Bacterial type I DNA restriction/modification (RM) enzymes are capable of restricting horizontal gene transfer in pathogenic bacteria. This is particularly significant, as such enzymes prevent the spread of antibiotic resistance genes, such as MRSA. Employing electron microscopy and molecular modeling integrated with an array of available biochemical data, Dryden and colleagues analyze the structures of the EcoKI and EcoR124 enzymes and shed light on their mechanism of action. Particular insight is provided into the large-scale conformational changes undergone upon DNA sequence recognition and the subsequent long-distance bidirectional translocation to their cleavage sites.
Keywords: DNA restriction enzymes, DNA translocation, electron microscopy, small-angle scattering, molecular modeling, DNA translocases
Abstract
Type I DNA restriction/modification (RM) enzymes are molecular machines found in the majority of bacterial species. Their early discovery paved the way for the development of genetic engineering. They control (restrict) the influx of foreign DNA via horizontal gene transfer into the bacterium while maintaining sequence-specific methylation (modification) of host DNA. The endonuclease reaction of these enzymes on unmethylated DNA is preceded by bidirectional translocation of thousands of base pairs of DNA toward the enzyme. We present the structures of two type I RM enzymes, EcoKI and EcoR124I, derived using electron microscopy (EM), small-angle scattering (neutron and X-ray), and detailed molecular modeling. DNA binding triggers a large contraction of the open form of the enzyme to a compact form. The path followed by DNA through the complexes is revealed by using a DNA mimic anti-restriction protein. The structures reveal an evolutionary link between type I RM enzymes and type II RM enzymes.
An important goal in understanding large molecular machines is determination of their molecular architecture, as this provides the foundation for understanding their mechanism. It is >40 years since the ∼440-kDa multifunctional type I DNA restriction/modification (RM) enzymes were first purified (Linn and Arber 1968; Meselson and Yuan 1968). Their discovery, and particularly their ability to cut dsDNA, led researchers onto the track of the site-specific type II restriction endonucleases and helped to usher in the era of genetic engineering (Loenen 2003).
These type I RM enzymes are found in over half of bacterial genomes, including many pathogens (Roberts et al. 2010), and play a role in controlling horizontal gene transfer (HGT) (Murray 2002; Waldron and Lindsay 2006). This is of special importance in the spread of antibiotic resistance in pathogens, such as multidrug-resistant Staphylococcus aureus (MRSA), where the Sau1 type I RM system appears to regulate HGT (Waldron and Lindsay 2006). The barrier to HGT provided by the numerous variants of Sau1 appears to define the clonal structure of S. aureus populations around the world.
So important is the apparent role of type I RM enzymes as a barrier to HGT that mobile genetic elements, including phage and conjugative transposons and plasmids, have developed an extensive range of anti-restriction measures to overcome the RM barrier (Tock and Dryden 2005). Anti-restriction measures directed against type II RM systems are relatively simple, involving a reduction in the number of target sequences on their mobile genome or the incorporation of modified bases. Anti-restriction measures against the type I RM systems are much more diverse and sophisticated and include the production of a range of specialized anti-restriction proteins and enzymes, including mimics of DNA structure (Walkinshaw et al. 2002; McMahon et al. 2009). These DNA mimics are highly charged and elongated proteins capable of binding more tightly to the type I RM enzyme than even its DNA target sequence.
In order to achieve a better understanding of fundamental aspects of HGT, further knowledge of the structure and mechanism of type I RM systems is essential. In contrast to the situation with type II restriction enzymes, it is only very recently that partial atomic structures for type I RM subunits have emerged (Calisto et al. 2005; Kim et al. 2005; Obarska et al. 2006; Obarska-Kosinska et al. 2008; Kennaway et al. 2009; Lapkouski et al. 2009; Uyen et al. 2009; Taylor et al. 2010; Gao et al. 2011), and their mechanisms, which most dramatically involve the translocation of thousands of base pairs of DNA with the formation of supercoiled loops (Yuan et al.1980; Endlich and Linn 1985; Studier and Bandyopadhyay 1988; García and Molineux 1999), still present many questions. The type I RM enzymes are complex structures with two HsdR restriction (R) subunits, two HsdM modification (M) subunits, and one HsdS sequence specificity (S) subunit (Murray 2000; Loenen 2003), each with a number of domains (Supplemental Fig. S1).
The HsdS contain two DNA target recognition domains (TRDs), one for each part of the bipartite recognition sequence, joined by two α helices in an antiparallel arrangement. The HsdS structure displays pseudo twofold symmetry (Calisto et al. 2005; Kennaway et al. 2009; Kim et al. 2005; Gao et al. 2011), and this symmetry is imposed on the whole structure of these enzymes (Kneale 1994; Dryden et al. 1995; Davies et al. 1999; Kennaway et al. 2009). The two HsdM lie on either side of the HsdS to form the M2S1 methyltransferase (MTase) core. The HsdM contain a methyltransferase catalytic domain. The two HsdR are located on either side of the MTase core and contain a nuclease domain and an ATP-hydrolyzing DNA translocation “motor” domain (Powell et al. 1998; Davies et al. 1999).
The MTase methylates specific adenine bases in the target sequences on hemimethylated, newly replicated host DNA (Dryden 1999; Madhusoodanan and Rao 2010), while the HsdR are switched on only when unmodified target sequences are detected on incoming DNA. Although unmodified targets can appear on host DNA during times of cell stress, restriction of host DNA is prevented by “restriction alleviation” (Makovets et al. 2004). The restriction reaction appears to work by fixing the enzyme to the recognition sequence and initiating an extended period of ATPase-driven DNA translocation (Seidel et al. 2008), which pulls DNA in toward the enzyme from both sides of the target site. Loops of DNA are extruded, and when the translocation stalls, the DNA is cleaved at the stall site rather than the target sequence. As the enzyme tracks the DNA helix, supercoiling can build up in the extruded loops. How the enzyme can translocate for up to 50,000 base pairs (bp) (García and Molineux 1999) and at speeds of up to 1000 bp per second (Seidel et al. 2004) in the face of the torsional stress induced by this supercoiling is not known, although it seems that the enzyme makes many abortive attempts to initiate the translocation (McClelland et al. 2005).
We now present a structure of EcoKI (specificity sequence target AACNNNNNNGTGC), the archetypal type I RM enzyme found on the chromosome of Escherichia coli K12, and EcoR124I (specificity sequence target GAANNNNNNRTCG), found on a conjugative plasmid, as determined by reconstruction from electron microscopy (EM) and single-particle analysis together with small-angle X-ray scattering (SAXS) and small-angle neutron scattering (SANS). The structures show EcoKI bound to a DNA duplex or to a DNA mimic anti-restriction protein, and EcoR124I in the absence and presence of a DNA duplex or an anti-restriction protein. With the aid of numerous biochemical constraints, known crystallographic structures of type I RM subunits, the location of the HsdS and HsdM within the MTase at low resolution (Callow et al. 2007; Taylor et al. 2010), and our recent reconstruction of the core MTase structure (Kennaway et al. 2009), we can construct a unique arrangement of the subunits within the EM structures, rationalize all previous knowledge about these complex machines, and reveal an evolutionary link between type I and type II RM systems.
Results
We analyzed here a variety of complexes with EM, SANS, and SAXS; namely, EcoR124I with bound DNA (30 bp), EcoR124I without DNA, EcoKI with bound DNA (75 bp), and EcoR124I with bound Ocr anti-restriction protein. It is important to note that, in the absence of ATP hydrolysis, EcoKI is a stable assembly containing all five protein subunits (Dryden et al. 1997; Roberts et al. 2011). However, for EcoR124I, the binding constants for each of the two HsdR differ by over two orders of magnitude (0.6 nM and 200 nM, respectively), and the two forms of the enzyme, R1M2S1 and R2M2S1, are in dynamic equilibrium (Supplemental Material; Supplemental Fig. S3; Janscak et al. 1998; Mernagh et al. 1998). The R1M2S1 form is active in translocation but not DNA cleavage (Janscak et al. 1998). At the micromolar concentrations used for the SAXS and SANS experiments, dissociation of the R2M2S1 complex is minimal, both with and without bound DNA; however, at the ∼100 nM concentrations used for EM, there will be a considerable fraction of EcoR124I in the R1M2S1 form. In all experiments using 1:1 protein:DNA complexes, the concentrations are above the binding constants for DNA; hence, free enzyme and free DNA concentrations will be very low.
Negative stain EM of EcoR124I
Single-particle analysis of negative stain EM images showed large differences between DNA-bound (Fig. 1A; Supplemental Fig. S2A,B) and unbound EcoR124I enzymes (Fig. 1B; Supplemental Fig. 2C,D), with their longest dimensions being ∼18 nm versus ∼22–26 nm, respectively. (Smaller particles were the R1M2S1 form and were analyzed separately as described later.) We refer here to the form of EcoR124I with DNA as the “closed” form and the form without DNA as the “open” form, following the nomenclature used for the two states of the EcoR124I MTase (with and without bound DNA) on the basis of SAXS analysis (Taylor et al. 1994). Apparent twofold symmetry was visible in many image averages.
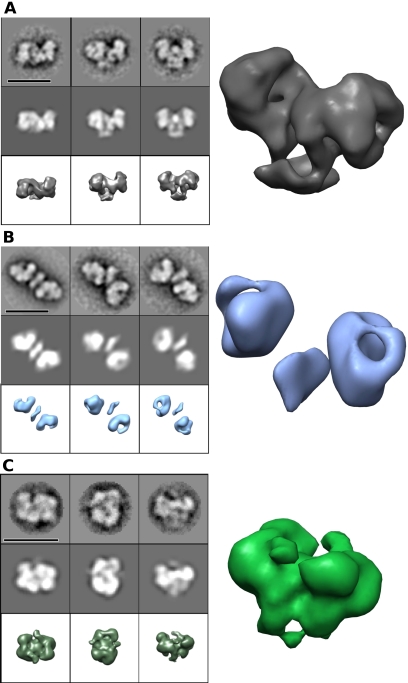
Figure 1.
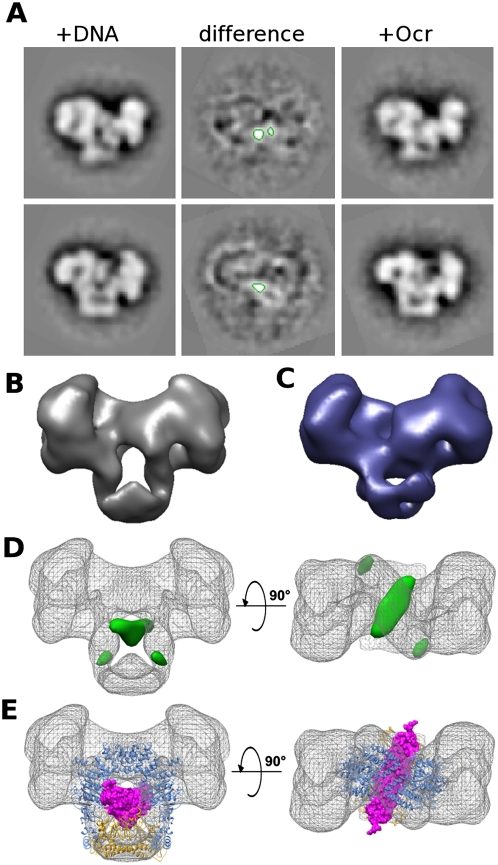
Gallery of type I RM structures and conformations determined by EM and single-particle analysis. (A) EcoR124I+DNA (closed state) negative stain EM. (B) EcoR124I without DNA (open state) negative stain EM. (C) EcoKI+DNA negative stain EM. For each 3 × 3 panel, the top rows are image averages, the middle rows are their corresponding reprojections, and the bottom rows are 3D surface views of the 3D reconstruction (bars, 200 Å); on the right is a larger 3D surface perspective view. Supporting EM data can be found in Supplemental Figure S2. Data on the assembly of the EcoR124I enzyme can be found in Supplemental Figure S3.
A three-dimensional (3D) reconstruction was generated (Fig. 1A; Supplemental Fig. S2A,B) of the EcoR124I RM enzyme bound to a 30-bp dsDNA fragment containing its unmethylated recognition site (Supplemental Material; Supplemental Fig. S3). This map has a resolution of ∼2.1 nm (by 0.5 Fourier shell correlation [FSC]) and is of good overall quality, due to it having a reasonable range of orientations on the grid (although still mostly limited to a single axis of rotation) and little or no flexibility. Twofold rotational symmetry was imposed during the refinement due to the apparent symmetry seen in the two-dimensional (2D) analysis and the twofold pseudosymmetry of the assembly imposed by previous experimental and theoretical work using sequence analysis (Kneale 1994; Dryden et al. 1995), domain-swapping experiments (Fuller-Pace et al. 1984; Gann et al. 1987; Cowan et al. 1989; Abadjieva et al. 1993; Meister et al. 1993), and three crystal structures of HsdS from different species (Calisto et al. 2005; Kim et al. 2005; Gao et al. 2011).
The DNA was not visible in our negative stain images. The same problem has recently been observed in images of the p53 protein in complex with DNA (Melero et al. 2011). This lack of DNA staining is often the case at stain depths typical for protein staining, as it is thought that the heavy metals bind in the DNA grooves, matching the negative contrast (Griffith 1978). However, sometimes the DNA is visible (Mayanagi et al. 2011), so the absence or presence seems to depend on the particular system investigated.
EcoR124I without DNA in negative stain was highly extended and more flexible with a very limited range of orientations on the carbon film (Fig. 1B; Supplemental Fig. S2C,D). Most particles (∼80%) appeared to have their twofold axis roughly normal to the plane of the carbon film. A low-resolution (∼3.5-nm) 3D reconstruction of the EcoR124I RM complex without DNA was generated. The angular range was limited due to the shape of the complex, and this limits the resolution of the open conformation. Very thin connections between the domains could be seen in some negative stain images, but these are not well resolved in the subsequent 3D map due to the low resolution. These thin linkers are likely pivot points for flexing to allow the enzyme to close up. A very small proportion of these particles (<5%) were seen to be folded up into the closed state, indicating a dynamic equilibrium between states in the absence of cognate DNA.
Comparing unbound “open” EcoR124I with the DNA-bound “closed” state shows a very large movement of the subunits within the assembly (Fig. 1A,B). It is possible that the carbon film affects the unbound open state and that it has a more variable conformation in solution. However, the twofold symmetry seen in many of the particles suggests that this open conformation is relatively stable, given that random conformational variability would break the symmetry. Opening and closing by type I RM enzymes in such a manner must allow entry of DNA substrate. SAXS and SANS data previously showed that the EcoR124I MTase collapsed from an extended structure to a more compact form in the presence of DNA (Taylor et al. 1994, 2010), as also shown by the almost complete protection from proteolysis when the MTase was bound to DNA (Webb et al. 1995). The recent EM analysis of the EcoKI MTase also suggested an opening and closing cycle for DNA binding but of lesser magnitude than for EcoR124I (Kennaway et al. 2009).
Negative stain EM of EcoKI
Negatively stained particles of EcoKI with DNA bound (Fig. 1C; Supplemental Fig. S2E,F) appeared smaller than EcoR124I with DNA (∼16 nm long), and the orientations observed on the carbon grids had little similarity to those of EcoR124I. Once again, the DNA was not visible in negative stain even though a larger 75-bp duplex was used. EcoKI particles showed some variability, particularly in peripheral regions. Approximate twofold symmetry was apparent in some views of EcoKI, and a wider range of views were seen. This may indicate a more rounded shape than EcoR124I, allowing adsorption of EcoKI to the carbon film in a variety of orientations. The 3D reconstruction of negatively stained EcoKI with a 75-bp fragment of dsDNA displayed a compact structure with many features similar to EcoR124I with DNA, including recognizable density for the five subunits in a matching arrangement, suggesting a common architecture for type I RM enzymes. EcoKI also adopted a compact form in absence of DNA when examined by EM (the particles appeared to be identical) and did not appear elongated, as seen for EcoR124I without DNA (data not shown). The dynamic equilibrium between open and closed forms apparently favors the closed form for EcoKI under the conditions used for EM.
Small-angle scattering experiments on EcoR124I and EcoKI
In order to determine the subunit organization of EcoR124I and EcoKI, SAXS and SANS were performed in the absence of DNA. The techniques are complementary, with SANS uniquely allowing the spatial location of subunits to be determined by matching out one or another of the components by specific perdeuteration, together with contrast variation using a range of H2O/D2O mixtures.
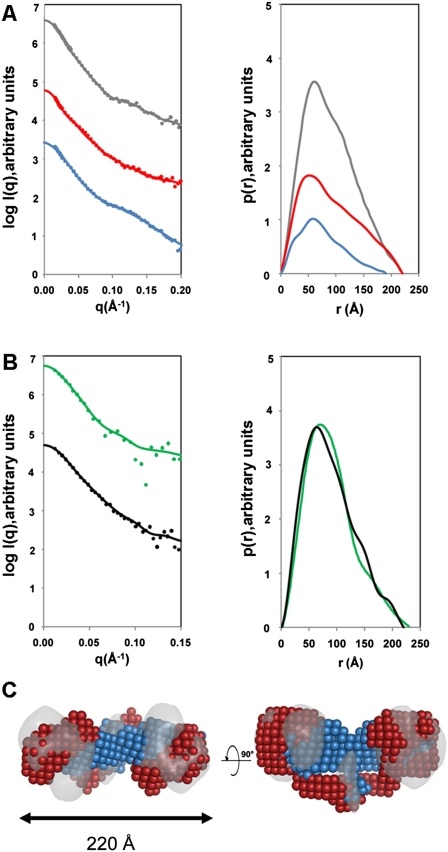
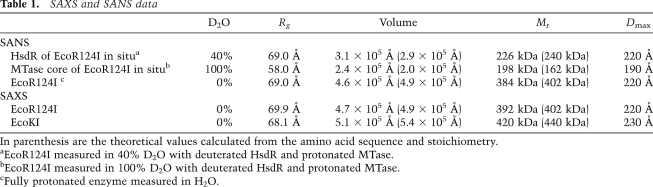
For SANS measurements, EcoR124I was prepared in either a fully protonated state or a perdeuterated state in which HsdR was deuterated and the MTase was protonated. The HsdR were deuterated to a level of 75%, such that the contrast match point of 100% D2O was achieved. SANS measurements were made in buffers containing 0%, 40%, and 100% D2O (Fig. 2A). The left panel of Figure 2A shows the scattering data, and the right panel of Figure 2A shows the derived pair distribution function, p(r). In 0% D2O, a radius of gyration (Rg) of 69 Å for the fully protonated EcoR124I was determined (Table 1). Fourier transformation of the scattering curve into a pair distribution function [p(r)] showed that the complex had a maximum dimension (Dmax) of 220 Å (right panel of Fig. 2A). Similar sets of measurements were carried out in 40% D2O for the perdeuterated complex in which the MTase core was matched out and only the scattering due to the two HsdR was visible. The Rg and Dmax were essentially unchanged, indicating that each HsdR must be positioned at either end of the RM enzyme, separated by the MTase core, consistent with DNA footprinting data (Mernagh et al. 1998; Powell et al. 1998). Finally, scattering data were collected in 100% D2O in which the HsdR were matched out, so only the scattering from the MTase was measured. An Rg of 58 Å and Dmax of 190 Å were determined for the MTase in situ within the body of the EcoR124I RM enzyme. Both the shape of the curve and values of Rg and Dmax for the MTase in situ were very similar to both SANS and SAXS results obtained for the pure EcoR124I MTase (Taylor et al. 1994, 2010). This indicates that the MTase in the absence of DNA has the same tertiary structure whether the HsdR are present or not.
Figure 2.
SANS and SAXS analyses. (A) SANS profiles of EcoR124I. The left panel shows the scattering data, and the right panel shows the pair distribution functions, p(r). (Gray) Protonated EcoR124I in 0% D2O; (blue) MTase core in situ within the RM enzyme (deuterated HsdR and protonated MTase measured in 100% D2O); (red) the two HsdR in situ in the RM enzyme (deuterated HsdR and protonated MTase measured in 40% D2O). (B) SAXS profiles of EcoR124I (black) and EcoKI (green). The panel on the left shows the scattering data, and the right panel shows the pair distribution functions, p(r). In both A and B, the solid lines in the scattering data represent the fits from the corresponding back-transformed distance distribution functions, p(r), in the panel on the right. (C) Multiphase ab initio modeling showing the location of the MTase core (blue) and the HsdR (red), superimposed on the EM map of EcoR124I from Figure 1B (gray). The panel on the right shows a 90° rotation about the long axis in the left panel. Data on the assembly of the EcoR124I enzyme can be found in Supplemental Figure S3.
Table 1.
SAXS and SANS data
SAXS data were also collected on the EcoKI and EcoR124I RM enzymes (Fig. 2B; Table 1). The scattering curves for both EcoKI and EcoR124I were essentially identical within error (left panel of Fig. 2B), indicating that the overall shapes of both RM enzymes in the absence of DNA are similar. From the pair distribution function (right panel of Fig. 2B), the Dmax of EcoKI (230 Å) was slightly larger than that determined for EcoR124I (220 Å), possibly due to the slightly larger overall mass of EcoKI. The Rg values of EcoKI and EcoR124I were 68 Å and 70 Å, respectively, in good agreement with the Rg obtained by SANS for EcoR124I.
Reconstruction of the EcoR124I structure from scattering data
An ab initio model of the EcoR124I RM enzyme, showing the location of HsdR and the MTase, can be constructed from the scattering experiments by both the simulated annealing procedure in DAMMIN (Svergun 1999) and multiphase bead modeling using MONSA (Svergun and Nierhaus 2000). Two phases were specified—one for the two HsdR, and the other for the MTase—while taking into account the Rg determined from the Guinier region. An overall symmetry of P2 was specified. The fit of the complete RM enzyme model to the 0% D2O SANS data was excellent, with a χ2 value of 0.98, while the MTase and HsdR fitted the 100% and 40% D2O SANS with χ2 values of 1.47 and 1.9, perhaps indicative of the flexible nature of the HsdR with respect to the MTase core. The MTase component was found to be located centrally within the envelope of the RM enzyme, while each HsdR was extended toward the outermost edge of the complex (Fig. 2C).
Location of HsdR in EcoR124I using EM
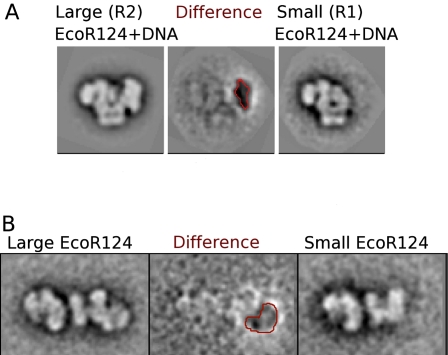
The SANS data clearly indicate that EcoR124I is an elongated structure with the two HsdR located toward the extreme ends on either side of the MTase core. The protein concentration used in the scattering experiments is substantially greater than the binding constant for the assembly of the R2M2S1 complex. However, at the lower protein concentration used for EM, a fraction of the enzyme will exist as the R1M2S1 form. In negative stain EM, the particles of EcoR124I with (Fig. 3A) or without (Fig. 3B) DNA were not 100% homogeneous, as smaller particles were noticed following alignment and classification. These smaller particles showed a single, strongly preferred orientation on the carbon, while the larger particles appeared to adopt a range of orientations. Some large particle views appeared to be in an orientation similar to those of the smaller particles, allowing a comparison to be made. Subtracting these similarly oriented images from each other to form difference images unambiguously showed a large missing region at the extremity of the small particles, which, consistent with the SANS analysis, must be the location of one of the HsdR. The observation of complexes with only one HsdR is consistent with previous biochemical data on EcoR124I (Janscak et al. 1998).
Figure 3.
2D difference images from EM data show the position of the HsdR in the EcoR124I complex. (A) Difference imaging between image averages of large (left) and small (right) particles in the EcoR124I+DNA negative stain EM data set reveals a large “negative density” region (red contour at −2.5 σ), consistent with a missing HsdR in the small particles. (B) Difference imaging of HsdR in the open state of EcoR124I (without DNA). Although the relative flexibility of the open complex gives rise to a less well-defined difference map, a region of negative density consistent with a missing HsdR is visible nevertheless (red contour).
Location of the DNA path in EcoR124I using a DNA mimic protein and determination of the orientation of the MTase in the RM enzyme
Having located the HsdR and the MTase core, ambiguities remain concerning the orientation of these units and the location of DNA within the overall structure. In particular, the lack of a clear position for the DNA would make defining the orientation of the MTase core ambiguous within the 3D map. Fortunately, we were able to use the Ocr DNA mimic protein, which binds very tightly to the DNA-binding site of the type I RM enzymes (Atanasiu et al. 2002; Walkinshaw et al. 2002), to infer the path of DNA through EcoR124I. Unlike the DNA, Ocr should be visible in EM experiments.
Complexes of EcoR124I with Ocr adopted a closed conformation in EM. The presence of Ocr also unexpectedly increased the proportion of complexes containing only one HsdR (one possibility is that the Ocr binds to free HsdR and thus lowers the proportion of the R2M2S1 complexes). However, using only those particles large enough to contain both HsdR, a 3D map was generated by projection matching onto the map for the EcoR124I+DNA complex (Fig. 4A–C). 2D difference images and 3D difference maps then revealed the position of a banana-shaped object running through the center of the RM enzyme but tilted at an angle relative to the long axis of the 3D map (Fig. 4D). The banana-like shape matches well with the structure of the Ocr protein (Fig. 4E; Walkinshaw et al. 2002). This orientation of Ocr in the EM map, coupled with the structural models of the MTase core of EcoKI (Kennaway et al. 2009) and of EcoR124I (based on that of EcoKI as described in the Supplemental Material; Supplemental Fig. S4A), allows only one possible orientation of the MTase atomic models within the EM envelope (Fig. 4E). The orientation chosen for the MTase exposes the double α-helical linker of the HsdS to the solvent. This is consistent with previous observations on the location of HsdS, which indicated that the linker could accommodate both small (Gubler and Bickle 1991) and large (Kannan et al. 1989) amino acid insertions and even a fusion with green fluorescent protein (Chen et al. 2010) without loss of function. Moreover, limited proteolysis showed that cleavage occurred preferentially in the α-helical region between the two TRDs of HsdS (Webb et al. 1995), and thus the linker must be highly accessible to solvent.
Figure 4.
2D and 3D difference mapping from EM data shows the route of DNA/Ocr through the EcoR124I complex. (A) Negative stain EM difference image averages of EcoR124I+DNA and EcoR124I+Ocr from two orientations show a smaller central area of positive difference (green contours at +4.5 σ), indicating the position of the DNA mimic Ocr, which excludes stain more effectively than DNA. (B) Surface view of the 3D reconstruction of EcoR124I+DNA. (C) Surface view of the 3D reconstruction of EcoR124I+Ocr shows the central hole is mostly occluded in the Ocr-containing complex when compared with the EcoR124I+DNA surface shown in B. (D) Two views of the DNA/Ocr 3D difference map (green surface, contoured at +4.5 σ) overlaid onto the EcoR124I+DNA map (gray mesh) showing the main positive difference densities. (E) Two views of the EcoR124I+DNA 3D map (gray mesh) with the EcoKI MTase core+Ocr atomic model (PDB code: 2y7C) docked in as a single rigid body. (Magenta spacefill) Ocr; (yellow ribbon) HsdS; (blue ribbon) 2× HsdM. The path of Ocr, as predicted from the MTase structure, matches well to the difference density.
Atomic modeling of the complete RM enzymes
The above data provide some of the constraints necessary for construction of atomic models. This procedure comprises two parts: first, completion of known crystallographic structures of the subunits by modeling of domains not resolved by crystallography, and second, insertion of the complete structures into the 3D envelopes provided by EM and scattering analyses. This latter part also benefits from the availability of published biochemical data on these enzymes as further constraints.
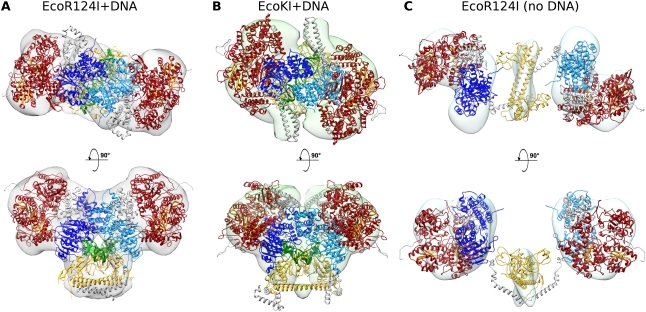
Atomic models of complete HsdR for EcoKI and EcoR124I were constructed based on known crystal structures (Supplemental Material; Supplemental Fig. S4B). The missing C terminus (amino acid residues 893–1038) of the HsdR of EcoR124I was modeled de novo using the program Rosetta (Supplemental Material). Although the positions of the central MTase core region and the HsdR are well defined, as described above, the orientation of the HsdR in the EM map of both EcoR124I and EcoKI was ambiguous, as the density was rather ring-like in shape. We can, however, propose a model that fits the data and gives the location and directionality of the DNA motor domains by aligning the RecA-like motor domains of the crystal structure of the dsDNA-bound SWI2/SNF2 chromatin remodeling translocase from Sulfolobus solfataricus (Protein Data Bank [PDB] code: 1z63) (Dürr et al. 2005) with those of HsdR (Lapkouski et al. 2009). As the direction of DNA translocation is defined in the chromatin remodeling translocase, it imposes a similar directionality on each HsdR, and since these have to pull DNA in toward the MTase core of the type I RM enzyme, the orientation of each HsdR relative to the MTase core becomes defined. Assuming that the DNA path between the DNA bound to the HsdR and the DNA bound to the core MTase must not be any longer than ∼40 bp, as determined by DNA footprinting experiments (Mernagh et al. 1998; Powell et al. 1998) and the minimum length of 45 bp of DNA required for ATP hydrolysis (Roberts et al. 2011), then the locations of the RecA-like motor domains of the HsdR are forced to be as shown, so that their DNA-binding sites are close to the DNA-binding site of the MTase core. Placement of the HsdR on either side of the MTase and interacting directly with DNA is further supported by the length of the structure of the ArdA anti-restriction DNA mimic protein (Nekrasov et al. 2007; McMahon et al. 2009), which occupies the entire DNA-binding site on type I RM enzymes. This then allows the complete structures for the closed forms of EcoR124I and EcoKI to be constructed as shown in Figure 5, A and B.
Figure 5.
Atomic models of EcoR124I+DNA, EcoR124I, and EcoKI+DNA docked into the EM map densities. (A) Two views of the EcoR124I+DNA model showing the MTase core closed around DNA (green; DNA bound to each HsdR is not shown for clarity). Adenine bases are flipped out into the active sites of each of the two HsdM (light and dark blue), induced by an ∼45° bend in the DNA. The HsdS is in yellow, and the two HsdR are shown in red, with the β sheets of the recA-like motor domains colored orange. Residues missing from the crystal structures (the 44 and 152 C-terminal residues of HsdM and HsdR, respectively) were modeled de novo and are shown in gray. The C-terminal regions of HsdM extend down to bind at the coiled coil of HsdS, and the HsdR C-terminal domains fill some empty density next to the N terminus of HsdM. (B) A model for the second type I RM enzyme, EcoKI bound to DNA. Colors are as in A, with residues modeled de novo shown in gray. The HsdS and HsdM from the MTase structure (PDB code: 2y2C) were docked in as a single rigid body. The HsdR modeled on those from EcoR124I (PDB code: 2w00), as described in the Supplemental Material, were placed in a position analogous to the EcoR124I model. (C) The model of EcoR124I in the open conformation (i.e., without DNA). Colors are as in A, with residues modeled de novo shown in gray. Although the EM map is at a lower resolution, a full atomic model can be built, aided by the EcoR124I+DNA model, SANS data, and 2D difference imaging. The HsdM and HsdR swing out as a unit away from HsdS. The predicted hinge regions in the C termini of the HsdM (modeled in gray) and their connections to HsdS are not well resolved.
Placement of the HsdR forces two large kinks in the DNA to allow the DNA to thread through the MTase core (Fig. 5A,B). This kinked path, which effectively shortens the through-space end-to-end distance of a duplex bound to the RM enzyme by ∼10 nm, is supported by atomic force microscopy (AFM) measurements of complexes of EcoR124I on DNA, which showed that binding of the nuclease shortened the length of a long linear DNA molecule by ∼11 nm (van Noort et al. 2004). AFM measurements of EcoKI bound to DNA also showed a pronounced kink (Walkinshaw et al. 2002; Neaves et al. 2009), and circular dichroism analysis of EcoR124I also indicated a large structural distortion to the DNA when bound (Taylor et al. 1994).
A fit of subunits into the EcoKI EM density map (Fig. 5B) corresponds closely to that of the EcoR124I in the closed state. The thin protrusions at either side of the EM envelope for EcoKI can fit the long coiled-coil N-terminal extensions predicted in the HsdR of EcoKI but absent in EcoR124I (Fig. 5B; Supplemental Material; Supplemental Figs. S1, S4B). These extensions appear to be quite variable in the EM analysis and are seen in both the presence and absence of DNA. The function of these extensions is unknown. Significant sequence differences exist between the two enzymes (Supplemental Figs. S1, S4), and this may account for other structural differences, although the overall architecture remains unchanged.
Fitting of subunits into the lower-resolution open EcoR124I map (Fig. 5C) was aided by the SANS data, 2D difference imaging (Fig. 3B), and the closed atomic model (Fig. 5A). An optimal fit was obtained by moving and rotating each HsdM–HsdR pair as a single rigid body away from HsdS. A relatively simple ∼90° rotation and an ∼80° twist around a pivot point near the C terminus of HsdM are sufficient to move between open and closed states. It has previously been shown that HsdR and HsdM can form a complex (Dryden et al. 1997), supporting movement of the two subunits as a rigid body. The C-terminal residues of the EcoKI HsdM are disordered in the crystal (PDB code: 2ar0) (Kennaway et al. 2009) and are proteolytically sensitive (Cooper and Dryden 1994), and so could play the role of the flexible linker proposed here. Proteolytic removal of this region precludes assembly of the EcoKI RM enzyme (Powell et al. 2003). A comparison of the atomic model of the open form of EcoR124I with that of the SAXS data using the program CRYSOL revealed a good fit with a χ2 value of 1.127. In addition, a comparison of the EM model with the SAXS data, obtained by first filling the EM envelope with dummy atoms, showed a comparably good fit with a χ2 value of 1.28. This comparison of the EM data, small-angle scattering data, and atomic models provides confidence in the atomic models presented.
Discussion
The data presented show how type I RM enzymes are assembled and how they bind and distort DNA prior to the initiation of DNA translocation driven by ATP hydrolysis. The EM and small-angle scattering structures are of low resolution; hence, the placement of atomic models within the structural envelopes could be ambiguous in the absence of data obtained using other methods. However, the type I RM enzymes have been extensively studied biochemically, biophysically, and genetically (Murray 2000, 2002; Loenen 2003; Tock and Dryden 2005), which provides several further constraints on the subunit orientations and gives confidence in the atomic models shown in Figure 5.
The structures shown in Figure 5 make it clear that there is an equilibrium between open and closed forms of the type I RM enzymes, with the equilibrium constant depending on the particular enzyme and the presence or absence of DNA (and presumably the cofactors S-adenosylmethionine and ATP, although these were not specifically examined in this study). EcoKI prefers to be closed whether DNA is present or not, and must therefore transiently open up to allow DNA access to the MTase core. EcoR124I appears to prefer an open form in the absence of DNA but is closed with DNA bound.
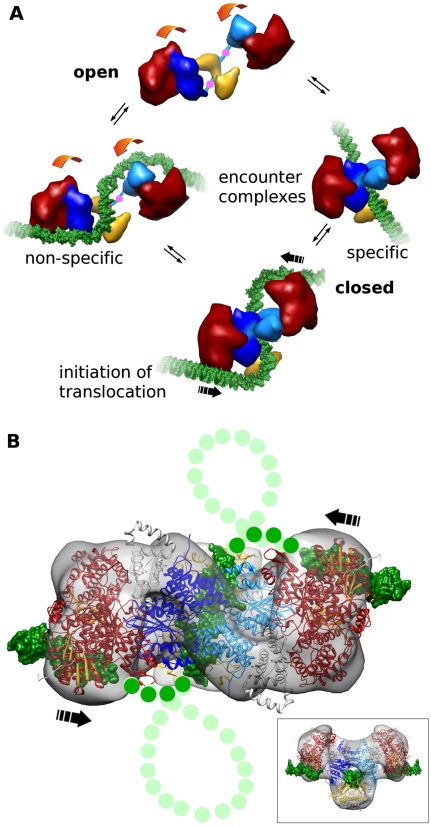
It would appear possible for the type I RM enzymes to reach the closed “initiation” complex with the S-shaped DNA path (Fig. 6A) via different routes. The open form can bind DNA nonspecifically using HsdR (left side of Fig. 6A) and diffuse along the DNA until the MTase core recognizes a target sequence or dissociates. The “trigger” for closing and formation of the initiation complex is most likely the recognition of the target sequence by the MTase core. Alternatively, the closed form of the enzyme must open up transiently to allow DNA to enter the MTase core, followed by closing of the core around the DNA (right side of Fig. 6A) and diffusion of the enzyme on the DNA until it either recognizes its DNA target sequence or reopens and dissociates. Starting the process of target sequence location and recognition via this pathway means that the motor domains of the HsdR will have to rely on the inherent flexibility of DNA for them to grasp it and force it into the S-like shape shown in the initiation complex.
Figure 6.
Schematic of large-scale conformational change and initiation of DNA looping and translocation. (A) Type I RM enzymes exist in a dynamic equilibrium between open and closed states (movement is shown by orange arrows, and pivot points in C-terminal regions of HsdM are indicated by pink dots). DNA (green) binding to form encounter complexes can occur nonspecifically to the HsdR (red) or via the target sequence to the MTase core (HsdM is in light and dark blue, and HsdS is in yellow). Complete closure of the enzyme and bending of the DNA around the HsdR produces the initiation complex for DNA translocation. (B) The predicted complete path of the DNA (green dots) through the atomic model of EcoR124I with segments of bound DNA. This is the proposed initiation complex (from Fig. 5A). During active translocation, the DNA would then form expanding loops from each side (light-green dots for DNA, and the direction of translocation is shown by black arrows). The inset shows the initiation complex turned 90° to the main panel.
The introduction of sharp bends in the DNA would require considerable energy to be expended by the enzyme. This may come from the transition between open and closed forms of the RM enzyme, but it may also require the hydrolysis of ATP by the HsdR. The models suggest that once the enzyme has closed around DNA and the motor of an HsdR subunit has a good “grip” on a segment of DNA, further hydrolysis of ATP would push the segment bound to the motor toward the central MTase core, as indicated by large arrows in Figure 6. Since the MTase core is also tightly bound to the DNA target sequence, DNA at the bend between the segments bound to the motor and to the MTase core would twist and perhaps even buckle, forming the small loop shown in Figure 6B. Formation of this highly strained loop is certain to be energetically unfavorable, in agreement with translocation measurements for type I RM enzymes in which it appears that much ATP is used in abortive attempts to initiate translocation (Seidel et al. 2008). Once the loop has formed, further DNA translocation would occur as the motors pump DNA toward the MTase core. Single-molecule experiments make it clear that the motors can work independently (Seidel et al. 2004, 2008), perhaps explaining why early EM studies showed both single- and double-looped structures (Yuan et al. 1980; Endlich and Linn 1985). In light of the large changes occurring upon DNA binding, it is possible that the actively translocating enzyme undergoes further changes in structure (e.g., in the presence of ATP). One may speculate that this great flexibility would allow the RM enzyme to accommodate the stresses built up during the extensive DNA translocation periods observed for these molecular machines. It is noteworthy, in this respect, that a process of deassembly of the enzymes occurs after DNA cleavage, and some of the subunits—although not all and depending on the particular type I RM enzyme—can be reused (Roberts et al. 2011; Simons and Szczelkun 2011).
Lapkouski et al. (2009) proposed a more speculative atomic model of EcoR124I using their structure of HsdR, a postulated DNA path across the subunit, and an early, incomplete model of the MTase core (Obarska et al. 2006). Although the current models and their model share much in common, there are two main differences; namely, the orientation of the HsdR with respect to the MTase core, and the path taken by the DNA. Previously (Lapkouski et al. 2009), the interface of HsdR with the MTase core was not defined when compared with the models presented here. More importantly, the DNA was proposed to bend across the motor domains of HsdR, so that it came near to the endonuclease domain in the same HsdR and could be cleaved. This model would suggest that the partially assembled R1M2S1 form of EcoR124I would be able to cleave DNA. However, no cleavage was observed for this partially assembled enzyme (Janscak et al. 1998), and thus the Lapkouski et al. (2009) model cannot be entirely correct. The current model suggests that the endonuclease domain of one HsdR is in proximity to DNA translocated by the other HsdR (Fig. 6B). This would explain the absence of DNA cleavage by partially assembled R1M2S1 forms of EcoR124I, despite the fact that such an assembly translocates DNA effectively (Janscak et al. 1998; Seidel et al. 2004, 2008). Thus, the current models are a significant improvement on the previously published models (Davies et al. 1999; Lapkouski et al. 2009).
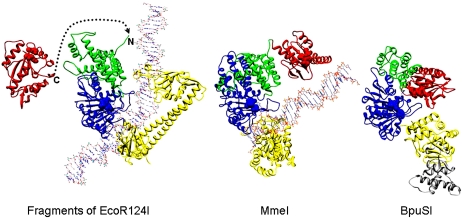
Last, the structural models presented can be compared with the structures of complex type II RM enzymes in groups IIB and IIG (Roberts et al. 2003), which recognize a target sequence but cleave at defined distances on either side of the target (IIB) or on one side of the target (IIG). These classes appear to contain structural domains in common with the type I RM enzymes; namely, endonuclease domains, an HsdM-like subunit, and TRDs, but no motor domains (Dryden 1999; Nakonieczna et al. 2009; Shen et al. 2011). In these enzymes, the motor domains of HsdR are missing and the endonuclease domain is directly fused to the HsdM-like subunit. In type IIB RM enzymes, DNA recognition is performed by an HsdS-like subunit with two TRDs, but for the type IIG restriction enzyme BpuSI, only one TRD with an inserted amino acid sequence is present. The type IIB RM enzyme is effectively a dimer of a type IIG RM enzyme. Thus, a type IIB RM enzyme is a “motor-less” type I RM system, and a type IIG system is half of a “motor-less” type I RM enzyme. Figure 7 compares the relative locations of one endonuclease domain, one HsdM, and the HsdS from the closed form of EcoR124I with the structures of the type IIG enzymes MmeI and BpuSI (Nakonieczna et al. 2009; Shen et al. 2011). MmeI recognizes the sequence TCCRAC and cuts downstream at N20/N18 or N21/N19. BpuSI recognizes the sequence GGGAC and cuts downstream at N10/N14. It can be seen how fusion of the endonuclease domain from HsdR to the start of HsdM in EcoR124I would move it to the same location as observed in the type IIG restriction enzymes and lead to cleavage downstream from the target sequence. Thus, the recently proposed role of gene fusions in the evolution of different groups of type II RM enzyme systems (Mokrishcheva et al. 2011) can be extended to include the evolution of the type I RM enzyme systems.
Figure 7.
Structural evolution of type IIG RM enzymes from a type I RM enzyme undergoing fusion of the C terminus of an endonuclease domain from HsdR, via deletion of the motor domains, to the N terminus of HsdM. The structure on the left shows part of EcoR124I, with one endonuclease domain from HsdR (in red), one HsdM (N-terminal domain is in green, and the MTase catalytic domain is in blue), and the HsdS (in yellow) (two TRDs). DNA bound to the MTase core is shown, but DNA bound to HsdR is omitted for clarity. The dashed line shows how the end of the endonuclease domain could join with the N terminus of HsdM to form a structure similar to the type IIG structures shown on the right. The catalytic motifs in the endonuclease domain and HsdM are shown in spacefill. The middle structure shows the structural model of MmeI with bound DNA with the same coloring used for equivalent domains (endonuclease domain, N-terminal domain, MTase catalytic domain, and TRD) (Nakonieczna et al. 2009; coordinates from ftp://genesilico.pl/iamb/models/RM.MmeI). The structure on the right shows the crystallographic structure of BpuSI (PDB code: 3s1s) with the same coloring of domains as in the other structures and with an inserted extra domain shown in gray (Shen et al. 2011). DNA is absent in this structure, and one can see that the endonuclease domain would be blocking the DNA-binding site on the TRD. Shen et al. (2011) proposed that the endonuclease domain would twist away to allow DNA sequence recognition.
Materials and methods
Full details are given in the Supplemental Material.
Protein expression and purification
EcoR124I MTase and EcoR124I HsdR (Taylor et al. 1992; Obarska-Kosinska et al. 2008), EcoKI (Dryden et al. 1997), and the Ocr anti-restriction protein (Stephanou et al. 2009) were expressed and purified as described previously.
Formation of protein and protein–DNA complexes
The R1M2S1 and R2M2S1 complexes of EcoR124I were formed by incubation of purified HsdR and EcoR124I MTase at 1:1 and 1:2 molar ratios, respectively. Formation of the protein–DNA and protein–Ocr complexes used a 1:1.5 molar ratio of protein to DNA or protein to Ocr dimer for EM studies and a 1:1 ratio of protein to DNA for scattering studies. The oligonucleotides used are described in the Supplemental Material.
EM and image processing
Negative stain grids were prepared by placing solutions of the protein complexes (∼40 μg/mL; i.e., ∼100 nM) onto UV-treated (Walker et al. 1985) continuous carbon-coated copper grids, then stained with 1% uranyl acetate solution. EcoR124I+DNA samples were prepared with a 30-bp fragment containing a centrally located recognition sequence, while EcoKI+DNA samples were prepared with a 75-bp fragment containing a centrally located recognition sequence. Grids were viewed in a Jeol 1200EX electron microscope fitted with a LaB6 (for EcoKI) or tungsten (for EcoR124I) electron source operating at 80 kV. Negatives (Kodak SO63) were recorded at 40,000× magnification with defocus ranging from ∼250 nm to ∼850 nm and were digitized at 15 μm (EcoR124I) or 20 μm (EcoKI) step size using an Imacon scanner. There were 3806 and 2647 negative stain particles in the EcoR124I±DNA data sets, respectively, and 2330 +Ocr particles. EcoKI negative stain data sets had 8300 unbound and 5910 +DNA particles.
EM maps have been submitted to Electron Microscopy Data Bank (EMDB) with the following accession codes: EcoR124+DNA 1890: EcoR124I (no DNA) 1891: EcoR124I+Ocr 1892: EcoKI+DNA 1893.
SAXS
SAXS measurements were performed on a Bruker Nanostar instrument using Cu Kα radiation with a wavelength of 1.54 Å. For EcoKI, 16 1-h measurements were taken of the sample (0.85 mg/mL) in 50 mM Tris-HCl (pH 8.0), 100 mM NaCl, and 1 mM Na2EDTA. For EcoR124I, two 1-h measurements were taken of the sample (2.2 mg/mL) in 20 mM Tris-HCl (pH 8.0), 100 mM NaCl, and 1 mM Na2EDTA. The sample-to-detector distance was 700 mm, providing a q-range of 0.08–0.32 Å−1. Samples were maintained at 10°C. No radiation damage was detected when successive data sets were overlaid.
SANS
The HsdR of EcoR124I were deuterated by expression in E. coli BL21 (DE3) cells using Enfors minimal medium containing 85% D2O with hydrogenated glycerol as the carbon source. Endonuclease complexes were formed by adding HsdR to the MTase, then dialyzing into 50 mM Tris-HCl (pH 8.0), 100 mM NaCl, and 1 mM Na2EDTA in varying H2O/D2O ratios. The complexes were further characterized by dynamic light scattering (data not shown). The protein concentration was between 2 and 4 mg/mL in these experiments (5–10 μM endonuclease). SANS data were collected using the D22 diffractometer at the ILL using two detector distances, 2 and 10 m, for 15 and 30 min, respectively, at 10°C, covering a scattering vector (q) range of 0.008–0.35 Å−1 with a wavelength of 6 Å. A 96 × 96-cm detector with a pixel size of 7.5 × 7.5 mm was used. Data for water were collected at a detector distance of 4 m, allowing the data to be placed on an absolute scale.
Construction of atomic models for type I RM complexes
Previous models of HsdS from EcoR124I (Obarska et al. 2006) and EcoKI MTase (Kennaway et al. 2009) and crystal structures of HsdM (3lkd) and HsdR (2w00) provided the starting points for constructing atomic models of EcoR124I and EcoKI. The open form of EcoR124I was constructed by independent fitting of two copies of the R1M1 subcomplex and the S subunit taken from the closed conformation. Model coordinates are available from us and at ftp://genesilico.pl/iamb/models/RM.typeI.
Acknowledgments
We dedicate this paper to the memory of Professor Noreen E. Murray (1935–2011). We thank Dr. Robert Knott for help in performing small-angle X-ray measurements at the Bragg Institute, ANSTO, Australia. D.T.F.D. would particularly like to acknowledge many years of support and encouragement from Professors Noreen and Kenneth Murray. We acknowledge funding from the Biotechnology and Biological Sciences Research Council (BB/D001870/1 to J.T. and D.T.F.D.), the Wellcome Trust (080304/Z/06/Z to G.G.K.), and the Foundation for Polish Science (TEAM/2009-4/2 to J.M.B.). The EM work on EcoKI was initiated as a result of the Isaac Newton Institute for Mathematical Sciences Workshop on “Statistical Mechanics of Molecular and Cellular Biological Systems,” January–July 2004.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.179085.111.
References
- Abadjieva A, Patel J, Webb M, Zinkevich V, Firman K. 1993. A deletion mutant of the type IC restriction endonuclease EcoR1241 expressing a novel DNA specificity. Nucleic Acids Res 21: 4435–4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanasiu C, Su TJ, Sturrock SS, Dryden DTF. 2002. Interaction of the Ocr gene 0.3 protein of bacteriophage T7 with EcoKI restriction/modification enzyme. Nucleic Acids Res 30: 3936–3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calisto BM, Pich OQ, Piñol J, Fita I, Querol E, Carpena X. 2005. Crystal structure of a putative type I restriction-modification S subunit from Mycoplasma genitalium. J Mol Biol 351: 749–762. [DOI] [PubMed] [Google Scholar]
- Callow P, Sukhodub A, Taylor J, Kneale G. 2007. Shape and subunit organisation of the DNA methyltransferase M.AhdI. J Mol Biol 369: 177–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Roberts GA, Stephanou AS, Cooper LP, White JH, Dryden DTF. 2010. Fusion of GFP to the M.EcoKI DNA methyltransferase produces a new probe of type I DNA restriction and modification enzymes. Biochem Biophys Res Commun 398: 254–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper LP, Dryden DTF. 1994. The domains of a type I DNA methyltransferase Interactions and role in recognition of DNA methylation. J Mol Biol 236: 1011–1021. [DOI] [PubMed] [Google Scholar]
- Cowan GM, Gann AA, Murray NE. 1989. Conservation of complex DNA recognition domains between families of restriction enzymes. Cell 56: 103–109. [DOI] [PubMed] [Google Scholar]
- Davies GP, Martin I, Sturrock SS, Cronshaw A, Murray NE, Dryden DTF. 1999. On the structure and operation of type I DNA restriction enzymes. J Mol Biol 290: 565–579. [DOI] [PubMed] [Google Scholar]
- Dryden DTF. 1999. Bacterial DNA methyltransferases. In S-adenosylmethionine-dependent methyltransferases: Structures and functions (ed. Cheng X, Blumenthal RM), pp. 283–340. World Scientific Publishing, Singapore. [Google Scholar]
- Dryden DTF, Sturrock SS, Winter M. 1995. Structural modelling of a type I DNA methyltransferase. Nat Struct Biol 2: 632–635. [DOI] [PubMed] [Google Scholar]
- Dryden DTF, Cooper LP, Thorpe PH, Byron O. 1997. The in vitro assembly of the EcoKI type I DNA restriction/modification enzyme and its in vivo implications. Biochemistry 36: 1065–1076. [DOI] [PubMed] [Google Scholar]
- Dürr H, Korner C, Muller M, Hickmann V, Hopfner KP. 2005. X-ray structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase core and its complex with DNA. Cell 121: 363–373. [DOI] [PubMed] [Google Scholar]
- Endlich B, Linn S. 1985. The DNA restriction endonuclease of Escherichia coli B. I. Studies of the DNA translocation and the ATPase activities. J Biol Chem 260: 5720–5728. [PubMed] [Google Scholar]
- Fuller-Pace FV, Bullas LR, Delius H, Murray NE. 1984. Genetic recombination can generate altered restriction specificity. Proc Natl Acad Sci 81: 6095–6099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gann AA, Campbell AJB, Collins JF, Coulson AF, Murray NE. 1987. Reassortment of DNA recognition domains and the evolution of new specificities. Mol Microbiol 1: 13–22. [DOI] [PubMed] [Google Scholar]
- García LR, Molineux IJ. 1999. Translocation and specific cleavage of bacteriophage T7 DNA in vivo by EcoKI. Proc Natl Acad Sci 96: 12430–12435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Tang Q, An X, Yan X, Liang D. 2011. Structure of HsdS subunit from Thermoanaerobacter tengcongensis sheds lights on mechanism of dynamic opening and closing of type I methyltransferase. PLoS ONE 6: e17346. doi: 101371/journalpone0017346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith JD. 1978. DNA structure: Evidence from electron microscopy. Science 201: 525–527. [DOI] [PubMed] [Google Scholar]
- Gubler M, Bickle TA. 1991. Increased protein flexibility leads to promiscuous protein–DNA interactions in type IC restriction-modification systems. EMBO J 10: 951–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janscak P, Dryden DTF, Firman K. 1998. Analysis of the subunit assembly of the type IC restriction–modification enzyme EcoR124I. Nucleic Acids Res 26: 4439–4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan P, Cowan GM, Daniel AS, Gann AA, Murray NE. 1989. Conservation of organization in the specificity polypeptides of two families of type I restriction enzymes. J Mol Biol 209: 335–344. [DOI] [PubMed] [Google Scholar]
- Kennaway CK, Obarska-Kosinska A, White JH, Tuszynska I, Cooper LP, Bujnicki JM, Trinick J, Dryden DTF. 2009. The structure of M.EcoKI type I DNA methyltransferase with a DNA mimic antirestriction protein. Nucleic Acids Res 37: 762–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS, De Giovanni A, Jancarik J, Adams PD, Yokota H, Kim R, Kim SH. 2005. Crystal structure of DNA sequence specificity subunit of a type I restriction-modification enzyme and its functional implications. Proc Natl Acad Sci 102: 3248–3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneale GG. 1994. A symmetrical model for the domain structure of type I DNA methyltransferases. J Mol Biol 243: 1–5. [DOI] [PubMed] [Google Scholar]
- Lapkouski M, Panjikar S, Janscak P, Smatanova IK, Carey J, Ettrich R, Csefalvay E. 2009. Structure of the motor subunit of type I restriction–modification complex EcoR124I. Nat Struct Mol Biol 16: 94–95. [DOI] [PubMed] [Google Scholar]
- Linn S, Arber W. 1968. Host specificity of DNA produced by Escherichia coli. X. In vitro restriction of phage fd replicative form. Proc Natl Acad Sci 59: 1300–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loenen WA. 2003. Tracking EcoKI and DNA fifty years on: A golden story full of surprises. Nucleic Acids Res 31: 7059–7069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhusoodanan UK, Rao DN. 2010. Diversity of DNA methyltransferases that recognize asymmetric target sequences. Crit Rev Biochem Mol Biol 45: 125–145. [DOI] [PubMed] [Google Scholar]
- Makovets S, Powell LM, Titheradge AJB, Blakely GW, Murray NE. 2004. Is modification sufficient to protect a bacterial chromosome from a resident restriction endonuclease? Mol Microbiol 51: 135–147. [DOI] [PubMed] [Google Scholar]
- Mayanagi K, Kiyonari S, Nishida H, Saito M, Kohda D, Ishino Y, Shirai T, Morikawa K. 2011. Architecture of the DNA polymerase B-proliferating cell nuclear antigen (PCNA)–DNA ternary complex. Proc Natl Acad Sci 108: 1845–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland SE, Dryden DTF, Szczelkun MD. 2005. Continuous assays for DNA translocation using fluorescent triplex dissociation: Application to type I restriction endonucleases. J Mol Biol 348: 895–915. [DOI] [PubMed] [Google Scholar]
- McMahon SA, Roberts GA, Johnson KA, Cooper LP, Liu H, White JH, Carter LG, Sanghvi B, Oke M, Walkinshaw MD, et al. 2009. Extensive DNA mimicry by the ArdA anti-restriction protein and its role in the spread of antibiotic resistance. Nucleic Acids Res 37: 4887–4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister J, MacWilliams M, Hübner P, Jütte H, Skrzypek E, Piekarowicz A, Bickle TA. 1993. Macroevolution by transposition: Drastic modification of DNA recognition by a type I restriction enzyme following Tn5 transposition. EMBO J 12: 4585–4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melero R, Rajagopalan S, Lázaro M, Joerger AC, Brandt T, Veprintsev DB, Lasso G, Gil D, Scheres SH, Carazo JM, et al. 2011. Electron microscopy studies on the quaternary structure of p53 reveal different binding modes for p53 tetramers in complex with DNA. Proc Natl Acad Sci 108: 557–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mernagh DR, Janscak P, Firman K, Kneale GG. 1998. Protein–protein and protein–DNA interactions in the type I restriction endonuclease R.EcoR124I. Biol Chem 379: 497–503. [DOI] [PubMed] [Google Scholar]
- Meselson M, Yuan R. 1968. DNA restriction enzyme from E. coli. Nature 217: 1110–1114. [DOI] [PubMed] [Google Scholar]
- Mokrishcheva ML, Solonin AS, Nikitin DV. 2011. Fused eco29kIR- and M genes coding for a fully functional hybrid polypeptide as a model of molecular evolution of restriction–modification systems. BMC Evol Biol 11: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray NE. 2000. Type I restriction systems: Sophisticated molecular machines (a legacy of Bertani and Weigle). Microbiol Mol Biol Rev 64: 412–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray NE. 2002. 2001 Fred Griffith review lecture Immigration control of DNA in bacteria: Self versus non-self. Microbiology 148: 3–20. [DOI] [PubMed] [Google Scholar]
- Nakonieczna J, Kaczorowski T, Obarska-Kosinska A, Bujnicki JM. 2009. Functional analysis of MmeI from methanol utilizer Methylophilus methylotrophus, a subtype IIC restriction–modification enzyme related to type I enzymes. Appl Environ Microbiol 75: 212–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neaves KJ, Cooper LP, White JH, Carnally SM, Dryden DTF, Edwardson JM, Henderson RM. 2009. Atomic force microscopy of the EcoKI type I DNA restriction enzyme bound to DNA shows enzyme dimerization and DNA looping. Nucleic Acids Res 37: 2053–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov SV, Agafonova OV, Belogurova NG, Delver EP, Belogurov AA. 2007. Plasmid-encoded antirestriction protein ArdA can discriminate between type I methyltransferase and complete restriction–modification system. J Mol Biol 365: 284–297. [DOI] [PubMed] [Google Scholar]
- Obarska A, Blundell A, Feder M, Vejsadova S, Sisakova E, Weiserova M, Bujnicki JM, Firman K. 2006. Structural model for the multisubunit type IC restriction-modification DNA methyltransferase MEcoR124I in complex with DNA. Nucleic Acids Res 34: 1992–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obarska-Kosinska A, Taylor JEA, Callow P, Orlowski J, Bujnicki JM, Kneale GG. 2008. HsdR subunit of the type I restriction-modification enzyme EcoR124I: Biophysical characterisation and structural modelling. J Mol Biol 376: 438–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell LM, Dryden DTF, Murray NE. 1998. Sequence-specific DNA binding by EcoKI a type IA DNA restriction enzyme. J Mol Biol 283: 963–976. [DOI] [PubMed] [Google Scholar]
- Powell LM, Lejeune E, Hussain FS, Cronshaw AD, Kelly SM, Price NC, Dryden DTF. 2003. Assembly of EcoKI DNA methyltransferase requires the C-terminal region of the HsdM modification subunit. Biophys Chem 103: 129–137. [DOI] [PubMed] [Google Scholar]
- Roberts RJ, Belfort M, Bestor T, Bhagwat AS, Bickle TA, Bitinaite J, Blumenthal RM, Degtyarev SKh, Dryden DTF, Dybvig K, et al. 2003. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res 31: 1805–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RJ, Vincze T, Posfai J, Macelis D. 2010. REBASE—a database for DNA restriction and modification: Enzymes genes and genomes. Nucleic Acids Res 38: D234–D236. doi: 10.1093/nar/gkp874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts GA, Cooper LP, White JH, Su TJ, Zipprich JT, Geary P, Kennedy C, Dryden DTF. 2011. An investigation of the structural requirements for ATP hydrolysis and DNA cleavage by the EcoKI type I DNA restriction and modification enzyme. Nucleic Acids Res 39: 7667–7676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel R, van Noort J, van der Scheer C, Bloom JGP, Dekker NH, Dutta CF, Blundell A, Robinson T, Firman K, Dekker C. 2004. Real-time observation of DNA translocation by the type I restriction modification enzyme EcoR124I. Nat Struct Mol Biol 11: 838–843. [DOI] [PubMed] [Google Scholar]
- Seidel R, Bloom JG, Dekker C, Szczelkun MD. 2008. Motor step size and ATP coupling efficiency of the dsDNA translocase. EcoR124I. EMBO J 27: 1388–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen BW, Xu D, Chan SH, Zheng Y, Zhu Z, Xu SY, Stoddard BL. 2011. Characterization and crystal structure of the type IIG restriction endonuclease RM.BpuSI. Nucleic Acids Res 39: 8223–8236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons M, Szczelkun MD. 2011. Recycling of protein subunits during DNA translocation and cleavage by type I restriction-modification enzymes. Nucleic Acids Res 39: 7656–7666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephanou AS, Roberts GA, Tock MR, Pritchard EH, Turkington R, Nutley M, Cooper A, Dryden DTF. 2009. A mutational analysis of DNA mimicry by Ocr the gene 0.3 antirestriction protein of bacteriophage T7. Biochem Biophys Res Commun 378: 129–132. [DOI] [PubMed] [Google Scholar]
- Studier FW, Bandyopadhyay PK. 1988. Model for how type I restriction enzymes select cleavage sites in DNA. Proc Natl Acad Sci 85: 4677–4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun DI. 1999. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys J 76: 2879–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun DI, Nierhaus KH. 2000. A map of protein–rRNA distribution in the 70 S Escherichia coli ribosome. J Biol Chem 275: 14432–14439. [DOI] [PubMed] [Google Scholar]
- Taylor IA, Patel J, Firman K, Kneale GG. 1992. Purification and biochemical-characterization of the EcoR124 type-I modification methylase. Nucleic Acids Res 20: 179–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor IA, Davis KG, Watts D, Kneale GG. 1994. DNA-binding induces a major structural transition in a type-I methyltransferase. EMBO J 13: 5772–5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JE, Callow P, Swiderska A, Kneale GG. 2010. Structural and functional analysis of the engineered type I DNA methyltransferase EcoR124INT. J Mol Biol 398: 391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tock MR, Dryden DTF. 2005. The biology of restriction and anti-restriction. Curr Opin Microbiol 8: 466–472. [DOI] [PubMed] [Google Scholar]
- Uyen NT, Park S, Choi J, Lee HJ, Nishi K, Kim JS. 2009. The fragment structure of a putative HsdR subunit of a type I restriction enzyme from Vibrio vulnificus YJ016: Implications for DNA restriction and translocation activity. Nucleic Acids Res 37: 6960–6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Noort J, van der Heijden T, Dutta CF, Firman K, Dekker C. 2004. Initiation of translocation by type I restriction-modification enzymes is associated with a short DNA extrusion. Nucleic Acids Res 32: 6540–6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldron DE, Lindsay JA. 2006. Sau1: A novel lineage-specific type I restriction-modification system that blocks horizontal gene transfer into Staphylococcus aureus and between S. aureus isolates of different lineages. J Bacteriol 188: 5578–5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker M, Knight P, Trinick J. 1985. Negative staining of myosin molecules. J Mol Biol 184: 535–542. [DOI] [PubMed] [Google Scholar]
- Walkinshaw MD, Taylor P, Sturrock SS, Atanasiu C, Berge T, Henderson RM, Edwardson JM, Dryden DTF. 2002. Structure of Ocr from bacteriophage T7 a protein that mimics B-form DNA. Mol Cell 9: 187–194. [DOI] [PubMed] [Google Scholar]
- Webb M, Taylor IA, Firman K, Kneale GG. 1995. Probing the domain structure of the type IC DNA methyltransferase M.EcoR124I by limited proteolysis. J Mol Biol 250: 181–190. [DOI] [PubMed] [Google Scholar]
- Yuan R, Hamilton DL, Burckhardt J. 1980. DNA translocation by the restriction enzyme from E. coli K. Cell 20: 237–244. [DOI] [PubMed] [Google Scholar]