Abstract
Genome-wide association studies have identified common variation in the CHRNA5–CHRNA3–CHRNB4 and CHRNA6–CHRNB3 gene clusters that contribute to nicotine dependence. However, the role of rare variation in risk for nicotine dependence in these nicotinic receptor genes has not been studied. We undertook pooled sequencing of the coding regions and flanking sequence of the CHRNA5, CHRNA3, CHRNB4, CHRNA6 and CHRNB3 genes in African American and European American nicotine-dependent smokers and smokers without symptoms of dependence. Carrier status of individuals harboring rare missense variants at conserved sites in each of these genes was then compared in cases and controls to test for an association with nicotine dependence. Missense variants at conserved residues in CHRNB4 are associated with lower risk for nicotine dependence in African Americans and European Americans (AA P = 0.0025, odds-ratio (OR) = 0.31, 95% confidence-interval (CI) = 0.31–0.72; EA P = 0.023, OR = 0.69, 95% CI = 0.50–0.95). Furthermore, these individuals were found to smoke fewer cigarettes per day than non-carriers (AA P = 6.6 × 10−5, EA P = 0.021). Given the possibility of stochastic differences in rare allele frequencies between groups replication of this association is necessary to confirm these findings. The functional effects of the two CHRNB4 variants contributing most to this association (T375I and T91I) and a missense variant in CHRNA3 (R37H) in strong linkage disequilibrium with T91I were examined in vitro. The minor allele of each polymorphism increased cellular response to nicotine (T375I P = 0.01, T91I P = 0.02, R37H P = 0.003), but the largest effect on in vitro receptor activity was seen in the presence of both CHRNB4 T91I and CHRNA3 R37H (P = 2 × 10−6).
INTRODUCTION
Genetic variation in the cholinergic nicotinic receptor genes (CHRNs) has repeatedly been found to be highly associated with nicotine dependence (reviewed in 1). Multiple independent variants have been identified that contribute to smoking-related phenotypes. Among these, the most strongly associated single nucleotide polymorphism (SNP) in several genome-wide association studies (GWAS) of nicotine dependence and correlated traits is a common non-synonymous change (rs16969968/D398N) in CHRNA5 (2). Despite big differences in the frequency of this SNP across populations, it shows a similar effect on risk, with odds ratios 1.3–2.00 in European, African American and Asian populations (3,4). In vitro functional studies have demonstrated that receptors including the amino acid encoded by the minor allele, which is the risk allele, show a reduced response to the nicotine agonist, epibatidine (5). Human carriers of the minor allele show decreased intrinsic resting connectivity strength in the dorsal anterior cingulate–ventral striatum/extended amygdala circuit (6). Additionally, a group of highly correlated variants, including the SNP rs588765 near CHRNA5, were shown to increase CHRNA5 mRNA expression and increase risk of nicotine dependence independently of rs16969968 (7,8). Common variants at the CHRNA6–CHRNB3 gene cluster were also recently shown to contribute to smoking quantity (2). Together, these results strongly suggest multiple mechanisms connecting nicotinic receptor function to nicotine dependence. Despite these findings, only a small proportion of the variance (∼5%) in nicotine dependence-related traits has been explained by these variants (7).
Neuronal nicotinic acetylcholine receptors are pentameric ion channels produced from various combinations of α and β receptor subunits and are the main target for nicotine in the brain. The vast majority of neuronally expressed nicotinic receptors contain the α4 and β2 subunits, but many other combinations are formed and have distinct properties and expression patterns. As such, it is thought that the various aspects of nicotine dependence (craving, withdrawal, tolerance, sensitivity, etc.) may be due to these distinct characteristics. For example, recent studies have shown that mice lacking Chrna5 fail to display the adverse effects of high doses of nicotine and mice lacking Chrnb4 or Chrna5 show reduced somatic signs of withdrawal after ceasing long-term nicotine treatment (9,10). Additionally, mice over-expressing Chrnb4 were recently shown to display a strong aversion to nicotine that can be reversed by virally mediated expression of α5 D398N variant in the medial habenula (11). This work suggests that aversion of nicotine in the mouse is regulated by the balanced activity of β4 and α5 nicotinic receptor subunits in the medial habenula. A recent association study of smoking cessation also suggests that variants in CHRNB4 may decrease craving and withdrawal symptoms (12). These findings suggest that multiple nicotinic receptor subunit genes likely play a role in the development and maintenance of nicotine dependence and that variants in these genes may have different and in some cases opposing functional effects.
Rare genetic variants have recently been shown to contribute to a number of common human diseases (reviewed in 13). Specifically, multiple rare variants have been reported in genes previously shown to harbor common variants associated with common diseases (14–16). Additionally, studies have demonstrated associations between rare variation in nicotinic receptor genes and nicotine dependence (17,18). As a class, rare SNPs, copy-number variants and small insertion/deletion polymorphisms (indels) constitute the majority of human genetic variation and thus may hold the key to understanding part of the missing heritability of complex traits unaccounted for by recent GWAS. Sequencing is one of the few methods by which these variants can be investigated; however, the cost of sequencing hundreds or thousands of individuals for multiple genes remains high despite advances in sequencing technologies. An efficient alternative approach is to carry out DNA sequencing using DNA pooled from multiple individuals. To further characterize the role of rare genetic variation in the nicotinic acetylcholine receptors on nicotine dependence, we have employed this approach to identify rare variants in the CHRNA5, CHRNA3, CHRNB4, CHRNA6 and CHRNB3 genes in individuals with and without nicotine dependence. We report that rare (<5%) missense variants at conserved sites in CHRNB4 are associated with decreased risk of developing nicotine dependence and decrease the number of cigarettes consumed daily by smokers. These findings suggest that nicotine dependence is mediated in part by rare genetic variation and by variation in CHRNB4 specifically.
RESULTS
In order to identify novel rare variants associated with nicotine dependence, we sequenced 752 individuals from the extremes of the population distribution of Fagerström Test of Nicotine Dependence (FTND) scores in 6 pools. The protein coding regions of the five human cholinergic nicotinic receptor subunit genes constituting the gene clusters previously reported to harbor common variants associated with nicotine dependence (CHRNA5–CHRNA3–CHRNB4 and CHRNA6–CHRNB3) (Fig. 1) were sequenced on an Illumina GAIIx and analyzed using the short indel prediction by large deviation inference and nonlinear true frequency estimation by recursion (SPLINTER) algorithm (19). We obtained >30-fold coverage per allele at all positions within the 28 amplicons designed to cover the protein coding exons of CHRNA3, CHRNA5, CHRNA6, CHRNB3 and CHRNB4 and validated 24 (9 novel) missense variants (Fig. 2 and Table 1). We only attempted to validate missense variants as these are more likely to have a functional impact than non-coding or synonymous coding changes. As the SPLINTER algorithm produces a P-value for the probability that a predicted variant is a true positive, we also genotyped a number of variants below our cut-off for significance in order to determine the positive predictive value of the algorithm. A total of 42 variants were genotyped in the sequenced individuals. Of these, 14 were selected for having probabilities of being true positives just below our cut-off value. Only 1 of these 14 variants was validated, while 24 of 28 SNPs predicted to be true positives were validated. This produced a positive predictive value of 86%. Additionally, all variants included as part of the amplified positive control vector were found upon achieving >30-fold coverage at mutated sites (sensitivity = 100%) and only ∼80 sites in the 1908 bp negative control vector were predicted to be polymorphic (specificity = ∼95%). The majority of validated variants, as expected, are rare (22/24 = 92% have minor allele frequency < 5%) in African Americans or Europeans. In fact, many are present in only one sequenced individual (7/24 = 29% are singletons). Additionally, some of the missense variants present in these genes were found at less conserved sites (9/25; phyloP score < 2), suggesting that they are less likely to have a functional impact. Altogether, we identified 15 variants (4 novel) at conserved sites and used these for further analysis. Of these, 12 were also predicted to be damaging by Polyphen, SIFT or both (Table 1 and Supplementary Material, Table S1) (20,21).
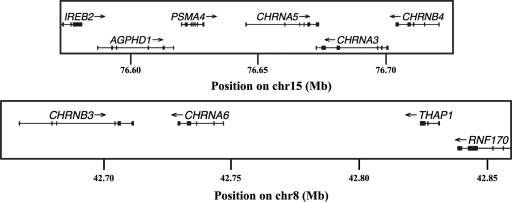
Figure 1.
Genomic context of sequenced genes. Positions are from human genome reference assembly build 36.1 (hg18). Direction of transcription is designated with an arrow. Exons are shown as squares.
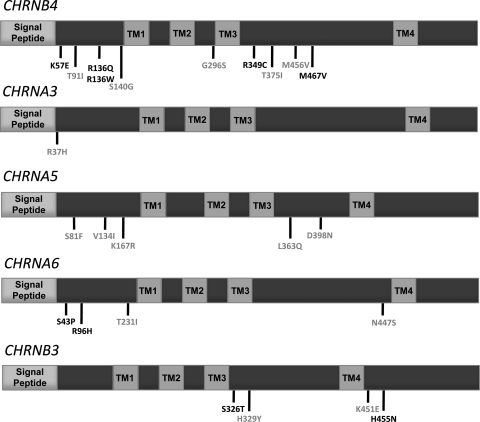
Figure 2.
Schematics of the CHRN genes studied and the locations of missense variants identified in sequenced individuals from the COGEND study. Each protein consists of a signal peptide, two extracellular domains, four transmembrane domains (TM1–4) and three intracellular domains. Conserved sites (phyloP44Vertebrate score ≥ 2) are shown in grey, while all others are shown in black.
Table 1.
Demographic and phenotypic characteristics of COGEND African Americans and European Americans
| CHRNB4 | African Americans |
European Americans |
||||
|---|---|---|---|---|---|---|
| Non-carriers | Carriers | P-value | Non-carriers | Carriers | P-value | |
| n | 681 | 29 | 1857 | 178 | ||
| Age | 37 ± 6 | 37 ± 7 | 0.64 | 37 ± 5 | 36 ± 6 | 0.72 |
| Women/men | 250/431 | 11/18 | 1.00 | 727/1151 | 67/110 | 0.87 |
| Nicotine cases/controls | 450/231 | 11/18 | 0.0025 | 974/883 | 77/101 | 0.023 |
| CPD | 17 ± 13 | 9 ± 6 | 6.6 × 10−5 | 19 ± 17 | 16 ± 14 | 0.021 |
| FTND Score | 4.0 ± 3.2 | 2.1 ± 2.8 | 0.0019 | 3.4 ± 3.5 | 2.7 ± 3.3 | 0.008 |
| Withdrawal symptom count | 3.7 ± 2.4 | 2.4 ± 2.9 | 0.012 | 3.4 ± 2.5 | 3.2 ± 2.4 | 0.42 |
Carriers are individuals who possess at least one of the following: T91I, T375I, G296S or M456V. ND cases are maximum lifetime FTND ≥ 4, while controls are maximum lifetime FTND ≤ 1. CPD values are mean ± SD and represent the average number of cigarettes smoked per day when the individual was smoking the most. FTND score is maximum lifetime FTND score. P-values were calculated using a two-sided Fisher's exact test or two-sided Wilcoxon rank-sum test.
P-values in bold are those less than 0.05.
We determined the frequency of each missense variant validated from sequencing in individuals from the Collaborative Study on the Genetics of Nicotine Dependence (COGEND) (710 African Americans and 2055 European Americans) using Sequenom. We then compared the frequency of each missense SNP as well as the number of carriers of conserved missense variants in cases and controls for each of the genes sequenced. To ensure novel findings, we excluded the SNP rs16969968 from analyses in European Americans as this would overshadow the effects of other variants in the CHRNA5 gene and because it is common in European Americans (frequency = 0.35). We find that two SNPs in CHRNB4 (T91I and T375I) and one SNP in CHRNA3 (R37H) are associated with decreased risk of nicotine dependence either in African Americans or in European Americans (Supplementary Material, Table S1). Notably, the T91I and R37H variants are in high linkage disequilibrium (LD) in European Americans (r2 = 0.89, n = 2035) and African Americans (r2 = 0.59, n = 710), making it difficult to determine the relative contribution of the two variants to the observed association. We do not find an association between rare variants, conserved or otherwise, and nicotine dependence at the other genes studied (CHRNB3, CHRNA6 and CHRNA5) in African Americans or European Americans.
We find that carriers of rare missense variants at conserved sites in CHRNB4 were less frequently nicotine dependent (ND) compared with non-carriers, both in African Americans and European Americans (African Americans P = 0.0025, odds-ratio (OR) = 0.31, 95% confidence-interval (CI) = 0.31–0.72; European Americans P = 0.023, OR = 0.69, 95% CI = 0.50–0.95) (Table 1). This is significant for African Americans after Bonferroni correction for multiple tests assuming one test for each gene studied (P < 0.01). As we are not able to distinguish between the effects of the T91I and R37H variants, carriers of missense variants at conserved site in either CHRNA3 or CHRNB4 are also less frequently ND compared with non-carriers (African American P = 0.0035; European American P = 0.012). To determine whether the observed association could be explained by the CHRNA3 R37H alone, we performed logistic regression including CHRNA3 R37H genotype as a covariate. CHRNB4 carrier status remained associated with nicotine dependence status in African Americans (P = 0.0025), but not in European Americans (P = 0.37). This is most likely because rs61737499 (CHRNB4 T375I), one of the major contributors to the association SNP in African Americans, is monomorphic in European Americans. Additionally, we investigated the possibility that one of the four comorbid diagnostic and statistical manual of mental disorders IV (DSM IV) defined substance dependence phenotypes measured in our data set (alcohol, cocaine, marijuana and opiates) was contributing to the observed association at CHRNB4. None of these comorbid phenotypes significantly altered the association results observed between nicotine dependence and CHRNB4 carrier status in either African Americans or European Americans when included as a covariate in a logistic regression. These findings suggest that variants in CHRNB4 and possibly CHRNA3 protect against nicotine dependence and that this is not due to a correlation between nicotine dependence and other comorbid phenotypes.
Two groups of common genetic variants within the CHRNA5–CHRNA3–CHRNB4 gene cluster (tagged by rs16969968 and rs588765, respectively) were previously shown to affect nicotine dependence risk and represent the two strongest associations between SNPs in this region and nicotine dependence in European Americans and African Americans (2,7,22). To determine whether our findings could be due to LD with either of these variants, we performed logistic regression including each as a covariate. CHRNB4 carrier status remained associated with nicotine dependence status in African Americans (P = 0.0048), but not in European Americans (P = 0.13) when rs16969968 genotype was included as a covariate. However, we find that CHRNB4 carrier status is associated with nicotine dependence among carriers of the minor allele of rs16969968 (Fisher's exact test P = 0.03, OR = 0.55) in European Americans. CHRNB4 carrier status was associated with nicotine dependence status in African Americans (P = 0.0031) and in European Americans (P = 0.043) when the rs588765 genotype was included as a covariate. These findings suggest that our observed associations are not due to a correlation between rare variants in CHRNB4 and previously described common variant associations in the region.
To ensure that our findings within the African American portion of the COGEND sample is not due to population stratification, we calculated principal components using EIGENSTRAT (23). We then performed logistic regression to test the association between CHRNB4 carrier status at conserved sites and nicotine dependence, including the first two principal components (PC1 and PC2) as covariates. CHRNB4 carrier status remained associated with nicotine dependence status in African Americans (P = 0.0038). Additionally, we calculated local admixture using local ancestry in admixed populations (24). In the 100 kb encompassing the CHRNA5–CHRNA3–CHRNB4 gene cluster, all African American individuals were found to have local ancestry estimates of 0, 50 or 100% European admixture, while all European Americans had local ancestry estimates of 100% European ancestry. The average European admixture of African Americans in this region was 21.8%, consistent with previous findings (25). CHRNB4 carrier status remained significantly associated with nicotine dependence when we performed logistic regression, including local admixture estimates from the 100 kb encompassing the CHRNA5–CHRNA3–CHRNB4 gene cluster as a covariate in African Americans (P = 0.0029). These findings suggest that the observed association is not due to population stratification, either on a genome scale or on a local scale.
In order to examine which aspects of nicotine dependence are affected by variants in CHRNB4, we tested the association between several nicotine-related phenotypes in addition to case status. When we compared the distribution of cigarettes per day (CPD) in CHRNB4 conserved SNP carriers and non-carriers, we observed that carriers also have a lower mean CPD than non-carriers (African American P = 6.6 × 10−5, European American P = 0.021). FTND as a quantitative trait was also lower in carriers compared with non-carriers (African American P = 0.0019, European American P = 0.008) (Table 1). Using DSM-IV criteria to assign nicotine dependence case status, we find carriers to have reduced risk of becoming ND in African Americans (P = 0.03) and European Americans (P = 0.02). As Chrnb4 knockout mice exhibit reduced withdrawal symptoms upon cessation of prolonged nicotine exposure (10,26), we investigated whether conserved missense variants in CHRNB4 contribute to withdrawal symptoms among individuals who attempt cessation. We find that carriers of conserved missense variants in CHRNB4 show fewer signs of withdrawal as measured by the number of withdrawal symptoms endorsed in African Americans but not European Americans (African American P = 0.012, European American P = 0.42) (Table 1).
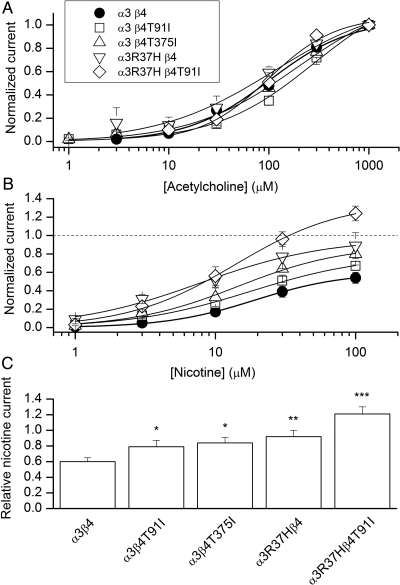
To determine whether the three major contributors to this association (CHRNB4 T375I, CHRNB4 T91I and CHRNA3 R37H) affect α3β4 receptor function, we heterologously expressed the variant forms of α3 and β4 in HEK293 cells. We examined the effect of the CHRNB4 T91I variant alone as well as in combination with CHRNA3 R37H because these two variants are in LD (EA r2 = 0.89, AA r2 = 0.59). We elicited responses with acetylcholine (the endogenous transmitter) or nicotine. The concentration–response relationships did not show significant differences in terms of the concentration producing a half-maximal response, for either acetylcholine or nicotine (Fig. 3A and B; t-test P > 0.2). However, each of these variants showed increased maximal response to nicotine relative to the maximal response to acetylcholine (Fig. 3B and C). Both CHRNB4 variants, the CHRNA3 R37H variant alone and the combination of CHRNA3 R37H and CHRNB4 T91I variants, showed significantly increased relative maximal response to nicotine compared with normal (t-test P ≤ 0.02). The maximal response produced from receptors containing CHRNB4 T91I and CHRNA3 R37H was also greater (>1.4-fold) than the response seen for CHRNA3 R37H alone, further suggesting an effect of CHRNB4 T91I independent of CHRNA3 R37H (t-test P = 0.02).
Figure 3.
Functional effects of the mutations to the α3 and β4 subunits. (A) The ACh concentration–response curves were obtained by fitting the data recorded at 1–1000 μm ACh to the Hill equation. The data are normalized to 1 mm ACh. Each data point is mean ± SEM from 4 to 9 cells. The fitting results are given in Supplementary Material, Table S2. (B) The nicotine concentration–response curves were obtained by fitting the data recorded at 1–100 μm nicotine to the Hill equation. Each cell was additionally exposed to 1 mm ACh to which the nicotine responses are normalized. Each data point is mean ± SEM from 3 to 9 cells. (C) The relative nicotine current is calculated as the ratio of the peak responses to 100 μm nicotine and 1 mm ACh from the same cell. The data are mean ± SEM from 6 to 13 cells. Statistical significance was assessed by t-test and shows comparison to the normal α3β4 receptor. *P < 0.05; **P = 0.003; ***P = 2 × 10−6.
DISCUSSION
We find that rare missense variants at evolutionarily conserved sites in CHRNB4 and CHRNA3 are associated with reduced risk of nicotine dependence in African Americans and European Americans. Our in vitro data suggest that this is likely due to the tendency of these variants to increase cellular response to nicotine, although the exact mechanism is unclear. We do not find an association between rare variants, conserved or otherwise, and nicotine dependence at the other genes studied (CHRNB3, CHRNA6 and CHRNA5) in African Americans or European Americans. This may be due to insufficient power resulting from low minor allele frequencies even after collapsing genotypes within genes. For instance, the frequency of carriers of missense variants at conserved loci for CHRNA5 in European Americans is 0.015, while the frequency of such carriers for CHRNB4 is 0.055. This increased carrier frequency for CHRNB4 was in large part due to the combined frequency of the two variants (T375I and T91I) which is why we chose to study these SNPs in vitro. It is important to note that rare variant associations are more dependent on stochastic events, namely the sampling of rare allele carriers from a population, than studies of common genetic variants. It is possible that our observed associations may be due to chance fluctuations in rare variant allele.
When considering genetic associations in admixed populations, it is important to ensure that findings are not due to subtle population stratification, either on a genome scale or locally at associated loci. As a result, we calculated both global and local measures of admixture for each of the African American individuals in COGEND. We find that neither global nor local admixture is able to account for the association we observe between rare missense variants at evolutionarily conserved sites in CHRNB4 and nicotine dependence.
In addition to nicotine dependence, we examined the effect of conserved missense variants in CHRNB4 on several phenotypes designed to tease apart various aspects of dependence. Specifically, we find that African American and European American rare variant carriers smoke fewer CPD and that African American carriers experience fewer withdrawal symptoms when compared with non-carriers. These data suggest that these CHRNB4 variants are likely contributing to the risk of developing nicotine dependence by allowing carriers to quit more easily, possibly by allowing them to smoke fewer CPD to combat withdrawal symptoms. It is difficult to determine with confidence which aspect of nicotine dependence variants in CHRNB4 are affecting, however, as each of the phenotypes analyzed are highly correlated in our sample (r2 from 0.32–0.85) and because our sample was selected to contain the extremes of the population distribution of FTND score.
To determine the functional consequence of changes in CHRNB4, we characterized two variants, as well as a conserved CHRNA3 missense variant correlated with CHRNB4 T91I. Both CHRNB4 variants were found to increase maximal nicotine response relative to response to acetylcholine (T375I P = 0.01, T91I P = 0.02). Additionally, the CHRNA3 missense variant alone and in combination with CHRNB4 T91I showed significant increases in the relative maximal response to nicotine (P = 0.003 and 2 × 10−6). The maximal response produced from receptors containing CHRNB4 T91I and CHRNA3 R37H was also greater (>1.4-fold) than the response seen for either CHRNB4 T91I or CHRNA3 R37H alone, further suggesting an effect of CHRNB4 T91I independent of CHRNA3 R37H (P = 0.02). (Table 2) These results are consistent with findings regarding the in vitro function of the previously described CHRNA5 D398N variant (5,27). Increased maximal response to nicotine here confers protection against nicotine dependence, in contrast to a decreased maximal response to epibatidine and an increased risk for nicotine dependence seen for the D398N variant in CHRNA5. Additionally, assuming increased agonist response is functionally similar to over-expression, our results are consistent with the increased aversion to nicotine seen in mice with targeted overexpression of Chrnb4 (11). We speculate, therefore, that one of the mechanisms by which vulnerability to nicotine dependence is conferred is through decreased sensitivity and thereby decreased aversion to nicotine.
Table 2.
Missense variants in sequenced nicotinic receptor genes present in the COGEND sample
| Gene | Amino acid position | rs number | CHR | hg18 position | Allele 1 | Allele 2 | PhyloP score | Sift prediction | Frequency in AA cases | Frequency in AA controls | Frequency in EA cases | Frequency in EA controls |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B3 | S326T | – | 8 | 42706583 | T | A | 0.67 | Tolerated | 0.002 | 0.002 | 0 | 0 |
| B3 | H329Y | – | 8 | 42706592 | C | T | 3.98 | Damaging | 0 | 0 | 0.0009 | 0.0030 |
| B3 | K451E | rs35327613 | 8 | 42710892 | G | A | 2.35 | Tolerated | 0.058 | 0.045 | 0.0009 | 0.0005 |
| B3 | H455N | – | 8 | 42710904 | C | A | 0.97 | Tolerated | 0 | 0.001 | 0 | 0 |
| A6 | N447S | rs16891583 | 8 | 42730159 | C | T | 4.96 | Damaging | 0.001 | 0 | 0 | 0 |
| A6 | T231I | – | 8 | 42730807 | G | A | 6.91 | Tolerated | 0 | 0 | 0 | 0.0005 |
| A6 | R96H | – | 8 | 42731315 | C | T | 0.94 | Damaging | 0 | 0.002 | 0 | 0 |
| A6 | S43P | – | 8 | 42739457 | A | G | 1.23 | Damaging | 0.011 | 0.018 | 0 | 0.0005 |
| A5 | S81F | – | 15 | 76660343 | C | T | 5.35 | Damaging | 0.001 | 0 | 0 | 0 |
| A5 | V134I | rs2229961 | 15 | 76667807 | A | G | 6.72 | Damaging | 0.002 | 0 | 0.0171 | 0.0122 |
| A5 | K167R | – | 15 | 76669288 | A | G | 5.40 | Damaging | 0.015 | 0.016 | 0 | 0.0005 |
| A5 | L363Q | rs79109919 | 15 | 76669876 | T | A | 4.77 | Damaging | 0.061 | 0.054 | 0.0009 | 0 |
| A5 | D398N | rs16969968 | 15 | 76669980 | A | G | 3.77 | Tolerated | 0.063 | 0.032 | 0.3874 | 0.3114 |
| A3 | R37H | rs8192475 | 15 | 76698285 | C | T | 2.67 | Damaging | 0.008 | 0.010 | 0.0430 | 0.0579 |
| B4 | M467V | rs61737502 | 15 | 76704628 | T | C | 1.58 | Tolerated | 0.083 | 0.068 | 0 | 0 |
| B4 | M456V | – | 15 | 76704661 | T | C | 4.71 | Damaging | 0 | 0 | 0 | 0.0005 |
| B4 | T375I | rs61737499 | 15 | 76708578 | G | A | 2.26 | Tolerated | 0.008 | 0.026 | 0 | 0 |
| B4 | R349C | – | 15 | 76708657 | A | G | 1.96 | Damaging | 0.001 | 0 | 0.0072 | 0.0072 |
| B4 | G296S | – | 15 | 76708816 | C | T | 5.69 | Damaging | 0 | 0.001 | 0 | 0 |
| B4 | S140G | rs56218866 | 15 | 76709284 | C | T | 2.27 | Tolerated | 0.047 | 0.040 | 0.0028 | 0.0051 |
| B4 | R136Q | rs56095004 | 15 | 76709295 | C | T | 0.41 | Tolerated | 0.003 | 0 | 0.0122 | 0.0106 |
| B4 | R136W | – | 15 | 76709296 | G | A | 0.33 | Tolerated | 0.012 | 0.012 | 0 | 0 |
| B4 | T91I | rs12914008 | 15 | 76710560 | A | G | 2.16 | Tolerated | 0.005 | 0.008 | 0.0378 | 0.0536 |
| B4 | K57E | – | 15 | 76714871 | T | C | 1.16 | Tolerated | 0 | 0.001 | 0 | 0 |
Cases are individuals with FTND score ≥ 4 and controls are individuals with FTND scores ≤ 1. PhyloP score is the basewise vertebrate conservation score. Positions are from human genome reference assembly build 36.1 (hg18).
In conclusion, we find an excess of conserved missense variants in CHRNB4 in smokers who are not ND compared with smokers who are ND. Moreover, the two major genetic contributors to this association lead to increased functional response to nicotine in vitro. We hypothesize that these variants act by increasing sensitivity to nicotine and that these changes lead to our overall finding of decreased risk for nicotine dependence. Further sequencing of individuals for whom detailed dependence-related information has been obtained is necessary to confirm and elaborate on these findings.
MATERIALS AND METHODS
Sample selection
DNA samples were collected as part of the Collaborative Genetic Study of Nicotine Dependence (COGEND). All members of the COGEND sample underwent a semi-structured interview, which assessed smoking behavior, other substance use and comorbid psychiatric conditions. The COGEND sample includes 710 African Americans [461 ND cases and 249 smokers with no symptoms of dependence (controls)] and 2055 European Americans (1062 ND cases and 993 controls). As these individuals were ascertained to study nicotine dependence, FTND scores (range = 1–10) were required to be ≤1 for controls and ≥4 for cases. In all cases, lifetime maximum FTND score was used. Additionally, to ensure that all individuals in the study had been exposed to nicotine, all members were required to have smoked at least 100 cigarettes in their lifetime. Among individuals with FTND scores ≥4, 96% (1458/1523) were also ND using DSM-IV nicotine dependence criteria. A total of 352 African Americans (176 ND cases and 176 smoking controls) and 400 European Americans (200 ND cases and 200 smoking controls) were sequenced. For African Americans, pools of 88 individuals were used (two case pools and two control pools). For European Americans, pools of 200 were used (one case pool and one control pool). The case pools include individuals with FTND scores ≥ 5, whereas the control pools include individuals with FTND scores ≤ 1. Individuals for case pools were chosen to select the most severely ND subjects in our sample. Follow-up genotyping of SNPs identified and validated in the sequenced individuals was done in the remaining portion of the COGEND sample (310 African Americans and 1662 European Americans).
Pooled sequencing
Pooled DNA sequencing was performed as previously described (28). The concentrations of individual DNA samples were first measured using Quant-iT™ PicoGreen reagent and pooled in equimolar amounts. Each pooled DNA sample was then used as the template for the amplification of each protein coding exon of the CHRNA3, CHRNA5, CHRNA6, CHRNB3 and CHRNB4 genes. Primers for the amplification of each exon were designed using Primer3 and reference sequences were taken from the human genome reference assembly build 36.1 (hg18) (Supplementary Material, Table S1). In order to ensure complete coverage of each desired exon, a minimum of 50 bp of flanking sequence on each side was required for each amplicon. We used Pfu high-fidelity DNA polymerase in all polymerase chain reactions (PCRs) to reduce the identification of SNPs generated as a result of the PCR (false positive SNPs). After PCR amplification of desired genomic regions, PCR products were cleaned using QIAquick PCR purification kits, quantified using Quant-iT PicoGreen reagent and ligated in equimolar amounts using T4 Ligase and T4 Polynucleotide Kinase. At this stage, positive and negative control vectors were amplified and added to each pool to serve as internal quality standards and to be used in data analysis. After ligation, concatenated PCR products were randomly sheared by sonication and prepared for Illumina sequencing on an Illumina Genome Analyzer IIx (GAIIx) according to the manufacturer's specifications. As previously shown by Vallania et al. (19), an average coverage of 30-fold per allele per pool was shown to correlate to optimal positive predictive value for the SNP-calling algorithm and was, therefore, the target level of coverage in this study. In order to obtain sufficient coverage per allele per pool, we obtained two lanes of Illumina GAIIx sequencing per pool. Coverage per amplicon was calculated for each pool after the first lane of sequence was obtained and only those amplicons not reaching 30-fold coverage per allele were included for the second lane of sequencing.
Sequencing analysis
For analysis, sequencing reads (36 bp reads) were aligned using an alignment algorithm developed by Vallania et al. (19) which aligns sequences allowing for two mismatches or indels of up to 4 bp. In order to quantify the specificity and sensitivity of this method, positive and negative control DNA were introduced as PCR products in the pooled sequencing protocol outlined above. As a positive control to estimate sensitivity for variant calling, a pool of 10 plasmids with a 72 bp insert was generated. One plasmid acts as the ‘wild-type’ insert, while the remaining nine plasmids contain one or two synthetically engineered mutations. All 10 plasmids were combined such that each the allele frequency of each known mutation would mimic either the allele frequency of a single allelic variant or 10 allelic variants in our human DNA pool. Following mixing of the vectors, PCR amplification across the insert sequence was performed and the PCR product was added to the normalized pool of human target PCR reactions during sequencing library preparation. We then obtained more coverage for each amplicon than that which was required to detect all variants in this amplified positive control vector pool in order to ensure a minimal SNP detection false negative rate. As a negative control and to model the sequencing error rate, 1908 bp of the pCMV6-XL5 plasmid was also amplified and included. One half of the pCMV6-XL5 plasmid sequence was used to train the SNP finding algorithm and the other to test the error model. Finally, to identify variants in the pooled sequence data, we used the SPLINTER algorithm (19). The SPLINTER algorithm produces a P-value for the probability that a predicted variant is a true positive. This probability is produced using large deviation theory to quantify the difference between observed allele frequencies within the sequence data to the background error rate for the same type of sequence changes seen in the amplified pCMV6-XL5 plasmid. A P-value cut-off value for each lane of Illumina sequencing was defined as the value at which all positive controls were identified. Only those variants falling below this cut-off value were considered ‘predicted’ by SPLINTER. All protein coding or splice site variants predicted by SPLINTER were then validated by individual Sequenom genotyping in each person from the source DNA pools. SNPs validated in the sequenced individuals were then genotyped in all members of the COGEND sample. All individual genotyping was performed using the Sequenom platform as described previously (8).
Association analysis
Each missense variant as well as the number of carriers of missense variants at conserved positions for each gene in the genes surveyed was tested for association with nicotine dependence in African Americans and European Americans using Fisher's exact test. Conservation was determined by basewise vertebrate conservation using PhyloP score (29). A site was called conserved when its phyloP score was ≥2, corresponding to a P-value of 0.01.The SNP rs56218866 (CHRNB4 S140G) was excluded from analyses as it had been shown previously to have no measurable effect on receptor function and did not effect the significance of the association between CHRNB4 carrier status and either nicotine dependence or CPD when taken as a covariate (30). For analysis of CPD, FTND score and withdrawal symptom count in carriers and non-carriers, Wilcoxon rank-sum tests were performed.
In vitro functional assay
Whole-cell voltage-clamp was performed as previously described to measure intracellular ion concentrations in response to exposure to nicotine (31). For each variant, we co-transfected HEK293 cells with plasmids expressing normal α3 and either normal β4 or variant β4, or variant α3 with normal or variant β4. Additionally, we co-transfected HEK293T cells with plasmids expressing the minor allele of α3 R37H and β4 T91I, because these SNPs are in very high LD and thus frequently co-occur in human populations. Concentration–response curves were produced for each subunit combination. EC50 values maximal response to agonist were each compared with normal using a two-tailed t-test.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. L.J.B. and A.M.G. are listed as inventors on the patent ‘Markers for Addiction’ (US 20070258898) covering the use of certain SNPs in determining the diagnosis, prognosis and treatment of addiction. L.J.B. acted as a consultant for Pfizer, Inc. in 2008.
FUNDING
The COGEND project is a collaborative research group and part of the National Institute on Drug Abuse (NIDA) Genetics Consortium. Subject collection was supported by NIH grant P01 CA89392 (L.J.B.). Phenotypic and genotypic data are stored in the NIDA Center for Genetic Studies (NCGS) at http://zork.wustl.edu/ under NIDA Contract HHSN271200477451C. Electrophysiological studies supported by NIH grant R21 DA26918 (G.A.).
Supplementary Material
ACKNOWLEDGEMENTS
In memory of Theodore Reich, founding Principal Investigator of COGEND, we are indebted to his leadership in the establishment and nurturing of COGEND and acknowledge with great admiration his seminal scientific contributions to the field. Lead investigators directing data collection are L.B., N.B., D.H. and E.J. The authors thank Heidi Kromrei and Tracey Richmond for their assistance in data collection. We thank the Genome Technology Access Center in the Department of Genetics at Washington University School of Medicine for help with genomic analysis. The Center is partially supported by NCI Cancer Center Support Grant #P30 CA91842 to the Siteman Cancer Center and by ICTS/CTSA Grant# UL1RR024992 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH), and NIH Roadmap for Medical Research. This publication is solely the responsibility of the authors and does not necessarily represent the official view of NCRR or NIH.
REFERENCES
- 1.Bierut L.J. Genetic vulnerability and susceptibility to substance dependence. Neuron. 2011;69:618–627. doi: 10.1016/j.neuron.2011.02.015. doi:10.1016/j.neuron.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thorgeirsson T.E., Gudbjartsson D.F., Surakka I., Vink J.M., Amin N., Geller F., Sulem P., Rafnar T., Esko T., Walter S., et al. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat. Genet. 2010;42:448–453. doi: 10.1038/ng.573. doi:10.1038/ng.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saccone N.L., Wang J.C., Breslau N., Johnson E.O., Hatsukami D., Saccone S.F., Grucza R.A., Sun L., Duan W., Budde J., et al. The CHRNA5-CHRNA3-CHRNB4 nicotinic receptor subunit gene cluster affects risk for nicotine dependence in African-Americans and in European-Americans. Cancer Res. 2009;69:6848–6856. doi: 10.1158/0008-5472.CAN-09-0786. doi:10.1158/0008-5472.CAN-09-0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li M.D., Yoon D., Lee J.Y., Han B.G., Niu T., Payne T.J., Ma J.Z., Park T. Associations of variants in CHRNA5/A3/B4 gene cluster with smoking behaviors in a Korean population. PLoS ONE. 2010;5:e12183. doi: 10.1371/journal.pone.0012183. doi:10.1371/journal.pone.0012183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bierut L.J., Stitzel J.A., Wang J.C., Hinrichs A.L., Grucza R.A., Xuei X., Saccone N.L., Saccone S.F., Bertelsen S., Fox L., et al. Variants in nicotinic receptors and risk for nicotine dependence. Am. J. Psychiatry. 2008;165:1163–1171. doi: 10.1176/appi.ajp.2008.07111711. doi:10.1176/appi.ajp.2008.07111711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hong L.E., Hodgkinson C.A., Yang Y., Sampath H., Ross T.J., Buchholz B., Salmeron B.J., Srivastava V., Thaker G.K., Goldman D., et al. A genetically modulated, intrinsic cingulate circuit supports human nicotine addiction. Proc. Natl Acad. Sci. USA. 2010;107:13509–13514. doi: 10.1073/pnas.1004745107. doi:10.1073/pnas.1004745107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saccone N.L., Culverhouse R.C., Schwantes-An T.H., Cannon D.S., Chen X., Cichon S., Giegling I., Han S., Han Y., Keskitalo-Vuokko K., et al. Multiple independent loci at chromosome 15q25.1 affect smoking quantity: a meta-analysis and comparison with lung cancer and COPD. PLoS Genet. 2010;6:e1001053. doi: 10.1371/journal.pgen.1001053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang J.C., Grucza R., Cruchaga C., Hinrichs A.L., Bertelsen S., Budde J.P., Fox L., Goldstein E., Reyes O., Saccone N., et al. Genetic variation in the CHRNA5 gene affects mRNA levels and is associated with risk for alcohol dependence. Mol. Psychiatry. 2009;14:501–510. doi: 10.1038/mp.2008.42. doi:10.1038/mp.2008.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fowler C.D., Lu Q., Johnson P.M., Marks M.J., Kenny P.J. Habenular alpha5 nicotinic receptor subunit signalling controls nicotine intake. Nature. 2011;471:597–601. doi: 10.1038/nature09797. doi:10.1038/nature09797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salas R., Pieri F., De Biasi M. Decreased signs of nicotine withdrawal in mice null for the beta4 nicotinic acetylcholine receptor subunit. J. Neurosci. 2004;24:10035–10039. doi: 10.1523/JNEUROSCI.1939-04.2004. doi:10.1523/JNEUROSCI.1939-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frahm S., Slimak M.A., Ferrarese L., Santos-Torres J., Antolin-Fontes B., Auer S., Filkin S., Pons S., Fontaine J.F., Tsetlin V., et al. Aversion to nicotine is regulated by the balanced activity of beta4 and alpha5 nicotinic receptor subunits in the medial habenula. Neuron. 2011;70:522–535. doi: 10.1016/j.neuron.2011.04.013. doi:10.1016/j.neuron.2011.04.013. [DOI] [PubMed] [Google Scholar]
- 12.Sarginson J.E., Killen J.D., Lazzeroni L.C., Fortmann S.P., Ryan H.S., Schatzberg A.F., Murphy G.M., Jr Markers in the 15q24 nicotinic receptor subunit gene cluster (CHRNA5-A3-B4) predict severity of nicotine addiction and response to smoking cessation therapy. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2011;156:275–284. doi: 10.1002/ajmg.b.31155. [DOI] [PubMed] [Google Scholar]
- 13.Schork N.J., Murray S.S., Frazer K.A., Topol E.J. Common vs. rare allele hypotheses for complex diseases. Curr. Opin. Genet. Dev. 2009;19:212–219. doi: 10.1016/j.gde.2009.04.010. doi:10.1016/j.gde.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johansen C.T., Wang J., Lanktree M.B., Cao H., McIntyre A.D., Ban M.R., Martins R.A., Kennedy B.A., Hassell R.G., Visser M.E., et al. Excess of rare variants in genes identified by genome-wide association study of hypertriglyceridemia. Nat. Genet. 2010;42:684–687. doi: 10.1038/ng.628. doi:10.1038/ng.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haller G., Torgerson D.G., Ober C., Thompson E.E. Sequencing the IL4 locus in African Americans implicates rare noncoding variants in asthma susceptibility. J. Allergy Clin. Immunol. 2009;124:1204–1209. doi: 10.1016/j.jaci.2009.09.013. e1209 doi:10.1016/j.jaci.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Momozawa Y., Mni M., Nakamura K., Coppieters W., Almer S., Amininejad L., Cleynen I., Colombel J.F., de Rijk P., Dewit O., et al. Resequencing of positional candidates identifies low frequency IL23R coding variants protecting against inflammatory bowel disease. Nat. Genet. 2011;43:43–47. doi: 10.1038/ng.733. doi:10.1038/ng.733. [DOI] [PubMed] [Google Scholar]
- 17.Wessel J., McDonald S.M., Hinds D.A., Stokowski R.P., Javitz H.S., Kennemer M., Krasnow R., Dirks W., Hardin J., Pitts S.J., et al. Resequencing of nicotinic acetylcholine receptor genes and association of common and rare variants with the Fagerstrom test for nicotine dependence. Neuropsychopharmacology. 2010;35:2392–2402. doi: 10.1038/npp.2010.120. doi:10.1038/npp.2010.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie P., Kranzler H.R., Krauthammer M., Cosgrove K.P., Oslin D., Anton R.F., Farrer L.A., Picciotto M.R., Krystal J.H., Zhao H., et al. Rare nonsynonymous variants in alpha-4 nicotinic acetylcholine receptor gene protect against nicotine dependence. Biol. Psychiatry. 2011;70:528–536. doi: 10.1016/j.biopsych.2011.04.017. doi:10.1016/j.biopsych.2011.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vallania F.L., Druley T.E., Ramos E., Wang J., Borecki I., Province M., Mitra R.D. High-throughput discovery of rare insertions and deletions in large cohorts. Genome Res. 2010;20:1711–1718. doi: 10.1101/gr.109157.110. doi:10.1101/gr.109157.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. doi:10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ng P.C., Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–874. doi: 10.1101/gr.176601. doi:10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saccone S.F., Hinrichs A.L., Saccone N.L., Chase G.A., Konvicka K., Madden P.A., Breslau N., Johnson E.O., Hatsukami D., Pomerleau O., et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum. Mol. Genet. 2007;16:36–49. doi: 10.1093/hmg/ddl438. doi:10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. doi:10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 24.Sankararaman S., Sridhar S., Kimmel G., Halperin E. Estimating local ancestry in admixed populations. Am. J. Hum. Genet. 2008;82:290–303. doi: 10.1016/j.ajhg.2007.09.022. doi:10.1016/j.ajhg.2007.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bryc K., Auton A., Nelson M.R., Oksenberg J.R., Hauser S.L., Williams S., Froment A., Bodo J.M., Wambebe C., Tishkoff S.A., et al. Genome-wide patterns of population structure and admixture in West Africans and African Americans. Proc. Natl Acad. Sci. USA. 2010;107:786–791. doi: 10.1073/pnas.0909559107. doi:10.1073/pnas.0909559107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jackson K.J., Martin B.R., Changeux J.P., Damaj M.I. Differential role of nicotinic acetylcholine receptor subunits in physical and affective nicotine withdrawal signs. J. Pharmacol. Exp. Ther. 2008;325:302–312. doi: 10.1124/jpet.107.132977. doi:10.1124/jpet.107.132977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuryatov A., Berrettini W., Lindstrom J. Acetylcholine receptor (AChR) alpha5 subunit variant associated with risk for nicotine dependence and lung cancer reduces (alpha4beta2)alpha5 AChR function. Mol. Pharmacol. 2011;79:119–125. doi: 10.1124/mol.110.066357. doi:10.1124/mol.110.066357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Druley T.E., Vallania F.L., Wegner D.J., Varley K.E., Knowles O.L., Bonds J.A., Robison S.W., Doniger S.W., Hamvas A., Cole F.S., et al. Quantification of rare allelic variants from pooled genomic DNA. Nat. Methods. 2009;6:263–265. doi: 10.1038/nmeth.1307. doi:10.1038/nmeth.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pollard K.S., Hubisz M.J., Rosenbloom K.R., Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–121. doi: 10.1101/gr.097857.109. doi:10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang Y., Salas R., Marubio L., Bercovich D., De Biasi M., Beaudet A.L., Dani J.A. Functional polymorphisms in the human beta4 subunit of nicotinic acetylcholine receptors. Neurogenetics. 2005;6:37–44. doi: 10.1007/s10048-004-0199-7. doi:10.1007/s10048-004-0199-7. [DOI] [PubMed] [Google Scholar]
- 31.Akk G., Li P., Bracamontes J., Steinbach J.H. Activation and modulation of concatemeric GABA-A receptors expressed in human embryonic kidney cells. Mol. Pharmacol. 2009;75:1400–1411. doi: 10.1124/mol.108.054510. doi:10.1124/mol.108.054510. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.