Telomerase recruitment is still a poorly understood step of telomere maintenance. Working with S. pombe, a model organism with telomere biology very similar to humans, Webb and Zakian now address this issue by investigating the molecular mechanism of Est1 function. They find that the Est1 14-3-3-like domain forms two distinct interactions with the TER1 telomerase RNA, and also with the Ccq1 protein. The latter interaction facilitates telomerase recruitment to the telomeres. This study dissects Est1's pivotal role in telomerase recruitment and telomere maintenance.
Keywords: 14-3-3-like, Ccq1, Est1, telomerase RNA
Abstract
The telomerase protein Est1 exists in multiple organisms, including Schizosaccharomyces pombe, humans, and Saccharomyces cerevisiae, but its function has only been closely examined in S. cerevisiae, where it is a recruiter/activator of telomerase. Here, we demonstrate that S. pombe Est1 was required for the telomere association of the telomerase holoenzyme, suggesting that it too has a recruitment role. Its association with telomeres was dependent on Trt1, the catalytic subunit, and Ccq1, a telomeric protein. Surprisingly, Est1 telomere binding was only partially dependent on TER1, the telomerase RNA, even though Est1 bound nucleotides 415–507 of TER1. A ter1-Δ415–507 strain had short telomeres and very low Est1 and Trt1 telomere association in late S phase but did not senesce. An unbiased search for mutations that reduced Est1–TER1 interaction identified mutations only in the Est1 14-3-3-like domain, a phosphoserine-binding motif, the first example of a 14-3-3-like domain with RNA-binding activity. These mutations also reduced Est1–Ccq1 binding. One such mutant prevented Est1 telomere association and caused telomere loss and slow senescence, similar to ccq1Δ. We propose that the Est1–Ccq1 interaction is critical for telomerase recruitment, while the Est1–TER1 interaction acts downstream from Ccq1-mediated recruitment to stabilize the holoenzyme at the telomere.
Telomerase is a specialized reverse transcriptase that solves the end replication problem by extending the 3′ ends of telomeres in the absence of a DNA template. This activity requires that the telomerase holoenzyme be positioned precisely at the 3′ hydroxyl end of the chromosome. The primary function of telomerase RNA is to template telomeric repeats added by the catalytic subunit to the end of the chromosome. However, other regions of telomerase RNAs distinct from the templating core also perform functions critical for telomere maintenance. Template boundary elements (Tzfati et al. 2000; Webb and Zakian 2008), pseudoknots, and the three-way junction motif (Brown et al. 2007) each influence the catalytic activity of the telomerase holoenzyme. Other RNA sequences, such as the Sm site in yeast (Seto et al. 1999), the BoxH/ACA in humans (Mitchell et al. 1999), and the ciliate loop IV (Robart et al. 2010), promote RNA and holoenzyme stability. In addition to these specialized functions, the RNA performs a more general structural role as a binding platform for different components of the telomerase ribonucleoprotein particle (RNP) that keeps these components in flexible proximity to each other.
The biological significance of this scaffold-like function as an organizer of the interactions of telomerase RNA with its protein-binding partners has been most closely examined in Saccharomyces cerevisiae (Sc). The three arms of its mature telomerase RNA, TLC1 (Dandjinou et al. 2004; Zappulla and Cech 2004), are each bound by different proteins: ScEst1 (Seto et al. 2002), Ku70/80 (Peterson et al. 2001), and the Sm ring (Seto et al. 1999), while the ScEst2 catalytic subunit binds to the central core of the RNA (Livengood et al. 2002). The long arms of TLC1 are quasihelical, with internal loops and bulges that provide flexibility to the RNA. The flexibility of the arms with distinct binding sites for proteins toward the ends of each arm led to the “beads on a string” model to describe the relationship of the RNP components (Zappulla and Cech 2004, 2006).
In S. cerevisiae, the telomere association of the catalytic subunit ScEst2 is biphasic, with peak interactions at G1/early S phase and again in late S/G2 phase (Taggart et al. 2002). The late S/G2-phase peak of ScEst2 binding coincides with telomerase activity and requires TLC1–ScEst1 bound to the telomere via the ScEst1–Cdc13 interaction (Chan et al. 2008). In contrast, the G1/early S-phase association depends on the interaction between TLC1 and Yku80 and is not required for telomere elongation (Fisher et al. 2004). These results reveal that TLC1 is important not just for its effects on telomerase activity, but also for telomerase–telomere association. In addition to ScEst2 telomere recruitment during late S/G2 phase, ScEst1 performs a telomerase activation role (Evans and Lundblad 1999).
The known components of the Schizosaccharomyces pombe telomerase RNP are the catalytic subunit Trt1 (Nakamura et al. 1997), the Est1 subunit (Beernink et al. 2003), the Sm ring (Leonardi et al. 2008), and the TER1 RNA (Leonardi et al. 2008; Webb and Zakian 2008). Trt1 and Est1 are both TER1-associated in vivo (Leonardi et al. 2008; Webb and Zakian 2008), and their association with each other requires TER1 (Leonardi et al. 2008).
Due to the many differences in telomere biology between S. cerevisiae versus S. pombe and humans, telomerase regulation is likely to be different in S. pombe and humans than in S. cerevisiae. For example, the C-terminal region of Est1 in both S. pombe and humans lacks the RNA recognition motif (RRM) that is required for ScEst1 interaction with TLC1 RNA (Zhou et al. 2000; Beernink et al. 2003; Reichenbach et al. 2003). Furthermore, the N-terminal domain of the human Est1 homologs (EST1A and EST1B) and S. pombe Est1 each contain a 14-3-3-like domain, which was originally identified in SMG7/hEST1C and binds phosphoserine (Fukuhara et al. 2005). More canonical 14-3-3 domains, which also bind phosphoserine, function in diverse processes such as signal transduction and cell cycle progress (Yaffe 2002). Although there is a region in the N terminus of ScEst1 with similarity to the 14-3-3-like domain, it contains only two of the five residues involved in phophoserine binding (Fukuhara et al. 2005). Because S. pombe TER1 does not interact with Pku80 (Webb and Zakian 2008), an interaction that brings S. cerevisiae Est2 to telomeres in G1 phase (Fisher et al. 2004), it is probably not surprising that S. pombe Trt1 is telomere-associated only during S phase (Moser et al. 2009), as is mammalian telomerase (Tomlinson et al. 2006). Furthermore, S. pombe and mammals lack an identifiable ScEst3-like subunit, which is essential for S. cerevisiae telomerase activity in vivo (Lendvay et al. 1996), and its recruitment by ScEst1 is proposed to provide the activating function of ScEst1 (Tuzon et al. 2011). An activation function via Est3 has also been proposed for Candida albicans Est1 (Hsu et al. 2007). Therefore, a different Est1 mechanism of action must be present in organisms that lack Est3. Finally, the ends of S. pombe telomeres are coated in a six-member complex that forms a molecular bridge between the internal dsDNA and the distal single-stranded G overhang (Miyoshi et al. 2008). This multisubunit complex is similar to the shelterin complex found at human telomeres (de Lange 2010). One of these S. pombe shelterin-like components, Ccq1, is required to recruit Trt1 to telomeres (Tomita and Cooper 2008).
In this study, we investigate the molecular mechanism of S. pombe Est1 function in telomere maintenance. We found that Est1 bound telomeres in late S phase, and this association was Ccq1- and Trt1-dependent but only partially dependent on TER1. In contrast, Trt1 association was completely dependent on Est1 and TER1. We determined the regions of both Est1 and TER1 that are required for their interaction. Mutations in the Est1 14-3-3-like domain eliminated its interaction with both TER1 RNA and Ccq1. Further analysis of one such mutant revealed that it had a ccq1Δ-like phenotype that was more severe than that of a ter1-Δ415–507 mutation that eliminated only the Est1–TER1 interaction. We propose that the Ccq1–Est1 interaction recruits telomerase to telomeres, while the Est1–TER1 interaction functions later to stabilize telomere-bound telomerase.
Results
Est1 binds telomeres in a Trt1- and Ccq1-dependent manner, but binding is only partially reduced in ter1Δ cells
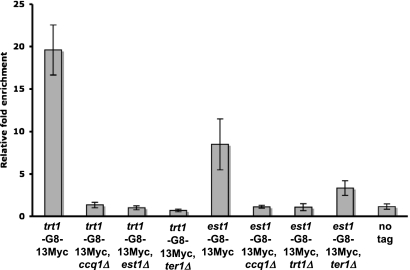
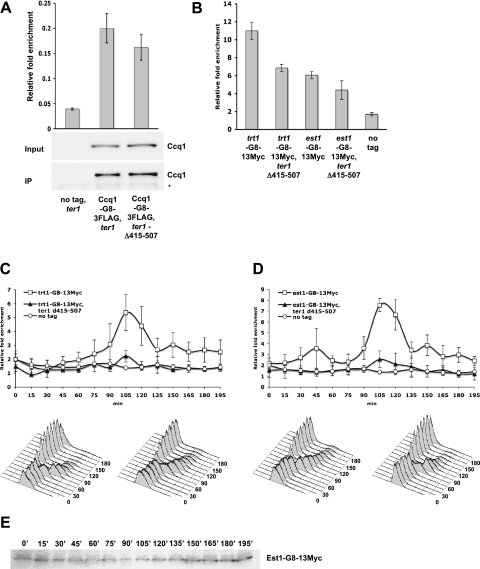
Est1 orthologs are found in S. cerevisiae, S. pombe, and humans, yet the mechanism of Est1 action has only been examined in detail in S. cerevisiae, and only ScEst1 has been shown to be telomere-associated (Taggart et al. 2002). We used chromatin immunoprecipitation (ChIP) and quantitative PCR (qPCR) to examine Est1 telomere association in wild-type and mutant cells. In cycling cells, Est1-G8-13Myc bound robustly to telomeres (Fig. 1). This binding was lost in both ccq1Δ and trt1Δ cells (Fig. 1), even though Est1-Myc was stable in both backgrounds (Supplemental Fig. 1; Leonardi et al. 2008; Webb and Zakian 2008; data not shown). However, Est1-Myc telomere binding still occurred in ter1Δ cells, albeit at reduced levels (Fig. 1). We also examined Trt1-G8-13Myc binding to telomeres. As shown previously, Trt1-Myc binding was Ccq1-dependent (Tomita and Cooper 2008). Trt1-Myc telomere binding was completely Est1- and TER1-dependent (Fig. 1). Thus, Est1-Myc binds telomeres, and this association requires Ccq1, Trt1, and, to a lesser extent, TER1.
Figure 1.
Trt1 and Est1 are dependent on Ccq1 and each component of the holoenzyme for normal levels of telomere association. Levels of Trt1 and Est1 telomere association in asynchronous culture in the presence and absence of Ccq1, TER1, and either Est1 or Trt1, respectively, were determined by ChIP. Trt1 and Est1 are tagged with a G8 linker and 13Myc epitopes. Relative fold enrichment compares telomere (STE1) immunoprecipitation (IP)/input signal with act1 IP/input signal for each holoenzyme component. Error bars indicate standard deviation for three independent experiments. Only trt1-G8-13Myc (P = 0.0079), est1-G8-13Myc (P = 0.015), and est1-G8-13Myc, ter1Δ (P = 0.0087) were significantly (P < 0.05) different from the no tag signal.
Est1 binds TER1 nucleotides 415–507
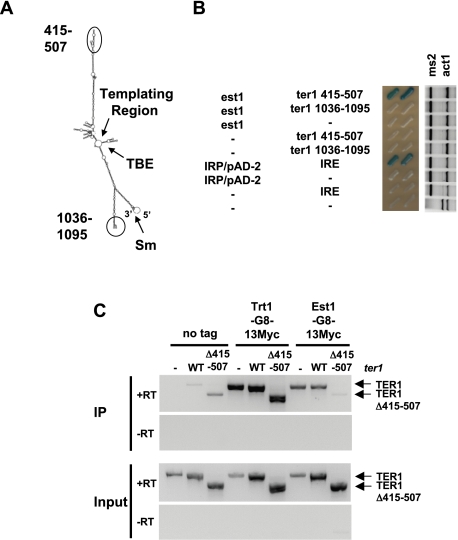
Because Est1 and Trt1 interaction depends on TER1 (Leonardi et al. 2008), and Est1 is partially dependent on TER1 for telomere association (Fig. 1), we identified the region of TER1 that interacts with Est1. We used the RNA three-hybrid system in S. cerevisiae (SenGupta et al. 1996) to screen for interactions between Est1, Trt1, and Ccq1 with the uncharacterized distal regions of the two TER1 arms (Webb and Zakian 2008), comprised of nucleotides 415–507 (hereafter called TER1-415–507) and 1036–1095 (hereafter called TER1-1036–1095) (Fig. 2A). Est1 interacted robustly with nucleotides 415–507. This binding was specific, as Est1 did not bind to TER1-1036–1095 or the empty hybrid ms2 RNA (Fig. 2B). Moreover, neither Trt1 nor Ccq1 interacted with TER1-415–507 (or with either of the other two RNAs) (Supplemental Fig. 2). The lack of Trt1 and Ccq1 binding to nucleotides 415–507 of the TER1 RNA provides additional evidence that the Est1interaction with this region is specific.
Figure 2.
TER1-415–507 is required for Est1 interaction in vivo. (A) Schematic of mature TER1 secondary structure determined by Vienna RNA Secondary Structure package. The previously identified templating sequence, template boundary element (TBE), and Sm-binding site (Sm) are depicted (Webb and Zakian 2008). An oval and circle mark nucleotides 415–507 and 1036–1095, respectively. (B) RNA three-hybrid screening of TER1 nucleotides 415–507 and 1036–1095 against Est1. (Left) Interaction strength between protein and RNA is determined by activation of a LacZ reporter and visualized by a colorimetric plate assay. IRP–IRE (iron regulatory protein–iron response element) interaction and activation of the reporter are positive controls. (Right) RT–PCR assay. Expression of RNAs as determined by RT–PCR of the constant ms2 region and compared with expression from the endogenous act1 locus. RT–PCR without added reverse transcriptase (−RT) demonstrates that the RNA preparations were free of contaminating DNA (data not shown). (C) Coimmunoprecipitation of TER1 and TER1-Δ415–507 in vivo with Trt1-G8-13Myc and Est1-G8-13Myc. Plasmid expression of TER1 and TER1-Δ415–507 are indicated. Levels of RNA association in immunoprecipitate (IP) after RNA immunoprecipitation and input were detected by RT–PCR. RT–PCR without added reverse transcriptase demonstrates removal of contaminating DNA.
Next, we performed RNA coimmunoprecipitation assays to examine Est1-Myc and Trt1-Myc interaction with TER1-Δ415–507. The ability of Est1-Myc to interact with endogenous TER1 or wild-type TER1 expressed from a plasmid was determined by the amount of RNA in the immunoprecipitates and then compared with the amount of TER1-Δ415–507, which was expressed from a plasmid. In comparison with wild-type RNA, the amount of TER1-Δ415–507 in the Est1-Myc immunoprecipitate was severely reduced (Fig. 2C, rightmost lanes) no greater than the amount of TER1-Δ415–507 RNA in the no tag control (Fig. 2C, leftmost lanes). In contrast, TER1-Δ415–507 RNA was precipitated as well as wild-type TER1 RNA in a strain expressing Trt1-Myc (Fig. 2C, middle lanes). Thus, we conclude that nucleotides 415–507 are essential for Est1–TER1 interaction but not for Trt1–TER1 association.
Nucleotides 415–507 of TER1 are required for wild-type telomere length
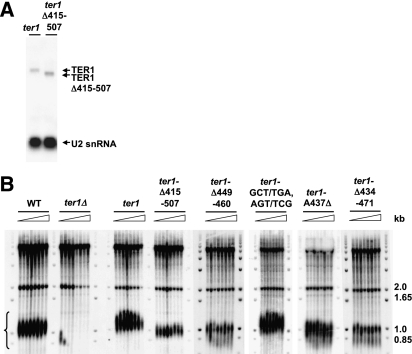
To determine whether interaction between Est1 and TER1 is required for telomere maintenance, we generated strains with the ter1-Δ415–507 allele or ter1 expressed from the leu1 locus in a ter1Δ strain. A Northern blot of total RNA from ter1-Δ415–507 and ter1 wild-type cells demonstrated that the steady-state levels of the mutant and wild-type RNAs were similar (Fig. 3A). Southern blot analysis of DNA from ter1-Δ415–507 cells showed that telomeres were ∼150 base pairs (bp) shorter than wild type, but were stably maintained over seven successive streaks (Fig. 3B).
Figure 3.
TER1-Δ415–507 is stably expressed and is necessary for telomere maintenance. (A) Northern blot of total RNA harvested from strains harboring a disruption of the endogenous ter1 locus and expressing wild-type TER1 (ter1) or TER1-Δ415–507 (ter1-Δ415–507) from the leu1 locus. The same TER1 probe is used to detect wild-type and mutant RNAs. U2 snRNA serves as a loading control. (B) Telomere blots. In all but the wild-type (WT) strain, TER1 RNA (or mutants thereof) was expressed from the leu1 locus in a ter1Δ background. Lanes contain EcoRI-digested DNA from wild-type S. pombe (WT), ter1Δ (ter1Δ), wild-type TER1 (ter1), TER1-Δ415–507 (ter1-Δ415–507), TER1-Δ449–460 (ter1-Δ449–460), TER1-GCT/TGA, AGT/TCG (ter1-GCT/TGA, AGT/TCG), TER1-A437Δ (ter1-A437Δ), or TER1-Δ434–471 (ter1-Δ434–471). Each lane depicts DNA prepared from a meiotic spore clone of the indicated genotype that was streaked seven times, harvested, and hybridized to a telomeric probe. A bracket marks telomeres and molecular markers are labeled.
A bulged adenosine within the nucleotide 415–507 region of TER1 is important for Est1 interaction and telomere maintenance
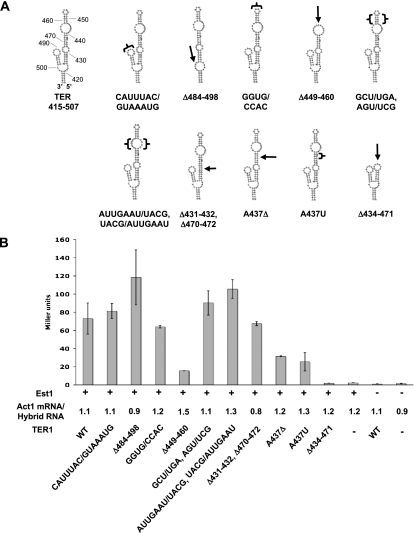
A series of mutant RNAs were constructed to identify regions within TER1-415–507 that are necessary for Est1 interaction using the RNA three-hybrid assay to monitor association. We first analyzed the contribution of the short 4-bp helix (nucleotides 484–498) within TER1-415–507 (Fig. 4A). Changing the sequence of the terminal loop from CAUUUAC to GUAAAUG (CAUUUAC/GUAAAUG) (Fig. 4B), or even the complete removal of the helix plus the terminal loop (Δ484–498), did not reduce Est1 interaction with the RNA (Fig. 4A,B). Thus, this short helix is not required for Est1 interaction.
Figure 4.
Identification of TER1-415–507 nucleotides that are important for Est1 interaction. (A) Predicted secondary structures of TER1-415–507 and the deletion and substitution mutants thereof that were analyzed in an Est1–TER1 interaction assay. Deletions are indicated by black arrows, and substituted regions are denoted by brackets. Deleted nucleotides and substituted sequences are noted below each structure. (B) Est1 interaction with TER1-415–507 mutants determined by quantitative liquid β-galactosidase RNA three-hybrid assay. Error bars depict standard deviation from three independent assays. The ratio of Act1 mRNA to hybrid RNA demonstrates that TER1 mutant RNA expression was at or near wild-type levels.
Three TER1 mutants were generated to test the contribution of the most distal region to Est1 interaction. Substitution of the terminal loop from GGUG to CCAC did not affect Est1 binding (Fig. 4B). However, the Δ449–460 allele, which deleted the entire end of the long helix, reduced Est1 interaction with the remaining RNA to ∼20% of wild-type levels, demonstrating that the terminal helix of the arm is critical for Est1 binding (Fig. 4B). When the ter1-Δ449–460 allele was integrated into the genome, the cells had short telomeres, which were only slightly longer than telomeres in ter1-Δ415–507 cells (Fig. 3B). The GCT/TGA, AGT/TCG allele was used to determine whether the structure or the sequence of the distal helix is important. Although this mutation changed the sequence of the distal helix, it still bound Est1 at normal levels (Fig. 4B), and telomere length was normal in ter1-GCT/TGA, AGT/TCG cells (Fig. 3B). These results demonstrate that the presence but not the sequence of the distal helix is important for Est1–TER1 interaction and normal telomere maintenance. Two unpaired regions (431–432 and 470–472, and 441–447 and 462–465) in the long helix were not needed for Est1–TER1 interaction, as deletion of the first unpaired region (Δ431–432 and Δ470–472) or altering the sequence of the second (AUUGAAU/UACG and UACG/AUUGAAU) did not reduce the ability of Est1 to interact with the RNA.
The TER1 structure predicts a bulged adenosine (A437) 2 nucleotides (nt) 5′ of the large unpaired region of the S. pombe TER1 Est1-binding arm. Deletion (A437Δ) and substitution (A437U) mutants of the A437 bulge each reduced Est1 interaction with the mutant RNA to ∼50%, demonstrating that both the bulge and its nucleotide identity are important. Cells expressing either ter1-A437Δ (Fig. 3B) or ter1-A437T (data not shown) had short telomeres but did not senesce even after seven streaks. Finally, removal of both the distal helix (449–460) and the bulged adenosine (A437) in TER1-Δ434–471 (Fig. 4A) reduced Est1 association (Fig. 4B) and maintained the short telomere phenotype seen in ter1-415–507 cells (Fig. 3B). Therefore, we define the Est1-binding region of TER1 as the most distal 37-nt structure in the longest arm.
TER1 nucleotides 415–507 are not required for TER1–Ccq1 association or telomerase catalytic activity
To understand the molecular defect that caused the ter1-Δ415–507 short telomere phenotype, we carried out a quantitative coimmunoprecipitation experiment to determine whether the 415–507 region in TER1 is required for the TER1–Ccq1 interaction. We immunoprecipitated Ccq1-3xFlag from both wild-type and TER1-Δ415–507 cells and quantified the amount of TER1 RNA and Ade6 RNA in the immunoprecipitate (Ade6 is an abundant mRNA that served as a specificity control) (Fig. 5A). Ccq1-Flag precipitated similar amounts of wild-type and TER1-Δ415–507 RNA. This interaction was specific, as the more abundant Ade6 mRNA was not preferentially immunoprecipitated (Fig. 5A; Supplemental Fig. 3). Thus, the Est1–TER1 interaction is not necessary for Ccq1–TER1 association either before or after telomerase associates with the telomere.
Figure 5.
TER1 nucleotides 415–507 are not required for TER1–Ccq1 interaction, but are necessary for peak late S-phase association of Trt1 and Est1 with the telomere. (A) RNA immunoprecipitation was performed with strains in which endogenous ter1 was disrupted and wild-type TER1 or TER1-Δ415–507 was expressed from the leu1 locus in a ccq1-G8-3Flag strain background. The no tag control strain was isogenic to the wild-type TER1-expressing strain. Data were analyzed by qRT–PCR. Relative fold enrichment is the ratio of amplification of a common region of TER1 IP/input to Ade6 mRNA IP/input. Error bars represent standard error for three independent reactions. Ccq1-G8-3Flag levels in the IP and input were visualized by α-Flag Western blot analysis. The asterisk indicates a nonspecific band. (B) Effect of ter1-Δ415–507 on Trt1-G8-13Myc and Est1-G8-13Myc telomere association in cycling cells. ChIP samples were analyzed as described in Figure1. Application of the one-tailed Student's t-test demonstrated significant difference (P < 0.05) between trt1-G8-13Myc and trt1-G8-13Myc, ter1-Δ415-507 (P = 0.016) and between est1-G8-13Myc and est1-G8-13Myc, ter1-Δ415-507 (P = 0.048). (C) Temperature-sensitive cdc25-22 cells expressing Trt1-G8-13Myc or lacking an epitope-tagged protein (no tag) were arrested in late G2 phase by growth at 37°C and then released to synchronous cell cycle by temperature shift to 24°C. Cells were harvested at 15-min intervals for ChIP and FACS analysis during growth at 24°C. In these strains, TER1-Δ415–507 RNA is expressed from the leu1 locus, and the endogenous ter1 locus is disrupted. ChIP samples were analyzed as described in Figure 1. Synchronicity of the strains was confirmed by FACS analysis. A similar profile was observed for the no tag control (data not shown) (n = 3). (D) Synchronous telomere-association ChIP profiles of cells expressing Est1-G8-13Myc or no epitope-tagged protein (no tag) were analyzed as described in C. (E) Levels of Est1-G8-13Myc were monitored during the cell cycle by α-Myc Western blotting of synchronous ChIP input samples.
We also performed an in vitro telomerase assay with immunoprecipitated Trt1-Myc to determine whether nucleotides 415–507 of TER1 affected the catalytic activity of telomerase. This possibility is not unprecedented, as telomerase RNA sequences distant from the templating region (e.g., pseudoknots) (Brown et al. 2007) influence telomerase catalytic activity in other systems. However, the in vitro telomerase activity was indistinguishable over a 90-min time course for extracts made from wild-type and ter1-Δ415–507 strains (Supplemental Fig. 4). Thus, nucleotides 415–507 of TER1 are not required for in vitro catalysis.
Est1 binds telomeres in late S phase and Trt1, and Est1 binding is reduced in ter1-Δ415–507 cells
Using ChIP, we found that both Trt1 and Est1 telomere binding were compromised in cycling ter1-Δ415–507 cells (Fig. 5B). To get a more accurate view of the impact of the ter1-Δ415–507 mutant on telomerase binding to telomeres, we carried out ChIP in cdc25-22 cells synchronized by arrest in late G2 phase by growth at high temperature. After release to the permissive temperature, cdc25-22 cells progressed into the next cell cycle, as shown by FACS analyses; consistent with other studies (Booher et al. 1989; Moreno et al. 1989), 70%–80% of the cells moved synchronously through the cell cycle (Fig. 5C,D bottom panels). We examined telomere binding of both Trt1-Myc and Est1-Myc in ter1 and ter1-Δ415–507 cells. As shown previously, Trt1-Myc binding occurred mainly in late S phase (Moser et al. 2009). Although this binding was reduced to ∼20% of wild-type levels in ter1-Δ415–507 cells, its timing was not affected (Fig. 5C). Peak Est1-Myc telomere association also occurred during late S phase, ∼105 min after release from the late G2 arrest (Fig. 5D), and, as with Trt1-Myc, this association was reduced to ∼15% in ter1-Δ415–507 cells without a change in timing. In wild-type cells, lower levels of Trt1-Myc and Est1-Myc were detected at telomeres at times other than late S phase (Fig. 5 C,D), which we attribute to incomplete synchrony. For example, the Est1-Myc peak in G1 phase was not significant and was not seen in all experiments. Western analysis of the synchronous input ChIP extracts demonstrated that Est1 function was not regulated by protein abundance (Fig. 5D). We conclude that the association, but not the timing, of Trt1 and Est1 with the telomere is highly compromised in ter1-Δ415–507 cells.
Est1 interacts with TER1 via the N-terminal 14-3-3-like domain
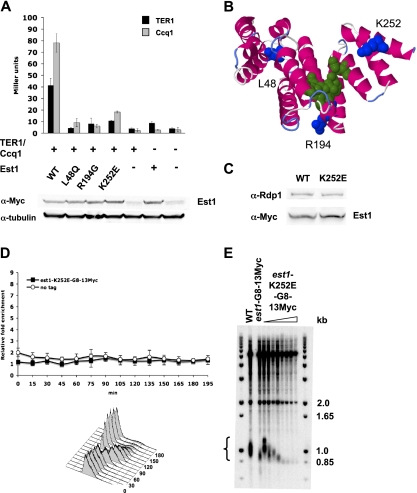
Because Est1 does not have an RNA-binding motif (Beernink et al. 2003), it is not clear from its sequence how it interacts with TER1. We used mutagenic PCR to generate mutations within the est1 ORF and tested their effects on the Est1–TER1 interaction with the RNA three-hybrid assay. Fourteen point mutants were identified that failed to interact with nucleotides 415–507 of TER1. Eleven of these alleles mapped to the N-terminal 14-3-3-like phophoserine-binding domain. Of the 14 mutants, only L48Q, R194G, and K252E produced wild-type levels of protein (Supplemental Fig. 5; data not shown). By the three-hybrid assay, each of the three mutants in the 14-3-3-like domain was severely impaired for TER1 binding (reduced to 10%–25% of wild-type levels) (Fig. 6A, black bars). Another group has also identified mutations in the Est1 14-3-3-like domain that do not interact with TER1 (T Nakamura, pers. comm.).
Figure 6.
Est1 telomere maintenance and binding requires the 14-3-3-like domain. (A) Quantification of TER1 and Ccq1 interaction with Est1 mutants by RNA three-hybrid and two-hybrid analysis. Wild-type Est1 and mutations in the 14-3-3-like domain of Est1 were generated by error-prone PCR. Wild-type and Est1 mutants were expressed in the pACT2 (three-hybrid analysis) and pGBKT7 (two-hybrid analysis) vectors. RNA three-hybrid analysis was used to monitor Est1 wild-type or mutant protein interaction with TER1, and two-hybrid analysis was used to monitor Est1 wild-type or mutant protein interaction with Ccq1. Nucleotides 415–507 of TER1 were expressed from the pIII/ms2-2 vector, and Ccq1 was expressed from pACT2. Quantification of interaction strengths in Miller units was determined by a β-galactosidase assay for both three-hybrid and two-hybrid analyses. Est1–TER1 interactions are depicted in black, and Est1–Ccq1 interactions are depicted in gray. Error bars represent standard deviation for n = 3. Stability of the mutant est1 proteins was demonstrated by Western blotting of lysates from two-hybrid analysis. α-tubulin levels are a loading control. (B) Predicted protein structure of the Est1 14-3-3-like domain (amino acids 1–260) generated by Phyre2 using normal modeling mode (Kelley and Sternberg 2009). Amino acids L48, R194, and K252 are colored blue. Conserved residues (R79, R180, Y181, and Q215) that are predicted to be involved in phosphoserine binding (Fukuhara et al. 2005) are colored green. (C) Western blotting demonstrates stability of Est1-G8-13Myc and Est1-K252E-G8-13Myc. (D) Est1-K252E-G8-13Myc does not associate with telomeres. Synchronous ChIP was performed on cells expressing Est1-K252E-G8-13Myc as described in Figure 5 (n = 3). (E) est1-K252E-G8-13Myc causes rapid telomere loss. Telomere blot of successive streaks of cells expressing Est1-K252E-G8-13Myc compared with wild-type telomere length and telomeres in cells expressing Est1-G8-13Myc.
The 14-3-3-like domain of Est1 also interacts with Ccq1. Using the two-hybrid assay, we found that each of the three Est1 mutants (L48Q, R194G, and K252E) were also defective for their interaction with Ccq1, with binding reduced to 10%–25% of wild-type levels (Fig. 6A, gray bars). We conclude that Est1 residues L48, R194, and K252 are necessary for full Est1–TER1 and Est1–Ccq1 interactions.
The Est1 K252E mutant does not bind telomeres and is telomerase-deficient
A strain with an integrated est1-K252E-G8-13Myc allele was used to determine the in vivo effects of this mutation. By Western blotting, Est1-K252E-Myc was stable in S. pombe (Fig. 6C). By ChIP, Est1-K252E-Myc did not bind telomeres (Fig. 6D). As in ccq1Δ cells (Tomita and Cooper 2008), telomeres were very short in est1-K252E-Myc cells and senesced by streak 7 (Fig. 6E; data not shown). Thus, the est1-K252E-Myc allele, which disrupts Est1 interaction with both TER1 and Ccq1, has a more severe phenotype than ter1-Δ415–507, which disrupts only the Est1–TER1 interaction.
Discussion
Here we show that Est1, a telomerase subunit that is essential for telomerase-mediated telomere lengthening, bound telomeres in late S phase, the same time as Trt1 binding (Moser et al. 2009). This cell cycle-dependent binding was not due to regulated Est1 abundance (Fig. 5E), as occurs in S. cerevisiae (Taggart et al. 2002; Osterhage et al. 2006). Est1 telomere association was completely dependent on the shelterin-like component Ccq1, occurring at background levels in ccq1Δ cells (Fig. 1). Likewise, Trt1 telomere binding is Ccq1-dependent (Fig. 1; Tomita and Cooper 2008). These results demonstrate that Ccq1is a component of the only pathway that recruits both subunits of the holoenzyme to the telomere.
Ccq1–Est1 binding is almost surely direct, as Ccq1 and Est1 interacted in S. cerevisiae (Fig. 6A), which is unlikely to express a hypothetical bridging protein. As this study was being reviewed, another group reported that Est1 and Ccq1 interact and showed that this interaction requires Ccq1 phosphorylation (Moser et al. 2011). Their results are consistent with our finding that the Ccq1–Est1 interaction was disrupted by mutations in the Est1 14-3-3-like domain (Fig. 6A), a domain that interacts with phosphorylated serine in other contexts (Fukuhara et al. 2005). Because 14-3-3-like domains occur in other Est1 family proteins, including human Est1, this region may also connect telomerase to telomeres in multicellular organisms.
Unlike ScEst1, S. pombe and human Est1 lack a canonical RNA interaction motif. Nonetheless, Est1 interacted specifically with TER1 RNA (Fig. 2). Again, because this interaction occurred in S. cerevisiae, as well as in S. pombe (Fig. 2), it is likely direct, although we cannot exclude the possibility of a heterologous bridging protein. Est1 associated with nucleotides 415–507, which form the end of one of the long quasihelical arms in the predicted TER1 secondary structure (Fig. 2A). In addition to being required for the Est1–TER1 interaction, the Est1 14-3-3-like domain was also required for the Est1–Ccq1interaction (Fig. 6A). Indeed, the same mutations that disrupted Est1–TER1 binding also disrupted Est1–Ccq1 interaction. To our knowledge, this is the first example of a 14-3-3-like domain-binding RNA. Thus, the 14-3-3-like motif mediates RNA–protein and protein–protein interactions. This finding suggests that 14-3-3-like domains in other proteins, such as the nonsense-mediated mRNA decay proteins SMG5, SMG7, and dual function SMG6/EST1A, may also interact with RNA.
The est1-K252E allele that eliminated Est1 interaction with both TER1 and Ccq1 had short telomeres, and cells eventually senesced. However, the senescence rate in the est1-K252E cells was slower than senescence in trt1Δ, est1Δ, or ter1Δ cultures (Fig. 6E; Nakamura et al. 1997; Beernink et al. 2003; Leonardi et al. 2008; Webb and Zakian 2008) but similar to the rate in ccq1Δ cells (Tomita and Cooper 2008). Since eliminating the Est1–TER1 interaction with the ter1-Δ415–507 allele did not cause senescence, we hypothesize that the most likely explanation for the telomerase-deficient behavior of est1-K252E cells is the loss of the Est1–Ccq1 interaction. Thus, when Ccq1 is telomere-associated but unable to interact with Est1, the holoenzyme no longer binds telomeres, which can explain the telomerase-deficient phenotype of est1-K252E. This finding suggests that it is the Ccq1–Est1 interaction, not the Ccq1–Trt1 association, that is essential for telomerase–telomere interaction. This interpretation is consistent with the observation that Est1 still associated with telomeres in ter1Δ cells, but Trt1 did not (Fig. 1).
Budding and fission yeast Est1 contacts telomerase RNA by very different protein motifs: the RRM domain in ScEst1 (Zhou et al. 2000) and, as shown here, the 14-3-3-like domain in S. pombe (Fig. 6). Given this difference, it is surprising that there is considerable conservation between the two distantly related yeasts in the RNA secondary structures that are important for Est1 binding. In S. pombe, Est1 interacts with the terminal stem–loop at the tip of a long arm (449–460 nt) in a structural, rather than sequence-dependent, manner (Fig. 4). The presence and specific sequence of a bulged adenosine (A437), which is predicted to lie 2 nt 5′ of the large unpaired region of the S. pombe TER1, was also necessary for full Est1 binding (Fig. 4). A similarly positioned bulge region is critical for ScEst1–TLC1 interaction (Seto et al. 2002).
In synchronous ter1-Δ415–507 cells, Est1 association in S phase was only 15% of wild-type levels (Fig. 5D). Trt1 binding was similarly reduced (Fig. 5C), and telomeres were very short (Fig. 3B). Given these findings, it is surprising the ter1-Δ415–507 cells did not senesce. In contrast, disrupting the TLC1–ScEst1 interaction results in telomere loss and senescence (Seto et al. 2002). The low telomerase binding in ter1-Δ415–507 cells was not due to reduced Ccq1–TER1 binding (Fig. 5A; Supplemental Fig. 3). Because the Trt1–Ccq1 association is TER1 independent (Tomita and Cooper 2008), and Trt1 interacted normally with TER1-Δ415–507 RNA (Fig. 2C), defects in these interactions also do not explain our results. Thus, the ter1-Δ415–507 allele is a separation-of-function allele that eliminates the physical connection between Est1 and the catalytic subunit, which only interact through TER1 (Leonardi et al. 2008), but retains the Est1–Ccq1 direct interaction and the Trt1–Ccq1 association.
We favor the following model to explain our data. Telomerase recruitment to telomeres is mediated by the interaction of the Est1 14-3-3-like domain with Ccq1. We hypothesize that this interaction is required for the subsequent association of Trt1 with the telomere. Consistent with this model, Ccq1 T93 phosphorylation by Tel1 (ATM) and/or Rad3 (ATR) is a critical first step in telomerase–telomere association, as this phosphorylation is required for Ccq1–Est1 interaction (Moser et al. 2011). After the telomerase holoenzyme is recruited to the telomere, we suggest that the TER1–Est1 interaction tethers Trt1 and Est1 to the telomere. Upon holoenzyme recruitment in late S phase, a conformational change in the shelterin-like complex occurs that makes the end of the chromosome accessible to telomeric repeat addition by Trt1. It is during this rearrangement, which transforms the end of the telomere from a protected to unprotected telomerase-receptive state, that the telomeric coassociation of Est1 and Trt1 by TER1 is required. Unlike Trt1, Est1 telomere association is not completely dependent on TER1 (Fig. 1). However, in ter1-Δ415–507 cells, Trt1 associates with TER1 (Fig. 2), but not Est1, and associates with telomeres to the same suboptimal levels as Est1 (Fig. 5C,D). Therefore, TER1-mediated Est1–TER1–Trt1 tethering promotes proper holoenzyme telomere association and telomere length. Our model is consistent with the possibility that the Est1–Ccq1 and Est1–TER1 interactions are mutually exclusive, with the initial Ccq1–Est1 telomerase recruitment separated from the latter Est1–Trt1 tethering function by switching from Est1–Ccq1 to Est1–TER1 binding. We note that in the predicted structure for the Est1 14-3-3-like domain, the residues identified in this study (L48, R194, and K252) surround the putative phosphoserine-binding pocket (Fig. 6B), which is consistent with an RNA interaction that blocks protein–protein interaction. Conceivably, the Est1 switch may be part of a checkpoint to verify the proximity of telomerase before the telomere is deprotected to allow repeat addition.
Materials and methods
Yeast strains
Standard molecular genetic techniques were used to generate the fission yeast strains used in this study. The pJK148-integrating vector was used to express wild-type ter1 and ter1 alleles from the leu1 locus using the endogenous ter1 promoter. Integration of alleles was confirmed by Southern blotting.
Nucleic acid manipulation
Plasmids used in RNA three-hybrid analysis included pACT2 to express protein partners and pIII/MS2-2 (SenGupta et al. 1996) to express the hybrid TER1 RNA fragments. RT–PCR amplification and subcloning were used to create est1/pACT2. Standard PCR and overlapping PCR were used to create the pIII/MS2-2/ter1-1036–1095 and pIII/MS2-2/ter1-415–507 wild-type and allele plasmids for expression in the L40 RNA three-hybrid strain (Zhang et al. 1999). Plasmid inserts were confirmed by sequencing. ccq1/pACT2 and est1/pGBKT7 vectors were generated for two-hybrid analysis. Telomeric DNA Southern blotting and Northern blotting were performed as previously described (Webb and Zakian 2008).
RNA three-hybrid and protein–protein two-hybrid analyses
RNA three-hybrid analysis was performed in the L40 S. cerevisiae strain (MATa ura3-52 leu2-3,112 his3200 trp11 ade2 LYS2::(lexAop)-HIS3 ura3::(lexAop)-lacZ) (Zhang et al. 1999). Identification of Est1 TER1-415–507 binding-deficient mutants by the RNA three-hybrid system was performed by screening ∼7000 transformants generated by gap repair of error-prone PCR (Cirino et al. 2003) templated from est1 cDNA (Roche). Plasmid DNA was recovered and sequenced from independent transformants that exhibited reduced lacZ reporter activation. Eighty percent (11) of the mutations that failed to activate the reporter mapped to the N-terminal 14-3-3-like domain of Est1. Of these 11 alleles, the protein products of three (L48Q, R194G, and K252E) were stably expressed. The pJ69-A strain (MATa trp1-901 leu2-3,1112 ura3-52 his3-200 gal4Δ gal80Δ GAL2-ADE2 LYS2::GAL1-HIS3 MET2::GAL7-LacZ) was used in two-hybrid analysis (James et al. 1996).
ChIP
ChIP was performed as described (Taggart et al. 2002) with the following modifications: Fifty milliliters of cells was cross-linked with 1% formaldehyde, washed two times with TBS (20 mM Tris-HCl at pH 7.5, 150 mM NaCl), and stored as a pellet at −80°C after collection. Pellets were resuspended in ChIP lysis buffer (50 mM Hepes/KOH at pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% Na-deoxycholate, 2 mM PMSF, Complete Mini EDTA-free protease inhibitor [Roche]) and lysed with glass beads in a FastPrep-24 machine (MP Biomedicals) until >95% of cells were lysed. Immunoprecipitation and bead capture incubations were performed for 2 h. After bead capture, each washing step was performed twice. est1-G8-13Myc and trt1-G8-13Myc epitope-tagged alleles used in these assays were previously described (Webb and Zakian 2008). The primers used to amplify the telomere-adjacent region STE1 and act1 gene by real-time PCR analysis were as described in Tomita and Cooper (2008). The presence of telomeres in senescing strains was confirmed by Southern blotting.
RNA immunoprecipitation
Coimmunoprecipitation of wild-type TER1 or Δ415–507 mutant RNAs with Ccq1-G8-3xFlag was performed as previously described (Webb and Zakian 2008) with minor modifications. Immunoprecipitation was performed in TMG300 buffer (10 mM Tris-HCl at pH 8.0, 1 mM MgCl2, 10% [v/v] glycerol, 1 mM EDTA, 100 mM NaCl, 0.1 mM DTT) in the presence of Complete Mini EDTA-free protease inhibitors (Roche), 2 mM PMSF, RNasin (Promega), and SUPERasein (Ambion). Extract was precleared with α-Myc agarose-conjugated beads (Sigma) for 1 h at 4°C before α-Flag agarose-conjugated bead (Sigma) incubation for 1 h at 4°C. Recovered coimmunoprecipitated RNAs were analyzed by qRT–PCR using the iScript One-Step RT–PCR kit with SYBR Green (Bio-Rad) as directed.
In vitro telomerase activity assay
These assays were performed as described (Beernink et al. 2003), with minor modifications. Instead of a mortar and pestle, cells were ground in a 6750 Freezer Mill (SPEX CertiPrep) five times for 2 min each with 2-min rest intervals. A total of 6.75 mg of extract was used in the assays. Washing was performed with a Nutator (BD Clay Adams) for 5 min.
Acknowledgments
We thank T.M. Nakamura for sharing results before publication, C.J. DeCoste for assistance in flow cytometry and cell counter, and M.L. Bochman, N. Sabouri, and Y. Wu for comments on the manuscript. This work was supported by U.S. National Institutes of Health (NIH) grants GM043265 and ARRA supplement. C.J.W. was supported by a U.S. National Research Service Award and American Cancer Society Post-doctoral Fellowship.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.181826.111.
References
- Beernink HT, Miller K, Deshpande A, Bucher P, Cooper JP 2003. Telomere maintenance in fission yeast requires an Est1 ortholog. Curr Biol 13: 575–580 [DOI] [PubMed] [Google Scholar]
- Booher RN, Alfa CE, Hyams JS, Beach DH 1989. The fission yeast cdc2/cdc13/suc1 protein kinase: Regulation of catalytic activity and nuclear localization. Cell 58: 485–497 [DOI] [PubMed] [Google Scholar]
- Brown Y, Abraham M, Pearl S, Kabaha MM, Elboher E, Tzfati Y 2007. A critical three-way junction is conserved in budding yeast and vertebrate telomerase RNAs. Nucleic Acids Res 35: 6280–6289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan A, Boule JB, Zakian VA 2008. Two pathways recruit telomerase to Saccharomyces cerevisiae telomeres. PLoS Genet 4: e1000236 doi: 10.1371/journal.pgen.1000236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirino PC, Mayer KM, Umeno D 2003. Generating mutant libraries using error-prone PCR. Methods Mol Biol 231: 3–9 [DOI] [PubMed] [Google Scholar]
- Dandjinou AT, Levesque N, Larose S, Lucier JF, Abou Elela S, Wellinger RJ 2004. A phylogenetically based secondary structure for the yeast telomerase RNA. Curr Biol 14: 1148–1158 [DOI] [PubMed] [Google Scholar]
- de Lange T 2010. How shelterin solves the telomere end-protection problem. Cold Spring Harb Symp Quant Biol 75: 167–177 [DOI] [PubMed] [Google Scholar]
- Evans SK, Lundblad V 1999. Est1 and Cdc13 as comediators of telomerase access. Science 286: 117–120 [DOI] [PubMed] [Google Scholar]
- Fisher TS, Taggart AK, Zakian VA 2004. Cell cycle-dependent regulation of yeast telomerase by Ku. Nat Struct Mol Biol 11: 1198–1205 [DOI] [PubMed] [Google Scholar]
- Fukuhara N, Ebert J, Unterholzner L, Lindner D, Izaurralde E, Conti E 2005. SMG7 is a 14-3-3-like adaptor in the nonsense-mediated mRNA decay pathway. Mol Cell 17: 537–547 [DOI] [PubMed] [Google Scholar]
- Hsu M, Yu EY, Singh SM, Lue NF 2007. Mutual dependence of Candida albicans Est1p and Est3p in telomerase assembly and activation. Eukaryot Cell 6: 1330–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144: 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJ 2009. Protein structure prediction on the Web: A case study using the Phyre server. Nat Protoc 4: 363–371 [DOI] [PubMed] [Google Scholar]
- Lendvay TS, Morris DK, Sah J, Balasubramanian B, Lundblad V 1996. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics 144: 1399–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardi J, Box JA, Bunch JT, Baumann P 2008. TER1, the RNA subunit of fission yeast telomerase. Nat Struct Mol Biol 15: 26–33 [DOI] [PubMed] [Google Scholar]
- Livengood AJ, Zaug AJ, Cech TR 2002. Essential regions of Saccharomyces cerevisiae telomerase RNA: Separate elements for Est1p and Est2p interaction. Mol Cell Biol 22: 2366–2374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell JR, Cheng J, Collins K 1999. A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol Cell Biol 19: 567–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi T, Kanoh J, Saito M, Ishikawa F 2008. Fission yeast Pot1-Tpp1 protects telomeres and regulates telomere length. Science 320: 1341–1344 [DOI] [PubMed] [Google Scholar]
- Moreno S, Hayles J, Nurse P 1989. Regulation of p34cdc2 protein kinase during mitosis. Cell 58: 361–372 [DOI] [PubMed] [Google Scholar]
- Moser BA, Subramanian L, Chang YT, Noguchi C, Noguchi E, Nakamura TM 2009. Differential arrival of leading and lagging strand DNA polymerases at fission yeast telomeres. EMBO J 28: 810–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser BA, Chang YT, Kosti J, Nakamura TM 2011. Tel1ATM and Rad3ATR kinases promote Ccq1–Est1 interaction to maintain telomeres in fission yeast. Nat Struct Mol Biol 18: 1408–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura TM, Morin GB, Chapman KB, Weinrich SL, Andrews WH, Lingner J, Harley CB, Cech TR 1997. Telomerase catalytic subunit homologs from fission yeast and human. Science 277: 955–959 [DOI] [PubMed] [Google Scholar]
- Osterhage JL, Talley JM, Friedman KL 2006. Proteasome-dependent degradation of Est1p regulates the cell cycle-restricted assembly of telomerase in Saccharomyces cerevisiae. Nat Struct Mol Biol 13: 720–728 [DOI] [PubMed] [Google Scholar]
- Peterson SE, Stellwagen AE, Diede SJ, Singer MS, Haimberger ZW, Johnson CO, Tzoneva M, Gottschling DE 2001. The function of a stem-loop in telomerase RNA is linked to the DNA repair protein Ku. Nat Genet 27: 64–67 [DOI] [PubMed] [Google Scholar]
- Reichenbach P, Hoss M, Azzalin CM, Nabholz M, Bucher P, Lingner J 2003. A human homolog of yeast Est1 associates with telomerase and uncaps chromosome ends when overexpressed. Curr Biol 13: 568–574 [DOI] [PubMed] [Google Scholar]
- Robart AR, O'Connor CM, Collins K 2010. Ciliate telomerase RNA loop IV nucleotides promote hierarchical RNP assembly and holoenzyme stability. RNA 16: 563–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- SenGupta DJ, Zhang B, Kraemer B, Pochart P, Fields S, Wickens M 1996. A three-hybrid system to detect RNA-protein interactions in vivo. Proc Natl Acad Sci 93: 8496–8501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto AG, Zaug AJ, Sobel SG, Wolin SL, Cech TR 1999. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature 401: 177–180 [DOI] [PubMed] [Google Scholar]
- Seto AG, Livengood AJ, Tzfati Y, Blackburn EH, Cech TR 2002. A bulged stem tethers Est1p to telomerase RNA in budding yeast. Genes Dev 16: 2800–2812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taggart AK, Teng SC, Zakian VA 2002. Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science 297: 1023–1026 [DOI] [PubMed] [Google Scholar]
- Tomita K, Cooper JP 2008. Fission yeast Ccq1 is telomerase recruiter and local checkpoint controller. Genes Dev 22: 3461–3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson RL, Ziegler TD, Supakorndej T, Terns RM, Terns MP 2006. Cell cycle-regulated trafficking of human telomerase to telomeres. Mol Biol Cell 17: 955–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuzon CT, Wu Y, Chan A, Zakian VA 2011. The Saccharomyces cerevisiae telomerase subunit Est3 binds telomeres in a cell cycle- and Est1-dependent manner and interacts directly with Est1 in vitro. PLoS Genet 7: e1002060 doi: 10.1371/journal.pgen.1002060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfati Y, Fulton TB, Roy J, Blackburn EH 2000. Template boundary in a yeast telomerase specified by RNA structure. Science 288: 863–867 [DOI] [PubMed] [Google Scholar]
- Webb CJ, Zakian VA 2008. Identification and characterization of the Schizosaccharomyces pombe TER1 telomerase RNA. Nat Struct Mol Biol 15: 34–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe MB 2002. How do 14-3-3 proteins work?—Gatekeeper phosphorylation and the molecular anvil hypothesis. FEBS Lett 513: 53–57 [DOI] [PubMed] [Google Scholar]
- Zappulla DC, Cech TR 2004. Yeast telomerase RNA: A flexible scaffold for protein subunits. Proc Natl Acad Sci 101: 10024–10029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappulla DC, Cech TR 2006. RNA as a flexible scaffold for proteins: Yeast telomerase and beyond. Cold Spring Harb Symp Quant Biol 71: 217–224 [DOI] [PubMed] [Google Scholar]
- Zhang B, Kraemer B, SenGupta D, Fields S, Wickens M 1999. Yeast three-hybrid system to detect and analyze interactions between RNA and protein. Methods Enzymol 306: 93–113 [DOI] [PubMed] [Google Scholar]
- Zhou J, Hidaka K, Futcher B 2000. The Est1 subunit of yeast telomerase binds the Tlc1 telomerase RNA. Mol Cell Biol 20: 1947–1955 [DOI] [PMC free article] [PubMed] [Google Scholar]