Abstract
Height is a complex trait under strong genetic influence. To date, numerous genetic loci have been associated with height in individuals of European ancestry. However, few large-scale discovery genome-wide association studies (GWAS) of height in minority populations have been conducted and thus information about population-specific height regulation is limited. We conducted a GWA analysis of height in 8149 African-American (AA) women from the Women's Health Initiative. Genetic variants with P< 5 × 10−5 (n = 169) were followed up in a replication data set (n = 20 809) and meta-analyzed in a total of 28 958 AAs and African-descent individuals. Twelve single-nucleotide polymorphisms (SNPs) representing 7 independent loci were significantly associated with height at P < 5 × 10−8. We identified novel SNPs in 17q23 (TMEM100/PCTP) and Xp22.3 (ARSE) reflecting population-specific regulation of height in AAs and replicated five loci previously reported in European-descent populations [4p15/LCORL, 11q13/SERPINH1, 12q14/HMGA2, 17q23/MAP3K3 (mitogen-activated protein kinase3) and 18q21/DYM]. In addition, we performed an admixture mapping analysis of height which is both complementary and supportive to the GWA analysis and suggests potential associations between ancestry and height on chromosomes 4 (4q21), 15 (15q26) and 17 (17q23). Our findings provide insight into the genetic architecture of height and support the investigation of non-European-descent populations for identifying genetic factors associated with complex traits. Specifically, we identify new loci that may reflect population-specific regulation of height and report several known height loci that are important in determining height in African-descent populations.
INTRODUCTION
Adult body height is a quantitative trait under strong genetic influence, with heritability estimates ranging from 0.69–0.98 (1–3). Yet, prior to 2007, candidate gene and genome-wide linkage studies had limited success in identifying genes consistently associated with height (4). In recent years, genome-wide association studies (GWAS) conducted primarily in populations of European descent have identified new common genetic variants associated with height (5–10). A number of genes involved in skeletal growth and development, construction of extra-cellular matrix proteins as well as cell cycle regulation have been implicated (4). A recent study of 183 727 European-descent individuals reported 180 loci influencing adult height (11). Yet, genetic variation at these loci explains only ∼10% of the phenotypic variation in height, though the authors estimate that unidentified variants could account for up to 16% of the variation. Some of the remaining heritable contribution to height may be explained by small effects not detectable using current methods or by incomplete linkage disequilibrium (LD) between genotyped and causal single-nucleotide polymorphisms (SNPs) (12). Race-specific variants may also contribute to height, and a better understanding of the genetic factors associated with height in minority populations could provide insight into the genetic architecture of height and other complex traits. However, to date, few GWAS of height have been conducted in non-European-descent populations with the exception of studies in Asian populations, including Chinese (13), Japanese (14), Korean (15,16) and Filipino (17) individuals. One study of approximately 1930 African-descent individuals found 14 loci to be associated with height, though none reached genome-wide statistical significance (18); a limitation of this study was its low power to detect variants of moderate effect. As an admixed population, AAs have a genetic profile reflecting the mixing of two ancestral, genetically distinct populations. Because this population admixture increases the genomic diversity, it has important implications for complex traits (19,20) and can be used to identify regions of the genome where African and European ancestral alleles are associated with height. Therefore, we conducted discovery GWA and admixture mapping analyses of height in AA women from the Women's Health Initiative (WHI) SNP Health Association Resource (SHARe). We then replicated our top GWA findings from WHI in a large data set of AA and African-descent individuals.
RESULTS
Discovery GWA results in WHI AAs
The 8149 AA women included in the WHI SHARe height GWA had a mean (SD) age of 62 (7) years. At baseline, ∼5 and 53% of the women self-reported having a doctor-diagnosis of osteoporosis and osteoarthritis, respectively. Body height ranged from 146.2 to 177.0 cm and was approximately normally distributed with a mean (SD) of 162.5 (6.2) cm. Height was negatively correlated with age in the age range of the WHI population (50–79 years) and hence analyses are adjusted for age. Although osteoarthritis was not associated with body height in the WHI population, osteoporosis was significantly associated with modest decreases in height, beta (SD) = −0.70 (0.33) cm, P = 0.03; women with osteoporosis were excluded in sensitivity analyses.
The WHI discovery GWA included 855 034 genotyped SNPs. The genomic control inflation factor λ for the association analyses was 1.087 after controlling for four eigenvectors, suggesting limited residual population substructure or cryptic relatedness (Supplementary Material, Fig. S1). For polygenic traits such as height, elevated λ might be expected (21). WHI SHARe GWA findings are shown in Supplementary Material, Figure S2. In sensitivity analyses, we attempted to replicate GWAS findings (14 SNPs which did not reach genome-wide significance) from one study of height in AAs (18); none of these SNPs was significant at P = 0.05/14 in our WHI AA data set (data not shown).
Replication and combined meta-analysis of GWA findings
The replication data set included up to 20 809 AAs and African-descent individuals. Adding WHI SHARe to these numbers, we had a total sample size of 28 958 individuals for the meta-analysis (Supplementary Material, Table S1), though it varied by SNP. A total of 169 SNPs with P < 5 × 10−5 from the WHI-SHARe GWA discovery phase were carried forward to the replication phase. Of these, 12 were significant in the combined WHI discovery + replication meta-analysis at P < 5 × 10−8 (Table 1). Of the 12 significant SNPs, 9 were in regions previously reported to be associated with height in largely European-descent populations (4p15, 11q13, 12q14, 17q23/MAP3K3, 17q24/PRKCA and 18q21) and the remaining 3 SNPs appear to reflect novel regions (17q23/TMEM100-PCTP and Xp22) in AAs.
Table 1.
Significant findings from the replication meta-analysis in AAs
| SNP | Locus | Positiona | Closest gene(s) | Allele | CEU AFb | YRI AFb | WHI |
Replication data set |
Meta-analysis P-valued | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AF | n | Beta ± SEc | P-valuec | AF | n | Beta ± SEc | P-valuec | ||||||||
| Regions not previously associated with height | |||||||||||||||
| rs1549519e | 17q23 | 51 112 786 | TMEM100/PCTP | T | 0.79 | 0.05 | 0.22 | 8046 | −0.095 ± 0.020 | 2.19 × 10−6 | 0.20 | 20 413 | −0.048 ± 0.012 | 7.99 × 10−5 | 5.80 × 10−9 |
| rs1426745e | 17q23 | 51 131 494 | TMEM100/PCTP | G | 0.21 | 0.93 | 0.76 | 8100 | 0.092 ± 0.019 | 2.09 × 10−6 | 0.78 | 20 347 | 0.041 ± 0.012 | 4.72 × 10−4 | 4.82 × 10−8 |
| rs12393627 | Xp22 | 2 895 723 | ARSE | G | <0.01 | 0.43 | 0.36 | 8146 | 0.079 ± 0.016 | 1.46 × 10−6 | 0.37 | 7584 | 0.093 ± 0.019 | 6.35 × 10−7 | 4.96 × 10−12 |
| Regions previously associated with height | |||||||||||||||
| rs925098 | 4p15 | 17 528 909 | LCORL | G | 0.35 | 0.39 | 0.35 | 8101 | 0.084 ± 0.016 | 3.31 × 10−7 | 0.35 | 20 451 | 0.056 ± 0.010 | 5.55 × 10−9 | 3.05 × 10−14 |
| rs2320299 | 4p15 | 17 581 470 | LCORL | G | 0.35 | 0.47 | 0.37 | 8140 | 0.076 ± 0.016 | 2.54 × 10−6 | 0.37 | 20 496 | 0.049 ± 0.010 | 2.61 × 10−7 | 8.50 × 10−12 |
| rs2011603 | 4p15 | 17 634 582 | LCORL | T | 0.64 | 0.62 | 0.66 | 8147 | −0.096 ± 0.016 | 6.51 × 10−9 | 0.66 | 20 495 | −0.039 ± 0.010 | 1.12 × 10−4 | 2.71 × 10−10 |
| rs606452 | 11q13 | 74 953 826 | SERPINH1 | C | 0.84 | 0.55 | 0.65 | 8081 | −0.077 ± 0.017 | 3.47 × 10−6 | 0.64 | 20 474 | −0.043 ± 0.010 | 1.96 × 10−5 | 1.56 × 10−9 |
| rs7968682 | 12q14 | 64 658 147 | HMGA2 | T | 0.52 | 0.48 | 0.58 | 8144 | −0.073 ± 0.016 | 5.49 × 10−6 | 0.58 | 20 484 | −0.043 ± 0.009 | 4.68 × 10−6 | 4.22 × 10−10 |
| rs11658329 | 17q23 | 59 116 763 | MAP3K3 | G | 0.72 | 0.15 | 0.31 | 8140 | −0.072 ± 0.017 | 2.90 × 10−5 | 0.30 | 20 415 | −0.049 ± 0.010 | 2.07 × 10−6 | 4.89 × 10−10 |
| rs3889237 | 17q24 | 62 209 892 | PRKCA | C | 0.58 | <0.01 | 0.15 | 8015 | −0.096 ± 0.023 | 3.40 × 10−5 | 0.14 | 19 096 | −0.056 ± 0.014 | 7.92 × 10−5 | 3.25 × 10−8 |
| rs357897e | 18q21 | 44 838 182 | DYMf | T | 0.86 | 0.23 | 0.39 | 8139 | 0.071 ± 0.016 | 1.69 × 10−5 | 0.36 | 20 496 | 0.044 ± 0.010 | 5.96 × 10−6 | 1.09 × 10−9 |
| rs1787200e | 18q21 | 44 841 652 | DYMf | T | 0.89 | 0.23 | 0.40 | 8146 | 0.074 ± 0.016 | 6.00 × 10−6 | 0.37 | 20 478 | 0.044 ± 0.010 | 8.04 × 10−6 | 7.37 × 10−10 |
Chr, chromosome; AF, coded allele frequency; SE, standard error.
aChromosome position (Build 36).
bHapMap CEU and YRI allele frequencies.
cBeta coefficient representing the difference in mean height Z-score associated with each additional allele, and corresponding P-value.
dMeta-analysis of height Z-scores with inverse variance weighting.
eThese SNPs are in high LD with each other in WHI AAs.
fAlthough the DYM region has been previously associated with height in European Americans, these SNPs are not in LD with the reported SNPs.
Our top novel finding was rs12393627 (P = 5 × 10−12) located at Xp22, which has not been reported previously. The top two SNPs at 18q21 are located in the dymeclin (DYM) gene and are in high LD with each other in the WHI AA, r2 > 0.95 (Fig. 1). Another DYM SNP, rs8099594, has been previously associated with height in European-descent populations (4). In conditional analyses, adjustment for rs8099594 did not substantially change findings for either of our top DYM SNPs (Table 2). DYM is a large gene spanning almost 400 kb. Our DYM SNPs, rs357897 and rs1787200, are located >370 kb from the previously reported European height-associated locus and are in low LD with it in the WHI AA population (r2 < 0.3) as well as in HapMap CEU (r2 < 0.2) and YRI (r2 < 0.3). rs8099594 was not significantly associated with height in the WHI AA population, P = 0.208.
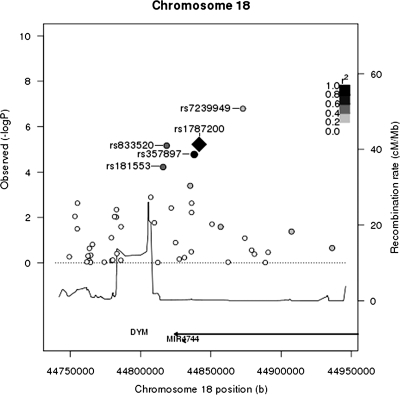
Figure 1.
Height association P-value and LD plot of the chromosome 18 DYM region P-values for WHI-only GWA results for SNPs within 150 kb of the top SNP from the replication, rs1787200, in the DYM region are shown. LD between rs1787200 and other SNPs in WHI is indicated using a gray scale, with black indicating the highest LD and white, the lowest LD. rs1787200 (black diamond) is in high LD with the other top DYM SNP from the replication meta-analysis, rs357897 (black circle). Top SNPs for the WHI-only analysis (rs7239949, rs833520 and rs181553) are also labeled.
Table 2.
Conditional analysis for significant loci
| Locus | Target SNP | Adjustment SNP | n (WHI only) | Target SNP marginal P-valuea | Target SNP conditional P-value |
|---|---|---|---|---|---|
| 17q23/TMEM100 | rs1549519 | rs11658329/MAP3K3 | 8037 | 1.95 × 10−6 | 4.57 × 10−5 |
| 17q23/TMEM100 | rs1426745 | rs11658329/MAP3K3 | 8091 | 1.87 × 10−6 | 4.13 × 10−5 |
| 17q24/PRKCA | rs3889237 | rs11658329/MAP3K3 | 8006 | 3.67 × 10−5 | 1.27 × 10−3 |
| 18q21/DYM | rs357897 | rs8099594/DYM | 8137 | 1.85 × 10−5 | 1.83 × 10−5 |
| 18q21/DYM | rs1787200 | rs8099594/DYM | 8140 | 6.88 × 10−6 | 6.47 × 10−6 |
aP-values may differ from those in Table 1 because only individuals with complete data for both the target and adjustment SNPs were included in the models.
Within 17q23–24, we have evidence both confirming previously identified loci and for a novel finding. The 17q23 SNP with the lowest P-value = 5 × 10−10, rs11658329, is located in an intron of the MAP3K3 gene. In CEU, it is in strong LD (r2 = 0.96) with rs2665838, an SNP in the chorionic somatomammotropin hormone 1 (CSH1)/growth hormone 1 (GH1) locus which has been associated with height in European-descent populations (11). rs11658329 is also in strong LD (r2 > 0.99) with rs8081612, an SNP in MAP3K3 previously associated with height in European-descent individuals and confirmed in a multi-ethnic cohort (22), which included small numbers of WHI AAs. We also identified two SNPs in the 17q23 locus, rs1549519 and rs1426745, which are located in the 3′ flanking region of the TMEM100 gene and are in high LD with each other (r2 = 0.89) (Fig. 2). Additionally, we identified rs3889237 in the nearby 17q24 locus, which is located in an intron of the PRKCA gene. In a conditional analysis, adjustment of the two 17q23 TMEM100 SNPs for the MAP3K3 SNP did not substantially affect their significance (Table 2). However, in spite of the large distance between the PRKCA SNP rs3889237 and the MAP3K3 SNP, results from the conditional analysis suggest that much of the rs3889237 association is explained by the MAP3K3 SNP. For this reason, we classify the 17q24/PRKCA finding as a region previously associated with height in Table 1, whereas the TMEM100 SNPs appear to be novel. In summary, we find evidence for two independent height-associated loci in chromosome 17q23–24 region: a locus near TMEM100-PCTP newly reported in AAs, and a second locus near the previously reported CSH1-GH1 European height region that, in AAs, is characterized by extended LD and encompasses MAP3K3 and PRCKA.
Figure 2.
Height association P-value and LD plot of the chromosome 17 TMEM100-PCTP region P-values for WHI-only GWA results for SNPs within 100 kb of the top hit in the TMEM100-PCTP region are shown. LD between rs1549519 and other SNPs in this region in the study population is indicated using a gray scale, with black indicating the highest LD and white, the lowest LD. rs1549519 (black diamond) is in high LD with the other top SNP from the replication meta-analysis, rs1426745.
For the other known regions, variants at 4p15, 11q23 and 12q24 were in moderate-to-strong LD (r2 > 0.7) in CEU with at least one SNP previously identified in European populations. The LD pattern for the 4p15 region is shown in Figure 3. The SNP with the lowest P-value in the replication, rs925098, is located in an intron of the ligand-dependent nuclear receptor co-repressor-like (LCORL) gene. It is in high LD, r2 > 0.7, with the other top SNPs that we identified in this region, rs2011603 and rs2320299. rs925098 is not in LD (r2 ≤ 0.1) in HapMap CEU with the previously reported LCORL SNPs: rs6449353 (11), rs6830062 (7,10), rs7678436 (14) or rs16896068 (5); however, it is in moderate-to-strong LD with rs6842303 (7) (r2 = 0.76) and rs2724475 (11) (r2 = 0.96), indicating that it is likely not a novel finding. Figure 3 also shows LD and association data in the WHI data set for the three previously reported LCORL SNPs that were genotyped in WHI (rs6830062, rs6842303 and rs16896068).
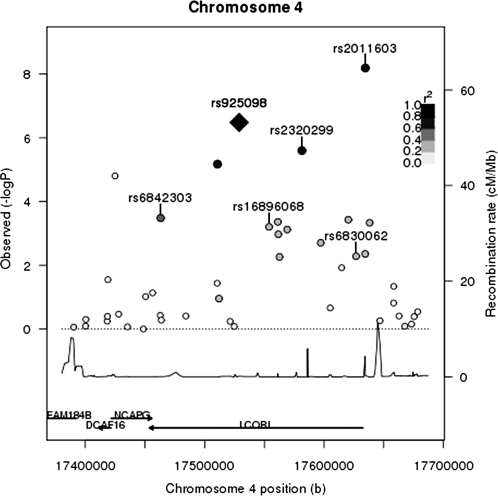
Figure 3.
Height association P-value and LD plot of the chromosome 4 LCORL region P-values for WHI-only GWA results within 150 kb of the top SNP identified in the replication meta-analysis, rs925098, in the LCORL region are shown. rs925098 is labeled with a black diamond. LD between rs925098 and other SNPs in this region in the WHI study population is indicated using a gray scale, with black indicating the highest LD and white, the lowest LD. SNPs in the region with modestly significant associations with height and in high LD (r2 > 0.6) with rs925098 include rs232099 and rs2011603. These data suggest that these three SNPs may not be independent. Other SNPs previously reported to be associated with height in European-descent populations (rs6842303, rs16896068 and rs6830062) are in lower LD with rs925098.
In an additional sensitivity analysis, we used publically available height GWAS results in approximately 130 000 European-descent individuals available from the GIANT consortium (11) to look up our top 11 findings from the autosomes; X chromosome results were not available. Of the 11 autosomal SNPs, 8 SNPs from the 4p15, 11q13, 12q14, 17q23 and 18q21 regions also reached genome-wide significance in the European-descent GIANT population (Supplementary Material, Table S3). However, the two novel 17q23 (TMEM100/PCTP) SNPs were not significant in GIANT (with P-values ranging from 0.14 to 0.18); nor was the 17q24 PRKCA SNP, rs3889237.
Admixture mapping results in WHI AAs
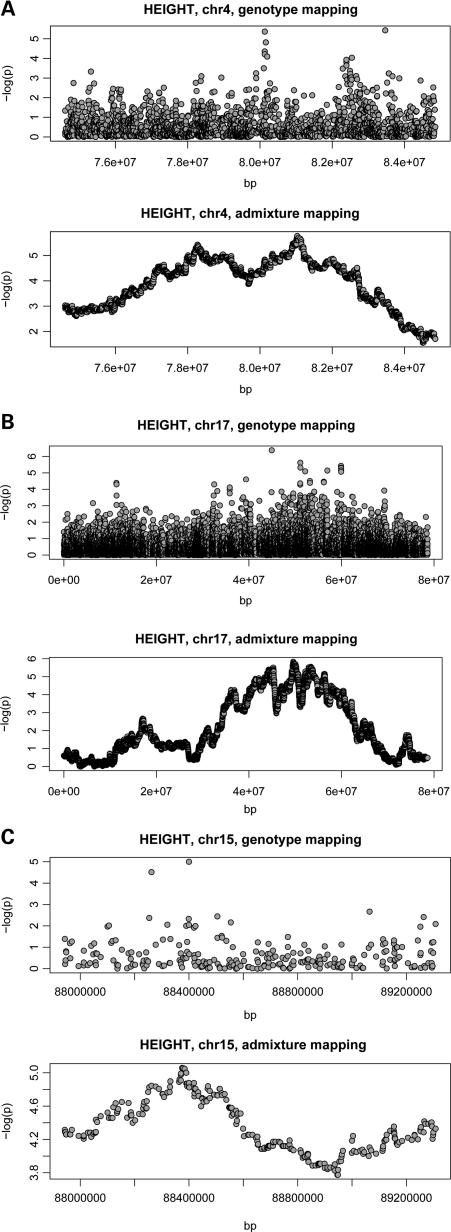
Admixture mapping identifies regions of the genome where the ancestry of a chromosomal region is associated with the trait of interest, after adjusting for the genome-wide average ancestry proportions. Results from the height admixture mapping analyses in WHI are shown in Figure 4. Low P-values meeting the genome-wide significance threshold in the admixture analysis (which is P < 1 × 10−5) on chromosomes 4, 12, 15 and 17 suggest that these regions may harbor variants that both influence height and have large allele frequency differences between the European and African ancestral populations. The strongest signal with the lowest P-values (P = 1–2 × 10−6) was found on chromosome 4 between 80 and 82 Mb (4q21). Within this region, several SNPs show suggestive genotype associations and thus could explain the admixture mapping signal. For example, rs899900 is moderately associated with height in WHI, P = 6.7 × 10−7. In conditional analyses, adjustment for rs899900 explains the majority of the admixture mapping signal. Although this chromosome 4 region does not overlap with the WHI GWA findings which were centered at 4p15, we found some moderate height associations (5 × 10−5< P < 1 × 10−8) on either end of this admixture region (Fig. 5A). On chromosome 17, we saw evidence for ancestral associations with height in the region between 40 and 60 Mb; these results are consistent with our SNP associations in this region (Fig. 5B). In fact, the majority of this ancestry signal is explained by an SNP near the TMEM100 gene, rs1549519. Results for chromosome 15 (15q26) were marginally significant, but again consistent for the SNP associations in this region (Fig. 5C). The modest peak on chromosome 12 is centered at the P-terminus close to the end of the chromosome where the local ancestry estimates tend to be noisier, and should be interpreted cautiously.
Figure 4.
WHI admixture mapping results. A plot of admixture mapping −log10 P-value results in the WHI data set on chromosomes 1–22 and X is shown. Much like a Manhattan plot, P-values are plotted in order of SNP position on the chromosome. The tall peaks reaching P-values of <1 × 10−5 suggest that SNPs indicative of African ancestry on chromosomes 4, 12 and 17 are associated with height.
Figure 5.
Comparison of GWA and admixture results in the WHI data set for chromosomal regions 4 (A), 17 (B) and 15 (C). In the WHI data set, GWA results for height are contrasted with height admixture mapping results for the same regions on the chromosome. Locations on the respective chromosome (in base pairs) are shown on the X-axes and −log10 P-values are indicated on the Y-axes.
DISCUSSION
In this discovery GWAS of height in AA women and replication population including a combined sample of more than 28 000 African-descent individuals, we identified new loci on chromosomes 17 (TMEM100/PCTP), 18 (DYM) and X (ARSE) significantly associated with height in AAs and replicated several height loci previously reported in European-descent individuals (4p15/LCORL, 11q13/SERPINH1, 12q14/HMGA2 and 17q23/MAP3K3). Using admixture mapping as a complementary approach to detect association signals in AAs, we identified a region on chromosome 4, independent of the LCORL locus, and regions on chromosomes 15 and 17, which potentially harbor population-specific variants that influence height, though these findings require further confirmation. Height is a trait that is influenced by many loci, some appear to be population specific, such as our GWA findings for chromosomes 17q23 and Xp22.3 and admixture findings for chromosome 4, whereas others are similar across European- and African-descent populations.
Of the novel findings, we identified a significant SNP in the Xp22.3 locus, rs12393627, which is located ∼3 kb upstream of the ARSE or arylsulfatase E (chondrodysplasia punctata 1) gene, though other arylsulfatase genes (ARSH, ARSD, ARSF) are also found in this region. The arylsulfatase family of proteins direct composition of bone and cartilage matrix. Rare mutations in the ARSE gene have been associated with short stature, among other features (23). Our SNP does not appear to be in high LD with any of the known functional variants in ARSE, but association data for the X chromosome are currently limited in the literature and in publically available databases, particularly for AAs. To our knowledge, we are the first to report the association between the Xp22 locus and common variation in body height using population data.
The DYM (18q21) region has previously been associated with height, yet our significant DYM SNPs were in a different part of the gene and were in low LD with the previously identified SNPs in the HapMap CEU, HapMap YRI and the WHI AA populations, which suggests that our SNPs may represent a distinct height locus in DYM in AA or perhaps are more highly correlated with the AA causal variant in this region. In our sensitivity analyses looking up GIANT results, these SNPs reached GWAS significance in European-descent populations also, which implies that the signal in this region may not be population specific.
The two SNPs that we identified at 17q23 (closest genes TMEM100 and PCTP) appear to be independent of MAP3K3 locus in WHI. Furthermore, they are located ∼1 Mb from the closest locus (NOG) associated with height in the large Lango Allen et al. study (11). Given their distance from NOG and lack of LD with the NOG SNP in the WHI population or in HapMap CEU (r2 < 0.01 in both populations), we believe that these SNPs are likely independent of the region identified by Lango Allen et al.
Although we identified novel loci associated with height, we also replicated loci previously associated with height in studies of European-descent populations, such as LCORL (5,7,10,11). The LCORL locus was also replicated in a height GWAS of Japanese individuals (14). Although the reported SNPs from all studies differ from our top LCORL SNPs, three were genotyped in our study. We replicated these associations and furthermore identified LCORL SNPs more robustly associated with height in AAs. Similarly, our 11q13 SNP located in an intron of the SERPINH1 gene and the 12q14 SNP which is downstream of HMGA2 gene are each in high LD in CEU (r2 > 0.93) with previously reported SNPs in these loci (11,22,24). The 17q22–24 region is rich in genes, including the growth hormone gene cluster encompassing a distance of ∼65 kb (25). Variation in the GH1 gene has been associated with height in both population (11,26) and family (27) studies. Our significant finding in the 17q24 locus, rs3889237, is located in an intron of the protein kinase C alpha (PRKCA) gene. PRKCA has been previously associated with BMI in females based on findings from a linkage study in Costa Rican families (28); however, it has not previously been associated with height. Results from conditional analyses in WHI data suggest that the PRKCA SNP is not independent from the MAP3K3 locus. Furthermore, this SNP was not significant in the European-descent GIANT consortium. This finding may reflect extended LD in this region as a result of admixture. Finally, our MAP3K3 SNP rs11658329 is in high LD in CEU with rs2665838, an SNP in the CSH1/GH1 locus previously associated with height in Lango Allen et al. (11) and also with an MAP3K3 SNP reported in Lanktree et al. (22).
Comparison of allele frequencies between HapMap CEU and YRI populations (Table 1) provides some clues as to why we were likely able to replicate previous findings and identify novel loci. For many of the previously reported SNPs, allele frequencies for CEU and YRI do not substantially differ from those in AA; we might expect associations to be consistent across races. Indeed, population allele frequencies were similar for the LCORL locus. In contrast, the allele for the Xp22 SNP associated with height in AA is essentially not present (or very rare in HapMap CEU), which demonstrates the advantage of studying a non-European population in which the allele frequency is higher and thus more likely to be identified through the GWAS approach. This X chromosome region did not pass the significance threshold in the admixture analyses, perhaps due to attenuated admixture associations as a result of its location at the P-terminus close to the end of the chromosome where the local ancestry estimates tend to be noisier. Again, focusing on allele frequency differences between the ancestral populations, admixture mapping provided findings at 17q23 consistent with the GWA and identified additional loci on chromosomes 4 (4q21) and 15 (15q26) associated with height. Although the lead SNPs driving these admixture signals at 4q21 and 15q26 were modestly associated with height in the WHI-GWAS, they were not significant in the replication. Although this may cast doubt on whether they are true findings, a full replication of these results would require an admixture replication data set, not just a GWAS replication.
The discovery GWA was conducted in WHI postmenopausal women ranging in age from 50 to 79. Adult height decreases with age due to conditions such as osteoporosis and osteoarthritis (10). Thus, height measurements in this older population may not reflect peak adult height (29). Self-reported osteoarthritis was not associated with height in the WHI AAs, but osteoporosis, though less prevalent, was associated with height. Excluding women with osteoporosis in the WHI analyses did not have appreciable effects on results for the top 12 SNPs from the replication (data not shown). Although osteoarthritis was prevalent in the WHI population, and in spite of reasonable power, we did not replicate an osteoarthritis-associated locus (GDF5-UQC) on chromosome 20 reported to contribute to height variation in largely European-descent populations (8); the P-value for rs6060369 was 0.07 in the WHI AA population. Importantly, we did not test rs143383 because it was not available in the WHI cohort; rs143383 was reported to be more strongly associated with height in AA perhaps due to LD differences at this locus.
Our replication data set included several cohorts, varying in age, sex, geographic location and in some cases, disease status. The use of normalized Z-scores in the meta-analysis allowed us to compare results across the varied populations while accounting for differences in the distributions of height. However, there was some evidence of heterogeneity in the meta-analysis. This heterogeneity is likely a reflection of the relatively large and varying Z-score beta values between WHI and the replication data set which included both AA and African subjects. However, in general, coded allele frequencies were consistent between the WHI and the replication data set, suggesting that major allele frequency differences between the populations are not of concern. An additional limitation pertains to the complexities of studying an admixed population. Although the Affymetrix 6.0 chip provides dense SNP coverage including over 900 000 SNPs, a recent analysis of 76 genes found that only ∼45–55% of SNPs, depending on the method employed, were tagged (r2 > 0.8) in Yoruban samples (30). Coverage for CEU was better, but these results suggest that association analyses using the Affymetrix 6.0 chip in AA populations may miss potentially important variants (20).
In spite of these limitations, our study was conducted in a large, well-characterized population with high-quality genetic data and measured body height. We are the first to report a discovery GWAS and admixture analysis of height in AA women. We pursued and replicated GWAS findings of interest using a large replication data set consisting of AAs and one small African cohort—identifying novel variants associated with height and generalizing findings from European-descent populations to AAs. Our synthesis of European-descent and AA results for chromosomes 4 and 18, in particular, may help to target future research in the LCORL and DYM genes. In addition, we describe the relationship between local ancestry and height—identifying potential associations between ancestry and height on chromosomes 4, 15 and 17. Overall, our findings provide insight into the genetic architecture of height and support investigation of non-European-descent populations for identifying genetic factors associated with complex traits.
MATERIALS AND METHODS
Study population
The WHI is a national health study focusing on strategies for preventing chronic diseases in postmenopausal women. A total of 161 808 women aged 50–79 years old were recruited from 40 clinical centers in the USA between 1993 and 1998 (31). The WHI cohort consists of an observational study, two clinical trials of postmenopausal hormone therapy (estrogen alone and estrogen plus progestin), a clinical trial of calcium and vitamin D supplements and a dietary modification trial. Study recruitment and exclusion criteria have been described previously (31). Study protocols and consent forms were approved by the institutional review boards at all participating institutions. The WHI SHARe minority cohort includes 8515 self-identified AA women from WHI who provided written informed consent for study participation and DNA analysis. Body height (cm) was measured at baseline using wall-mounted stadiometers. A total of 47 women did not have baseline height. We used height data collected at follow-up visits (all within 3 years) on 41 of these women; 6 women without any height data were excluded. Body height measurements were truncated for the 1st and 99th percentiles (n = 157). This truncation was requested by the Fred Hutchinson Cancer Research Center Institutional Review Board to preserve participant anonymity upon dbGAP submission; we retained these truncated values in the analyses. Research was approved by the Fred Hutchinson Cancer Research Center Institutional Review Board.
Genotyping
Genetic data were obtained from genome-wide scans using the Genome-wide Human SNP Array 6.0 (Affymetrix, Santa Clara, CA, USA, www.affymetrix.com) containing 909622 SNPs and following the manufacturer's protocol. Genotyping quality control included examination of concordance rates for blinded and un-blinded duplicates. Approximately 1% of SNPs failed genotyping and SNPs with call rates <95% or concordance rates <98%, minor allele frequency (MAF) <1% were excluded. After these exclusions, 855034 SNPs on chromosomes 1–22 and X were available for association analysis. Genotyping failed in 99 samples. Additional participants were excluded based on low call rates <95%, and sex or race/ethnicity discrepancy. First- or second-degree relatedness was assessed as described in Thornton and McPeek (32). For the related individuals, the first- or second-degree relative with the highest call rate was retained and other family members were excluded. After applying all exclusions, a total of 8149 AA women with available height data were included in the analysis.
Population stratification correction
A principal-component (PC) analysis of all samples was performed using EIGENSTRAT (33). The first 10 PCs were calculated for each individual and evaluated for their contribution to ancestral variation. Because most of the ancestral variation was explained by the first four PCs, only these were included as covariates in the analyses.
Association analysis
Linear regression models were used to assess associations between each SNP and baseline body height (continuous). Models were adjusted for age and ancestry PCs 1–4. Using additive genetic models, each SNP was coded as a count of the variant alleles (0, 1 or 2). Results for SNPs with minor allele frequencies ≥0.01 are reported. We inspected quantile–quantile (QQ) plots to determine whether the distribution of the observed SNP association P-values was consistent with the null distribution. We calculated lambda (λ), an indicator of over-dispersion due to potential population stratification by dividing the mean of the test statistics by the mean of the expected values from a χ2 distribution with 1 degree of freedom. Using λ, we investigated correction of P-values using genomic control to account for potential residual confounding by genetic ancestry (34,35). Analyses were conducted using the R software (version 2.11.1).
Plots were prepared in R using International HapMap3 data for Yoruba in Ibadan, Nigeria (YRI), and Utah residents with ancestry from northern and western Europe (CEU) (http://hapmap.ncbi.nlm.nih.gov/). HapMap population LD estimates were obtained from the SNP Annotation and Proxy Search (SNAP) software (36). Power calculations were conducted assuming additive genetic models using Quanto v1.2 (37,38).
Replication
SNPs that achieved genome-wide significance of P<5×10−8 or that had highly suggestive P-values (5×10−8<P<5×10−5) in the WHI data were tested in an independent AA- and African-descent population of approximately 20000 men and women (39). This replication data set includes data on AAs from the Candidate-Gene Association Resource (CARe) Consortium (40) including Atherosclerosis Risk in Communities (ARIC), Coronary Artery Risk Development in young Adults (CARDIA), Cleveland Family Study (CFS), Jackson Heart Study (JHS) and the Multi-Ethnic Study of Atherosclerosis (MESA); the African Americans Breast Cancer Cohort (AABC) consisting of nine studies (in preparation), the African Americans Prostate Cancer Cohort (AAPC) consisting of 10 studies (Nat. Genet., in press), the Maywood cohort and one African cohort recruited from Igbo-Ora and Ibadan in southwest Nigeria (18). All participants were genotyped on the Affymetrix 6.0 platform; additional details about the height consortium are described in N'Diaye et al. (39). Stratifying on sex and case–control status (if applicable), height was regressed on age and other appropriate covariates (e.g. recruitment center), after which the height residuals were normalized into height Z-scores. Height Z-scores from the different sub-data sets were combined and associations between genotyped or imputed SNPs and height Z-scores were tested using linear regression models with adjustment for ancestry. SNPs were coded assuming additive genetic models. Association results from all studies were combined in a meta-analysis using the inverse variance meta-analytic method as implemented in the software METAL (41).
Meta-analysis
To facilitate meta-analyses using WHI SHARe results and results from the height replication data set, we re-analyzed the replication SNPs in WHI SHARe data using height Z-scores as the outcome. Briefly, we regressed age on height and normalized the height residuals. In linear regression models using these normalized residuals or height Z-scores as the outcome, we tested for associations with each SNP and included PCs for ancestry adjustment. We then combined these WHI Z-score results with the height replication data set Z-score results and conducted an inverse variance-weighted meta-analysis using METAL (41).
Within WHI alone, we estimated our power to detect associations with a small 1.0 cm change in height to be ∼20% for SNPs with MAF = 0.05 and α = 5 × 10−8, increasing to >77% for SNPs with MAF ≥ 0.10. Although our initial power to detect small differences in height associated with rare alleles was low, our power to detect small changes in height in the replication meta-analysis for rare alleles (MAF ∼0.02) was ∼50% increasing to >99% for alleles with MAF ≥ 0.05.
Admixture analysis
For each AA individual in the sample, locus-specific ancestry was estimated using a higher order extension (up to order 15) of the Markov-hidden Markov model (42). Briefly, this method uses a graphical model approach to adaptively capture local haplotype structure within each ancestral population, and thereby more accurately accounts for background LD (Johnson et al., in preparation). In simulation studies, this method compares favorably with existing methods in terms of the accuracy and computational efficiency; the error rate was <1.5%. In the current analysis, phased haplotype data from the HapMap3 CEU and YRI individuals were augmented as the reference panels. Local ancestry data were coded as 0 = two European alleles, 1 = one European/one African allele, 2 = two African alleles at each locus. Using the local ancestry estimates, we performed an admixture mapping analysis to detect variants that are present at different frequencies among the European and AA ancestral populations. In this analysis, we regressed height on locus-specific ancestry, adjusting for age and genome-wide ancestry proportions using the first four PCs (43). Given the correlation between SNPs along the genome, the number of independent tests is reduced compared with the GWAS and hence significance level was assessed at the more liberal threshold of P < 1 × 10−5 (44).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
This work was supported by the National Heart, Lung, and Blood Institute, National Institutes of Health, and the US Department of Health and Human Services through contracts N01WH22110, 24152, 32100-2, 32105-6, 32108-9, 32111-13, 32115, 32118-32119, 32122, 42107-26, 42129-32 and 44221. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript. Funding information for the replication data set is provided in Supplementary Material.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the WHI investigators and staff for their dedication, and the study participants for making the program possible. A full listing of WHI investigators is available at http://www.whiscience.org/publications/WHI_investigators_shortlist.pdf.
REFERENCES
- 1.Perola M., Sammalisto S., Hiekkalinna T., Martin N.G., Visscher P.M., Montgomery G.W., Benyamin B., Harris J.R., Boomsma D., Willemsen G., et al. Combined genome scans for body stature in 6,602 European twins: evidence for common Caucasian loci. PLoS Genet. 2007;3:e97. doi: 10.1371/journal.pgen.0030097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sale M.M., Freedman B.I., Hicks P.J., Williams A.H., Langefeld C.D., Gallagher C.J., Bowden D.W., Rich S.S. Loci contributing to adult height and body mass index in African American families ascertained for type 2 diabetes. Ann. Hum. Genet. 2005;69:517–527. doi: 10.1046/j.1529-8817.2005.00176.x. [DOI] [PubMed] [Google Scholar]
- 3.Hirschhorn J.N., Lindgren C.M., Daly M.J., Kirby A., Schaffner S.F., Burtt N.P., Altshuler D., Parker A., Rioux J.D., Platko J., et al. Genomewide linkage analysis of stature in multiple populations reveals several regions with evidence of linkage to adult height. Am. J. Hum. Genet. 2001;69:106–116. doi: 10.1086/321287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weedon M.N., Frayling T.M. Reaching new heights: insights into the genetics of human stature. Trends Genet. 2008;24:595–603. doi: 10.1016/j.tig.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 5.Weedon M.N., Lango H., Lindgren C.M., Wallace C., Evans D.M., Mangino M., Freathy R.M., Perry J.R., Stevens S., Hall A.S., et al. Genome-wide association analysis identifies 20 loci that influence adult height. Nat. Genet. 2008;40:575–583. doi: 10.1038/ng.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lettre G., Jackson A.U., Gieger C., Schumacher F.R., Berndt S.I., Sanna S., Eyheramendy S., Voight B.F., Butler J.L., Guiducci C., et al. Identification of ten loci associated with height highlights new biological pathways in human growth. Nat. Genet. 2008;40:584–591. doi: 10.1038/ng.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gudbjartsson D.F., Walters G.B., Thorleifsson G., Stefansson H., Halldorsson B.V., Zusmanovich P., Sulem P., Thorlacius S., Gylfason A., Steinberg S., et al. Many sequence variants affecting diversity of adult human height. Nat. Genet. 2008;40:609–615. doi: 10.1038/ng.122. [DOI] [PubMed] [Google Scholar]
- 8.Sanna S., Jackson A.U., Nagaraja R., Willer C.J., Chen W.M., Bonnycastle L.L., Shen H., Timpson N., Lettre G., Usala G., et al. Common variants in the GDF5-UQCC region are associated with variation in human height. Nat. Genet. 2008;40:198–203. doi: 10.1038/ng.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Estrada K., Krawczak M., Schreiber S., van Duijn K., Stolk L., van Meurs J.B., Liu F., Penninx B.W., Smit J.H., Vogelzangs N., et al. A genome-wide association study of northwestern Europeans involves the C-type natriuretic peptide signaling pathway in the etiology of human height variation. Hum. Mol. Genet. 2009;18:3516–3524. doi: 10.1093/hmg/ddp296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soranzo N., Rivadeneira F., Chinappen-Horsley U., Malkina I., Richards J.B., Hammond N., Stolk L., Nica A., Inouye M., Hofman A., et al. Meta-analysis of genome-wide scans for human adult stature identifies novel loci and associations with measures of skeletal frame size. PLoS Genet. 2009;5:e1000445. doi: 10.1371/journal.pgen.1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lango Allen H., Estrada K., Lettre G., Berndt S.I., Weedon M.N., Rivadeneira F., Willer C.J., Jackson A.U., Vedantam S., Raychaudhuri S., et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang J., Benyamin B., McEvoy B.P., Gordon S., Henders A.K., Nyholt D.R., Madden P.A., Heath A.C., Martin N.G., Montgomery G.W., et al. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lei S.F., Yang T.L., Tan L.J., Chen X.D., Guo Y., Guo Y.F., Zhang L., Liu X.G., Yan H., Pan F., et al. Genome-wide association scan for stature in Chinese: evidence for ethnic specific loci. Hum. Genet. 2009;125:1–9. doi: 10.1007/s00439-008-0590-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okada Y., Kamatani Y., Takahashi A., Matsuda K., Hosono N., Ohmiya H., Daigo Y., Yamamoto K., Kubo M., Nakamura Y., et al. A genome-wide association study in 19 633 Japanese subjects identified LHX3-QSOX2 and IGF1 as adult height loci. Hum. Mol. Genet. 2010;19:2303–2312. doi: 10.1093/hmg/ddq091. [DOI] [PubMed] [Google Scholar]
- 15.Cho Y.S., Go M.J., Kim Y.J., Heo J.Y., Oh J.H., Ban H.J., Yoon D., Lee M.H., Kim D.J., Park M., et al. A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat. Genet. 2009;41:527–534. doi: 10.1038/ng.357. [DOI] [PubMed] [Google Scholar]
- 16.Kim J.J., Lee H.I., Park T., Kim K., Lee J.E., Cho N.H., Shin C., Cho Y.S., Lee J.Y., Han B.G., et al. Identification of 15 loci influencing height in a Korean population. J. Hum. Genet. 2010;55:27–31. doi: 10.1038/jhg.2009.116. [DOI] [PubMed] [Google Scholar]
- 17.Croteau-Chonka D.C., Marvelle A.F., Lange E.M., Lee N.R., Adair L.S., Lange L.A., Mohlke K.L. Genome-wide association study of anthropometric traits and evidence of interactions with age and study year in Filipino women. Obesity. 2011;19:1019–1027. doi: 10.1038/oby.2010.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang S.J., Chiang C.W., Palmer C.D., Tayo B.O., Lettre G., Butler J.L., Hackett R., Adeyemo A.A., Guiducci C., Berzins I., et al. Genome-wide association of anthropometric traits in African- and African-derived populations. Hum. Mol. Genet. 2010;19:2725–2738. doi: 10.1093/hmg/ddq154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patterson N., Petersen D.C., van der Ross R.E., Sudoyo H., Glashoff R.H., Marzuki S., Reich D., Hayes V.M. Genetic structure of a unique admixed population: implications for medical research. Hum. Mol. Genet. 2010;19:411–419. doi: 10.1093/hmg/ddp505. [DOI] [PubMed] [Google Scholar]
- 20.Cooper R.S., Tayo B., Zhu X. Genome-wide association studies: implications for multiethnic samples. Hum. Mol. Genet. 2008;17:R151–R155. doi: 10.1093/hmg/ddn263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang J., Weedon M.N., Purcell S., Lettre G., Estrada K., Willer C.J., Smith A.V., Ingelsson E., O'Connell J.R., Mangino M., et al. Genomic inflation factors under polygenic inheritance. Eur. J. Hum. Genet. 2011;19:807–812. doi: 10.1038/ejhg.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lanktree M.B., Guo Y., Murtaza M., Glessner J.T., Bailey S.D., Onland-Moret N.C., Lettre G., Ongen H., Rajagopalan R., Johnson T., et al. Meta-analysis of dense genecentric association studies reveals common and uncommon variants associated with height. Am. J. Hum. Genet. 2011;88:6–18. doi: 10.1016/j.ajhg.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Casarin A., Rusalen F., Doimo M., Trevisson E., Carraro S., Clementi M., Tenconi R., Baraldi E., Salviati L. X-linked brachytelephalangic chondrodysplasia punctata: a simple trait that is not so simple. Am. J. Med. Genet. A. 2009;149A:2464–2468. doi: 10.1002/ajmg.a.33039. [DOI] [PubMed] [Google Scholar]
- 24.Weedon M.N., Lettre G., Freathy R.M., Lindgren C.M., Voight B.F., Perry J.R., Elliott K.S., Hackett R., Guiducci C., Shields B., et al. A common variant of HMGA2 is associated with adult and childhood height in the general population. Nat. Genet. 2007;39:1245–1250. doi: 10.1038/ng2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.George D.L., Phillips J.A., III, Francke U., Seeburg P.H. The genes for growth hormone and chorionic somatomammotropin are on the long arm of human chromosome 17 in region q21 to qter. Hum. Genet. 1981;57:138–141. doi: 10.1007/BF00282009. [DOI] [PubMed] [Google Scholar]
- 26.Millar D.S., Lewis M.D., Horan M., Newsway V., Easter T.E., Gregory J.W., Fryklund L., Norin M., Crowne E.C., Davies S.J., et al. Novel mutations of the growth hormone 1 (GH1) gene disclosed by modulation of the clinical selection criteria for individuals with short stature. Hum. Mutat. 2003;21:424–440. doi: 10.1002/humu.10168. [DOI] [PubMed] [Google Scholar]
- 27.Wagner J.K., Eble A., Hindmarsh P.C., Mullis P.E. Prevalence of human GH-1 gene alterations in patients with isolated growth hormone deficiency. Pediatr. Res. 1998;43:105–110. doi: 10.1203/00006450-199801000-00016. [DOI] [PubMed] [Google Scholar]
- 28.Murphy A., Tantisira K.G., Soto-Quiros M.E., Avila L., Klanderman B.J., Lake S., Weiss S.T., Celedon J.C. PRKCA: a positional candidate gene for body mass index and asthma. Am. J. Hum. Genet. 2009;85:87–96. doi: 10.1016/j.ajhg.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sorkin J.D., Muller D.C., Andres R. Longitudinal change in height of men and women: implications for interpretation of the body mass index: the Baltimore Longitudinal Study of Aging. Am. J. Epidemiol. 1999;150:969–977. doi: 10.1093/oxfordjournals.aje.a010106. [DOI] [PubMed] [Google Scholar]
- 30.Bhangale T.R., Rieder M.J., Nickerson D.A. Estimating coverage and power for genetic association studies using near-complete variation data. Nat. Genet. 2008;40:841–843. doi: 10.1038/ng.180. [DOI] [PubMed] [Google Scholar]
- 31.The Women's Health Initiative Study Group. Design of the Women's Health Initiative clinical trial and observational study. Control Clin. Trials. 1998;19:61–109. doi: 10.1016/s0197-2456(97)00078-0. [DOI] [PubMed] [Google Scholar]
- 32.Thornton T., McPeek M.S. ROADTRIPS: case-control association testing with partially or completely unknown population and pedigree structure. Am. J. Hum. Genet. 2010;86:172–184. doi: 10.1016/j.ajhg.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 34.Devlin B., Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 35.Bacanu S.A., Devlin B., Roeder K. Association studies for quantitative traits in structured populations. Genet. Epidemiol. 2002;22:78–93. doi: 10.1002/gepi.1045. [DOI] [PubMed] [Google Scholar]
- 36.Johnson A.D., Handsaker R.E., Pulit S.L., Nizzari M.M., O'Donnell C.J., de Bakker P.I. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gauderman W.J., Morrison J.M. QUANTO 1.1: a computer program for power and sample size calculations for genetic-epidemiology studies. 2006 [Google Scholar]
- 38.Gauderman W.J. Sample size requirements for association studies of gene-gene interaction. Am. J. Epidemiol. 2002;155:478–484. doi: 10.1093/aje/155.5.478. [DOI] [PubMed] [Google Scholar]
- 39.N'Diaye A., Chen G.K., Palmer C.D., Ge B., Tayo B., Mathias R.A., Ding J., Nalls M.A., Adeyemo A., et al. Identification, replication, and fine-mapping of loci associated with adult height in individuals of African ancestry. PLoS. Genet. 2011;7:e1002298. doi: 10.1371/journal.pgen.1002298. doi:10.1371/journal.pgen.1002298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lettre G., Palmer C.D., Young T., Ejebe K.G., Allayee H., Benjamin E.J., Bennett F., Bowden D.W., Chakravarti A., Dreisbach A., et al. Genome-wide association study of coronary heart disease and its risk factors in 8,090 African Americans: The NHLBI CARe Project. PLoS Genet. 2011;7:e1001300. doi: 10.1371/journal.pgen.1001300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tang H., Coram M., Wang P., Zhu X., Risch N. Reconstructing genetic ancestry blocks in admixed individuals. Am. J. Hum. Genet. 2006;79:1–12. doi: 10.1086/504302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Basu A., Tang H., Arnett D., Gu C.C., Mosley T., Kardia S., Luke A., Tayo B., Cooper R., Zhu X., et al. Admixture mapping of quantitative trait loci for BMI in African Americans: evidence for loci on chromosomes 3q, 5q, and 15q. Obesity. 2009;17:1226–1231. doi: 10.1038/oby.2009.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang H., Siegmund D.O., Johnson N.A., Romieu I., London S.J. Joint testing of genotype and ancestry association in admixed families. Genet. Epidemiol. 2010;34:783–791. doi: 10.1002/gepi.20520. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.