Abstract
While there has been significant progress in the development of engineering principles for synthetic biology, a substantial challenge is the construction of robust circuits in a noisy cellular environment. Such an environment leads to considerable intercellular variability in circuit behavior, which can hinder functionality at the colony level. Here, we engineer the synchronization of thousands of oscillating colony “biopixels” over centimetre length scales through the use of synergistic intercellular coupling involving quorum sensing within a colony and gas-phase redox signaling between colonies. We use this platform to construct an LCD-like macroscopic clock that can be used to sense arsenic via modulation of the oscillatory period. Given the repertoire of sensing capabilities of bacteria such as E. coli, the ability to coordinate their behavior over large length scales sets the stage for the construction of low cost genetic biosensors that are capable of detecting heavy metals and pathogens in the field.
Synthetic biology can be broadly parsed into the “top-down” synthesis of genomes1,2 and the “bottom-up” engineering of relatively small genetic circuits3–9. In the genetic circuits arena, toggle switches10 and oscillators11 have progressed into triggers12, counters13 and synchronized clocks14. Sensors have arisen as a major focus in the context of biotechnology7,15,16, while oscillators have provided insights into the basic-science functionality of cyclic regulatory processes17,18. A common theme is the concurrent development of mathematical modeling that can be used for experimental design and characterization, as in physics and the engineering disciplines.
The synchronization of genetic clocks provides a particularly attractive avenue for synthetic biology applications. Oscillations permeate science and technology in a number of disciplines, with familiar examples including AC power19, GPS20, and lasers21. These technologies have demonstrated that operating in the frequency domain can offer significant advantages over steady-state designs in terms of information gathering and transmission. In particular, oscillatory sensors confer a number of advantages to traditional ones22, since frequency is easily digitized and can be quickly updated with repeated measurements. For sensors that use optical reporters, measurements of frequency are less sensitive to experimental factors such as beam power and exposure time than intensity measurements which must be normalized and calibrated.
While the bottom-up approach to synthetic biology is increasingly benefiting from DNA synthesis technologies, the general design principles are still evolving. Within this context, a substantial challenge is the construction of robust circuits in a cellular environment that is governed by noisy processes such as random bursts of transcription and translation23–30. Such an environment leads to considerable intercellular variability in circuit behavior, which can hinder their functionality at the colony level. An ideal design strategy for reducing variability across a cellular population would involve both strong and long-range coupling that would instantaneously synchronize the response of millions of cells. Quorum sensing typically involves strong intercellular coupling over tens of microns9,14, yet the relatively slow diffusion time of molecular communication through cellular media leads to signaling delays over millimetre scales. Faster communication mechanisms, such as those mediated in the gas phase, may increase the length scale for instantaneous communication, but are comparatively weak and short-lived since the vapor species more readily disperse.
Synergistic Synchronization
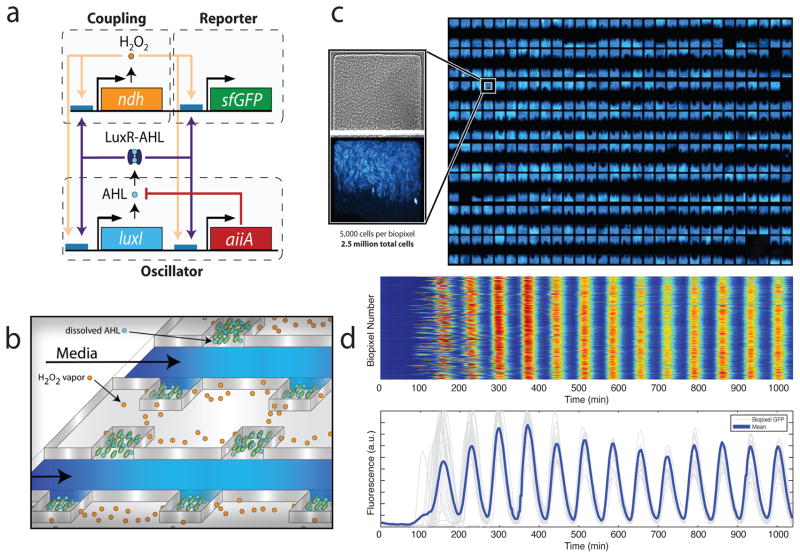
In order to develop a frequency modulated biosensor, we designed a gene network capable of synchronizing genetic oscillations across multiple scales (Fig. 1a and Suppl. Fig. 1). We constructed an LCD-like microfluidic array that allows many separate colonies of sensing bacteria to grow and communicate rapidly by gas exchange (Fig. 1b and Suppl. Fig. 9). Since previous work14 has demonstrated that coupling through quorum sensing leads to incoherent oscillations at the millimetre scale, this mode of cellular communication is too slow for the generation of synchronized oscillations at the macroscopic scale. However, the slower quorum sensing can be used to synchronize small local colonies, provided there is a second level of design that involves faster communication for coordination between the colonies. Therefore rather than attempting to engineer a sensor from a single large-colony oscillator, we wired together thousands of small oscillating colonies, or “biopixels”, in a microfluidic array. Coupling between biopixels involves redox signaling by hydrogen peroxide (H2O2) and the native redox sensing machineries of E. coli. The two coupling mechanisms act synergistically in the sense that the stronger, yet short-range, quorum sensing is necessary to coherently synchronize the weaker, yet long-range, redox signaling. Using this method we demonstrate synchronization of approximately 2.5 million cells across a distance of 5 mm, over 1,000 times the length of an individual cell (Fig. 1c–d). This degree of synchronization yields extremely consistent oscillations, with a temporal accuracy of about 2 minutes compared to 5 – 10 minutes for a single oscillator14 (Fig. 1d).
Fig. 1.
Sensing array of radically coupled genetic biopixels. (a) Network diagram. The luxI promoter drives expression of luxI, aiiA, ndh, and sfGFP in four identical transcription modules. The quorum-sensing genes luxI and aiiA generate synchronized oscillations within a colony via AHL. The ndh gene codes for NDH-2, an enzyme that generates H2O2 vapor which is an additional activator of the luxI promoter. H2O2 is capable of migrating between colonies and synchronizing them. (b) Conceptual design of the sensing array. AHL diffuses within colonies while H2O2 migrates between adjacent colonies through the PDMS. Arsenite-containing media is passed in through the parallel feeding channels. (c) Fluorescent image of an array of 500 E. coli biopixels containing about 2.5 million cells. Inset: brightfield and fluorescent images display a biopixel of 5,000 cells. (d) Heatmap and trajectories depicting time-lapse output of 500 individual biopixels undergoing rapid synchronization. Sampling time is 2 minutes.
The global synchronization mechanism is comprised of two modes of communication that work on different scales. The quorum-sensing machinery (LuxI, AiiA) uses an acyl-homoserine lactone (AHL) to mediate intracolony synchronization. In our device, the degree to which neighboring colonies are able to influence each other via AHL diffusion is negligible owing to the high media channel flow rates. Instead, we engineered the cells to communicate via gas exchange by placing a copy of the gene coding for NADH dehydrogenase II (NDH-2) under the control of an additional lux promoter. NDH-2 is a membrane-bound respiratory enzyme that produces low levels of H2O2 and superoxide (O2−)31. Since H2O2 vapor is able to pass through the 25 μm oxygen-permeable PDMS walls that separate adjacent colonies, periodic production of NDH-2 yields periodic migration of H2O2 between biopixels. When H2O2 enters the cell, it transiently changes its redox state, interacting with our synthetic circuit through the native aerobic response control systems, including arcAB which has a binding site in the lux promoter region32,33. Under normal conditions, ArcAB is partially active so lux is partially repressed. In contrast, oxidizing conditions triggered by H2O2 inactivate ArcAB, relieving this repression. Each oscillatory burst promotes firing in neighboring colonies by relieving repression on the lux promoter. This constitutes an additional positive feedback that rapidly synchronizes the population (Suppl. Fig. 2).
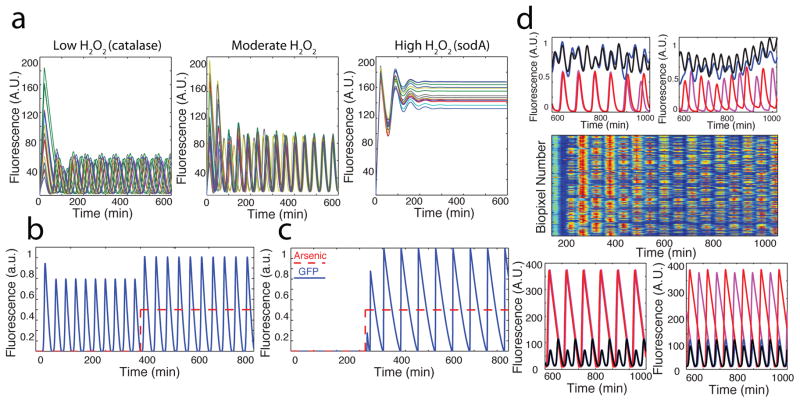
We investigated the effects of catalase and superoxide dismutase (SOD) to probe the nature of H2O2communication. When a population of synchronized colonies was exposed to a step increase of 200 U/ml catalase, an enzyme that rapidly degrades extracellular H2O234, synchronization was broken and colonies continued to oscillate individually (Suppl. Fig. 3). Since the cell membrane is impermeable to catalase, this confirms that communication between colonies depends on external H2O2 while oscillations within a colony do not. Conversely, when we enhanced the rate of superoxide conversion to H2O2 by expressing sodA35,36 from an additional lux promoter, colonies quickly fired in a spatial wave and failed to oscillate further despite no changes to growth rate or cell viability (Suppl. Fig. 4). Since H2O2 is produced internal to the cell, this confirms that H2O2 is capable of escaping the cell and activating lux-controlled genes in neighboring colonies via diffusion. The apparent higher output of H2O2 by SOD as compared to NDH-2 is likely due to its very high catalytic efficiency 37. Finally, we observed synchronization between arrays of traps even when they were fluidically isolated but held in close proximity (Suppl. Fig. 5). These devices share no common fluid sources or channels, making communication by dissolved molecules like AHL impossible. Taken together, these results confirm that gaseous H2O2 is the primary mode of communication between oscillating colonies.
Based on our understanding of the mechanism for global synchronization, we expected that we could simplify the circuitry by eliminating ndh and achieve the same effect with intermittent bursts of high-intensity blue light. In this design, the GFP molecule acts as a photosensitizer, releasing free radicals upon exposure that produce oxygen species (ROS) including H2O238. At the peak of oscillation, significant vapor-phase H2O2 is produced by exposing GFP-containing cells to fluorescent light. Conversely, at the trough of oscillation, cells contain almost no GFP, and therefore produce very little H2O2 upon fluorescing. Bursts of light thus generate bursts of H2O2 vapor whose concentration depends on the oscillating GFP level, just as periodic production of NDH-2 did previously. Indeed, this strategy was similarly able to synchronize our sensor array (Fig. 1d). Numerous controls were performed to ensure that synchronized oscillations did not occur at low fluorescence intensities (Suppl. Fig. 6 and Suppl. Movie 9).
To probe this mode of synchronization, we investigated the effects of thiourea and the antibiotics ampicillin and kanamycin. When a synchronized population of colonies was exposed to 35 mM thiourea, a potent radical quencher39,40, we observed sharply decaying synchronized oscillations while growth rate and cell viability were unaffected (Suppl. Fig. 7). This suggests that without O2−, oscillations cannot be produced. Next, we ran a series of experiments switching the antibiotic resistance genes on our plasmids. We noted that radical-producing antibiotics41, particularly Ampicillin, significantly reduced the degree of synchronization, showing that an excess of radical species such as O2− also hinders communication (Suppl. Fig. 8). Since our final constructs included a plasmid with kanamycin resistance, which was also found to produce some radicals, we used full (50 μg/ml) selection when growing up the cells but very low (5 μg/ml) selection during the experimental run. Persistence of oscillations, sequencing, and subsequent growth in full selection following the run confirmed the presence of all 3 plasmids despite this low experimental selection. Catalase and sodA results were identical to those with NDH-2 synchronization. These results show that fluorescence-mediated synchronization involves the production of radical species following fluorescence exposure and communication via H2O2.
Sensing Array of Biopixels
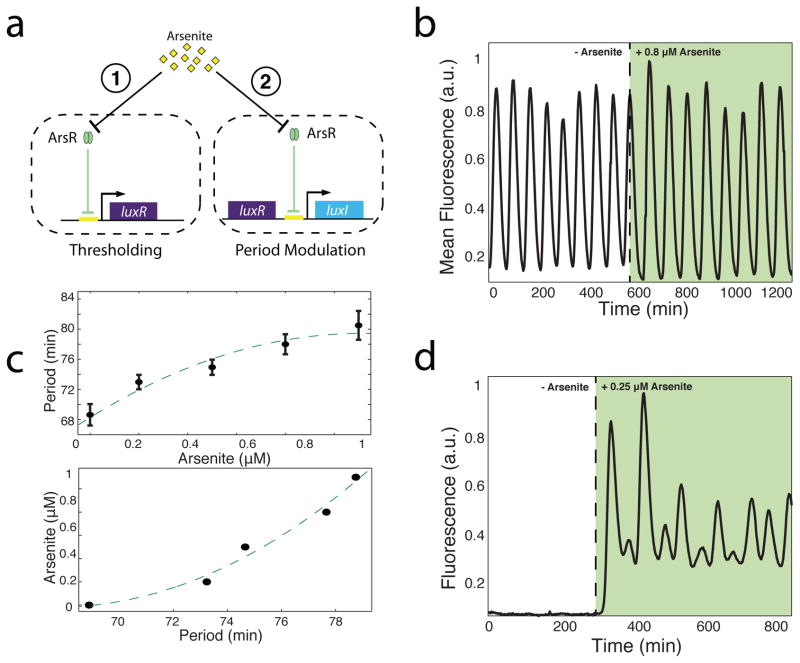
With a platform for generating consistent and readily-detectable oscillations, we sought to use the circuit to engineer an arsenic-sensing macroscopic biosensor. We rewired the network to include an extra copy of the positive-feedback element, the AHL-synthase LuxI, under the control of a native arsenite-responsive promoter which is repressed by ArsR in the absence of arsenite (Fig. 2a). When arsenite is not present in the media, supplemental luxI is not transcribed and the circuit functions normally, generating baseline oscillations. However, the addition of trace amounts of arsenite relieves this repression and allows supplemental luxI to be produced, increasing the oscillatory amplitude and period. Tuning the level of LuxI by varying arsenite concentration results in clear changes to the oscillatory period (Fig. 2b). To determine the range of detection, we swept arsenite concentrations from 0 to 1 μM and measured the oscillatory period (Fig. 2c, top). Using statistical methods (Suppl. Data Analysis), we generated a sensor calibration curve (Fig. 2c, bottom) that depicts the maximum possible arsenite concentration present (α = 95%) for a given measured period. This curve is an illustration of how data generated by our array would be used to measure arsenite concentrations in an unknown sample using our device. Our system was able to reliably quantify arsenite levels as low as 0.2 μM, below the 0.5 μM WHO-recommended level for developing nations42.
Fig. 2.
Frequency modulated genetic biosensor. (a) Network diagrams depicting two constructed sensing modules. In thresholding (1), the luxR gene is removed from the oscillator network and supplemented by a new copy driven by an arsenic-responsive promoter. In period modulation (2), a supplemental luxI gene tagged for increased degradation is driven by the arsenic-responsive promoter which affects the period of oscillation. (b) A sample period modulation sensor output following a step increase of 0.8 μM arsenite. Oscillatory period increases from 69 minutes to 79 minutes. (c) (Top) Period versus arsenite concentration for the sensor array. Error bars indicate ± 1 standard deviation averaged over 500 biopixel trajectories. Dotted line represents model-predicted curve. (Bottom) Sensor calibration curve generated from experimental data. Points indicate the maximum arsenite level with 95% certainty for a given measured period as determined statistically from experimental data. (d) Thresholder output following a step increase of 0.25 μM arsenite. A dramatic shift from rest to oscillatory behavior is observed within 20 minutes following the addition of arsenite.
As an alternative sensing strategy, we rewired the network to include a copy of the luxR gene controlled by an arsenic-responsive promoter while removing it from the rest of the circuit (Fig. 2a). Since the LuxR-AHL complex must be present to activate the lux promoter43, cells produce no LuxR when the media is free of arsenite, generating no fluorescence or oscillations. The addition of arsenite stimulates the production of LuxR, restoring circuit function and producing clear, synchronized oscillations (Fig. 2d). This ON/OFF detection system has a threshold of 0.25 μM, a detection limit that can be adjusted by changing the copy number, ribosome binding site (RBS) strength, or promoter strength of the sensing plasmid (Suppl. Plasmid Construction).
The sensing array is also capable of producing complex behaviors arising from the dynamic interaction of cellular colonies. By making modifications to the size, number, and arrangement of biopixels in the device, we are able to dramatically alter the output waveforms. For example, when we constructed a device in which trap separation distance is increased (45 μm versus 25 μm), we observed local anti-phase synchronization between neighboring colonies (Fig. 3d, top right). To explore this phenomenon on a larger scale, we constructed a device that contains an array of 416 traps constructed according to the specifications above. In these experiments, we observe initial global synchronization that gradually falls into local anti-phase synchronization across the array (Fig. 3d, middle and Suppl. Movie 4). Phase alignment is maintained over at least 48 hours, with patches of synchronization typically 3–6 colonies in size. Alternatively, by changing dimensions such that the array contains traps of two slightly different sizes, we observe a 1:2 resonance synchronization where larger traps pulse at double the frequency of smaller traps while maintaining synchronization (Fig. 3d, top). Finally, when LuxR is limited as in the thresholding scheme, we observe synchronized oscillations of alternating large and small peaks in both experiment and model (Suppl. Fig. 12). Our computational model (see Modeling Box) captures these effects (Fig. 3d, bottom and Suppl. Fig. 11,12) and indicates that further array manipulation will yield new, richer dynamics that could not be produced directly by changing circuit structure.
Fig. 3.
Computational modeling of radical synchronization and biosensing. (a) Time series of a population of biopixels producing varying amounts of H2O2 vapor. Synchronization occurs only for moderate levels while high levels lock ON and low levels oscillate asynchronously. (b) A typical time series for our period modulation sensor undergoing a step increase of arsenite. Oscillations increase in both amplitude and period. (c) A typical time series output for the thresholding sensor. Oscillations arise following the addition of arsenite. (d) Experimental and computational output depicting complex dynamic behaviors between neighboring traps. (Top 2 panels) 1:2 resonance and anti phase synchronization observed when trap size (left, black/blue = 95 μm depth and red/magenta = 85 μm depth) and separation distance (right, same colors) are modified experimentally, (Middle) Scaled-up array experimental data for increased trap separation experiments demonstrating anti phase synchronization, (Bottom) Computational model trajectories depicting 1:2 resonance and anti phase synchronization when trap size (same colors as experimental data) and separation distance are changed.
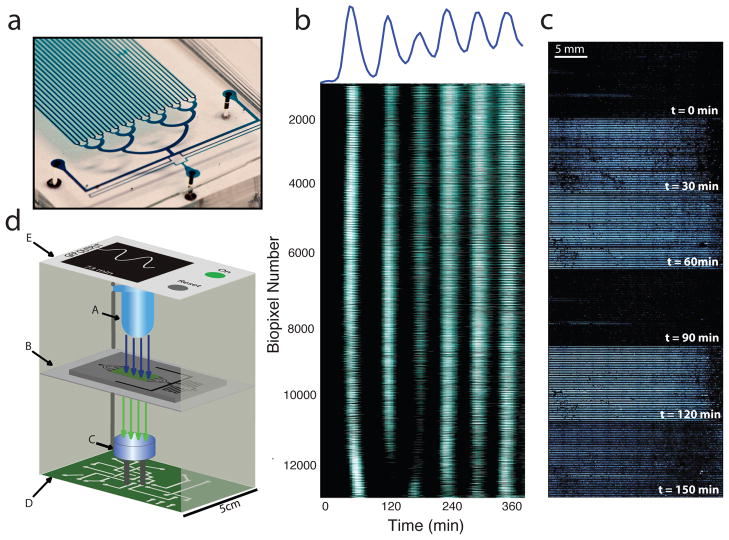
While our sensor array is capable of performing a variety of complex functions in the laboratory, adapting this technology to a real-world device will require the elimination of the expensive and bulky microscopy equipment. However, measuring genetic oscillations in the absence of any magnification or powerful illumination will require even further increased signal. Using this mechanism of global synchronization, we were able to scale up to a 24 mm × 12 mm array that houses over 12,000 communicating biopixels (Fig. 4a). Synchronization is maintained across the entire array, a distance over 5,000 times the length of an individual cell, using an inexpensive LED (Fig. 4b,c). The signal strength generated by the large number of cells in the array (about 50 million) will allow us to adapt the device to function as a handheld sensor. In our conceptual design (Fig. 4d), the sensor will continuously read the oscillatory frequency using off-the-shelf electronic components costing less than $50.
Fig. 4.
Radical synchronization on a macroscopic scale. (a) The scaled-up array is 24 mm × 12 mm and houses over 12,000 biopixels that contain approximately 50 million total cells when filled. (b) Global synchronization is maintained across the array. Heatmap of individual trajectories of all 12,224 oscillating biopixels. (c) Image series depicting global synchronization and oscillation for the macroscopic array. Each image is produced by stitching 72 fields of view imaged at 4X magnification. (d) Schematic diagram illustrating our design for a handheld device utilizing the sensing array. An LED (A) excites the array (B) and emitted light is collected by a photodetector (C), analyzed by an onboard processor (D), and displayed graphically (E).
There have been many examples of bacteria-based biosensors44–46, usually involving an optical reporter driven by a single promoter. Since optical intensity readings are sensitive to imaging conditions like beam power and exposure time, measurements must typically be normalized and calibrated. Measuring period of oscillation allows us to avoid these issues since peak-to-peak time does not depend on individual peak intensity. Secondly, oscillations produced at the colony level effectively decouple the signal from the growth state of individual cells, which can also affect fluorescence intensity. By using a dynamic readout that depends on communication between biopixels, we scan and tune potential output signals by changing device parameters rather than redesigning the underlying circuit. For example, we might design a new sensing scheme in which oscillations synchronize with the addition of some toxin and shift to anti phase or resonant synchronization when critical toxin levels are present.
Scaling Up Synthetic Biology
By nesting two modes of communication we are able to expand the scale over which individual cells are coordinated and increase the complexity of their interaction. Indeed, there are many familiar examples of hierarchical systems. Airline routes are often designed such that small airports are connected locally to larger hubs that are connected internationally. It would neither be feasible nor desirable to connect every airport together. Similarly, individual cells communicate locally by one method, generating impulses large enough to enable colonies to communicate globally by another. Nesting communication mechanisms in this way may allow us to better scale up synthetic circuits of different types, such as switches and logic gates, paving the way for the next generation of synthetic biology pursuits.
Modeling Box
Our model of the frequency modulated (FM) biosensor is based on a published model for the quorum-sensing synchronized oscillator14. In addition to the reactions reflected in that model, we include the arsenite-induced production and degradation of LuxI and/or LuxR. From the biochemical reactions, we derived a set of delay-differential equations to be used as our model. These delayed reactions mimic the complex cascade of processes (transcription, translation, maturation, etc.) leading to formation of functional proteins. As expected, our model predicts oscillations that change frequency when changes in arsenite occur (Fig. 2c and 3b). The amplitude and period of the oscillations both depend on the concentrations of the toxin. We then modified the model to describe the LuxR-based detection system. Our model predicts a marked transition from rest to oscillations upon addition of arsenite, consistent with experimental observations (Fig. 3c).
The multi-scale nature of communication in our array allows us to treat colony and array-level dynamics separately, where arsenite affects the quorum-sensing machinery of a colony, producing changes to oscillatory period that propagate between biopixels in the array. To quantitatively describe the mechanisms driving synchronization at the array-level, we treat each colony as a single oscillator that acts according to degrade-and-fire kinetics47. We also include the production of H2O2 and its interaction with neighboring colonies by two-dimensional diffusion. Using this model we identified three regimes that correlate well with experimental observations (Fig. 3a). When the effective production of H2O2 is low, as with catalase, we observe unsynchronized oscillations owing to constant, mild repression of the lux promoter via ArcA (Fig. 3a, left). In contrast, when H2O2 production is very high, neighboring colonies rapidly fire in succession and remain on due to the permanent activation of the lux promoter, consistent with the SOD experiment (Fig. 3a, right). Finally, at intermediate H2O2, we observe globally synchronized oscillations (Fig. 3a, middle). As colonies are moved further apart, synchronicity breaks due to slowed migration of H2O2 (Suppl. Fig. 10).
Supplementary Material
Acknowledgments
This work was supported by the National Institute of General Medical Sciences of the National Institutes of Health (GM069811), the Department of Defense National Defense Science and Engineering Graduate Fellowship (AP), and the National Science Foundation Graduate Research Fellowship (PS). LT was supported, in part, by ONR MURI grant N00014-07-0741. We would like to thank Jim Imlay, Sameh Ali, and Jim Collins for helpful discussions and Jen Hickman, Brooks Taylor, and Katie Lomax for their help with illustrations.
Footnotes
Supplementary information, including methods, supplementary figures and tables, is linked to the online version of the paper at www.nature.com/nature.
The authors declare no competing financial interests.
References
- 1.Gibson D, et al. Complete chemical synthesis, assembly, and cloning of a mycoplasma genitalium genome. Science. 2008;319:1215. doi: 10.1126/science.1151721. [DOI] [PubMed] [Google Scholar]
- 2.Gibson D, et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science. 2010;329:52. doi: 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
- 3.Hasty J, McMillen D, Collins JJ. Engineered gene circuits. Nature. 2002;420:224–30. doi: 10.1038/nature01257. [DOI] [PubMed] [Google Scholar]
- 4.Sprinzak D, Elowitz MB. Reconstruction of genetic circuits. Nature. 2005;438:443–8. doi: 10.1038/nature04335. [DOI] [PubMed] [Google Scholar]
- 5.Endy D. Foundations for engineering biology. Nature. 2005;438:449–53. doi: 10.1038/nature04342. [DOI] [PubMed] [Google Scholar]
- 6.Ellis T, Wang X, Collins J. Diversity-based, model-guided construction of synthetic gene networks with predicted functions. Nature biotechnology. 2009;27:465. doi: 10.1038/nbt.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kobayashi H, et al. Programmable cells: interfacing natural and engineered gene networks. Proc Natl Acad Sci U S A. 2004;101:8414–9. doi: 10.1073/pnas.0402940101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.You L, Cox I, RS, Weiss R, Arnold FH. Programmed population control by cell-cell communication and regulated killing. Nature. 2004;428:868–71. doi: 10.1038/nature02491. [DOI] [PubMed] [Google Scholar]
- 9.Basu S, Gerchman Y, Collins CH, Arnold FH, Weiss R. A synthetic multicellular system for programmed pattern formation. Nature. 2005;434:1130–4. doi: 10.1038/nature03461. [DOI] [PubMed] [Google Scholar]
- 10.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–42. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 11.Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–8. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- 12.Lu T, Collins J. Dispersing biofilms with engineered enzymatic bacteriophage. Proceedings of the National Academy of Sciences. 2007;104:11197. doi: 10.1073/pnas.0704624104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedland A, et al. Synthetic gene networks that count. Science. 2009;324:1199. doi: 10.1126/science.1172005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Danino T, Mondragon-Palomino O, Tsimring L, Hasty J. A synchronized quorum of genetic clocks. Nature. 2010;463:326–330. doi: 10.1038/nature08753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tabor J, et al. A synthetic genetic edge detection program. Cell. 2009;137:1272–1281. doi: 10.1016/j.cell.2009.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu T, Collins J. Engineered bacteriophage targeting gene networks as adjuvants for antibiotic therapy. Proceedings of the National Academy of Sciences. 2009;106:4629. doi: 10.1073/pnas.0800442106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stricker J, et al. A fast, robust and tunable synthetic gene oscillator. Nature. 2008;456:516–9. doi: 10.1038/nature07389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mondragon-Palomino O, Danino T, Selimkhanov J, Tsimring L, Hasty J. Entrainment of a population of synthetic genetic oscillators. Science. 2011;333 doi: 10.1126/science.1205369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Westinghouse G. System of electrical distribution. US Patent No. 1887;373:035. [Google Scholar]
- 20.Lewandowski W, Azoubib J, Klepczynski W. Gps: Primary tool for time transfer. Proceedings of the IEEE. 1999;87:163–172. [Google Scholar]
- 21.Vladimirov A, Kozyreff G, Mandel P. Synchronization of weakly stable oscillators and semiconductor laser arrays. EPL (Europhysics Letters) 2003;61:613. [Google Scholar]
- 22.Gast T. Sensors with oscillating elements. Journal of Physics E: Scientific Instruments. 1985;18:783. [Google Scholar]
- 23.Ozbudak EM, Thattai M, Kurtser I, Grossman AD, van Oudenaarden A. Regulation of noise in the expression of a single gene. Nat Genet. 2002;31:69–73. doi: 10.1038/ng869. [DOI] [PubMed] [Google Scholar]
- 24.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–6. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 25.Golding I, Paulsson J, Zawilski S, Cox E. Real-time kinetics of gene activity in individual bacteria. Cell. 2005;123:1025–1036. doi: 10.1016/j.cell.2005.09.031. [DOI] [PubMed] [Google Scholar]
- 26.Blake W, et al. Phenotypic consequences of promoter-mediated transcriptional noise. Molecular cell. 2006;24:853–865. doi: 10.1016/j.molcel.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 27.Huh D, Paulsson J. Non-genetic heterogeneity from stochastic partitioning at cell division. Nature genetics. 2010 doi: 10.1038/ng.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murphy K, Adams R, Wang X, Balázsi G, Collins J. Tuning and controlling gene expression noise in synthetic gene networks. Nucleic acids research. 2010;38:2712. doi: 10.1093/nar/gkq091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sevim V, Gong X, Socolar J. Reliability of transcriptional cycles and the yeast cell-cycle oscillator. PLoS Computational Biology. 2010;6:e1000842. doi: 10.1371/journal.pcbi.1000842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Austin D, et al. Gene network shaping of inherent noise spectra. Nature. 2006;439:608–611. doi: 10.1038/nature04194. [DOI] [PubMed] [Google Scholar]
- 31.Messner K, Imlay J. The identification of primary sites of superoxide and hydrogen peroxide formation in the aerobic respiratory chain and sulfite reductase complex of escherichia coli. Journal of Biological Chemistry. 1999;274:10119. doi: 10.1074/jbc.274.15.10119. [DOI] [PubMed] [Google Scholar]
- 32.Bose JL, et al. Bioluminescence in vibrio fischeri is controlled by the redox-responsive regulator arca. Molecular Microbiology. 2007;65:538–553. doi: 10.1111/j.1365-2958.2007.05809.x. [DOI] [PubMed] [Google Scholar]
- 33.Georgellis D, Kwon O, Lin E. Quinones as the redox signal for the arc two-component system of bacteria. Science. 2001;292:2314. doi: 10.1126/science.1059361. [DOI] [PubMed] [Google Scholar]
- 34.Seaver L, Imlay J. Hydrogen peroxide fluxes and compartmentalization inside growing escherichia coli. Journal of bacteriology. 2001;183:7182. doi: 10.1128/JB.183.24.7182-7189.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fridovich I. The biology of oxygen radicals. Science. 1978;201:875. doi: 10.1126/science.210504. [DOI] [PubMed] [Google Scholar]
- 36.McCord J, Fridovich I. Superoxide dismutase. Journal of Biological Chemistry. 1969;244:6049. [PubMed] [Google Scholar]
- 37.Berg J, Tymoczko JL, Stryer L. Biochemistry. W.H. Freeman; 2006. [Google Scholar]
- 38.Remington S. Fluorescent proteins: maturation, photochemistry and photophysics. Current opinion in structural biology. 2006;16:714–721. doi: 10.1016/j.sbi.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 39.Kelner M, Bagnell R, Welch K. Thioureas react with superoxide radicals to yield a sulfhydryl compound. explanation for protective effect against paraquat. Journal of Biological Chemistry. 1990;265:1306. [PubMed] [Google Scholar]
- 40.Touati D, Jacques M, Tardat B, Bouchard L, Despied S. Lethal oxidative damage and mutagenesis are generated by iron in delta fur mutants of escherichia coli: protective role of superoxide dismutase. Journal of bacteriology. 1995;177:2305. doi: 10.1128/jb.177.9.2305-2314.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kohanski MA, DePristo MA, Collins JJ. Sublethal antibiotic treatment leads to multidrug resistance via radical-induced mutagenesis. Molecular Cell. 2010;37:311–320. doi: 10.1016/j.molcel.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nordstrom D. Worldwide occurrences of arsenic in ground water. Science. 2002;296:2143. doi: 10.1126/science.1072375. [DOI] [PubMed] [Google Scholar]
- 43.Waters C, Bassler B. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 44.van der Meer J, Belkin S. Where microbiology meets microengineering: design and applications of reporter bacteria. Nature Reviews Microbiology. 2010;8:511–522. doi: 10.1038/nrmicro2392. [DOI] [PubMed] [Google Scholar]
- 45.Daunert S, et al. Genetically engineered whole-cell sensing systems: coupling biological recognition with reporter genes. Chemical Reviews. 2000;100:2705–2738. doi: 10.1021/cr990115p. [DOI] [PubMed] [Google Scholar]
- 46.Leveau J, Lindow S. Bioreporters in microbial ecology. Current opinion in microbiology. 2002;5:259–265. doi: 10.1016/s1369-5274(02)00321-1. [DOI] [PubMed] [Google Scholar]
- 47.Mather W, Bennett M, Hasty J, Tsimring L. Delay-Induced Degrade-and-Fire Oscillations in Small Genetic Circuits. Biophys J Phys Rev Lett. 2009;102:068105. doi: 10.1103/PhysRevLett.102.068105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Quan J, Tian J. Circular polymerase extension cloning of complex gene libraries and pathways. PloS one. 2009;4:e6441. doi: 10.1371/journal.pone.0006441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stocker J, et al. Development of a set of simple bacterial biosensors for quantitative and rapid measurements of arsenite and arsenate in potable water. Environ Sci Technol. 2003;37:4743–4750. doi: 10.1021/es034258b. [DOI] [PubMed] [Google Scholar]
- 50.Keiler K, Waller P, Sauer R. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger rna. Science. 1996;271:990. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.