Nuclear organization can be studied using chromosome conformation capture (3C) technology, which is a biochemical method of analyzing contact frequencies between genomic sites in cells. New 3C-based technologies that provide higher-resolution results (such as 4C, 5C, HiC, and ChIA-PET) have been developed in the past 10 years. In this review, de Wit and de Laat discuss these novel technologies and their contribution to our current knowledge of genome structure.
Keywords: chromosome conformation capture, functional genomics, genome structure, nuclear organization
Abstract
Over the past 10 years, the development of chromosome conformation capture (3C) technology and the subsequent genomic variants thereof have enabled the analysis of nuclear organization at an unprecedented resolution and throughput. The technology relies on the original and, in hindsight, remarkably simple idea that digestion and religation of fixed chromatin in cells, followed by the quantification of ligation junctions, allows for the determination of DNA contact frequencies and insight into chromosome topology. Here we evaluate and compare the current 3C-based methods (including 4C [chromosome conformation capture-on-chip], 5C [chromosome conformation capture carbon copy], HiC, and ChIA-PET), summarize their contribution to our current understanding of genome structure, and discuss how shape influences genome function.
For more than a century, researchers have been making inquiries into the organization of the nucleus, the largest and most easily discernable organelle in the eukaryotic cell. Early in the 20th Century, Cajal (1903) identified subnuclear structures that were later named Cajal bodies. Twenty-five years later, Heitz (1928) observed differentially staining chromatin in interphase nuclei of mosses and described it as heterochromatin and euchromatin. Interest in nuclear structure was further fueled by the realization that the nucleus contains genetic material in the form of DNA fibers. In humans, when unwound, the DNA measures ∼2 m, ∼200,000 times the diameter of an average mammalian cell nucleus. Packing DNA inside the nucleus therefore imposes tremendous organizational challenges. While already conceptually interesting, the shape of the genome becomes even more fascinating when one realizes that it also relates to genome functioning. Although we are still far from understanding this exact relationship, breakthrough technologies are now available for the systematic and detailed analysis of nuclear organization.
Traditionally, nuclear organization is studied by microscopy, and thus it is appropriate to start by highlighting some important observations made under the microscope. However, the emphasis of this review is on novel genomics strategies that are based on chromosome conformation capture (3C) technology. Ten years ago, Dekker et al. (2002) developed 3C technology, a biochemical strategy to analyze contact frequencies between selected genomic sites in cell populations. Since then, various 3C-derived genomics methods have been developed. In comparison with microscopy, 3C-based methods enable more systematic DNA topology studies at a higher resolution: These technologies can put observations made on single genes in selected cells in the context of genomic behavior in cell populations. The generated DNA contact maps start teaching us the rules that dictate genome structure and functioning inside the cell. As we explain here, these rules are probabilistic, not deterministic, implying that they cannot predict the shape and functioning of the genome in individual cells. To investigate this, microscopy is indispensable. The combination of 3C-based genomics approaches and microscopy will therefore continue to be necessary to fully uncover the shape of the genome and its impact on the function of the living cell.
Microscopy observations into mammalian genome organization
Chromosomes occupy their own territory in the cell nucleus (Cremer et al. 1982; Bolzer et al. 2005; Cremer and Cremer 2006) and adopt a preferential radial position within the nucleus, with large chromosomes found more often at the nuclear periphery and small chromosomes found more interiorly. The spatial separation of chromosomes in the nucleus is not absolute: Chromosomal intermingling takes place at the periphery of the territories (Branco and Pombo 2006). A further division can be found within the chromosome territory, with gene-poor and gene-rich regions spatially separated (Shopland et al. 2006; Simonis et al. 2006).
The segregation of active and inactive chromatin inside the nucleus raises the possibility that nuclear positioning affects gene activity. This idea is supported by DNA fluorescence in situ hybridization (FISH) observations that certain genes (e.g., HoxB and uPA) loop out of their chromosome territory upon activation (Chambeyron and Bickmore 2004; Ferrai et al. 2010b). This is likely driven by regulatory DNA sequences, such as enhancers and locus control regions (LCRs) (Noordermeer et al. 2008). In addition, a correlation between expression status and positioning relative to the nuclear periphery and pericentromeric heterochromatin has been observed for some genes (most notably the imunnoglobulin and β-globin loci) (Kosak et al. 2002; Ragoczy et al. 2006). In these instances, the silent genes are found to be closer to these nuclear landmarks than the active ones (Meister et al. 2010). Again, evidence exists that this positioning can be controlled by transcription factors binding to regulatory DNA sequences (Lundgren et al. 2000).
The proximity of a gene relative to other genes has also been associated with its regulation. This suggestion came from FISH experiments showing that a number of erythoid-specific genes that are located far apart on a chromosome specifically colocalize with each other when they are actively transcribed (Osborne et al. 2004). While this may suggest that genes have a relatively unconstrained ability to move and search for preferred neighboring genes, live-cell imaging studies suggest otherwise. Tagging loci with arrays of bacterial operator sequences and expressing the cognate DNA-binding protein fused to a fluorescent protein allow for the spatial and temporal tracking of genes. This has revealed that genomic loci generally show constrained motion within a small subvolume of the mammalian cell nucleus (Tsukamoto et al. 2000; Janicki et al. 2004; Strickfaden et al. 2010). Targeting specific factors to these arrays can sometimes—but not always—induce repositioning, as was seen for lamin-associated proteins that can direct loci to the nuclear periphery (Finlan et al. 2008; Kumaran and Spector 2008; Reddy et al. 2008). This, again, sometimes—but not always—leads to reduced transcriptional output of genes surrounding the arrays on the linear chromosome.
The power of FISH and other microscopy methods lies in their ability to do single-cell analyses of gene positioning (for a more complete overview, see Ferrai et al. 2010a; Geyer et al. 2011). However, on a genomic and cell population scale, they are limited in throughput and resolution. It is therefore unclear whether they uncover general principles of nuclear organization or the peculiarities of individual genes.
Genomics methods are now available for investigating nuclear organization. DamID (van Steensel and Henikoff 2000) is one such method that, when directed to proteins of the nuclear lamina, provides maps of chromatin associated with the nuclear periphery (Pickersgill et al. 2006). In mice and in human cell lines, these lamina-associated domains (LADs) are megabase-sized regions from across the genome. They are generally gene-poor, transcriptionally inactive, and late replicating (Guelen et al. 2008; Peric-Hupkes et al. 2010). Interestingly, genes that are activated or poised for transcription can dissociate from the nuclear lamina (Peric-Hupkes et al. 2010).
Besides DamID, a series of other genomics approaches has been developed that measures DNA–DNA contact probabilities. These strategies enable the highly detailed uncovering of chromosome topology.
3C technology: toward three-dimensional (3D) genomics
The seminal study by Dekker et al. (2002) describing the 3C method has sparked the development of a large number of 3C-derived genomics methods. To understand their potential and limitations, we first look at the principles and applications of the underlying 3C technology.
The initial step in 3C and 3C-derived methods is to establish a representation of the 3D organization of the DNA. To this end, the chromatin is fixed using a fixative agent, most often formaldehyde (Dekker et al. 2002). Next, the fixed chromatin is cut with a restriction enzyme recognizing 6 base pairs (bp)—such as HindIII (Dekker et al. 2002), BglII (Tolhuis et al. 2002), SacI, BamHI (Tan-Wong et al. 2008), or EcoRI (Palstra et al. 2003)—or with more frequent cutters, such as AciI (Miele et al. 2009) or DpnII (Tan-Wong et al. 2008; Comet et al. 2011). In the subsequent step, the sticky ends of the cross-linked DNA fragments are religated under diluted conditions to promote intramolecular ligations (i.e., between cross-linked fragments). DNA fragments that are far away on the linear template, but colocalize in space, can, in this way, be ligated to each other. A template is thereby created that is, in effect, a one-dimensional (1D) cast of the 3D nuclear structure.
The way to establish the 3D conformation of a locus or chromosome is to measure the number of ligation events between nonneighboring sites. In 3C, this is done by semiquantitative (Dekker et al. 2002) or quantitative (Splinter et al. 2006; Wurtele and Chartrand 2006) PCR amplification of selected ligation junctions (Fig. 1). For this, primers are designed near and toward the ends of all restriction fragments of interest. By comparing the amplification efficiency of different primer combinations, a matrix of ligation frequencies is established that serves as proxies for pairwise interaction frequencies.
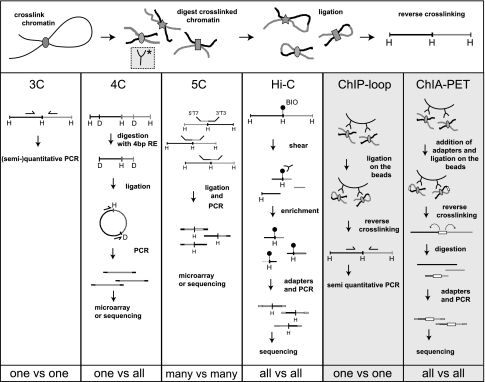
Figure 1.
Overview of 3C-derived methods. An overview of the 3C-derived methods that are discussed is given. The horizontal panel shows the cross-linking, digestion, and ligation steps common to all of the “C” methods. The vertical panels indicate the steps that are specific to separate methods.
In the original study by Dekker et al. (2002), from this matrix, the average 3D conformation of yeast chromosome III was determined, showing that it forms a contorted ring. The method was then adapted for the mammalian system and used to demonstrate that chromatin loops exist in vivo between regulatory DNA elements and their target genes. This was originally done in studies investigating the β-globin locus, where the upstream LCR was shown to physically interact with the active globin genes, thereby looping out the intervening 30–50 kb of chromatin fiber (Fig. 2A; Tolhuis et al. 2002). The term active chromatin hub (ACH) was introduced to describe such spatial clustering of genic sequences with surrounding regulatory sequences (Fig. 2B; Tolhuis et al. 2002). The composition of the β-globin ACH was demonstrated to dynamically follow the transcriptional changes that accompany development and differentiation (Palstra et al. 2003), and transcription factors were found to drive or stabilize its formation (Drissen et al. 2004; Vakoc et al. 2005; Splinter et al. 2006). 3C technology also demonstrated looping between regulatory sequences and genes at other loci, including the H19–Igf2 locus (Murrell et al. 2004), interleukin TH2 (Spilianakis et al. 2005), and the α-globin locus (Vernimmen et al. 2007).
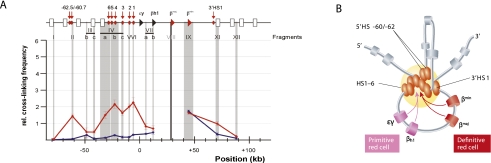
Figure 2.
3C data and the ACH. (A) Example of 3C data showing enhancer looping at the mouse β-globin locus (reproduced from Tolhuis et al. [2002] with permission from Elsevier, © 2002). The relative cross-linking frequency (Y-axis) is plotted for the β-major gene (the anchor point; thick black vertical line) with selected other restriction fragments (gray vertical lines) across the locus. The organization of the locus is shown at the top, with arrows pointing at regulatory sequences, the LCR being comprised of regulatory sequence 1–6, and the β-globin genes shown as triangles. In fetal livers (red line), where β-major is active and under the control of the LCR, loops are found between the gene and the LCR. In fetal brains (blue line), where the gene is silent, no such loops are observed. (B) Based on various 3C experiments in the β-globin locus, the ACH model was proposed. A schematic representation shows the conformation of the locus in its active conformation. Used with permission from Splinter and de Laat (2011).
With 3C, it is also possible to pick up enhancers that were previously unknown to regulate a gene. A survey of the spatial environment of the CFTR gene—which, when mutated, can cause cystic fibrosis—identified a number of cell type-specific interactions. Some of these sequences showed synergistic enhancer activity in a reporter assay, suggesting that they may activate CFTR expression (Gheldof et al. 2010).
Enhancer activity on gene expression can be blocked by insulator sequences (Wallace and Felsenfeld 2007). Insulator sequences are bound by proteins such as CTCF in mammals (Phillips and Corces 2009) or Su(Hw) in flies (Geyer and Corces 1992). 3C technology has been used to demonstrate that the function of certain insulators is dependent on spatial organization. In mammals, CTCF sites form chromatin loops by contacting each other in the β-globin locus (Splinter et al. 2006) and H19/Igf2 locus (Zhao et al. 2006). CTCF recruits additional factors to its binding sites, such as cohesin (Parelho et al. 2008; Wendt et al. 2008) and TAF3 (Wang et al. 2011), which may facilitate DNA loop formation (Hadjur et al. 2009). In Drosophila, a single gypsy insulator element was shown to block the repressive effects of an upstream Polycomb response element (PRE) on a downstream reporter gene. Interestingly, two gypsy insulator elements located between the repressive (PRE) and the transgene rescued the PRE-mediated gene silencing by forming a loop and bringing the PRE spatially closer to the gene (Comet et al. 2011).
In addition to enhancer and insulator loops, a number of recent studies have pointed to the existence of loops between the start and end of a gene. 3C experiments in mouse liver cells showed that ribosomal DNA promoters have an increased propensity to interact with terminator sequences and that these loops are associated with increased rDNA expression (Nemeth et al. 2008). A mechanistic explanation is that gene looping facilitates reloading of RNA polymerase and thereby increases expression throughput, forming a so-called “ribomotor” (Kempers-Veenstra et al. 1986). Also, promoter–terminator looping has been observed on RNA polymerase II transcribed sequences. In yeast, loops form on genes when they are either active or poised, but not when they are repressed (O'Sullivan et al. 2004). In human cells, loop formation between the two long terminal repeats (LTRs) of the HIV provirus has been shown to be dependent on gene expression (Perkins et al. 2008). It has also been proposed that gene loops have a role in transcriptional memory: Gene loops form after an initial round of (slow) gene activation and are essential for subsequent fast reactivation (Laine et al. 2009). Although promoter–terminator looping is generally associated with increased gene activity, for the BRCA1 gene, the looped conformation is actually associated with lower expression compared with the nonlooped conformation (Tan-Wong et al. 2008). In this case, the gene loop is thought to confer repression on the locus.
Various technical issues need to be considered when interpreting 3C data. For example, it is important to realize that any two sequences nearby on the linear chromosome are, by definition, close in space, and therefore sequences over hundreds of kilobases frequently cross-link and ligate to the anchor, independently of the chromatin's 3D conformation. Thus, in order to appreciate loops visualized by 3C-based technologies, one needs to find the anchor interacting with a distant sequence more frequently than with intervening sequences. Therefore, 3C methods intrinsically rely on quantitative rather than qualitative measurements. 3C technology uses PCR for the quantitative detection of a given ligation junction. The importance of this assessment is underscored by the following consideration: At most alleles, cross-linking will result in larger chromatin aggregates with many DNA fragments together (“hairballs”), within which all DNA ends compete with each other for ligation to the anchor fragment. Even a frequent and stable enhancer–promoter interaction will therefore only occasionally result in the corresponding ligation junction. Combined with the fact that every anchor fragment is only present twice in a diploid cell and that a single cell therefore contributes maximally two ligation junctions of interest, this implies that 3C PCR requires faithful and quantitative amplification of very rare ligation junctions from many genome equivalents. For this and other reasons, (semi)quantitative 3C PCR is notoriously difficult and requires strict controls and careful experimental design and data interpretation (Dekker 2006; Hagege et al. 2007; Simonis et al. 2007).
Despite its inherent difficulties, 3C has been instrumental in delineating chromatin loops between sites relatively close on the linear chromosome template. However, for sites separated over distances more than a few hundred kilobases, specific ligation products become too infrequent to be accurately quantified by 3C PCR. The advent of genome-scale methods such as microarrays and high-throughput sequencing has enabled the development of more unbiased methods that offer a solution for assessing the relative abundance of such long-range DNA–DNA contacts.
Chromosome conformation capture-on-chip (4C) technology
4C originally combined 3C technology with microarrays to analyze the contacts of a selected genomic site (or “viewpoint”) with all of the genomic fragments that are represented on the array (Simonis et al. 2006). 4C-seq refers to the same strategy, but uses next-generation sequencing (NGS) instead of microarrays to analyze contacting sequences (Splinter et al. 2011). 4C is also an acronym for circular chromosome conformation capture (Zhao et al. 2006), which uses a slightly different protocol. 4C is known as a “one versus all” strategy because, in it, a single viewpoint is defined, and the genome is screened for sequences that contact this selected site (Fullwood et al. 2009b).
The practical steps involved in 4C technology are outlined in Figure 1 and explained in detail in Simonis et al. (2006, 2007). In brief, in 4C technology, the ligated 3C template is processed with a second round of DNA digestion and ligation to create small DNA circles (some of which contain the 3C ligation junctions). Using viewpoint-specific primers, inverse PCR specifically amplifies all sequences contacting this chromosomal site. They can then be analyzed by microarrays or, nowadays, by NGS methods. The latter is cheaper, provides higher resolution, enables more accurate quantification of DNA interaction frequencies, and has a larger dynamic range (Fig. 3).
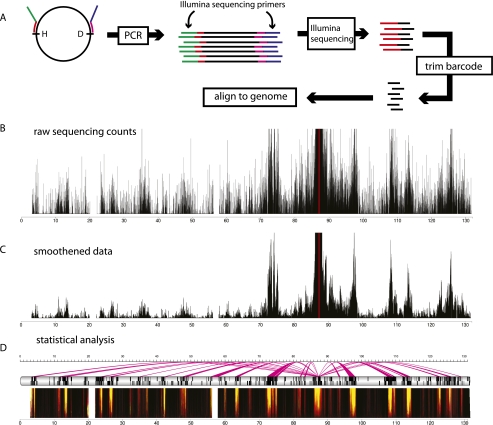
Figure 3.
4C-seq example data and analysis. (A) In 4C-seq, ligation junctions (or “captures”) of a given genomic site (“viewpoint” or “bait”) are PCR-amplified using viewpoint-specific primers. Because the 4C primers also contain the Illumina sequencing primers, the PCR products can be sequenced without further processing. The reads contain the primer and the ligation junction. After trimming the primer sequence, the remainder of the reads are aligned to the genome. (B) A chromosomal map displays the raw 4C-seq counts of the mouse Rad23a locus in neural precursor cells. The 4C profile shows the characteristic peak flanking the viewpoint. (C) With windowed approaches such as running mean or median, a low-pass filter is applied to the data, allowing a view of chromosomal interactions mostly free of noise. (D) Ultimately, high-level analyses, such as domainograms for visualization or false discovery rate (FDR)-based methods for identifying statistically significant interactions, can be employed (Splinter et al. 2011). Domainograms are multiscale representations for the enrichment of 4C captures in a given region. Lightly colored areas show genomic regions that are significantly enriched for 4C captures. An FDR-based method can be employed to identify regions that are significantly enriched. The magenta arcs show the interactions with the viewpoint across the chromosome.
4C technology was first applied to investigate the DNA interaction profiles of a tissue-specific gene (β-globin) embedded in an inactive chromosomal region and a housekeeping gene (Rad23a) present in an active gene-rich region (Simonis et al. 2006). Rad23a made contacts with active regions on its own chromosome and on other chromosomes. The profile of contacts was largely conserved between two tissues. The erythroid-specific β-globin locus, on the other hand, had a profoundly different profile of interactions, depending on its expression status. In erythroid cells, the locus contacts other active regions, whereas in fetal brains, where the locus is inactive, inactive regions are contacted. As such, 4C demonstrated the separation between active and inactive chromatin at a much higher resolution than appreciable by microscopy. It also showed that the activity of a single gene locus can have a marked effect on its steady-state nuclear position.
4C studies have also been used to address whether or not it is the act of transcription itself that dictates the position of a locus. A modified 4C approach to study the DNA interactions of the active β-globin and α-globin loci led to the conclusion that coregulated genes preferentially meet at dedicated transcription sites in the nucleus (Schoenfelder et al. 2010). A possible interpretation of these results is that genes dynamically move to specific nuclear locations for their transcription, rather than the transcription machinery moving to the genes. However, results from other 4C studies are at odds with this interpretation. Blocking elongation of RNA polymerase with α-amanitin or DRB did not seem to affect the contacts of active genes (Palstra et al. 2008), suggesting that chromosome conformation is not dependent on ongoing transcription. The reverse (i.e., activation of a large number of genes) also does not dramatically affect nuclear organization. This was concluded from a series of 4C experiments that addressed how chromosome conformation is affected by a stimulation of the glucocorticoid receptor (GR) (Hakim et al. 2011). The cellular response is very rapid and affects hundreds of genes (John et al. 2009). The effect on nuclear organization, however, was found to be modest and to mostly consist of the expansion of existing interacting regions, rather than a massive reorganization of chromosomes. No large-scale movement of GR-responsive genes was observed, not even toward each other (Hakim et al. 2011).
The relative stability of chromosome conformation in a given cell type was confirmed by a study that assessed the influence of an ectopic enhancer on 3D genome interactions (Noordermeer et al. 2011a). A human β-globin LCR ectopically placed in a cluster of mouse housekeeping genes was originally found by FISH to more often position this genomic region outside its chromosome territory (Noordermeer et al. 2008). 4C technology was then applied to determine where the LCR drags the cluster to. No new contacts were observed; rather, some of the pre-existing interchromosomal contacts were formed more frequently. Interestingly, this happened specifically at genes controlled by GATA-1 and EKLF, two transcription factors that also bind to the LCR (Noordermeer et al. 2011a). Collectively, these data suggest that chromosomal context dictates the nuclear subvolume that can be sampled by a genomic element (gene and enhancer). Within this predetermined space, the element may find preferred interaction partners (such as genes controlled by shared transcription factors).
The link between structure and transcription was further elucidated in a 4C study that focused on dosage compensation of the mammalian X chromosome. Using an allele-specific 4C strategy, it was shown that the inactive and active X chromosomes adopt distinct topologies. The noncoding RNA Xist drives X inactivation, but is not required to maintain gene silencing (Wutz et al. 2002). Intriguingly, upon conditional deletion of Xist, the inactive X chromosome did change its conformation to adopt a structure similar to the active X chromosome, despite the continued suppression of silenced genes on the inactive X chromosome. This indicates that the conformation of chromosomes is not critically dependent on the expression of the genes they are comprised of.
4C studies in Drosophila further uncovered the specification of preferred nuclear environments. Genes repressed by Polycomb group (PcG) proteins were found to preferentially cluster (Bantignies et al. 2011; Tolhuis et al. 2011), and contacts were mostly restricted to the same chromosome arm. When the 4C analysis was repeated on a chromosome arm that harbors an inversion across the centromere, specific contacts were only observed on the newly fused chromosome arm (Tolhuis et al. 2011). This is interesting from an evolutionary standpoint, because the vast majority of synteny breakpoints occur within the same chromosome arm (or Müller element) (Richards et al. 2005; Clark et al. 2007), suggesting that the observed Müller element-constrained chromosome organization is important during evolution. The clustering of PcG-repressed genes may also be of functional importance: Mutations in one target locus were found to (slightly) weaken the repression of an interacting PcG target locus (Bantignies et al. 2011).
4C technology is the preferred strategy to assess the DNA contact profile of individual genomic sites. As may be obvious from the above examples, 4C is currently limited to the description of long-range contacts with larger regions elsewhere on the chromosome (in cis) or on other chromosomes (in trans). Local interactions—for example, between a gene and its enhancer 50 kb away—are not yet readily picked up due to a lack of resolution. Most 4C strategies use restriction enzymes with a 6-nucleotide (nt) recognition sequence that cut, on average, once every few kilobases, creating fragments that are much larger than the average regulatory sequences (which are often not larger than several hundred base pairs). Increased resolution was obtained when a frequent cutter was used that recognizes 4 bp and theoretically cuts every 256 bp. Using this enzyme, a known contact between a regulatory sequence and a gene of the α-globin cluster was picked up (Lower et al. 2009). Whether or not this strategy is robust enough to also identify de novo regulatory interactions remains to be seen. One disadvantage of this published strategy is that no further processing of the very large DNA circles was included, which hampers efficient PCR amplification and may explain the apparent low data complexity. Clearly, though, 4C is the natural scheme to also pick up local interactions between enhancers, promoters, and other regulatory sequences. Further improvements on the technique are expected that will better allow robust screening for these important local interactions.
Chromosome conformation capture carbon copy (5C) technology
5C can be described as a “many versus many” technology. It allows concurrent determination of interactions between multiple sequences (Dostie et al. 2006). In 5C (Fig. 1), the 3C template is hybridized to a mix of oligonucleotides, each of which partially overlaps a different restriction site in the genomic region of interest. Pairs of oligonucleotides that correspond to interacting fragments are juxtaposed on the 3C template and can be ligated together. Since all 5C oligos carry one of two universal sequences at their 5′ ends, all ligation products can subsequently be amplified simultaneously in a multiplex PCR reaction. Readout of these junctions occurs either on a microarray or by high-throughput sequencing.
The resolution of the technique is determined by the spacing between neighboring oligonucleotides on the linear chromosome template. It can never reach the resolution of 4C, HiC, or ChIA-PET (discussed later), as not every unique end of a restriction fragment will allow the design of a 5C oligonucleotide. On the other hand, and different from 4C, 5C provides a matrix of interaction frequencies for many pairs of sites: This puts contacts between given DNA sites in the context of those between other pairs of sites. 5C and HiC are therefore used to reconstruct the (average) 3D conformation of larger genomic regions.
So far, 5C technology has been applied to the human β-globin locus (Dostie et al. 2006) the human α-globin locus (Fig. 4A; Bau et al. 2011), and the human HOXA–D gene clusters (Fraser et al. 2009; Ferraiuolo et al. 2010; Wang et al. 2011). At the globin loci, interactions between regulatory sequences and genes previously identified by 3C technology were readily picked up, showing that this technology is a medium-throughput alternative to 3C for the identification of enhancer–promoter interactions. It is not widely used for this purpose, though, likely because 3C is considered easier in design and data analysis. However, its increase in throughput and reduced bias in PCR amplification efficiency between pairs of sites theoretically makes 5C superior to 3C.
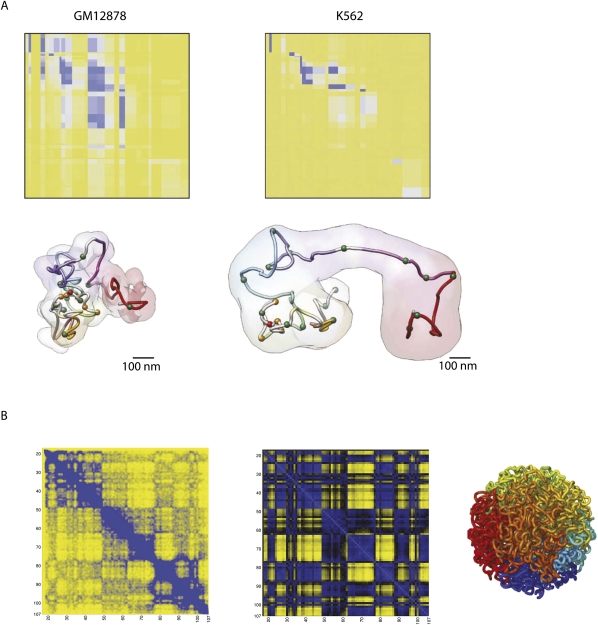
Figure 4.
5C and HiC offer matrices of interaction frequencies. (A) 5C results across 500 kb around the inactive and active α-globin locus (in GM12878 and K562 cells, respectively) (top) are modeled to show that the active locus adopts a more open conformation, whereas in the inactive state, the locus shows a closed conformation (bottom). Used with permission from Macmillan Publishers Ltd. (from Bau et al. 2011) (© 2011). (B) Based on HiC data, chromosome-wide matrices of interaction frequencies can be plotted. A contact map with 1-Mb resolution (with a step size of 100 kb) of human chromosome 14 is shown. The middle panel shows the plaid-like pattern, which is the ultimate result of the HiC analysis method. The right panel shows a fractal globule, a model for human chromosome organization that was postulated based on HiC data (Lieberman-Aiden et al. 2009) and theoretical analysis (Grosberg and et al. 1993). Plots are based on data from Gene Expression Omnibus entry GSE18199 (Lieberman-Aiden et al. 2009).
5C not only identifies interactions between specific pairs of sites, but also builds a matrix of contact frequencies across entire genomic regions and, as such, is the method of choice to start reconstructing their conformations (Fig. 4A). Invariably, the data show that when active, gene clusters adopt a looped but compact topology that facilitates local contacts between genomic elements; e.g., between regulatory sequences and genes. When the chromatin organization of the HOXA locus was assayed in two cell types expressing either the 5′HOXA genes or 3′HOXA gene, preferred long-range interactions were only observed within the active parts of the gene cluster, showing diametrically opposed HOXA interaction patterns between the two cell types (Wang et al. 2011). In vivo, the spatiotemporal expression of Hox genes is collinear with their chromosomal order in the gene clusters. It was recently shown that different contact domains form along the developing body axis, each separating the different genes that are active and inactive at the corresponding position of the anterior–posterior axis (Noordermeer et al. 2011b). In conclusion, 5C technology is the method of choice for understanding DNA contacts between specific sites in the context of other contacts made in a genomic region.
HiC
NGS methods have also led to the development of a number of “all versus all” methods, among which the first was the HiC method (Lieberman-Aiden et al. 2009). In HiC, the procedure for creating a 3C template is slightly adjusted (Fig. 1). Before ligation, the restriction ends are filled in with biotin-labeled nucleotides. Following a blunt end ligation, DNA is purified and sheared, and a biotin pull-down is performed to ensure that only ligation junctions are selected for further analysis. Reads are mapped back to the genome, and when a pair is found on two different restriction fragments, this is scored as an interaction between these two fragments. From this, a matrix of ligation frequencies between all fragments in the genome can be constructed. A variation (Duan et al. 2010) of the HiC method uses the 4C strategy to further digest with a secondary restriction enzyme and ligate to trim the 3C template to small circles, but is followed by a third round of digestion that uses the primary 3C restriction enzyme. The ends are filled in with a biotinylated adapter containing an EcoP15I restriction site, which is cut 25–27 nt away. Biotin pull-down and paired-end sequencing enables the detection of ligation junctions.
The resolution of the published HiC experiments in mammals, based on ∼10 million paired-end reads, was ∼1 Mb (Lieberman-Aiden et al. 2009). Because of the quadratic nature of “all versus all” data, an increase in resolution by 10-fold requires a 100-fold increase in sequence depth. The spatial separation of active and inactive regions, previously observed in 4C experiments (Simonis et al. 2006), was confirmed in a genome-wide manner by the HiC data. Gene-dense, presumably active regions cluster with other gene-dense regions. Conversely gene-poor, inactive regions cluster with other gene-poor regions. Furthermore, the HiC data revealed that nuclear organization was quite constant between two different cell lines (K562 and GM06990), hinting at a core organization present in most cell lines.
The two microenvironments that were picked up by the HiC method were different with respect to a number of features: The open chromatin compartment was enriched for genes, active histone marks, and DNase I hypersensitivity sites. On the other hand, the closed chromatin compartment was enriched for inactive histone marks, such as H3K27me3, and depleted for genes and DNase I hypersensitivity sites. More sequencing depth and perhaps even more sophisticated analysis methods (Yaffe and Tanay 2011) will determine whether there may be a further compartmentalization of these two microenvironments.
In both baker's and fission yeast, “all versus all” genome-wide chromosome conformation experiments were able to reach kilobase resolution owing to their much smaller genome sizes (both ∼12.4 Mb) and increased sequence depth. For Saccharomyces cerevisiae, the previously observed Rabl configuration (Jin et al. 2000; Bystricky et al. 2004; Zimmer and Fabre 2011) was confirmed, with the data showing clustering of the centromeres and clustering of the telomeres (Duan et al. 2010). As can be predicted from this structure, the short arms of chromosomes showed more frequent interactions. This was also observed in high-throughput in vivo imaging experiments that measured the nuclear position of genomic regions relative to nuclear landmarks (Therizols et al. 2010). The HiC data also showed that tRNA genes dispersed throughout the genome come together in two distinct nuclear clusters: one nucleolar cluster (Thompson et al. 2003), and the other cluster interacting with the centromere. In both S. cerevisiae and Schizosaccharomyces pombe, the HiC data indicate that the chromosomes are organized in chromosomal territories. However, different conclusions were drawn with respect to the clustering of functionally related genes. In S. cerevisiae, apart from tRNA genes, no other groups of genes were found significantly clustered (Duan et al. 2010). However, in S. pombe, cell cycle genes and other functionally related genes were claimed to come together (Tanizawa et al. 2010). Whether these are species-specific differences or are caused by different analysis methods requires further study.
ChIA-PET
ChIA-PET combines chromatin immunoprecipitation (ChIP) with 3C-type analysis and offers the possibility to direct analysis of chromatin interactions exclusively to those formed between sites bound by a given DNA- or chromatin-interacting protein (Fullwood et al. 2009a). As such, it is a genome-wide version of the ChIP-loop procedure (Horike et al. 2005). Essentially, the technique analyzes ligation junctions formed between DNA sites that are pulled down with an antibody against a protein of interest (Fig. 1). ChIA-PET has so far been applied to sites bound by the estrogen receptor α (ERα) (Fullwood et al. 2009a) and CTCF (Handoko et al. 2011). The interactome map of the ERα-binding sites consisted of a few thousand intrachromosomal loops, of which the most prominent ones were reproducible between experiments. Invariably, these were loops between highly enriched binding sites that were relatively close on the linear chromosome template (most were within 100 kb of each other) (Fullwood et al. 2009a). ChIA-PET cannot address whether the identified loops depend on the protein of interest: The technique strictly relies on pulling down DNA sites via their associated proteins and is therefore incompatible with protein knockout or knockdown. The technique also has a rather low signal-to-noise ratio; only a small percentage of the paired-end tags reveals a chromatin loop, the unambiguous identification of which, for example, requires that the two paired sequences both localize to a (different) binding site of the protein of interest. Therefore, a current limiting factor of ChIA-PET is that it only identifies contacts between DNA sites bound by the same factor. 4C, 5C, and HiC are not dependent on associated proteins and are not hampered by these limitations. However, these strategies will not discriminate between the bound and unbound versions of a given genomic site and will therefore miss chromatin loops formed in small subpopulations of cells that happen to have the factor of interest bound to these sites. Thus, ChIA-PET has both advantages and disadvantages over other 3C strategies. In the ERα study, 3C was used to test at a few locations the effect of ERα knockdown, where it led to the loss of loop formation.
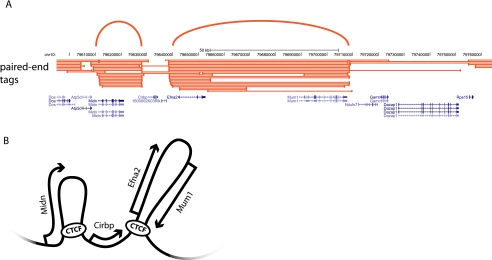
ChIA-PET was recently also applied to uncover the interactome mediated by CTCF in mouse embryonic stem cells (ESCs). Very deep sequencing identified chromatin loops between <10% of the CTCF-binding sites, most (1480) in cis between sites relatively nearby on the chromosome, but also an unexpectedly high number (336) of reproducible interactions in trans. Handoko et al. (2011) speculate that the current numbers are only the tip of the iceberg, as other CTCF sites may well also form chromatin loops that are undetectable with the current sequence depth and strategy. The identified loops formed by CTCF could separate enhancers from promoters and active chromatin from inactive chromatin and could therefore be important for its insulator function or facilitate communication between distal enhancers and genes (Fig. 5). The study highlights the central role of CTCF in the spatial regulation of gene expression (Handoko et al. 2011). It will be particularly interesting to understand the function of the interchromosomal CTCF interactions and find out what determines whether a CTCF site forms a chromatin loop or not.
Figure 5.
ChIA-PET offers insight into the chromosome interactome for the CTCF protein. (A) Results of three ChIA-PET libraries for the insulator protein CTCF. Paired-end tags show possible interactions between two protein-binding sites. Arcs highlight loops formed between genomic sites. (B) Schematic representation of a possible model for the folding of the locus shown in A.
Interpretation of chromosome capture experiments: further considerations
The resolution of all 3C-based methods is limited by the choice of the first restriction enzyme. For a six-cutter like HindIII, there are ∼800,000 HindIII sites in the mouse genome, and the average resolution throughout the genome will be ∼4 kb. However, this is only theoretical, as a number of factors can influence this resolution. Local distribution of restriction sites can vary between different genomic regions, resulting in different resolutions at different genomic locations. An additional factor that influences the results is the presence of repeats in the genome. For 3C, this is a relatively minor problem because one can be quite flexible in the selection of primers for PCR. For sequencing-based methods, this can be more challenging, especially in 4C-seq and 5C, which rely on the sequence directly adjacent to the restriction site. However, this can be partially circumvented by increasing the length of the sequencing reads, which gives higher mapping specificity. Additional factors that may influence HiC results were recently assessed and combined into a probabilistic modeling method that corrects systematic biases (Yaffe and Tanay 2011).
A key characteristic of 3C, 4C, 5C, HiC, and ChIA-PET experiments is the very high capture probability between neighboring fragments, in keeping with their close spatial proximity. Moving further away from a given fragment leads to exponential decrease of the capture probability until it reaches a baseline level (Lieberman-Aiden et al. 2009; Wijchers and de Laat 2011). The rapid decline in contact probability makes it so that specific ligation junctions between two given sites far apart on the chromosome, or on different chromosomes, will be rare. As discussed, this makes 3C (“one versus one”) unsuitable for the analysis of long-range contacts. As for the higher-order genomics versions of 3C, windowed approaches are necessary for the analysis of long-range chromatin contacts. Indeed, for far cis and trans DNA contacts, 4C and HiC data sets are not reproducible at the single-fragment resolution, but are highly reproducible over genomic windows. In such approaches, when a long-range interaction within or between chromosomes is described, this is often a statistical definition, meaning that two (multifragment) regions have a higher probability for making contacts compared with other regions at a similar distance on the same chromosome or elsewhere on other chromosomes. For a more thorough statistical definition of 4C, HiC, or ChIA-PET interactions, we refer to Lieberman-Aiden et al. (2009), Li et al. (2010), Splinter et al. (2011), and Yaffe and Tanay (2011).
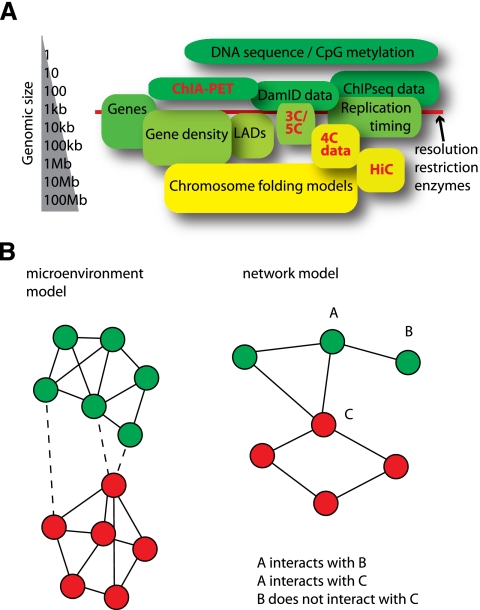
Knowing which regions of the genome are contacted by a gene of interest (4C) or interpreting the overall chromosomal conformation (HiC) is only interesting when some form of integrative genomics analysis is performed. However, because long-range contacts are generally formed between genomic regions measuring in the range of 100 kb to 1 Mb, there is often more than three orders of magnitude difference between the scale of the genomic interaction data and genomic data such as CpG methylation, ChIP, DNase I hypersensitivity, and expression data (Fig. 6A; Marti-Renom and Mirny 2011). This scale difference is further complicated by the fact that across larger chromosomal domains, many genomic variables correlate. For example, C/G nucleotide density correlates with gene density, which in turn correlates with the density of SINE repeats, which shows an inverse correlation with LINE repeat density. Furthermore, regions of high gene density also show a high density of transcription factor-binding sites, DNase I hypersensitivity sites, and certain post-translational histone modifications (Myers et al. 2011). Although these are only correlations, it is important to keep these potential confounding factors in mind when formulating hypotheses with regard to the underlying features of nuclear organization.
Figure 6.
Different levels at which methods that probe the 1D and 3D genome operate. (A) A length scale shows the resolution of the data that are obtained with various genome-wide methods. This is important for comparisons made between various data sets. (B) Graph representation of possible models of genome organization; circles identify loci in the genome, and colors represent different chromatin states. The microenvironment model describes strong separation between genomic regions that have different chromatin states, but assumes few preferences in contacts between genomic regions with similar chromatin. In the network model, on the other hand, such preferences do exist, and interactions between similarly typed chromatin regions can be mutually exclusive.
Proper statistical analysis is especially important when dealing with gene clusters. Since spatial interactions are often formed between larger regions covering multiple genes, an interacting region may spuriously overlap with one or more genes having the same function. Most tools for scoring enrichment of functional annotation, such as Database for Annotation, Visualization, and Integrated Discovery (DAVID) (Dennis et al. 2003) or FatiGO (Al-Shahrour et al. 2004), assume independent sampling, a prerequisite that is clearly not satisfied in studies of long-distance chromatin interactions. This limitation can be overcome by performing nonstandard statistical procedures, such as circular permutation of the gene order along the chromosome (de Wit et al. 2008) or the collapsing of gene clusters to single observations in the statistical analysis.
A further issue to consider is the number of contacts a given gene appears to have. In 4C, a single locus can be engaged in tens or hundreds of contacts (depending on the threshold applied to define contacts). These contacts are collected from many cells and will not all be present in the same cell. Likely, the large number of contacts reflects cell-to-cell differences in genome topology. It was previously argued that upon exit from mitosis, each chromosome probably adopts one of a limited number of energetically favored conformations that will position a given gene next to a few other genes (Simonis et al. 2006).
This is especially important to keep in mind when considering 3D modeling based on 3C data. 3D chromatin models based on 5C and HiC data offer a tantalizing first step toward visualizing abstract interaction frequencies (Lieberman-Aiden et al. 2009; Duan et al. 2010; Tanizawa et al. 2010; Bau et al. 2011), but need to be interpreted with caution (Langowski 2010). First, microscopy has shown that genome topology changes over time and is different between cells. The dynamics of chromatin structure and cell-to-cell variation is not appreciable by 3C-based methods, and it cannot be determined whether two different interactions of A with B and C occur simultaneously or sequentially and/or whether they are mutually exclusive. 3C and derivatives only provide steady-state conformations measured across a population of cells. The resulting average 3D genome models are therefore not likely to be found exactly as such in any given cell at a given time. This is different from protein-folding models, as these molecules generally form stable 3D configurations. Ultimately, DNA contacts need to be studied in single cells on single alleles to understand how the one interaction influences the other.
Insights into nuclear organization
Based on the results discussed above, we can start to theorize about models for nuclear organization and can envision two extremes: the compartmentalization or microenvironment model and the network model (Fig. 6B). In the microenvironment model, the genome is subdivided into two or more types of chromatin that segregate in the nucleus, forming distinct microenvironments that include genomic segments with the same chromatin type. An example of this would be euchromatin versus heterochromatin or peripheral versus interior chromatin. On the other hand, we can imagine a model of the nucleus where mutually exclusive interactions occur. For instance, locus A interacts with loci B and C, but not at the same time, because loci B and C do not interact. This is analogous to a protein network where a single factor may be part of multiple nonoverlapping complexes. Therefore, we call this the network model. Both models have their own implications with regard to the underlying factors that shape the 3D genome; e.g., self-organization versus active formation of contacts.
When we entertain a bird's eye view of the nucleus, the microenvironment model seems to be the most likely model for chromosome organization. Contact profiles of genes that apparently belong to the same chromatin type but are located on different ends of the chromosome are highly similar (Lieberman-Aiden et al. 2009; Splinter et al. 2011). When we zoom in and consider nearby contacts made by individual regulatory sequences, 3C and 5C data have shown that there can be more complex organization than just two different microenvironments (Tolhuis et al. 2002; Dostie et al. 2006; Bau et al. 2011). The most reasonable model for nuclear organization is probably a hybrid of the two models. The microenvironment model underlies the segregation of active and inactive chromatin and acts at the level that positions chromosomal domains relative to each other in cis and in trans. The network model, on the other hand, seems to underlie the shape within chromosomal domains, positioning genes and regulatory DNA elements relative to each other in the nuclear space.
Future directions
With dropping sequencing costs, rising throughput, and more laboratories being acquainted with 3C-based technologies, a flood of genome interaction profiles is expected to be generated in the decade to come. Likely, these data sets will teach us about the stability of genome conformations during development, identify the proteins that dictate folding, and uncover how structure influences the function of the genome and vice versa.
3C-derived methods fail to detect cell-to-cell variation and cannot, by definition, assess the dynamics in the system. Therefore, the ultimate challenge is the development of strategies that analyze structures in single cells. Live-cell imaging methods that can track a single locus must be employed (Robinett et al. 1996; Janicki et al. 2004), possibly in combination with single-molecule detection methods, in order to provide further insight into the dynamics of the nuclear structure (Dange et al. 2011). Integration of 3C/4C/5C/HiC/ChIA-PET data with high-resolution live-cell imaging of tagged loci and single-molecule tracking data will lead to a better understanding of the dynamics underlying chromosomal interaction data and the cell-to-cell variation of these interactions. Ultimately, the goal will be to understand the relationship between the structure and the function of the genome. Current functional genomics studies mainly focus on evolutionary conservation, the mapping of transcription factor-binding sites, and DNA and histone modifications by ChIP-seq, or the mapping of regulatory potential by, for example, DNase I profiling. Invariably, these approaches produce linear maps of genomic information. The systematic mapping of DNA contacts by high-resolution 3C-based genomics methods will identify the 3D contacts these sites are engaged in, which will enable the discrimination of the functional from nonfunctional sites and uncover the wiring of genes with regulatory sequences in the genome (Splinter and de Laat 2011). Thus, to really assign function to the genome, 3C-based technologies are needed to convert our linear information on regulatory potential in the genome into functional 3D regulatory DNA networks.
Acknowledgments
We thank Erik Splinter for preparing Figure 1, and members of the de Laat laboratory for discussion. This work was financially supported by grants from the Dutch Scientific Organization (NWO) to E.dW. (700.10.402, “Veni”) and W.dL. (91204082 and 935170621), InteGeR FP7 Marie Curie ITN (PITN-GA-2007-214902), and a European Research Council Starting Grant (209700, “4C”) to W.dL.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.179804.111.
References
- Al-Shahrour F, Diaz-Uriarte R, Dopazo J 2004. FatiGO: A Web tool for finding significant associations of gene ontology terms with groups of genes. Bioinformatics 20: 578–580 [DOI] [PubMed] [Google Scholar]
- Bantignies F, Roure V, Comet I, Leblanc B, Schuettengruber B, Bonnet J, Tixier V, Mas A, Cavalli G 2011. Polycomb-dependent regulatory contacts between distant Hox loci in Drosophila. Cell 144: 214–226 [DOI] [PubMed] [Google Scholar]
- Bau D, Sanyal A, Lajoie BR, Capriotti E, Byron M, Lawrence JB, Dekker J, Marti-Renom MA 2011. The three-dimensional folding of the α-globin gene domain reveals formation of chromatin globules. Nat Struct Mol Biol 18: 107–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolzer A, Kreth G, Solovei I, Koehler D, Saracoglu K, Fauth C, Muller S, Eils R, Cremer C, Speicher MR, et al. 2005. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol 3: e157 doi: 10.1371/journal.pbio.0030157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco MR, Pombo A 2006. Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS Biol 4: e138 doi: 10.1371/journal.pbio.0040138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bystricky K, Heun P, Gehlen L, Langowski J, Gasser SM 2004. Long-range compaction and flexibility of interphase chromatin in budding yeast analyzed by high-resolution imaging techniques. Proc Natl Acad Sci 101: 16495–16500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajal S 1903. Un sencillo metodo de coloracion selectiva del reticulo protoplasmico y sus efectos en los diversos organos nerviosos de vertebrados e invertebrados. Trab Lab Investig Biol Univ Madr 2: 129–221 [Google Scholar]
- Chambeyron S, Bickmore WA 2004. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev 18: 1119–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, Markow TA, Kaufman TC, Kellis M, Gelbart W, Iyer VN, et al. 2007. Evolution of genes and genomes on the Drosophila phylogeny. Nature 450: 203–218 [DOI] [PubMed] [Google Scholar]
- Comet I, Schuettengruber B, Sexton T, Cavalli G 2011. A chromatin insulator driving three-dimensional Polycomb response element (PRE) contacts and Polycomb association with the chromatin fiber. Proc Natl Acad Sci 108: 2294–2299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T, Cremer C 2006. Rise, fall, and resurrection of chromosome territories: A historical perspective. Part II. Fall and resurrection of chromosome territories during the 1950s to 1980s. Part III. Chromosome territories and the functional nuclear architecture: Experiments and models from the 1990s to the present. Eur J Histochem 50: 223–272 [PubMed] [Google Scholar]
- Cremer C, Cremer T, Gray JW 1982. Induction of chromosome damage by ultraviolet light and caffeine: Correlation of cytogenetic evaluation and flow karyotype. Cytometry 2: 287–290 [DOI] [PubMed] [Google Scholar]
- Dange T, Joseph A, Grunwald D 2011. A perspective of the dynamic structure of the nucleus explored at the single-molecule level. Chromosome Res 19: 117–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E, Braunschweig U, Greil F, Bussemaker HJ, van Steensel B 2008. Global chromatin domain organization of the Drosophila genome. PLoS Genet 4: e1000045 doi: 10.1371/journal.pgen.1000045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J 2006. The three ‘C’ s of chromosome conformation capture: Controls, controls, controls. Nat Methods 3: 17–21 [DOI] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N 2002. Capturing chromosome conformation. Science 295: 1306–1311 [DOI] [PubMed] [Google Scholar]
- Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA 2003. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 4: R60 doi: 10.1186/gb-2003-4-9-r60 [PubMed] [Google Scholar]
- Dostie J, Richmond TA, Arnaout RA, Selzer RR, Lee WL, Honan TA, Rubio ED, Krumm A, Lamb J, Nusbaum C, et al. 2006. Chromosome conformation capture carbon copy (5C): A massively parallel solution for mapping interactions between genomic elements. Genome Res 16: 1299–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drissen R, Palstra RJ, Gillemans N, Splinter E, Grosveld F, Philipsen S, de Laat W 2004. The active spatial organization of the β-globin locus requires the transcription factor EKLF. Genes Dev 18: 2485–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Z, Andronescu M, Schutz K, McIlwain S, Kim YJ, Lee C, Shendure J, Fields S, Blau CA, Noble WS 2010. A three-dimensional model of the yeast genome. Nature 465: 363–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrai C, de Castro IJ, Lavitas L, Chotalia M, Pombo A 2010a. Gene positioning. Cold Spring Harb Perspect Biol 2: a000588 doi: 10.1101/cshperspect.a000588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrai C, Xie SQ, Luraghi P, Munari D, Ramirez F, Branco MR, Pombo A, Crippa MP 2010b. Poised transcription factories prime silent uPA gene prior to activation. PLoS Biol 8: e1000270 doi: 10.1371/journal.pbio.1000270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraiuolo MA, Rousseau M, Miyamoto C, Shenker S, Wang XQ, Nadler M, Blanchette M, Dostie J 2010. The three-dimensional architecture of Hox cluster silencing. Nucleic Acids Res 38: 7472–7484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlan LE, Sproul D, Thomson I, Boyle S, Kerr E, Perry P, Ylstra B, Chubb JR, Bickmore WA 2008. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet 4: e1000039 doi: 10.1371/journal.pgen.1000039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser J, Rousseau M, Shenker S, Ferraiuolo MA, Hayashizaki Y, Blanchette M, Dostie J 2009. Chromatin conformation signatures of cellular differentiation. Genome Biol 10: R37 doi: 10.1186/gb-2009-10-4-r37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH, et al. 2009a. An oestrogen-receptor-α-bound human chromatin interactome. Nature 462: 58–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullwood MJ, Wei CL, Liu ET, Ruan Y 2009b. Next-generation DNA sequencing of paired-end tags (PET) for transcriptome and genome analyses. Genome Res 19: 521–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer PK, Corces VG 1992. DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev 6: 1865–1873 [DOI] [PubMed] [Google Scholar]
- Geyer PK, Vitalini MW, Wallrath LL 2011. Nuclear organization: Taking a position on gene expression. Curr Opin Cell Biol 23: 354–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheldof N, Smith EM, Tabuchi TM, Koch CM, Dunham I, Stamatoyannopoulos JA, Dekker J 2010. Cell-type-specific long-range looping interactions identify distant regulatory elements of the CFTR gene. Nucleic Acids Res 38: 4325–4336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosberg A, Rabin Y, Havlin S, Neer A 1993. Crumpled globule model of the three-dimensional structure of DNA. EPL 23: 373–378 [Google Scholar]
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, et al. 2008. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453: 948–951 [DOI] [PubMed] [Google Scholar]
- Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, Fraser P, Fisher AG, Merkenschlager M 2009. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature 460: 410–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagege H, Klous P, Braem C, Splinter E, Dekker J, Cathala G, de Laat W, Forne T 2007. Quantitative analysis of chromosome conformation capture assays (3C-qPCR). Nat Protoc 2: 1722–1733 [DOI] [PubMed] [Google Scholar]
- Hakim O, Sung MH, Voss TC, Splinter E, John S, Sabo PJ, Thurman RE, Stamatoyannopoulos JA, de Laat W, Hager GL 2011. Diverse gene reprogramming events occur in the same spatial clusters of distal regulatory elements. Genome Res 21: 697–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handoko L, Xu H, Li G, Ngan CY, Chew E, Schnapp M, Lee CW, Ye C, Ping JL, Mulawadi F, et al. 2011. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat Genet 43: 630–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitz E 1928. Das heterochromatin der moose. 1 Jahrb wiss Bot 69: 762–818 [Google Scholar]
- Horike S, Cai S, Miyano M, Cheng JF, Kohwi-Shigematsu T 2005. Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome. Nat Genet 37: 31–40 [DOI] [PubMed] [Google Scholar]
- Janicki SM, Tsukamoto T, Salghetti SE, Tansey WP, Sachidanandam R, Prasanth KV, Ried T, Shav-Tal Y, Bertrand E, Singer RH, et al. 2004. From silencing to gene expression: Real-time analysis in single cells. Cell 116: 683–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin QW, Fuchs J, Loidl J 2000. Centromere clustering is a major determinant of yeast interphase nuclear organization. J Cell Sci 113: 1903–1912 [DOI] [PubMed] [Google Scholar]
- John S, Johnson TA, Sung MH, Biddie SC, Trump S, Koch-Paiz CA, Davis SR, Walker R, Meltzer PS, Hager GL 2009. Kinetic complexity of the global response to glucocorticoid receptor action. Endocrinology 150: 1766–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempers-Veenstra AE, Oliemans J, Offenberg H, Dekker AF, Piper PW, Planta RJ, Klootwijk J 1986. 3′-End formation of transcripts from the yeast rRNA operon. EMBO J 5: 2703–2710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, Singh H 2002. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science 296: 158–162 [DOI] [PubMed] [Google Scholar]
- Kumaran RI, Spector DL 2008. A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. J Cell Biol 180: 51–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laine JP, Singh BN, Krishnamurthy S, Hampsey M 2009. A physiological role for gene loops in yeast. Genes Dev 23: 2604–2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langowski J 2010. Chromosome conformation by crosslinking: Polymer physics matters. Nucleus 1: 37–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Fullwood MJ, Xu H, Mulawadi FH, Velkov S, Vega V, Ariyaratne PN, Mohamed YB, Ooi HS, Tennakoon C, et al. 2010. ChIA-PET tool for comprehensive chromatin interaction analysis with paired-end tag sequencing. Genome Biol 11: R22 doi: 10.1186/gb-2010-11-2-r22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326: 289–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lower KM, Hughes JR, De Gobbi M, Henderson S, Viprakasit V, Fisher C, Goriely A, Ayyub H, Sloane-Stanley J, Vernimmen D, et al. 2009. Adventitious changes in long-range gene expression caused by polymorphic structural variation and promoter competition. Proc Natl Acad Sci 106: 21771–21776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundgren M, Chow CM, Sabbattini P, Georgiou A, Minaee S, Dillon N 2000. Transcription factor dosage affects changes in higher order chromatin structure associated with activation of a heterochromatic gene. Cell 103: 733–743 [DOI] [PubMed] [Google Scholar]
- Marti-Renom MA, Mirny LA 2011. Bridging the resolution gap in structural modeling of 3D genome organization. PLoS Comput Biol 7: e1002125 doi: 10.1371/journal.pcbi.1002125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister P, Towbin BD, Pike BL, Ponti A, Gasser SM 2010. The spatial dynamics of tissue-specific promoters during C. elegans development. Genes Dev 24: 766–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miele A, Bystricky K, Dekker J 2009. Yeast silent mating type loci form heterochromatic clusters through silencer protein-dependent long-range interactions. PLoS Genet 5: e1000478 doi: 10.1371/journal.pgen.1000478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell A, Heeson S, Reik W 2004. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat Genet 36: 889–893 [DOI] [PubMed] [Google Scholar]
- Myers RM, Stamatoyannopoulos J, Snyder M, Dunham I, Hardison RC, Bernstein BE, Gingeras TR, Kent WJ, Birney E, Wold B, et al. 2011. A user's guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol 9: e1001046 doi: 10.1371/journal.pbio.1001046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemeth A, Guibert S, Tiwari VK, Ohlsson R, Langst G 2008. Epigenetic regulation of TTF-I-mediated promoter-terminator interactions of rRNA genes. EMBO J 27: 1255–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noordermeer D, Branco MR, Splinter E, Klous P, van Ijcken W, Swagemakers S, Koutsourakis M, van der Spek P, Pombo A, de Laat W 2008. Transcription and chromatin organization of a housekeeping gene cluster containing an integrated β-globin locus control region. PLoS Genet 4: e1000016 doi: 10.1371/journal.pgen.1000016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noordermeer D, de Wit E, Klous P, van de Werken H, Simonis M, Lopez-Jones M, Eussen B, de Klein A, Singer RH, de Laat W 2011a. Variegated gene expression caused by cell-specific long-range DNA interactions. Nat Cell Biol 13: 944–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noordermeer D, Leleu M, Splinter E, Rougemont J, De Laat W, Duboule D 2011b. The dynamic architecture of Hox gene clusters. Science 334: 222–225 [DOI] [PubMed] [Google Scholar]
- Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W, et al. 2004. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet 36: 1065–1071 [DOI] [PubMed] [Google Scholar]
- O'Sullivan JM, Tan-Wong SM, Morillon A, Lee B, Coles J, Mellor J, Proudfoot NJ 2004. Gene loops juxtapose promoters and terminators in yeast. Nat Genet 36: 1014–1018 [DOI] [PubMed] [Google Scholar]
- Palstra RJ, Tolhuis B, Splinter E, Nijmeijer R, Grosveld F, de Laat W 2003. The β-globin nuclear compartment in development and erythroid differentiation. Nat Genet 35: 190–194 [DOI] [PubMed] [Google Scholar]
- Palstra RJ, Simonis M, Klous P, Brasset E, Eijkelkamp B, de Laat W 2008. Maintenance of long-range DNA interactions after inhibition of ongoing RNA polymerase II transcription. PLoS One 3: e1661 doi: 10.1371/journal.pone.0001661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, et al. 2008. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell 132: 422–433 [DOI] [PubMed] [Google Scholar]
- Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SW, Solovei I, Brugman W, Graf S, Flicek P, Kerkhoven RM, van Lohuizen M, et al. 2010. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell 38: 603–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins KJ, Lusic M, Mitar I, Giacca M, Proudfoot NJ 2008. Transcription-dependent gene looping of the HIV-1 provirus is dictated by recognition of pre-mRNA processing signals. Mol Cell 29: 56–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JE, Corces VG 2009. CTCF: Master weaver of the genome. Cell 137: 1194–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickersgill H, Kalverda B, de Wit E, Talhout W, Fornerod M, van Steensel B 2006. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat Genet 38: 1005–1014 [DOI] [PubMed] [Google Scholar]
- Ragoczy T, Bender MA, Telling A, Byron R, Groudine M 2006. The locus control region is required for association of the murine β-globin locus with engaged transcription factories during erythroid maturation. Genes Dev 20: 1447–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy KL, Zullo JM, Bertolino E, Singh H 2008. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature 452: 243–247 [DOI] [PubMed] [Google Scholar]
- Richards S, Liu Y, Bettencourt BR, Hradecky P, Letovsky S, Nielsen R, Thornton K, Hubisz MJ, Chen R, Meisel RP, et al. 2005. Comparative genome sequencing of Drosophila pseudoobscura: Chromosomal, gene, and cis-element evolution. Genome Res 15: 1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinett CC, Straight A, Li G, Willhelm C, Sudlow G, Murray A, Belmont AS 1996. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J Cell Biol 135: 1685–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, Andrews S, Kurukuti S, Mitchell JA, Umlauf D, Dimitrova DS, et al. 2010. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet 42: 53–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shopland LS, Lynch CR, Peterson KA, Thornton K, Kepper N, Hase J, Stein S, Vincent S, Molloy KR, Kreth G, et al. 2006. Folding and organization of a contiguous chromosome region according to the gene distribution pattern in primary genomic sequence. J Cell Biol 174: 27–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, de Laat W 2006. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat Genet 38: 1348–1354 [DOI] [PubMed] [Google Scholar]
- Simonis M, Kooren J, de Laat W 2007. An evaluation of 3C-based methods to capture DNA interactions. Nat Methods 4: 895–901 [DOI] [PubMed] [Google Scholar]
- Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA 2005. Interchromosomal associations between alternatively expressed loci. Nature 435: 637–645 [DOI] [PubMed] [Google Scholar]
- Splinter E, de Laat W 2011. The complex transcription regulatory landscape of our genome: Control in three dimensions. EMBO J 30: 4345–4355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splinter E, Heath H, Kooren J, Palstra RJ, Klous P, Grosveld F, Galjart N, de Laat W 2006. CTCF mediates long-range chromatin looping and local histone modification in the β-globin locus. Genes Dev 20: 2349–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splinter E, de Wit E, Nora EP, Klous P, van de Werken HJ, Zhu Y, Kaaij LJ, van Ijcken W, Gribnau J, Heard E, et al. 2011. The inactive X chromosome adopts a unique three-dimensional conformation that is dependent on Xist RNA. Genes Dev 25: 1371–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickfaden H, Zunhammer A, van Koningsbruggen S, Kohler D, Cremer T 2010. 4D chromatin dynamics in cycling cells: Theodor Boveri's hypotheses revisited. Nucleus 1: 284–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanizawa H, Iwasaki O, Tanaka A, Capizzi JR, Wickramasinghe P, Lee M, Fu Z, Noma K 2010. Mapping of long-range associations throughout the fission yeast genome reveals global genome organization linked to transcriptional regulation. Nucleic Acids Res 38: 8164–8177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan-Wong SM, French JD, Proudfoot NJ, Brown MA 2008. Dynamic interactions between the promoter and terminator regions of the mammalian BRCA1 gene. Proc Natl Acad Sci 105: 5160–5165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therizols P, Duong T, Dujon B, Zimmer C, Fabre E 2010. Chromosome arm length and nuclear constraints determine the dynamic relationship of yeast subtelomeres. Proc Natl Acad Sci 107: 2025–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson M, Haeusler RA, Good PD, Engelke DR 2003. Nucleolar clustering of dispersed tRNA genes. Science 302: 1399–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W 2002. Looping and interaction between hypersensitive sites in the active β-globin locus. Mol Cell 10: 1453–1465 [DOI] [PubMed] [Google Scholar]
- Tolhuis B, Blom M, Kerkhoven RM, Pagie L, Teunissen H, Nieuwland M, Simonis M, de Laat W, van Lohuizen M, van Steensel B 2011. Interactions among Polycomb domains are guided by chromosome architecture. PLoS Genet 7: e1001343 doi: 10.1371/journal.pgen.1001343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto T, Hashiguchi N, Janicki SM, Tumbar T, Belmont AS, Spector DL 2000. Visualization of gene activity in living cells. Nat Cell Biol 2: 871–878 [DOI] [PubMed] [Google Scholar]
- Vakoc CR, Letting DL, Gheldof N, Sawado T, Bender MA, Groudine M, Weiss MJ, Dekker J, Blobel GA 2005. Proximity among distant regulatory elements at the β-globin locus requires GATA-1 and FOG-1. Mol Cell 17: 453–462 [DOI] [PubMed] [Google Scholar]
- van Steensel B, Henikoff S 2000. Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat Biotechnol 18: 424–428 [DOI] [PubMed] [Google Scholar]
- Vernimmen D, De Gobbi M, Sloane-Stanley JA, Wood WG, Higgs DR 2007. Long-range chromosomal interactions regulate the timing of the transition between poised and active gene expression. EMBO J 26: 2041–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace JA, Felsenfeld G 2007. We gather together: Insulators and genome organization. Curr Opin Genet Dev 17: 400–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, et al. 2011. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472: 120–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, et al. 2008. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 451: 796–801 [DOI] [PubMed] [Google Scholar]
- Wijchers PJ, de Laat W 2011. Genome organization influences partner selection for chromosomal rearrangements. Trends Genet 27: 63–71 [DOI] [PubMed] [Google Scholar]
- Wurtele H, Chartrand P 2006. Genome-wide scanning of HoxB1-associated loci in mouse ES cells using an open-ended chromosome conformation capture methodology. Chromosome Res 14: 477–495 [DOI] [PubMed] [Google Scholar]
- Wutz A, Rasmussen TP, Jaenisch R 2002. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat Genet 30: 167–174 [DOI] [PubMed] [Google Scholar]
- Yaffe E, Tanay A 2011. Probabilistic modeling of Hi-C contact maps eliminates systematic biases to characterize global chromosomal architecture. Nat Genet 43: 1059–1065 [DOI] [PubMed] [Google Scholar]
- Zhao Z, Tavoosidana G, Sjolinder M, Gondor A, Mariano P, Wang S, Kanduri C, Lezcano M, Sandhu KS, Singh U, et al. 2006. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet 38: 1341–1347 [DOI] [PubMed] [Google Scholar]
- Zimmer C, Fabre E 2011. Principles of chromosomal organization: Lessons from yeast. J Cell Biol 192: 723–733 [DOI] [PMC free article] [PubMed] [Google Scholar]