The enteric nervous system (ENS) is crucial for many intestinal functions, and defects in the formation of the ENS cause Hirschsprung disease (HSCR). The molecular mechanisms that control collective cell migration of the ENS cells during embryogenesis are not well known. In this study, Niswander and colleagues find a novel regulator of enteric neural crest cell (ENCC) migration, Phactr4, and demonstrate that Phactr4 links PP1 with integrin signaling and cofilin activity to regulate the cytoskeleton and control directed cell migration. Thus, this study provides insight into a novel function for Phactr4 in controlling ENCC migration.
Keywords: directional cell migration, enteric neural crest cell, Hirschsprung disease, Phactr4, PP1, β1 integrin, cofilin
Abstract
Hirschsprung disease (HSCR) is caused by a reduction of enteric neural crest cells (ENCCs) in the gut and gastrointestinal blockage. Knowledge of the genetics underlying HSCR is incomplete, particularly genes that control cellular behaviors of ENCC migration. Here we report a novel regulator of ENCC migration in mice. Disruption of the Phactr4 gene causes an embryonic gastrointestinal defect due to colon hypoganglionosis, which resembles human HSCR. Time-lapse imaging of ENCCs within the embryonic gut demonstrates a collective cell migration defect. Mutant ENCCs show undirected cellular protrusions and disrupted directional and chain migration. Phactr4 acts cell-autonomously in ENCCs and colocalizes with integrin and cofilin at cell protrusions. Mechanistically, we show that Phactr4 negatively regulates integrin signaling through the RHO/ROCK pathway and coordinates protein phosphatase 1 (PP1) with cofilin activity to regulate cytoskeletal dynamics. Strikingly, lamellipodia formation and in vivo ENCC chain migration defects are rescued by inhibition of ROCK or integrin function. Our results demonstrate a previously unknown pathway in ENCC collective migration in vivo and provide new candidate genes for human genetic studies of HSCR.
The enteric nervous system (ENS) has been called the “second brain,” as it is comprised of 100 million neurons and plays an autonomous role in controlling many intestinal functions, including peristalsis, gastric and pancreatic secretion, and immune response (Heanue and Pachnis 2007). The ENS is composed of neural crest-derived neurons and glia that are organized into ganglia, and these ganglia interconnect to form an enteric plexus. Defects in ENS development in humans cause Hirschsprung disease (HSCR), a common congenital disorder occurring in 1:5000 live births. HSCR is characterized by an absence of enteric neurons in terminal regions of the gut due to an embryonic defect in ENS formation. The receptor tyrosine kinase RET and the G protein-coupled receptor endothelin receptor B (EDNRB) and their respective ligands, GDNF and EDN3, are critical in ENS development (Heanue and Pachnis 2007; Amiel et al. 2008). Mutations in c-Ret and EdnrB are responsible for ∼50% and 5% of HSCR cases, respectively (McCallion et al. 2003). However, the mechanisms responsible for many of the remaining HSCR cases are still unclear. Moreover, the complex inheritance pattern of HSCR indicates that mutations at additional loci contribute to the disease.
Mice provide an excellent animal model to study the genetics and mechanisms of ENS formation. During mouse embryogenesis, ENS progenitors derive from vagal and sacral neural crest cells (NCCs). At embryonic day 9.5 (E9.5), vagal NCCs emigrate from the neural tube and invade the foregut, then migrate along the entire gastrointestinal tract in a rostrocaudal direction (Young et al. 2004). Sacral NCCs make a small contribution of neurons and glial cells by colonizing the hindgut at E15.5 (Druckenbrod and Epstein 2005). Different cellular processes such as neural crest specification, proliferation, differentiation, and migration are important for complete innervation of the gut (Asai et al. 2006; Simpson et al. 2007; Okamura and Saga 2008; Wallace et al. 2009). The study of enteric NCC (ENCC) migration has revealed complex cellular behaviors at the migratory wave front. Close to the wave front, there are a few solitary ENCCs, and these help direct the forward migration of ENCCs that follow as chains of cells, which then spread out to form an interconnected network within the gut (Young et al. 2004; Druckenbrod and Epstein 2005). In terms of ENCC migration, only a few genes are known to be important, and these include L1 cell adhesion molecule, β1-integrin, Ret, and matrix metalloprotease-2 (MMP-2) (Anderson et al. 2006; Asai et al. 2006; Breau et al. 2009; Anderson 2010).
Directed cell migration is crucial for complete colonization of the gut by the ENS. To migrate toward a chemoattractant, a cell must become polarized and form a single dominant lamellipodial protrusion at its leading edge in the direction of migration. Directional migration is a result of spatially restricted stable and persistent lamellipodium. Lamellipodial protrusion requires reorganization of the actin cytoskeleton, and this process is mediated by multiple signals, including external cues, the intracellular polarity machinery, and adhesion receptors. Integrins are cell surface transmembrane receptors clustered in the leading edge and at cell–matrix adhesions, where they connect the extracellular matrix (ECM) to the actin cytoskeleton network. When activated, integrins undergo conformational change to recruit signaling molecules such as focal adhesion kinase (FAK), SRC, and ERK to the cell–matrix adhesions. This serves to control the temporal and spatial activities of downstream small RhoGTPases to direct local cell membrane dynamics (Geiger et al. 2001). The actin-severing protein cofilin is a downstream target of RhoGTPases, and the action of cofilin on actin polymerization helps generate propulsive force and directional protrusions at the leading edge (Bernstein and Bamburg 2010). Small RhoGTPase can activate LIM kinase, which phosphorylates cofilin at Ser3 to inhibit its activity (Arber et al. 1998; Maekawa et al. 1999), whereas general serine/threonine phosphatases such as protein phosphatase 1 (PP1) can activate cofilin by dephosphorylation (Ambach et al. 2000; Larsen et al. 2003; Oleinik et al. 2010). Cofilin does not regulate lamellipodia formation, but is required for directional migration by controlling actin reorganization to generate polarized lamellipodia at the leading edge (Dawe et al. 2003; Ghosh et al. 2004). When cofilin activity is decreased in fibroblasts by β1 integrin-triggered phosphorylation of cofilin on Ser3 through the RhoA–ROCK1 pathway, cells undergo random intrinsic migration (Danen et al. 2005). Thus, lamellipodial dynamics controlled by cofilin and integrin signaling are crucial factors in dictating directional cell migration.
Genetic mutants can provide significant new insights into the molecular mechanisms of cell migration. Starting from a forward genetic screen in mice, we describe here a novel function of the Phactr4 gene in ENCC migration. Phactr4 belongs to a small group of proteins with predicted PP1- and actin-interacting regulatory domains (Allen et al. 2004). Little is known of the in vivo functions of the Phactr family. Our previous studies identified a missense mutation of Phactr4 (called Phactr4humdy) within the PP1-binding domain that disrupts PP1, but not actin, binding and causes a misregulation of PP1 activity. During murine neural tube closure and eye development, Phactr4 through PP1 controls Rb phosphorylation and proliferation (Kim et al. 2007). Here we report a HSCR-like defect in ENS development in Phactr4humdy mutant mouse embryos due to defective collective cell migration of ENCCs. This results in greatly reduced numbers of ENCCs in the caudal gut, with abnormal accumulation of material in the gut. Time-lapse live imaging of ENCC migration from the neural tube and within the gut indicates that both Phactr4 and PP1 are required for directed ENCC migration. Mutant ENCCs display random protrusions and undirected migration, and Phactr4 acts cell-autonomously in the regulation of cytoskeletal dynamics. Phactr4 protein colocalizes with β1 integrin and cofilin at the tips of lamellipodia. Biochemical studies show that Phactr4 is required to negatively regulate integrin signaling, and disrupted integrin signaling through RHO/ROCK leads to misregulation of cofilin phosphorylation. Lamellipodia formation and ENCC chain migration defects can be rescued in vivo by inhibition of integrin signaling or by activation of cofilin. Thus, Phactr4 regulates actin cytoskeleton dynamics through cofilin activity that is controlled by PP1 and integrin signaling during ENCC migration. These data suggest Phactr4 and PP1 be considered as candidate genes in the etiology of human HSCR.
Results
Phactr4humdy embryos exhibit intestinal hypoganglionic phenotype
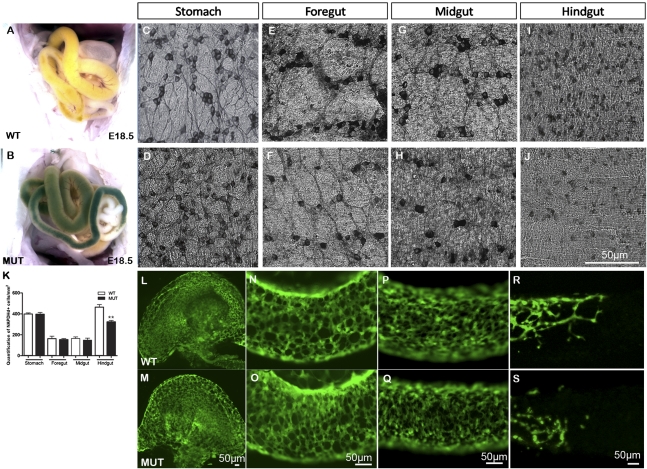
Phactr4humdy/humdy mutant embryos displayed an intestinal blockage phenotype with an abnormal accumulation of material in the gut. Normally, intestines of wild-type embryos at E18.5 were white and/or yellow in color, but the intestines of mutant embryos were green and/or dark red, indicating a gastrointestinal tract problem (Fig. 1A,B). Histological sections showed retention of meconium in E18.5 mutant intestine (Supplemental Fig. S1A,B). To characterize ENCCs in the gut, we analyzed the expression of nicotinamide adenine dinucleotide phosphate (NADPH) diaphorase, which highlights the major neuronal population in the myenteric plexus at E18.5 (Fig. 1C–J). Wild-type enteric neurons were clustered in myenteric ganglia interconnected by neurites in the stomach, foregut, and midgut (Fig. 1C,E,G). However, mutant neurons were largely individually localized and the neurite pattern was disorganized (Fig. 1D,F,H). The number of NADPH diaphorase-positive cells was normal in the stomach, foregut, and midgut but showed a statistically significant decrease in the mutant hindgut compared with wild-type (30.2% decrease in the mutant) (Fig. 1K). Ganglia hypotrophy was clearly observed in the mutant hindgut (Fig. 1I,J).
Figure 1.
Colonic hypoganglionosis in Phactr4humdy/humdy embryos. (A,B) Material is retained in the intestines of E18.5 mutants (B) presenting as green intestines versus the normal white or orange intestines of wild-type embryos (A). (C–J) Whole-mount NADPH diaphorase staining of E18.5 wild-type (top panels) and Phactr4humdy (bottom panels) gut regions. (K) Statistically significant 30.2% decrease in NADPH diaphorase-stained cells in the mutant hindgut, compared with wild type (E18.5). Data are expressed as mean ± standard deviation (SD) in three independent experiments. (**) P < 0.01, Student's t-test. (L–S) Confocal images of whole-mount gut preparations from E12.5 wild-type;RetTGM/+ (top panels) and Phactr4humdy/humdy;RetTGM/+ (bottom panels) embryos. (L,M) Stomach. (N,O) Foregut. (P,Q) Midgut. (R,S) Hindgut.
The defect in ENCC number and organization was observed early in development. RetTGM/+ (Enomoto et al. 2001) was used to visualize ENCCs that are marked by GFP expression under the control of the Ret promoter. By E12.5, wild-type and mutant ENCCs colonized the gut to post-caecum hindgut level (Fig. 1L–S). However, the number of mutant ENCCs was reduced, and these cells were more randomly distributed and often appeared as individualized cells relative to the organized network in wild type (Fig. 1R,S). Phactr4humdy/humdy mutants die at or before birth, and hence it was not possible to examine whether megacolon is present in Phactr4 mutants after birth, similar to that observed in HSCR patients, although this would be expected based on the embryonic hypoganglionic phenotype and retention of meconium.
Neural crest specification and ENCC proliferation and differentiation are normal in Phactr4humdy mutants
ENCC colonization of the gut is dependent on proper neural crest specification, proliferation, and differentiation. Molecular analyses of these processes showed no apparent alteration. Neural crest specification appeared normal, as revealed by neural crest markers AP2α and Sox10 (Supplemetnal Fig. S2A–D). Our previous studies showed Phactr4 regulates the cell cycle through PP1, Rb, and E2F1. Although in the neural tube and retina the humdy mutation causes excess cell proliferation (Kim et al. 2007), analysis of phospho-histone 3 staining of E9.5, E10.5, and E12.5 Phactr4humdy/humdy;RetTGM/+ embryonic ENCC in the gut (Supplemental Fig. S2E) showed no significant difference between wild type and mutant. Importantly, although loss of E2F1 function can rescue exencephaly, coloboma, and abnormal proliferation of neural progenitors in humdy embryos (Kim et al. 2007), it could not rescue the ENS defects (Supplemental Fig.S1), indicating a role for Phactr4 in the ENS that is independent of the Rb–E2F1-regulated cell cycle. Mutant ENCCs also underwent differentiation, as revealed by neuron and glia markers Tuj1, PGP9.5, and GFAP (Supplemental Fig. S2G–L). TUNEL staining for apoptotic ENCCs showed no significant difference at E9.5, E10.5, and E12.5 (Supplemental Fig. S2F). However, after ENCC migration and colonization of the gut, increased apoptosis was detected in mutant ENCCs at E14.5. By E18.5, no apoptosis was observed in wild-type or mutant ENS. Taken together, these results indicate that the decreased number and incomplete innervation by mutant ENCCs is not due to an alteration in cell specification, proliferation, or differentiation. The reduced number of ENCCs at E12.5 cannot be explained by cell death; however, the cell death seen at E14.5 may contribute to the overall phenotype of hypoganglionosis.
Phactr4 is required for ENCC migration
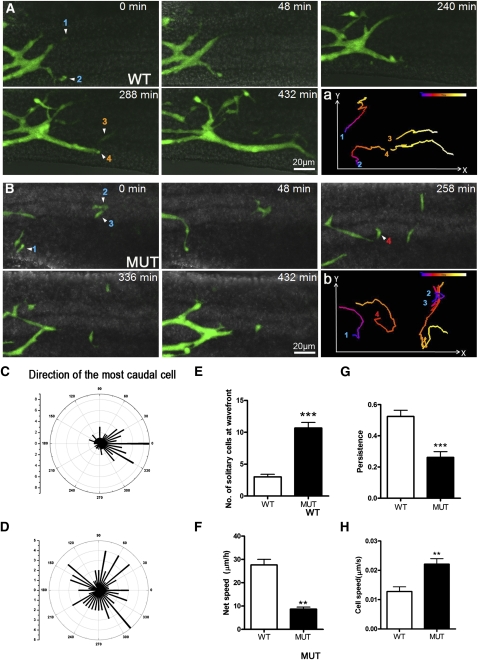
Another parameter critical to ENCC colonization of the gut is directed cell migration. To explore the in vivo migratory behavior of ENCCs within the gut, we used time-lapse live imaging. RetTGM/+ cells in organ explants of E12.5 hindgut from wild-type and Phactr4humdy/humdy embryos were imaged for up to 8 h (Fig. 2A,B; Supplemental Movies S1, S2). At the migration wave front, wild-type ENCC chain migration was readily observed and cell trajectories were in a relatively straight line from rostral to caudal (Fig. 2A [panel a], C; Supplemental Movie S1). There were very few solitary ENCCs (Fig. 2E), and individual cells were quickly joined by more rostral ENCCs to form small and dynamic groups of cells as either aggregates or chains, which efficiently invaded the hindgut. This pattern of migration and net speed (27.67 ± 2.33 μm/h) (Fig. 2F) is consistent with previous observations of wild-type ENCCs (Young et al. 2004). In contrast, in the mutant, directionality of ENCCs at the wave front was much more erratic (cell tracking was performed on three wild-type and three mutant gut explants) (Fig. 2B [panel b], D; Supplemental Movie S2), indicating that directional ENCC migration is disrupted by loss of Phactr4 function. Moreover, there was a large number of solitary ENCCs detached from the population and located away from the wave front chains (Fig. 2B). Quantification of the number of solitary cells at the wave front showed a 3.6-fold increase in the mutant (Fig. 2E), suggesting that Phactr4 is required to retain cell–cell adhesion at the migratory wave front. Some solitary ENCCs rejoined the rostral chains, whereas some eventually rounded up and underwent cell death. This latter subset was small and was detected by live imaging but not in fixed TUNEL-stained samples. The net speed and persistence of leading cell movements were significantly reduced in the mutant, although the speed of individual mutant cells was increased (Fig. 2F–H). Taken together, these observations indicate that Phactr4 is required to retain ENCCs in a chain at the migratory wave front and provide directional migration to allow complete innervation of the gut.
Figure 2.
Live imaging of ENCC migration along the gut. (A,B) Still images from time-lapse movies of the hindgut of E12.5 wild-type;RetTGM/+ (A) and Phactr4humdy/humdy;RetTGM/+ mutant (B) whole-mount gut showing the caudal progression of ENCCs. (A, panel a) Wild-type ENCCs migrated as streams of cells in interconnected chains with a strongly rostral-to-caudal trajectory (left to right, respectively). (B, panel b) In contrast, Phactr4humdy ENCCs were largely solitary at or close to the migratory wave front with a random trajectory. (A, panel a; B, panel b) Numbers indicate four different cells tracked over 8 h at 5-min intervals, and the migration track is color-coded to indicate the relative time point. (C,D) Polar histograms represent the trajectories of the most caudal cell at 15-min intervals in three explants of E12.5 hindgut. The trajectories were determined by drawing a straight line from the position of the most caudal cell with its position 15 min previously, with 0 degree being the rostrocaudal axis of the gut. The number shown on the inner arcs represent the frequency with which the cell was detected at that angle. (E,F) Quantification of the number of solitary cells at the migration wave front (E) and net speed at which the migratory wave front of ENCCs migrated caudally along the gut (F). The net speed was determined by measuring the distance between the location of the wave front at the beginning of the sequence and its location at the end of the sequence (a minimum of 8 h later). (G,H) Analysis of persistence (ratio of the direct length from start to end divided by the total track length) (G) and speed (H) of migrating ENCCs. Data are expressed as mean ± SD in three independent experiments. (***) P < 0.001; (**) P < 0.01; (*) P < 0.05, Student's t-test.
Phacrtr4 acts cell-autonomously to regulate directed cell migration
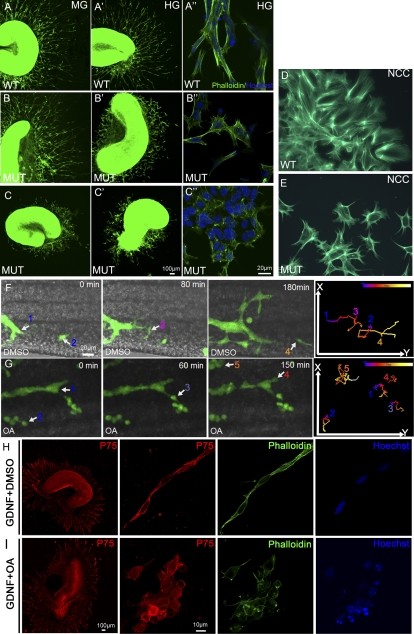
Phactr4 is expressed in ENCCs, as shown by PCR from FACs-sorted ENCCs (Supplemental Fig. S3A). Therefore, we next asked whether Phactr4 acts cell-autonomously to regulate ENCC migration. To study this, we dissected gut segments from E13.5 wild-type or Phactr4humdy/humdy embryos and cultured them in 3D collagen gels with GDNF to stimulate ENCC migration out of the explant (ENCCs visualized by immunostaining with p75NTR) (data not shown). Wild-type ENCCs showed extensive migration out of the explant (Fig. 3A,A′). Mutant ENCCs showed limited migration, wherein ∼85% of explants showed limited migration from the hindgut (Fig. 3B′) and ∼15% displayed severe migration defects from both the midgut and hindgut (Fig. 3C,C′). Higher magnification of wild-type ENCCs showed chains of elongated and polarized cells, whereas mutant ENCCs were largely individual and displayed an altered cell shape with multiple random protrusions (Fig. 3A″,B″,C″).
Figure 3.
Phactr4 and PP1 act cell-autonomously to control directed migration. (A–C) Segments of E13.5 proximal midgut (MG) (A–C) and distal hindgut (HG) (A′–C′) were cultured in 3D collagen matrix with GDNF for 3 d and stained with phalloidin (green) to detect the cytoskeleton and with Hoechst (blue) for nuclei. Explants of wild-type gut showed extensive migration of ENCCs out of both gut segments (A,A′). Phactr4humdy ENCCs are responsive to GDNF, but their migration out of the explant is variable. (C,C′) In severe cases, cells from the midgut and hindgut displayed limited migration. (B,B′) In mild cases, only cells from hindgut displayed a migration defect. (A″–C″) Higher magnification of wild-type ENCCs showed elongated cells that migrate together in chains (A″), while Phactr4humdy ENCCs had altered cell shape with random protrusions (B″,C″). (D,E) Vagal NCCs labeled with phalloidin 48 h after migrating from vagal neural tube explant. Wild-type NCCs are elongated and polarized (D), while Phactr4humdy NCCs had aberrant cell shape with random protrusions (E). (F,G) Still images from time-lapse movies of E12.5 wild-type;RetTGM/+ hindgut explants treated with DMSO (F) or 100 nM OA (G) for 3 h, then OA was removed and fresh medium was added, followed by time-lapse imaging over 8 h. Time is noted in minutes. Following OA treatment, cells displayed undirected cell protrusions and random cell movements. Numbers indicate different cells tracked over 8 h at 5-min intervals, and the tracks shown in the right panels are color-coded to indicate the relative time point. (H,I) Explants of wild-type gut cultured with GDNF for 2 d and then treated with DMSO (H) or 20 nM OA (I) for 1 h. (H) In the control explant, ENCCs were polarized and maintained long chains. (I) Inhibition of PP1 activity with OA resulted in altered cell shape with random protrusions. Red is anti-p75NTR antibody detecting the ENCC, green is phalloidin detecting the cytoskeleton, and blue is Hoechst detecting nuclei.
The variable migration defect was also evident in vivo. At E12.5, wild-type ENCCs have colonized the caecum completely and reached the hindgut (Supplemental Fig. S3B,D). In contrast, the mutant phenotype ranged from the most severe but rare cases where almost no ENCCs were detected throughout the gut (Supplemental Fig. S3C), likely reflecting a vagal neural crest emigration defect, to cases in which mutant ENCCs were reduced in the caecum or hindgut, even at E13.5 (Supplemental Fig. S3E,G).
Live imaging of ENCC migration from E13.5 gut explants showed properties similar to those seen in vivo. Wild-type ENCCs migrated out of the explants in chains, with a largely straight trajectory (Supplemental Fig. S4A,C; Supplemental Movie S3). In contrast, mutant ENCCs were more solitary and moved very rapidly (2.75-fold increase in ENCC speed compared with wild type) (Supplemental Fig. S4E) but without specific direction, as shown by the meandering trajectories and decreased persistence (Supplemental Fig. S4B,D,E; Supplemental Movie S4). The directionality of cell protrusions was also much more erratic in the mutant than wild type (Supplemental Fig. S4F,G). Together, these data indicate that Phactr4 is required cell-autonomously to regulate ENCC migration.
Phactr4 is also required cell-autonomously for vagal NCC emigration from explants of E9.0 neural tube (visualized with anti-p75NTR antibody). Wild-type NCCs migrated extensively from the neural tube, whereas there was a 32% decrease in NCCs that migrated from the humdy neural tube (Supplemental Fig. S5A–C). Actin cytoskeleton visualization by phalloidin staining showed that wild-type NCCs were elongated and polarized, with a single dominant lamellipodia at the leading edge (Fig. 3D). In contrast, humdy NCCs were less elongated, with multiple lateral lamellipodium, and more solitary cells were observed (Fig. 3E). In vivo migration to the foregut was also defective in E9.5 Phactr4humdy/humdy; RetTGM/+ embryos, with reduced number of ENCCs in the foregut (37% decrease relative to wild type) (Supplemental Fig. S5D–F). Together, these data indicate that Phactr4 acts cell-autonomously to direct ENCCs throughout their migratory pathway from neural crest emigration to ENCC migration into the foregut and along the intestinal tract. These data also suggest that Phactr4 regulates maintenance of chain formation and cell polarization, perhaps by affecting cytoskeletal dynamics.
PP1 is also required for ENCC migration along the gut
The Phactr4humdy mutation disrupts PP1 binding, and therefore we tested whether Phactr4 acts through PP1 in the regulation of ENCC migration. PP1 activity was inhibited with a pharmacological inhibitor, okadaic acid (OA), and the migratory wave front was visualized by dynamic imaging of wild-type E12.5 RetTGM/+ hindgut organ cultures. In DMSO control treatment, ENCCs at the migratory wave front migrated in long chains and efficiently colonized the gut (Fig. 3F; Supplemental Movie S5). In contrast, treatment with 100 nM OA caused individualization of ENCCs and they showed limited and undirected migration (Fig. 3G; Supplemental Movies S6–S8). PP1 functions cell-autonomously in the ENCCs, as shown by OA treatment following GDNF stimulation of ENCC migration from wild-type hindgut explants. OA addition resulted in chain dissociation, and the ENCCs were individualized, no longer polarized, and showed random protrusions (Fig. 3, cf. I and DMSO control in H). These studies serve to tie together Phactr4 and PP1 function in the control of ENCC migration.
Phactr4 mutation disrupts the actin cytoskeleton and lamellipodium formation through regulation of cofilin activity
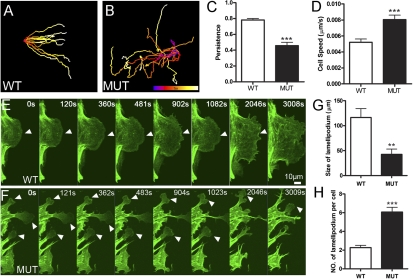
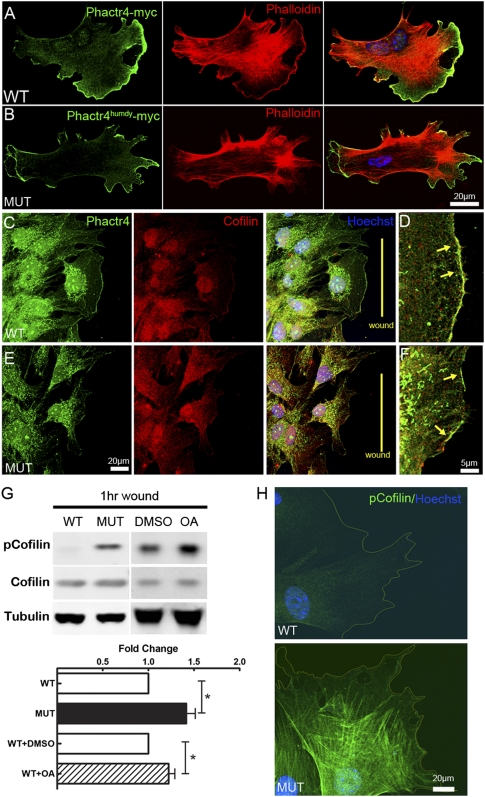
To explore further the molecular mechanism underlying Phactr4 regulation of directional migration, we established an in vitro system of wild-type and Phactr4humdy/humdy mouse embryonic fibroblasts (MEFs). In a wound healing assay, wild-type MEFs moved in a persistent fashion to close the wound, whereas the mutant MEFs moved randomly with erratic trajectories, mimicking the in vivo migration defects (Fig. 4A–D; Supplemental Movies S9, S10). To examine actin dynamics, we transfected MEFs with Lifeact-EGFP (Riedl et al. 2008) and followed their migration during wound healing. Wild-type cells at the wound front had one large fan-like lamellipodia, whereas mutant cells displayed multiple small lamellipodium and retraction fibers (Fig. 4E,F; Supplemental Movies S11, S12, a single cell is shown for wild type and mutant). Quantitative analysis revealed smaller size but increased number of lamellipodium in mutant MEFs (Fig. 4G,H).
Figure 4.
Altered actin dynamics in Phactr4humdy MEFs. (A,B) Trajectories of migratory wild-type MEFs (A) or mutant MEFs (B) in a wound healing assay. Cells tracked over 15 h at 3-min intervals. The track is color-coded to indicate relative time point. (C,D) Quantification of persistence (C) and speed (D) of migrating cells in a wound healing assay. (E,F) MEFs transfected with Lifeact-EGFP construct. Wild-type cells show one predominant lamellipodia (E), whereas mutant cells display increased numbers of small random lamellipodium and retraction fibers (F). Arrowheads show the lamellipodia. (G,H) Quantification of size (G) and number (H) of lamellipodium. Data are expressed as mean ± SD in three independent experiments. (**) P < 0.01; (***) P < 0.001, Student's t-test.
To determine the subcellular localization of Phactr4, Myc-tagged Phactr4wt or Phactr4humdy constructs were transfected into wild-type or mutant MEFs. This showed the concentration of Phactr4 at the tips of lamellipodium. Moreover, the Phactr4humdy mutation did not affect its localization (Fig. 5A,B), consistent with the ability of both wild-type and mutant protein to bind to actin (Kim et al. 2007). Endogenous Phactr4 also localized to the lamellipodia, as visualized with a Phactr4-specific antibody (Fig. 5C,E). However, the actin cytoskeletal network was disrupted in mutant cells, as phalloidin staining showed multiple small lamellipodium as well as multiple retraction fibers on all sides of the mutant cell (Fig. 4F, 5B). Normally, polarized lamellipodia are a result of cytoskeletal remodeling directed by small RhoGTPases. Cofilin, one of actin-interacting proteins downstream from RhoGTPase, plays an important role in regulating actin dynamics by severing filamentous actin. Cofilin activity is important for directional migration by reorganizing actin protrusions in response to external guidance cues (Ghosh et al. 2004; Paavilainen et al. 2004; Bernstein and Bamburg 2010). PP1 can activate cofilin by dephosphorylation (Ambach et al. 2000; Larsen et al. 2003; Oleinik et al. 2010). Immunostaining of wild-type and mutant cells showed that Phactr4 colocalized with cofilin at the tip of the lamellipodium (Fig. 5C–F). The distinct localization of Phactr4, combined with the fact that the humdy mutation specifically disrupts PP1 but not actin binding, led us to hypothesize that Phactr4 serves as a novel scaffold protein to bridge PP1 to cofilin to coordinate actin cytoskeletal dynamics. The relative amount of cofilin phosphorylated on Ser3 in wounded cells was significantly higher in mutant MEFs by both Western blot and immunostaining (Fig. 5G,H). Increased phospho-Ser3 (pSer3)-cofilin was also observed when PP1 activity was inhibited in wild-type MEFs (Fig. 5G). Together, these data provide evidence that Phactr4 acts through cofilin to regulate actin dynamics.
Figure 5.
Cofilin activity is disturbed in Phactr4humdy MEFs. (A,B) Myc-tagged Phactr4 wild-type (A) or humdy (B) construct was transfected into wild-type or mutant MEFs, respectively. MEFs were fixed and stained with anti-Myc antibody, phalloidin, and Hoechst, showing Phactr4 localization to the lamellipodium. (C–F) Detection of endogenous Phactr4 protein, with an anti-Phactr4 antibody (green) showing colocalization with cofilin (red) at the leading edge of lamellipodium in wounded wild-type (D) or mutant (E) MEFs. The arrow shows colocalization at the lamellipodium. (G) MEFs were grown to confluency in a laminin-coated dish for 36 h and then wounded extensively (evenly spaced wounds, 500 μm apart). Cells were allowed to migrate into the wound for 1 h. Where indicated, 0.1 μM OA or DMSO was added to wild-type MEFs during the wound healing period. Western blot analysis of total lysates with the indicated antibodies is shown. Quantification of protein expression showed 46% increase of pSer-cofilin in mutant and 26% increase in OA-treated wild-type cells. n = 6; (*) P < 0.05. (H) Immunostaining of pSer3-cofilin in wounded wild-type (top panel) and mutant (bottom panel) MEFs.
Phactr4 colocalizes with β1 integrin at the tips of lamellipodia and regulates integrin signaling
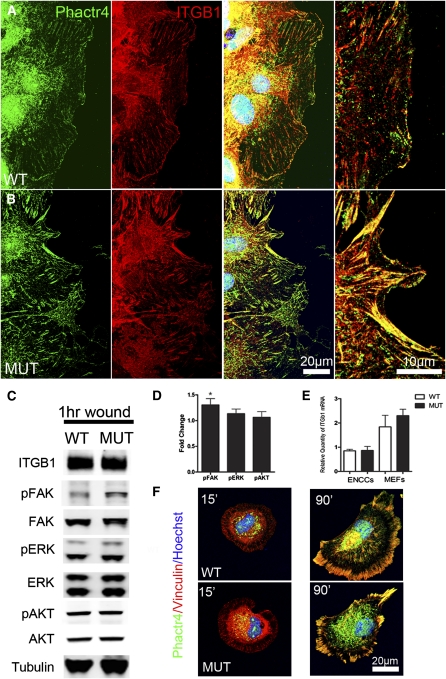
Integrins play key roles in controlling directional migration by regulating the actin cytoskeleton (Etienne-Manneville and Hall 2001; White et al. 2007; Legate et al. 2009). A conditional null mutation of β1 integrin in mouse ENCCs shows defects in cell–cell adhesion and directional migration (Breau et al. 2006, 2009). Moreover, β1 integrin tends to promote random migration through the Rho–ROCK–cofilin pathway (Danen et al. 2005). We therefore asked whether Phactr4 may interact with β1 integrin to coordinate the actin cytoskeleton. First, we examined the localization of Phactr4 protein in relation to β1 integrin (ITGB1) in the MEF wound healing assay. This showed a strong correlation between endogenous Phactr4 and ITGB1 at the tips of lamellipodium in both wild-type and mutant cells (Fig. 6A–B). Moreover, Phactr4 localizes to mature but not nascent focal adhesions (Fig. 6F), sites where integrin signaling activates one of its downstream targets, FAK (Legate et al. 2009).
Figure 6.
Phactr4 colocalizes with β1 integrin at the tip of lamellipodia and regulates integrin signaling. (A,B) Four hours after wounding, MEFs were fixed and stained with anti-Phactr4 (green) and anti-ITGB1 (red) antibodies and Hoechst (blue). Phactr4 colocalizes with ITGB1 at the leading edge of lamellipodium in wild type and mutant. (C) MEFs were grown on laminin and then wounded for 1 h. Western blot analysis of total lysates with the indicated antibodies is shown. (D) Quantification of protein expression based on experiments such as shown in C showed 30% increase of pFAK in mutant cells. n = 6; (*) P < 0.05. (E) ITGB1 mRNA level by quantitative RT–PCR of mRNAs isolated from MEFs or FACS-sorted ENCCs shows no significant change in RNA levels. n = 3. (F) MEFs were plated on a fibronectin-coated coverslip for 15 min and 90 min, and stained with antibody against Phactr4 (green), focal adhesion marker Vinculin (red), and nuclei marker Hoechst (blue). Phactr4 is localized to mature, but not nascent, focal adhesions in both wild-type and mutant cells.
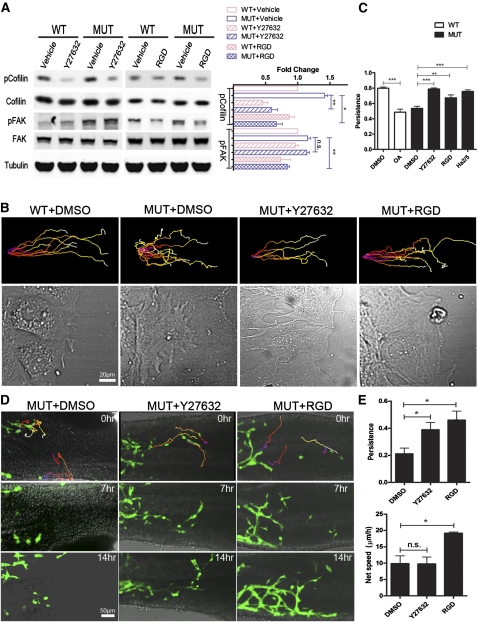
We then tested whether Phactr4 function is required for proper integrin signaling. Western blot analysis of wounded mutant MEFs, compared with wild type, showed an increase of phospho-FAK (pFAK) (Figure 6C,D), suggesting Phactr4 may serve as a negative regulator of integrin signaling. We also examined whether alteration in integrin signaling would modulate other signaling pathways such as ERK and AKT. However, this effect was specific, as no significant change in phospho-Erk (pERK) or phospho-Akt (pAKT) was observed (Figure 6C,D), nor was there a change in phospho-Numb (data not shown), another target of PP1(Nishimura and Kaibuchi 2007). The increase in integrin signaling is not due to up-regulation of integrin expression, as ITGB1 protein or mRNA level is not changed in either MEFs or sorted ENCCs (Fig. 6C,E). Furthermore, in a 1-h wound healing assay in the presence of GRGDTP peptide to block integrin activity or a ROCK inhibitor (Y27632), pSer3-cofilin levels were markedly decreased, indicating that the abnormal increase in phospho-cofilin in Phactr4humdy mutant cells is dependent on both β1 integrin and ROCK activity (Fig. 7A). RGD treatment, but not ROCK inhibitor, also decreased pFAK, suggesting that the increase in pFAK is due to increased integrin activity in mutant cells (Fig. 7A). Together, these results provide evidence that Phactr4 controls cofilin phosphorylation by down-regulating the activity of the β1 integrin–FAK–ROCK pathway.
Figure 7.
Rescue of random migration both in vitro and in vivo. (A) MEFs were grown on laminin and then wounded for 1 h. Where indicated, 5 μM Y27632, 100 μg/mL GRGDTP, or vehicle control was included during the wound healing period. Cells were lysed, and the cellular content of pSer3-cofilin, phospho-Y925-FAK, and β-tubulin relative to total cofilin and FAK was determined by Western blotting. Quantification of protein expression based on experiments such as shown. n = 3; (*) P < 0.05; (**) P < 0.01. (B) Confluent MEF monolayers were wounded, and the cells were allowed to migrate into the wound in the presence of 5 μM Y27632, 100 μg/mL GRGDTP, 10 μg/mL β1 integrin blocking antibody (Ha2/5 clone, Supplemental Movie S17), or vehicle control (DMSO). The cells were imaged every 3 min for 15 h, and then tracked by Imaris software. Representative trajectories of migrating cells (top panels) and selected phase-contrast images showing lamellipodia morphology of migrating cells (bottom panels). (C) Quantification of cell persistence. n > 300 track plots; (**) P < 0.01; (***) P < 0.001. Data are expressed as mean ± SD. (D) Still images from time-lapse movies of E12.5 Phactr4humdy/humdy;RetTGM/+ hindgut explants treated with DMSO, 20 μM Y27632, or 1 mg/mL GRGDTP and then imaged every 3 min for 16 h. Time is noted in hours. (Top panels) Cell trajectories were color-coded to indicate the relative time point. (E) Quantification of cell persistence and net speed. (*) P < 0.05. Data are expressed as mean ± SD in three independent experiments.
Rescue of ENCC directional migration by inhibition of integrin or ROCK activity
Given the mechanistic relationship defined above and the abnormal regulation of β1 integrin signaling and phospho-cofilin in Phactr4 mutant cells, we sought to directly determine whether the random migration and loss of persistent movement was a consequence of increased β1 integrin signaling. Indeed, treatment of mutant MEFs with GRGDTP peptide to block interaction of integrin with its ECM ligands, β1 integrin function-blocking antibody (clone Ha2/5), or the ROCK inhibitor restored persistent migration and rescued the formation of a single large lamellipodium (Fig. 7B,C; Supplemental Movies S13–S17). Moreover, cell–cell adhesion was also rescued with RGD treatment, as cells migrated collectively to close the wound (Fig. 7B; Supplemental Movie S16). We also tested whether ENCC migration in vivo could be rescued. Treatment of mutant hindgut organ cultures with ROCK inhibitor showed a normalization of directed cell movement and partial rescue of chain migration (Fig. 7D,E; Supplemental Movies S18–S20). Even more strikingly, GRGDTP peptide treatment strongly rescued mutant ENCC migration, resulting in restoration of chain migration and normalized persistence and net speed (Fig. 7D,E; Supplemental Movie S20), reminiscent of wild-type ENCC migration (Fig. 2A). Thus, the loss of Phactr4-dependent directional migration is due to an up-regulation of ITGB1 signaling, which promotes random migration. Collectively, these data reveal a novel role for Phactr4 in controlling directional migration and that Phactr4 acts at the lamellipodia to mediate integrin signaling through the ROCK–cofilin pathway.
Discussion
The coordinated migration of enteric neurons is essential for their correct positioning and proper integration to form the functional neuronal network of the mature ENS. However, relatively little is known about the molecular regulation of this collective cell migration. Here we identify Phactr4 as a novel regulator of ENCC migration. Moreover, we discovered a mechanistic link between Phactr4-mediated and integrin-dependent actin cytoskeleton dynamics (Supplemental Fig. S6). Phactr4humdy/humdy mutant embryos exhibit intestinal hypoganglionosis. This defect is independent of ENCC specification, proliferation, or differentiation, and instead is the result of a defect in ENCC migration. One of our key findings is that the Phactr4humdy mutation does not decrease the velocity of an individual cell, but does significantly reduce the persistence and directionality of migration, which results in disrupted chain migration both in vivo and in vitro. The Phactr4humdy mutation does not inhibit formation or retraction of cell protrusions, but the orientation and direction of the cell protrusions are significantly disrupted. Mechanistically, Phactr4 protein colocalizes with β1 integrin and cofilin at the protrusions and is found at mature focal adhesions, and loss of Phactr4 function results in increased integrin signaling and increased phosphorylation of FAK and cofilin. Thus, Phactr4 regulates integrin signaling and cofilin activity, and the coordination of these activities by Phactr4 controls polarized protrusion and directional migration. Most strikingly, the Phactr4humdy ENCC migration defects were rescued by inhibiting integrin function with an RGD peptide or by inhibiting ROCK activity, indicating that Phactr4 acts via integrin-mediated cofilin signaling and this functional relationship is essential for directed ENCC migration.
Our work provides in vivo evidence that Phactr4 regulates cytoskeletal remodeling. Wild-type cells have a highly polarized morphology, while Phactr4 mutant cell shape is greatly altered, with an increased number of random protrusions around the circumference of the cell. Studies in cultured cells of other Phactr family members support our in vivo results. Phactr3 (scapinin) enhances cell motility by interacting with the actin cytoskeleton (Sagara et al. 2009). Moreover, each Phactr family member, when overexpressed, leads to a change in cell shape and cell protrusions of variable length and direction (Favot et al. 2005). Our in vivo loss of Phactr4 function results also show dramatic changes in cell shape and cell protrusions, indicating that the level and/or localization of the Phactr proteins are critical in regulating the organization of the actin cytoskeleton. Indeed, Phactr4 is specifically localized to cell protrusions, where the actin cytoskeleton is actively remodeled during directed cell migration. Together, the data indicate that the Phactr family proteins have a common feature of modifying cell morphology to affect cell motility.
The yeast Phactr4 homolog is Afr1, and it also regulates actin dynamics. There is a specific Afr1 mutation that disrupts PP1 binding and results in abnormal budding versus polarized budding in wild-type budding yeast (Bharucha et al. 2008). In yeast, Afr1 brings PP1 to the septin cytoskeleton. Previously, we showed that mouse Phactr4 binds to actin and PP1, and the humdy mutation specifically disrupts interaction of Phactr4 with PP1 (Kim et al. 2007). This unique allele has served to reveal the in vivo functions of Phactr4 and PP1 in cell cycle regulation and cell migration. During neural and eye development, Phactr4 helps retain PP1 in the cytoplasm to control the activity of PP1 toward one of its targets, the retinoblastoma protein (Kim et al. 2007). During ENCC migration, we postulate that Phactr4 bridges PP1 and actin to regulate actin cytoskeletal dynamics during directional ENCC migration. Inability of Phactr4 to bring PP1 to actin, as well as pharmacological loss of PP1 function, impairs ENCC migration and causes undirected cell protrusions. It is intriguing to speculate that Phactr4, through its localization at lamellipodia and focal adhesions, serves to provide subcellular substrate specificity to PP1. We show that Phactr4 colocalizes with cofilin and cofilin phosphorylation increases dramatically when Phactr4 cannot bind PP1. Cofilin activity is important for directional cell migration by maintaining a polarized actin cytoskeleton (Dawe et al. 2003; Ghosh et al. 2004; Mouneimne et al. 2004). Here we show that the random migration of Phactr4humdy cells is associated with increased levels of inactive phosphorylated cofilin. Rho, Rac, and Cdc42 can activate LIM kinase, which phosphorylates cofilin at Ser3 to inhibit its activity (Arber et al. 1998; Maekawa et al. 1999), whereas protein phosphatases such as PP1 serve to dephosphorylate cofilin to enhance its activity. In our in vivo studies, the Rho/Rho kinase pathway is responsible for cofilin phosphorylation downstream from Phactr4 as mutant cells are rescued by ROCK inhibition, resulting in persistent migration with increased cofilin activity. Furthermore, inhibition of PP1 activity by OA stimulates an increase in cofilin Ser3 phosphorylation and causes random migration. Together, our findings provide a new pathway by which Phactr4 and PP1 act to regulate cofilin activity, which is required for directed collective cell migration in vivo.
The orientation of cell membrane protrusions determines the direction and behavior of a migrating cell. Intracellular signaling pathways at the leading edge that control actin cytoskeleton remodeling can therefore contribute to directional migration (Petrie et al. 2009). Integrins play a key role in sensing external cues, such as chemoattractants or wounds. Integrin signaling activates FAK as well as PI3K and MAPK pathways. In addition, integrin and its coreceptors can mediate adhesion formation, and the formation of new adhesions at the leading edge can contribute to directional migration, in part by modulating RhoGTPase activity to control protrusion formation (White et al. 2007). The specific localization of Phactr4 to membrane protrusions, coupled with the role of Phactr4 in regulating integrin signaling, allows the polarized activation of integrin signaling and adhesion formation at the leading edge of cells. Here we show that Phactr4 is a novel negative regulator of integrin signaling and propose that Phactr4 bridges external signals with the regulation of actin dynamics to reshape the cell and direct its movement (Supplemental Fig. S6). It is interesting that loss of one of the integrin-interacting proteins, integrin-linked kinase (ILK), also shows defects similar to Phactr4 mutant. ILK forms a complex with α-parvin and β1 integrin at focal adhesion sites. The absence of α-parvin or mutations in the α-parvin-binding domain of ILK causes abnormal contraction and cells fail to extend a persistent leading edge, leading to random migration in smooth muscle cells and collecting duct epithelial cells. These defects are due to increased RhoA–ROCK activity that results in elevated myosin light chain phosphorylation (Lange et al. 2009; Montanez et al. 2009). Furthermore, inhibition of ROCK activity can also rescue the defects in smooth muscle cells. Phactr4 is detected at mature but not nascent focal adhesions and colocalizes with β1 integrin (Fig. 6F), similar to the ILK/α-parvin complex found at integrin adhesions (Zhang et al. 2002). Therefore, it is possible that Phactr4 may spatially control the function of ILK/α-parvin/β1 integrin signaling. In vitro studies using podocytes have also shown that the stability of the ILK/α-parvin complex depends on the phosphorylation status of α-parvin (Yang et al. 2005). It is possible that Phactr4 acts through PP1 to regulate ILK/α-parvin/β1 integrin complex stability. The precise molecular mechanism by which the Phactr4/PP1 complex regulates integrin signaling remains open for future research.
Phactr4 not only affects directional migration but also ENCC chain migration. Phactr4humdy mutants show an increased number of solitary cells both in vivo and in vitro, and Phactr4 acts cell-autonomously to maintain cohesive ENCC chain migration, suggesting that Phactr4 is involved in cell–cell interactions. Time-lapse microscopy has revealed the importance of intercellular contacts between migrating ENCCs (Young et al. 2004; Anderson et al. 2006). L1 is a cell adhesion molecule expressed by the developing ENS, and L1 inhibition causes ENCCs to separate from their chains and became solitary (Anderson et al. 2006). Very little is known about the molecular regulation of ENCC migration beyond the proteins L1, β1-integrin, and MMP-2, which are involved in cell–cell/cell–matrix interactions and GDNF/RET signaling. Here we connected the function of Phactr4 with integrin; future studies may reveal an interconnection with L1 and/or MMP-2. Together, our studies provide the first in vivo evidence for a key intracellular regulator of cytoskeletal rearrangements needed for ENCC collective cell migration to and along the gut. Our results demonstrate a previously unknown pathway in ENCC collective migration in vivo (Supplemental Fig. S6) and provide new candidate genes for human genetic studies of HSCR.
Materials and methods
Mouse strains and genotyping
Phactr4humdy mutant embryos were genotyped with SSLP markers (Kim et al. 2007), specifically D4ski4010L, D2ski4010R, D4ski55-50L, and D4ski55-50R (Supplemental Table S1). The Phactr4humdy mutation was maintained on C3H/HeNCrl background for >10 generations. RetTGM was crossed into C3H/HeN background for five generations before crossing with Phactr4humdy. RetTGM genotype was determined by PCR analysis (Enomoto et al. 2001).
Organ cultures and time-lapse imaging
For collagen gel gut explant cultures, E13.5 proximal hindgut segments were placed on a glass-bottomed dish (MatTek) coated with 1 mg/mL 3D collagen gel (R&D Systems) with 10 ng/mL GDNF (US Biological) as described (Young et al. 2001). To inhibit PP1, explants were cultured for 2 d, and then 20 nM OA (Sigma) was added to the explants for 1 h and then the explants were washed with PBS followed by addition of culture medium. For imaging, explants were placed in a heat- and humidity-controlled chamber (37°C, 5% CO2/95% air) on the stage of an inverted Zeiss LSM510 Meta confocal microscope. All images were acquired with a 10× lens c-Apochromat NA 1.2. Time intervals for live-image acquisition are denoted in the Supplemental Material in the figure and movie legends.
For ex vivo time-lapse imaging, E12.5 gut segments containing RetTGM GFP+ cells were prepared as suspended explants as described (Hearn et al. 1999) and cultured in a MatTek glass-bottomed dish in DMEM with 10% fetal bovine serum and 0.075% penicillin/streptomycin. The region to be imaged was suspended across a “V” cut in a piece of black Millipore filter paper and held in place by attaching the mesentery to filter paper. Up to six gut explants were imaged by time lapse using a Zeiss Axioskop motorized stage microscope equipped with a heated stage (Zeiss). Images were captured as described in the Supplemental Material in the figure and movie legends. To inhibit PP1, 100 nM OA was added to the culture medium for 1 h or 3 h, and then the medium was removed and fresh medium was added. GRGDTP peptide (1 mg/mL; Calbiochem) or 20 μM Y27632 (Calbiochem) were added to the culture medium throughout the period of live imaging.
For NCC emigration analysis, the vagal region of neural tubes from E9.0 embryos was dissected and digested in 2 mg/mL Dispase II (Roche) as described (Newgreen and Minichiello 1995). Explants were placed on a fibronectin-coated (50 μg/mL; Sigma) 14-mm coverslip and incubated in 150 μL of complete culture medium for 24 h.
MEFs were isolated from +/+ and Phactr4humdy/humdy E13.5 embryos as described (Abbondanzo et al. 1993).
Additional Materials and Methods are included in the Supplemental Material.
Acknowledgments
We thank Rytis Prekeris for helpful discussions, Heather Young for helpful suggestions, Helen McBride for advice on gut explant culture, and members of our laboratory, especially Jianfu Chen and Carsten Schnatwinkel, for suggestions throughout this work and for helpful comments on the manuscript. We thank Lori Bulwith for technical assistance, and Jessica Goodman for analysis of the Phactr4humdy;E2f1 embryos. This work was supported by the Department of Pediatrics, and L.N. is an investigator of the Howard Hughes Medical Institute. T.-H.K initiated the project and phenotypic characterization, Y.Z. performed phenotypic characterization, developed the live imaging and mechanistic pathway, and wrote the paper. L.N. oversaw the research design and data analyses and wrote the paper.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.179283.111.
References
- Abbondanzo SJ, Gadi I, Stewart CL 1993. Derivation of embryonic stem cell lines. Methods Enzymol 225: 803–823 [DOI] [PubMed] [Google Scholar]
- Allen PB, Greenfield AT, Svenningsson P, Haspeslagh DC, Greengard P 2004. Phactrs 1–4: A family of protein phosphatase 1 and actin regulatory proteins. Proc Natl Acad Sci 101: 7187–7192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambach A, Saunus J, Konstandin M, Wesselborg S, Meuer SC, Samstag Y 2000. The serine phosphatases PP1 and PP2A associate with and activate the actin-binding protein cofilin in human T lymphocytes. Eur J Immunol 30: 3422–3431 [DOI] [PubMed] [Google Scholar]
- Amiel J, Sproat-Emison E, Garcia-Barcelo M, Lantieri F, Burzynski G, Borrego S, Pelet A, Arnold S, Miao X, Griseri P, et al. 2008. Hirschsprung disease, associated syndromes and genetics: A review. J Med Genet 45: 1–14 [DOI] [PubMed] [Google Scholar]
- Anderson RB 2010. Matrix metalloproteinase-2 is involved in the migration and network formation of enteric neural crest-derived cells. Int J Dev Biol 54: 63–69 [DOI] [PubMed] [Google Scholar]
- Anderson RB, Turner KN, Nikonenko AG, Hemperly J, Schachner M, Young HM 2006. The cell adhesion molecule L1 is required for chain migration of neural crest cells in the developing mouse gut. Gastroenterology 130: 1221–1232 [DOI] [PubMed] [Google Scholar]
- Arber S, Barbayannis FA, Hanser H, Schneider C, Stanyon CA, Bernard O, Caroni P 1998. Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase. Nature 393: 805–809 [DOI] [PubMed] [Google Scholar]
- Asai N, Fukuda T, Wu ZQ, Enomoto A, Pachnis V, Takahashi M, Costantini F 2006. Targeted mutation of serine 697 in the Ret tyrosine kinase causes migration defect of enteric neural crest cells. Development 133: 4507–4516 [DOI] [PubMed] [Google Scholar]
- Bernstein BW, Bamburg JR 2010. ADF/cofilin: A functional node in cell biology. Trends Cell Biol 20: 187–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharucha JP, Larson JR, Konopka JB, Tatchell K 2008. Saccharomyces cerevisiae Afr1 protein is a protein phosphatase 1/Glc7-targeting subunit that regulates the septin cytoskeleton during mating. Eukaryot Cell 7: 1246–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breau MA, Pietri T, Eder O, Blanche M, Brakebusch C, Fassler R, Thiery JP, Dufour S 2006. Lack of β1 integrins in enteric neural crest cells leads to a Hirschsprung-like phenotype. Development 133: 1725–1734 [DOI] [PubMed] [Google Scholar]
- Breau MA, Dahmani A, Broders-Bondon F, Thiery JP, Dufour S 2009. β1 integrins are required for the invasion of the caecum and proximal hindgut by enteric neural crest cells. Development 136: 2791–2801 [DOI] [PubMed] [Google Scholar]
- Danen EH, van Rheenen J, Franken W, Huveneers S, Sonneveld P, Jalink K, Sonnenberg A 2005. Integrins control motile strategy through a Rho-cofilin pathway. J Cell Biol 169: 515–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe HR, Minamide LS, Bamburg JR, Cramer LP 2003. ADF/cofilin controls cell polarity during fibroblast migration. Curr Biol 13: 252–257 [DOI] [PubMed] [Google Scholar]
- Druckenbrod NR, Epstein ML 2005. The pattern of neural crest advance in the cecum and colon. Dev Biol 287: 125–133 [DOI] [PubMed] [Google Scholar]
- Enomoto H, Crawford PA, Gorodinsky A, Heuckeroth RO, Johnson EM, Milbrandt J 2001. RET signaling is essential for migration, axonal growth and axon guidance of developing sympathetic neurons. Development 128: 3963–3974 [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S, Hall A 2001. Integrin-mediated activation of Cdc42 controls cell polarity in migrating astrocytes through PKCzeta. Cell 106: 489–498 [DOI] [PubMed] [Google Scholar]
- Favot L, Gillingwater M, Scott C, Kemp PR 2005. Overexpression of a family of RPEL proteins modifies cell shape. FEBS Lett 579: 100–104 [DOI] [PubMed] [Google Scholar]
- Geiger B, Bershadsky A, Pankov R, Yamada KM 2001. Transmembrane crosstalk between the extracellular matrix–cytoskeleton crosstalk. Nat Rev Mol Cell Biol 2: 793–805 [DOI] [PubMed] [Google Scholar]
- Ghosh M, Song X, Mouneimne G, Sidani M, Lawrence DS, Condeelis JS 2004. Cofilin promotes actin polymerization and defines the direction of cell motility. Science 304: 743–746 [DOI] [PubMed] [Google Scholar]
- Heanue TA, Pachnis V 2007. Enteric nervous system development and Hirschsprung's disease: Advances in genetic and stem cell studies. Nat Rev Neurosci 8: 466–479 [DOI] [PubMed] [Google Scholar]
- Hearn CJ, Young HM, Ciampoli D, Lomax AE, Newgreen D 1999. Catenary cultures of embryonic gastrointestinal tract support organ morphogenesis, motility, neural crest cell migration, and cell differentiation. Dev Dyn 214: 239–247 [DOI] [PubMed] [Google Scholar]
- Kim TH, Goodman J, Anderson KV, Niswander L 2007. Phactr4 regulates neural tube and optic fissure closure by controlling PP1-, Rb-, and E2F1-regulated cell-cycle progression. Dev Cell 13: 87–102 [DOI] [PubMed] [Google Scholar]
- Lange A, Wickstrom SA, Jakobson M, Zent R, Sainio K, Fassler R 2009. Integrin-linked kinase is an adaptor with essential functions during mouse development. Nature 461: 1002–1006 [DOI] [PubMed] [Google Scholar]
- Larsen M, Tremblay ML, Yamada KM 2003. Phosphatases in cell–matrix adhesion and migration. Nat Rev Mol Cell Biol 4: 700–711 [DOI] [PubMed] [Google Scholar]
- Legate KR, Wickstrom SA, Fassler R 2009. Genetic and cell biological analysis of integrin outside-in signaling. Genes Dev 23: 397–418 [DOI] [PubMed] [Google Scholar]
- Maekawa M, Ishizaki T, Boku S, Watanabe N, Fujita A, Iwamatsu A, Obinata T, Ohashi K, Mizuno K, Narumiya S 1999. Signaling from Rho to the actin cytoskeleton through protein kinases ROCK and LIM-kinase. Science 285: 895–898 [DOI] [PubMed] [Google Scholar]
- McCallion AS, Emison ES, Kashuk CS, Bush RT, Kenton M, Carrasquillo MM, Jones KW, Kennedy GC, Portnoy ME, Green ED, et al. 2003. Genomic variation in multigenic traits: Hirschsprung disease. Cold Spring Harb Symp Quant Biol 68: 373–381 [DOI] [PubMed] [Google Scholar]
- Montanez E, Wickstrom SA, Altstatter J, Chu H, Fassler R 2009. α-Parvin controls vascular mural cell recruitment to vessel wall by regulating RhoA/ROCK signalling. EMBO J 28: 3132–3144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouneimne G, Soon L, DesMarais V, Sidani M, Song X, Yip SC, Ghosh M, Eddy R, Backer JM, Condeelis J 2004. Phospholipase C and cofilin are required for carcinoma cell directionality in response to EGF stimulation. J Cell Biol 166: 697–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newgreen DF, Minichiello J 1995. Control of epitheliomesenchymal transformation. I. Events in the onset of neural crest cell migration are separable and inducible by protein kinase inhibitors. Dev Biol 170: 91–101 [DOI] [PubMed] [Google Scholar]
- Nishimura T, Kaibuchi K 2007. Numb controls integrin endocytosis for directional cell migration with aPKC and PAR-3. Dev Cell 13: 15–28 [DOI] [PubMed] [Google Scholar]
- Okamura Y, Saga Y 2008. Notch signaling is required for the maintenance of enteric neural crest progenitors. Development 135: 3555–3565 [DOI] [PubMed] [Google Scholar]
- Oleinik NV, Krupenko NI, Krupenko SA 2010. ALDH1L1 inhibits cell motility via dephosphorylation of cofilin by PP1 and PP2A. Oncogene 29: 6233–6244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paavilainen VO, Bertling E, Falck S, Lappalainen P 2004. Regulation of cytoskeletal dynamics by actin-monomer-binding proteins. Trends Cell Biol 14: 386–394 [DOI] [PubMed] [Google Scholar]
- Petrie RJ, Doyle AD, Yamada KM 2009. Random versus directionally persistent cell migration. Nat Rev Mol Cell Biol 10: 538–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl J, Crevenna AH, Kessenbrock K, Yu JH, Neukirchen D, Bista M, Bradke F, Jenne D, Holak TA, Werb Z, et al. 2008. Lifeact: A versatile marker to visualize F-actin. Nat Methods 5: 605–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagara J, Arata T, Taniguchi S 2009. Scapinin, the protein phosphatase 1 binding protein, enhances cell spreading and motility by interacting with the actin cytoskeleton. PLoS ONE 4: e4247 doi: 10.1371/journal.pone.0004247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson MJ, Zhang DC, Mariani M, Landman KA, Newgreen DF 2007. Cell proliferation drives neural crest cell invasion of the intestine. Dev Biol 302: 553–568 [DOI] [PubMed] [Google Scholar]
- Wallace AS, Barlow AJ, Navaratne L, Delalande JM, Tauszig-Delamasure S, Corset V, Thapar N, Burns AJ 2009. Inhibition of cell death results in hyperganglionosis: Implications for enteric nervous system development. Neurogastroenterol Motil 21: 768–e49 doi: 10.1111/j.1365-2982.2009.01309.x [DOI] [PubMed] [Google Scholar]
- White DP, Caswell PT, Norman JC 2007. αvβ3 and α5β1 integrin recycling pathways dictate downstream Rho kinase signaling to regulate persistent cell migration. J Cell Biol 177: 515–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Guo L, Blattner SM, Mundel P, Kretzler M, Wu C 2005. Formation and phosphorylation of the PINCH-1–integrin linked kinase–α-parvin complex are important for regulation of renal glomerular podocyte adhesion, architecture, and survival. J Am Soc Nephrol 16: 1966–1976 [DOI] [PubMed] [Google Scholar]
- Young HM, Hearn CJ, Farlie PG, Canty AJ, Thomas PQ, Newgreen DF 2001. GDNF is a chemoattractant for enteric neural cells. Dev Biol 229: 503–516 [DOI] [PubMed] [Google Scholar]
- Young HM, Bergner AJ, Anderson RB, Enomoto H, Milbrandt J, Newgreen DF, Whitington PM 2004. Dynamics of neural crest-derived cell migration in the embryonic mouse gut. Dev Biol 270: 455–473 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Tu Y, Velyvis A, Yang Y, Qin J, Wu C 2002. Assembly of the PINCH–ILK–CH–ILKBP complex precedes and is essential for localization of each component to cell–matrix adhesion sites. J Cell Sci 115: 4777–4786 [DOI] [PubMed] [Google Scholar]