues was far from the value of AZIN2 in testes. The fact that AZIN2 mRNA is mostly found in testes, where a specific testicular antizyme (AZ3) is expressed in the haploid germinal cells (22, 23), together with our findings that AZ3 affects polyamine uptake and that AZIN2 may counteract this effect strongly suggests that both proteins may be important to achieve the severe control of intracellular polyamines, which appears to be required at the later stages of spermatogenesis (49).
Collectively, our previous results on the functional activity of AZIN2 regulating ODC activity, and the present results showing that this protein markedly stimulates polyamine uptake and is considerably expressed in testes and brain, suggest that it is quite likely that AZIN2 may be an important and novel player in polyamine metabolism in these tissues. The precise role of this protein in testicular and neuronal functions, as well as its possible implication in malignant growth, is a subject of ongoing investigations in our laboratory.
This work was supported in part by Grants 00466/PI/04 from The Seneca Foundation (Autonomous Community of Murcia) and BFU2005-09378-C02 from the Spanish Ministry of Education. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: ODC, ornithine decarboxylase; AZ, antizyme; RT, reverse transcription; DMEM, Dulbecco's modified Eagle's medium; ORF, open reading frame; GFP, green fluorescent protein; AZBE, antizyme binding element.
References
- 1.Cohen, S. S. (1998) A Guide to the Polyamines, Oxford University Press, New York
- 2.Thomas, T., and Thomas, T. J. (2001) Cell. Mol. Life Sci. 58 244–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wallace, H. M., Fraser, A. V., and Hughes, A. (2003) Biochem. J. 376 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Childs, A. C., Mehta, D. J., and Gerner, E. W. (2003) Cell. Mol. Life Sci. 60 1394–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Igarashi, K., and Kashiwagi, K. (2000) Biochem. Biophys. Res. Commun. 271 559–564 [DOI] [PubMed] [Google Scholar]
- 6.Johnson, T. D. (1996) Trends Pharmacol. Sci. 17 22–27 [DOI] [PubMed] [Google Scholar]
- 7.Coffino, P. (2001) Nat. Rev. Mol. Cell Biol. 2 188–194 [DOI] [PubMed] [Google Scholar]
- 8.Seiler, N. (2004) Amino Acids 26 217–233 [DOI] [PubMed] [Google Scholar]
- 9.Pegg, A. E. (1988) Cancer Res. 48 759–774 [PubMed] [Google Scholar]
- 10.Gerner, E. W., and Meyskens, F. L. (2004) Nat. Rev. Cancer 4 781–792 [DOI] [PubMed] [Google Scholar]
- 11.Casero, R. A., and Marton, L. J. (2007) Nat. Rev. Drug Discov. 6 373–390 [DOI] [PubMed] [Google Scholar]
- 12.Auvinen, M., Paasinen, A., Andersson, L. C., and Holtta, E. (1992) Nature 360 355–358 [DOI] [PubMed] [Google Scholar]
- 13.Pegg, A. E. (2006) J. Biol. Chem. 281 14529–14532 [DOI] [PubMed] [Google Scholar]
- 14.Seiler, N., Delcros, J. G., and Moulinoux, J. P. (1996) Int. J. Biochem. Cell Biol. 28 843–861 [DOI] [PubMed] [Google Scholar]
- 15.Igarashi, K., and Kashiwagi, K. (1999) Biochem. J. 344 633–642 [PMC free article] [PubMed] [Google Scholar]
- 16.Soulet, D., Covassin, L., Kaouass, M., Charest-Gaudreault, R., Audette, M., and Poulin, R. (2002) Biochem. J. 367 347–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mangold, U. (2005) IUBMB Life 57 671–676 [DOI] [PubMed] [Google Scholar]
- 18.Matsufuji, S., Matsufuji, T., Miyazaki, Y., Murakami, Y., Atkins, J. F., Gesteland, R. F., and Hayashi, S. (1995) Cell 80 51–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayashi, S., Murakami, Y., and Matsufuji, S. (1996) Trends Biochem. Sci. 21 27–30 [PubMed] [Google Scholar]
- 20.Mitchell, J. L. A., Judd, G. G., Bareyal-Leyser, A., and Ling, S. Y. (1994) Biochem. J. 299 19–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suzuki, T., He, Y., Kashiwagi, K., Murakami, Y., Hayashi, S., and Igarashi, K. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 8930–8934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ivanov, I. P., Rohrwasser, A., Terreros, D. A., Gesteland, R. F., and Atkins, J. F. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 4808–4813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tosaka, Y., Tanaka, H., Yano, Y., Masai, K., Nozaki, M., Yomogida, K., Otani, S., Nojima, H., and Nishimune, Y. (2000) Genes Cells 5 265–276 [DOI] [PubMed] [Google Scholar]
- 24.Kitani, T., and Fujisawa, H. (1989) Biochim. Biophys. Acta 991 44–49 [DOI] [PubMed] [Google Scholar]
- 25.Mangold, U. (2006) Cell. Mol. Life Sci. 63 2095–2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murakami, Y., Ichiba, T., Matsufuji, S., and Hayashi, S. (1996) J. Biol. Chem. 271 3340–3342 [DOI] [PubMed] [Google Scholar]
- 27.Keren-Paz, A., Bercovich, Z., Porat, Z., Erez, O., Brener, O., and Kahana, C. (2006) Oncogene 25 5163–5172 [DOI] [PubMed] [Google Scholar]
- 28.Zhu, M. Y., Iyo, A., Piletz, J., and Regunathan, S. (2004) Biochim. Biophys. Acta 1670 156–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lopez-Contreras, A. J., Lopez-Garcia, C., Jimenez-Cervantes, C., Cremades, A., and Penafiel, R. (2006) J. Biol. Chem. 281 30896–30906 [DOI] [PubMed] [Google Scholar]
- 30.Kanerva, K., Matikie, L. T., Pelander, A., Heiskala, M., and Andersson, L. C. (2008) Biochem. J. 409 187–192 [DOI] [PubMed] [Google Scholar]
- 31.Pitkanen, L. T., Heiskala, M., and Andersson, L. C. (2001) Biochem. Biophys. Res. Commun. 287 1051–1057 [DOI] [PubMed] [Google Scholar]
- 32.Mangold, U., and Leberer, E. (2005) Biochem. J. 385 21–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu, C., Lang, D. W., and Coffino, P. (1999) J. Biol. Chem. 274 26425–26430 [DOI] [PubMed] [Google Scholar]
- 34.Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 35.Bradford, M. M. (1976) Anal. Biochem. 72 248–254 [DOI] [PubMed] [Google Scholar]
- 36.Mitchell, J. L. A., Diveley, R. R., and Bareyal-Leyser, A. (1992) Biochem. Biophys. Res. Commun. 186 81–88 [DOI] [PubMed] [Google Scholar]
- 37.Li, X. Q., and Coffino, P. (1992) Mol. Cell. Biol. 12 3556–3562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bercovich, Z., and Kahana, C. (2004) J. Biol. Chem. 279 54097–54102 [DOI] [PubMed] [Google Scholar]
- 39.Kim, S. W., Mangold, U., Waghorne, C., Mobascher, A., Shantz, L., Banyard, J., and Zetter, B. R. (2006) J. Cell Sci. 119 2853–2861 [DOI] [PubMed] [Google Scholar]
- 40.Ivanov, I. P., Gesteland, R. F., and Atkins, J. F. (1998) Genomics 52 119–129 [DOI] [PubMed] [Google Scholar]
- 41.Mitchell, J. L. A., Simkus, C. L., Thane, T. K., Tokarz, P., Bonar, M. M., Frydman, B., Valasinas, A. L., Reddy, V. K., and Marton, L. J. (2004) Biochem. J. 384 271–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Snapir, Z., Keren-Paz, A., Bercovich, Z., and Kahana, C. (2008) Biochem. J. 410 613–619 [DOI] [PubMed] [Google Scholar]
- 43.Choi, K. S., Suh, Y. H., Kim, W. H., Lee, T. H., and Jung, M. H. (2005) Biochem. Biophys. Res. Commun. 328 206–212 [DOI] [PubMed] [Google Scholar]
- 44.Jung, M. H., Kim, S. C., Jeon, G. A., Kim, S. H., Kim, Y., Choi, K. S., Park, S. I., Joe, M. K., and Kimm, K. (2000) Genomics 69 281–286 [DOI] [PubMed] [Google Scholar]
- 45.Nilsson, J., Grahn, B., and Heby, O. (2000) Biochem. J. 346 699–704 [PMC free article] [PubMed] [Google Scholar]
- 46.Takahashi, Y., Li, L. L., Kamiryo, M., Asteriou, T., Moustakas, A., Yamashita, H., and Heldin, P. (2005) J. Biol. Chem. 280 24195–24204 [DOI] [PubMed] [Google Scholar]
- 47.Schaner, M. E., Davidson, B., Skrede, M., Reich, R., Florenes, V. A., Risberg, B., Berner, A., Goldberg, I., Givant-Horwitz, V., Trope, C. G., Kristensen, G. B., Nesland, J. M., and Borresen-Dale, A. L. (2005) Mol. Cancer 4 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Duin, M., van Marion, R., Vissers, K., Watson, J. E., van Weerden, W. M., Schroder, F. H., Hop, W. C. J., van der Kwast, T. H., Collins, C., and van Dekken, H. (2005) Genes Chromosomes Cancer 44 438–449 [DOI] [PubMed] [Google Scholar]
- 49.Hakovirta, H., Keiski, A., Toppari, J., Halmekyto, M., Alhonen, L., Janne, J., and Parvinen, M. (1993) Mol. Endocrinol. 7 1430–1436 [DOI] [PubMed] [Google Scholar]