Abstract
Background
The Three-amino acid-loop-extension (TALE) superfamily of homeodomain-containing transcription factors have been implicated in normal hematopoiesis and in leukemogenesis and are important survival, differentiation, and apoptosis pathway modulators. In this work, we determined the expression levels of TALE genes in leukemic-derived cell lines, in blood samples of patients with Acute lymphoblastic leukemia (ALL), and in the blood samples of healthy donors.
Results
Here we show increased expression of MEIS1, MEIS2, and PREP1 genes in leukemia-derived cell lines compared with blood normal cells. High levels of MEIS1 and PREP1, and low levels of PBX4 expression were also founded in samples of patients with ALL. Importantly, silencing of MEIS1 decreases the proliferation of leukemia-derived cells but increases their survival after etoposide treatment. Etoposide-induced apoptosis induces down-regulation of MEIS1 expression or PREP1 up-regulation in chemotherapy-resistant cells.
Conclusions
Our results indicate that up-regulation of MEIS1 is important for sustaining proliferation of leukemic cells and that down-regulation of MEIS1 or up-regulation of PREP1 and PBX genes could be implicated in the modulation of the cellular response to chemotherapeutic-induced apoptosis.
Keywords: TALE genes, leukemia, MEIS1, PREP1, PBX, Apoptosis
Background
Three-amino-acid loop extension (TALE) genes belong to the homeobox group and are distinguished by the presence of three extra amino acids in the loop binding the first to the second alpha helix of the homeodomain [1]. TALE proteins include subfamilies MEINOX and PBC. MEINOX is composed of the members MEIS1, MEIS2, the recently described MEIS3, PREP1, and PREP2 in humans [1,2]. The PBC subfamily contains PBX1, PBX2, PBX3, and PBX4 proteins [3,4]. Expression of TALE genes has been related with normal development, differentiation, survival, apoptosis, and with the hematopoietic process [5-10]. Indeed, some TALE genes are targets for viral insertion or for chromosome translocations during leukemogenesis. In this regard, MEIS1 has been characterized as a common proviral integration site in BXH-2 mice [11]; in these mice, leukemic tumors that contain a viral integration site at the MEIS1 locus frequently possess an additional co-integration site in some HOX genes [12], which suggests the required cooperative effect of MEIS and HOX during leukemogenesis. Over-expression of MEIS1 in CD34+ hematopoietic cells has been related with suppression of differentiation, promotion of proliferation, and self-renewal. Interestingly, high levels of MEIS1 in myeloid progenitors have been shown to regulate the cellular response to some cytokines, favoring self-renewal or differentiation. Moreover, in the murine myeloid cell line 32Dcl3, it has been observed that MEIS1 can block granulocytic differentiation in response to G-CSF [13]. MEIS1 has been also found over-expressed in human leukemic cells [14].
Other TALE proteins that have been also related with normal hematopoiesis and leukemogenesis comprise members of the PBX group. PBX proteins were first identified as HOX cofactors involved in developmental gene regulation [15,16]. PBX1 plays a role in the development of blood cell populations because hematopoietic stem cells from PBX1-/- embryos have reduced colony-forming activity and are unable to establish multilineage hematopoiesis in competitive reconstitution experiments [8]. PBX-PREP1 complexes are required for the production of normal CD4 and CD8 T-lymphocytes. Furthermore, PBX-MEIS complexes have been implicated in megakaryocyte differentiation, and PBX-PREP complexes have been also connected with the regulation of Interleukin (IL)-10 production in macrophages during the phagocytosis of apoptotic cells [17]. PREP proteins are also important during development; for instance, deletion of PREP1 in mice and zebrafish induces embryonic lethality [18,19]. Mice hypomorphic for PREP1 exhibit defects in T-cell development, with a decreased number of single-positive thymocytes, increased apoptosis of double-positive thymocytes, and abnormalities in the expression of αβ and γδ T-cell receptors [19]. Additionally, reduction in PREP1 expression directly affects the expression of MEIS and PBX and consequently, normal embryonic hematopoiesis [20].
In summary, TALE genes codify for important transcription factors involved in hematopoiesis and leukemogenesis and are important survival, differentiation, and apoptosis pathway modulators in hematopoietic cells. In this study, we analyzed the expression of TALE genes in leukemia-derived cell lines, in samples of patients with Acute lymphoblastic leukemia (ALL), and in samples of clinically healthy donors. We observed consistent up-regulation of MEIS1 and PREP1 and down-regulation of PBX4 in leukemic cell lines and in the samples of patients with ALL. Interestingly, RNA-mediated down-modulation of MEIS1 lowers leukemic cell proliferation. Additionally, chemotherapeutic treatment of lymphoblastic cell lines induces an increment in PREP1, PBX2, and PBX4 messenger RNA (mRNA) levels that could be related with a more resistant phenotype.
Results
Higher Expression Levels of MEIS1, MEIS2, and PREP1 Genes in Leukemia-derived Cell Lines Compared with Normal Cells
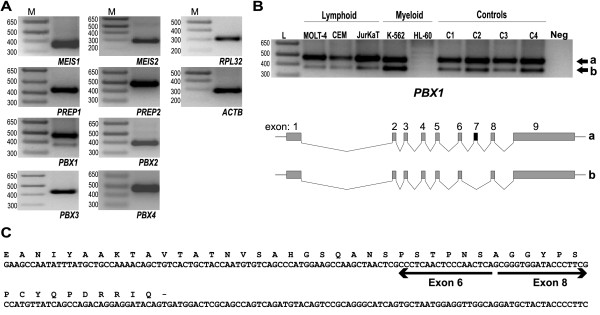
TALE genes are normally involved in the differentiation of hematopoietic cells and their expression has been related with deregulated differentiation. From this starting point, we wanted to determine the expression levels of TALE genes in cells with impaired differentiation. For this purpose, we choose leukemic-derived cell lines and normal differentiated cells as the study model. In order to analyze the expression of TALE genes, we first selected primer pairs to amplify these genes and proved these primers by conventional PCR reactions utilizing a pull of complementary DNA (cDNA) obtained from leukemia-derived cell lines (Table 1). All primer pairs were able to amplify a specific product corresponding to the expected size for each TALE gene (Figure 1A); however, for PBX1, we observed an additional band of lower molecular size. We carried out the same approach to analyze primer pairs designed to amplify the L32 ribosomal protein (RPL32) and Beta-Actin (ACTB) in order to have reference genes to measure relative gene expression (Figure 1A). Regarding PBX1, we also noticed the low-molecular-weight band existing in controls and in all leukemia derived cell lines (Figure 1B). By sequence analysis, we found that this corresponds to an isoform of PBX1 in which exon 7 is lost (Figures 1B and 1C). This indicated that our PCR primers were specific for PBX1, and also that they are able to amplify both PBX1 versions. Because we planned to quantify gene expression by real time PCR (qRT-PCR) utilizing the SYBR Green strategy, in the case of PBX1, the value obtained in quantification correspond to the sum of the expression of both isoforms.
Table 1.
Primer pairs used for PCR reactions
| GENE | ACCESION NUMBER | PRIMER SEQUENCE | Tm |
|---|---|---|---|
| MEIS1 | [GenBank:NM_002398] | F: CCC CAG CAC AGG TGA CGA TGA T R: TGC CCA TTC CAC TCA TAG GTC C |
60 |
| MEIS2 | [GenBank:NM_170677] | F: CCA TCG ACC TCG TCA TTG AT R: CCT CCT TTC TTC TGG CGT TTT T |
60 |
| PREP1 | [GenBank:NM_004571] | F: GGT TTT GGC CTG ATT CTA TTG C R: GTG GGG AGG GAG TGG TG |
65 |
| PREP2 | [GenBank:NM_022062] | F: GCC ACC AAT ATA ATG CGT TCT T R: GTG TTC CAA GCC CAG GTC |
65 |
| PBX1 | [GenBank:NM_002585] | F: CTA ACT CGC CCT CAA CTC C R: GTG TCC AGA TTG GCT GAA ATA G |
60 |
| PBX2 | [GenBank:NM_002586] | F: GGC GGC TCT TTC AAT CTC TCA R: GTC TCG TTA GGG AGG GGA TGA C |
65 |
| PBX3 | [GenBank:NM_006195] | F: CAA GGG TCC CAA GTC GG R: TGG CCT AAT TGG ATA AAG TGC T |
60 |
| PBX4 | [GenBank:NM_025245] | F: ATG GGG AAG TTT CAA GAA GAG G R: ATC TCG AGT CGC AGC AGA C |
65 |
| GAPDH | [GenBank:NM_002046] | F: CAC TGC CAC CCA GAA GAC TGT G R: TGT AGG CCA TGA GGT CCA CCA C |
60 |
| RPL32 | [GenBank:NM_000994] | F: GCA TTG ACA ACA GGG TTC GTA G R: ATT TAA ACA GAA AAC GTG CAC A |
60 |
| ACTB | [GenBank:NM_001101] | F: TCC GCA AAG ACC TGT ACG R: AAG AAA GGG TGT AAC GCA ACT A |
60 |
Figure 1.
Analysis of primers used for amplification of Three-amino-acid loop-extension (TALE) family member genes. A) A pull of cDNA obtained from leukemia-derived cell lines was utilized to test the specificity and efficiency of each set of primers in the amplification of TALE family genes. After 40 cycles of amplification by conventional PCR, the PCR products were separated into 2% agarose gels and visualized under Ultraviolet (UV) light. Amplification products of the reference genes employed (RPL32 and ACTB) are also included. The 1 Kb Plus DNA Ladder (Invitrogen, Life Science) is shown in the left line; B) Amplification of PBX1 in leukemia-derived cell lines and in healthy controls separated into 2% agarose gels and visualized under UV light (upper panel). Genome map of complete (a) and alternative (b) splicing of PBX1; C) Sequence of alternative splicing of PBX1 showing adjacent coding regions of the deleted exon.
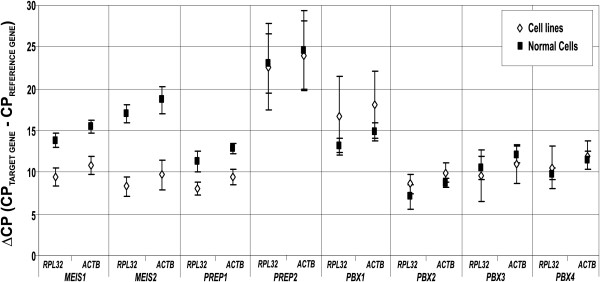
Next, we proceeded to analyze the expression of TALE genes by qRT-PCR in leukemia-derived cell lines. We employed five cell lines including Jurkat, CEM, MOLT-4, K562, and HL-60; the first three are lymphoblastic, and the latter two, myeloid. We determined the crossing point for each target gene and subsequently normalized this with the crossing point of an internal reference gene to calculate the ΔCP, which represents an absolute and more comparative value (see Materials and Methods). It is important to bear in mind that the ΔCP value is inversely proportional to gene expression. To obtain more consistent results, we use two different reference genes: RPL32, and ACTB. As can be observed in Figure 2, results obtained with RPL32 and ACTB follow the same tendency. In this regard, RPL32 and ACTB were selected as confident reference genes. Interestingly, when expression levels of TALE genes in leukemic cell lines and blood-derived normal cells were evaluated, significant differences were observed in the expression of MEIS1, MEIS2, and PREP1. Normal blood cells have greater ΔCP values for these three genes, thus lower expression (Figure 2). For PREP2 and all PBX members, we did not observe any variation. Additionally, on comparing ΔCP values we could note that in all cell lines and control cells, PREP2 possesses the lowest mRNA level.
Figure 2.
Baseline expression level of Three-amino-acid loop-extension (TALE) family genes (MEIS1, MEIS2, PREP1, PREP2, PBX1, PBX2, PBX3, and PBX4) in healthy cells vs. leukemia-derived cell lines. The graphics-display means and Standard deviation (SD) of ΔCP values obtained for the expression level of TALE genes. Values were calculated taking RPL32 or ACTB as reference genes. The squares and diamonds represent means ± SD of two independent experiments.
Up-regulation of MEIS1 and PREP1 and Down-regulation of PBX4 in ALL Samples vs. Those of Healthy Individuals
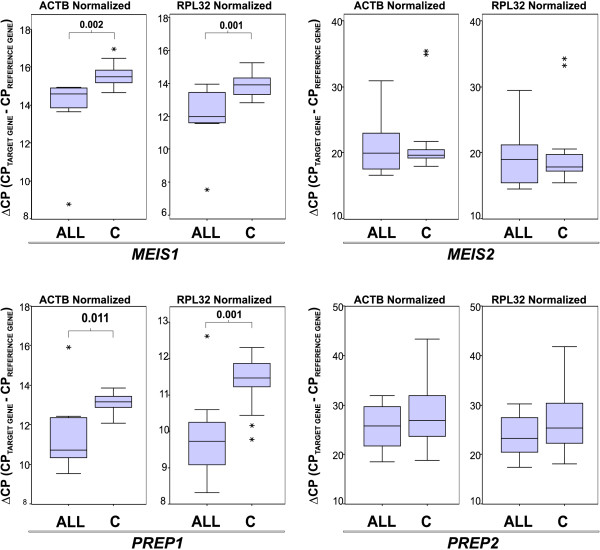
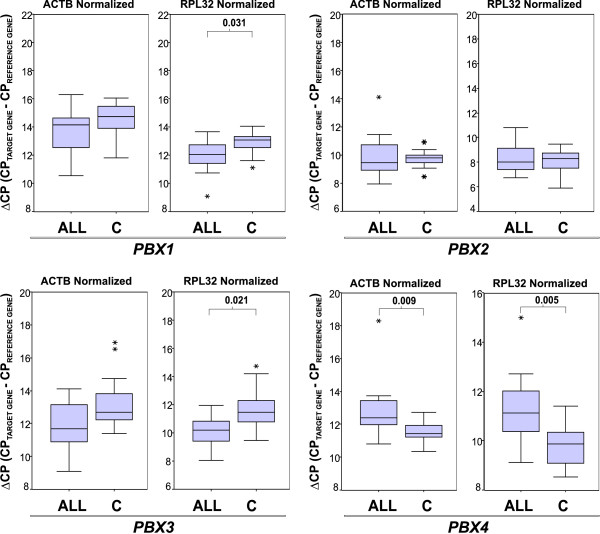
To confirm whether variations in TALE expression observed in cell lines were also observed in samples of patients with leukemia, we recruited 14 samples of patients diagnosed with Acute lymphoblastic leukemia (ALL) and 19 samples from clinically healthy volunteers (Table 2). We again analyzed the genetic expression of TALE genes by qRT-PCR employing the previously mentioned RPL32 and ACTB as reference genes to calculate ΔCP values. As can be observed in Figure 3, distribution of ΔCPs obtained for ALL samples were noticeably different from those obtained for control samples in the cases of MEIS1 and PREP1. Differences in ΔCP values for MEIS2 and PREP2 in patients compared with controls were not statistically significant. For the PBX group (see Figure 4), we observed that PBX1 and PBX3 were, to some extent, up-regulated in patients with ALL, but this difference was only statistically significant when we normalized with reference gene RPL32. PBX2 expression remained unchanged in patients and controls, and the sole member that clearly exhibited down-regulation in ALL samples was PBX4.
Table 2.
Overview of controls and patients
| Control ID | Gender | Age (years) | Patient ID | Gender | Age (years) | Diagnosis |
|---|---|---|---|---|---|---|
| 1 | M | 33 | 1 | M | 38 | ALL |
| 2 | M | 26 | 2 | M | 82 | ALL |
| 3 | F | 54 | 3 | M | 56 | ALL |
| 4 | F | 34 | 4 | F | 46 | ALL |
| 5 | F | 68 | 5 | F | 32 | ALL |
| 6 | M | 51 | 6 | F | 36 | ALL |
| 7 | F | 43 | 7 | F | 56 | ALL |
| 8 | F | 24 | 8 | M | 84 | ALL |
| 9 | F | 56 | 9 | M | 61 | ALL |
| 10 | M | 40 | 10 | M | 58 | ALL |
| 11 | F | 53 | 11 | F | 30 | ALL |
| 12 | F | 35 | 12 | M | 52 | ALL |
| 13 | F | 26 | 13 | F | 43 | ALL |
| 14 | M | 39 | 14 | M | 18 | ALL |
| 15 | M | 73 | ||||
| 16 | M | 45 | ||||
| 17 | F | 39 | ||||
| 18 | M | 40 | ||||
| 19 | M | 26 | ||||
ALL, Acute lymphoblastic leukemia; ID, identification; M, Masculine; F, Feminine.
Figure 3.
Levels of MEIS1-2 and PREP1-2 in healthy volunteers vs. patients with leukemia. Box plot graphics showing ΔCP values taking ACTB (left panel) or RPL32 (right panel) as reference genes. The graphics display median (dark lines), 25‒75th percentile (boxes), interquartile ranges (whiskers), and outliers (*) from the 14 patients with Acute lymphoblastic leukemia (ALL) and the 19 controls (C). Unpaired Student t test was used to compare ALL with Control group. Statistical significances are shown between groups only when p ≤ 0.05.
Figure 4.
Levels of PBX1-4 in healthy volunteers vs. patients with leukemia. Box plot graphics showing ΔCP values taking ACTB (left panel) or RPL32 (right panel) as reference genes. The graphics display median (dark lines), 25‒75th percentile (boxes), interquartile ranges (whiskers), and outliers (*) from the 14 patients with Acute lymphoblastic leukemia (ALL) and the 19 controls (C). Unpaired Student t test was used to compare ALL with Control group. Statistical significances are shown between groups only when p ≤ 0.05
MEIS1 Silencing Decreases the Proliferation Rate of Leukemic-derived Cell Lines
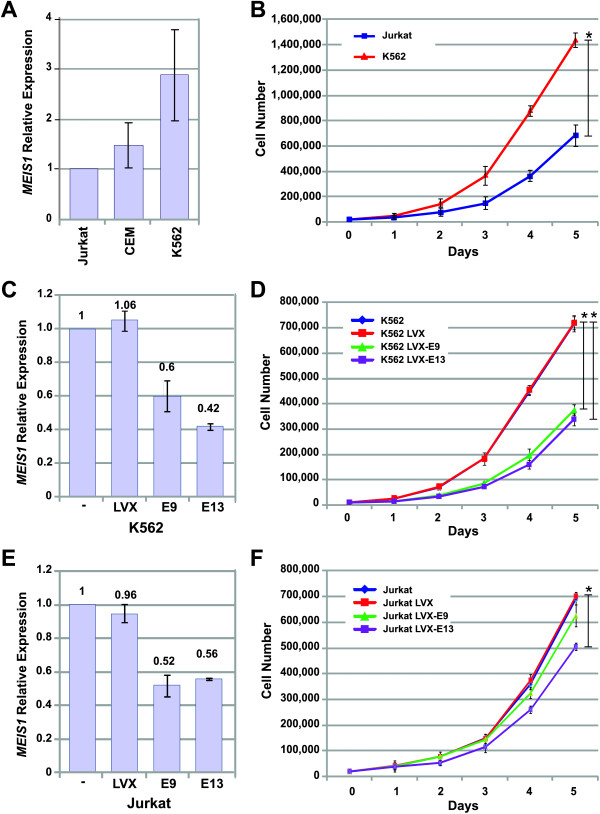
Because we determined a consistent up-regulation of MEIS1 and PREP1 in cell lines and in samples of patients with ALL, it was interesting to us to determine which type of advantage provides the high expression of these genes to leukemic cells. First we analyzed the role of MEIS1. The MEIS1 gene has been localized in chromosome 2 and it has been described that Jurkat cells are monosomic for this chromosome, CEM cells have two copies, and K562 cells are trisomic [21]; in this regard, expression of MEIS1 ought to be different in these cell lines. To test this hypothesis, we analyzed MEIS1 baseline expression in these cell lines by qRT-PCR (Figure 5A). As expected, Jurkat was the cell line with the lowest MEIS1 expression, followed by CEM and K562 expressing highest levels. Taking advantage of the existing different levels of MEIS1 in the cell lines, we utilized Jurkat and K562 cells to investigate whether high MEIS1 expression is related with increased proliferation. We observed that K562 have a higher proliferation rate than Jurkat cells (Figure 5B). To demonstrate the direct involvement of MEIS1 in this exacerbated proliferation, we performed silencing assays in both cell lines. We employed short hairpin RNAs shRNAs directed to two different regions of MEIS1 mRNA: one was directed to Exon 9 (E9), and the other to Exon 13 (E13). By using recombinant virus, we introduced these sequences into Jurkat and K562 cells. To assure that all infected cells were carrying the construction, a resistance gene to puromycin was also introduced and the infected cells were selected with this antibiotic. Additionally, we infected the cells with an empty virus (without shRNA) and selected them also with puromycin in order to posses a control for the selection and infection process. We then tested MEIS1 (mRNA) levels by qRT-PCR. As shown in Figures 5C and 5E, MEIS1 mRNA levels decrease with both shRNAs in both cell lines to nearly 50% of the initial expression. Employing the MEIS1-silenced cells, we then measured the proliferation rate and observed that proliferation was affected in all clones in which MEIS1 was silenced (Figures 5D and 5F). This effect was more obvious in K562 cells, which are the cells that express more MEIS1.
Figure 5.
Effect of MEIS1 expression on cell growth of leukemia-derived cell lines. A) Expression levels of MEIS1 were analyzed by qRT-PCR in Jurkat, CEM, and K562 cells; expression of RPL32 was also determined and used as reference gene to calculate relative expression; B) Cell proliferation analysis of K562 and Jurkat cells; C, E) Expression levels of MEIS1 in Jurkat and K562 cell lines infected with virus carrying shRNA-E9 or shRNA-E13. Values were obtained by qRT-PCR using RPL32 as reference gene; D, F) Proliferation of MEIS1-silenced cells. Jurkat and K562 cells were infected with an shRNA directed to exon 9 (LVX-E9) and an shRNA directed to exon 13 (LVX-E13). Cell growth was determined counting the cells daily for 5 days. Graphics show means ± Standard deviations (SD) of values obtained from three independent experiments. Statistical differences were calculated at the end point of proliferation curves using 2 way ANOVA analysis and Bonferroni posttest, (*) significances are shown between groups only when p ≤ 0.05.
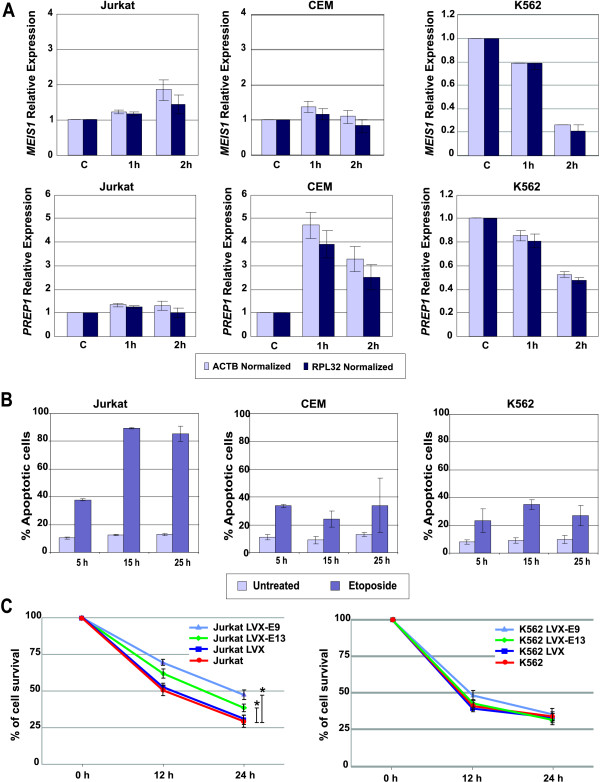
Expression of MEIS1 and PREP1 Is Modulated in Response to Apoptosis Induction by Etoposide
The other TALE member that we found up-regulated in leukemic cells was PREP1. Expression of this gene has been associated with resistance to apoptosis and it also has been described that PREP1 regulates MEIS1 expression [20,22]. In this respect, we subsequently analyzed whether the expression of PREP1 and MEIS1 was related with resistance to apoptosis induction by chemotherapeutic stimulus in leukemic cells. In order to assess this parameter, cultured cells were exposed to etoposide for 1 or 2 h; thereafter, variations in MEIS1 and PREP1 expression were analyzed by qRT-PCR. We observed that after etoposide treatment, Jurkat cells exhibit a tendency to increase MEIS1 expression, CEM cells remained unchanged, while diminishes K562 expression was noteworthy (Figure 6A). For PREP1, nearly no difference was observed in Jurkat cells; the response of CEM cells was more important because a notorious up-regulation was evidenced. Interestingly, K562 cells down-regulate PREP1 expression in response to etoposide (Figure 6A). To correlate these observations with phenotypic response, we measured the percentage of apoptotic cells after 5, 15, and 24 h of etoposide treatment. As can be observed in Figure 6B, Jurkat cells were the cells most sensitive to etoposide action; in contrast, CEM and K562 cells were the most resistant cells.
Figure 6.
Modulation of MEIS1 and PREP1 expression after etoposide treatment. A) Jurkat, CEM, and K562 cells were treated with 170 μM etoposide for 1 and 2 h; thereafter, total RNA was extracted and retrotranscribed. Real time-PCR assays were performed to determine the relative expression levels of MEIS1 and PREP1. Expression analysis was carried out by normalizing with non-treated cells and employing RPL32 as reference gene. The bars represent means ± Standard deviations (SD) of two independent experiments; B) Percentage of apoptotic cells induced by etoposide treatment. Jurkat, CEM, and K562 cells were treated with 170 μM etoposide; the percentage of apoptotic cells was measured using Annexin-V-FLUOS. The bars represent means ± Standard deviations (SD) of three independent experiments. C) MEIS1-silenced (LVX-E9 and -E13) cells were treated with 170 μM etoposide for 12 and 24 hours. Parental cells (Jurkat and K562) or empty vector-silenced cells (LVX) were also used. After etoposide treatment WST-1 was added to cell cultures and incubate for 3 additional hours. The percentage of cell survival was calculated measuring Optical density (OD) at 450 nm (OD of untreated cells was set as 100%). Statistical differences were calculated at the end point of the curves using 2 way ANOVA analysis and Bonferroni posttest, (*) significances are shown between groups only when p ≤ 0.05.
Given that K562 cells show a chemotherapeutic-resistant phenotype and that response of these cells to etoposide exposure is the down-modulation of MEIS1, and because we observed that Jurkat cells increased MEIS1 expression and were the most sensitive cells, we postulate that MEIS1 down-regulation could be a mechanism for resistance to etoposide-induced apoptosis. In this regard, Jurkat clones with MEIS1-silenced should be more resistant than Jurkat infected with the empty virus (pLVX) or with parental Jurkat cells. We tested this hypothesis exposing the cells to etoposide and measuring the percentage of surviving cells (Figure 6C). From this approach, we observed that Jurkat clones in which MEIS1 was silenced demonstrated a higher percentage of cell survival compared with pLVX infected cells or parental cells. MEIS1 silencing in K562 cells did not further increased the percentage of surviving cells.
Discussion
TALE genes are a particular group of homeobox genes that are important in the regulation of proliferation, apoptosis, and normal cell differentiation. Anomalous expression of these genes has been involved in the development of hematological malignancies [23]. In this work, we first analyzed variations in the expression of TALE genes in leukemia-derived cell lines compared with normal control cells. In that we observed dissimilar MEIS1, MEIS2, and PREP1 expression levels, we wished to confirm whether these changes were also observed in samples of patients with leukemia. Interestingly, we found variations in MEIS1, PREP1, and PBX4 expression. It has been reported that over-expression of MEIS1 blocks myeloid cell differentiation; thus, high levels of MEIS1 are required to maintain hematopoietic cells in an undifferentiated state [13]. We observed high levels of MEIS1 mRNA in leukemia-derived cell lines and also in the blood samples of patients with ALL; our results are in agreement with observations that lower levels of MEIS1 are unfavorable for cell life, because MEIS1 down-regulation has been related with decreased proliferation and poor survival of neuroblastoma and leukemia cell lines [24,25]. Moreover, Kawagoe et al. reported that down-regulation of MEIS1 is required to induce differentiation of hematopoietic cells [26]. Our findings support the notion that this gene plays an oncogenic role and that its expression is required to sustain proliferation and block differentiation in leukemia cells [24,27]. Controversially, it has been reported that high levels of this protein can also trigger apoptosis; we observed that high MEIS1-expressing K562 cells were more resistant to apoptosis induction than Jurkat cells, which exhibited lower levels of MEIS1; however, it is also well known that MEIS1 requires the presence of protein partners to achieve its different functions [16,28,29]; one explanation for the contradictory effects reported for MEIS1 could be that, regardless of higher MEIS1 expression, cells can regulate the action of this protein by modulating the expression of MEIS1 cofactors, such as HOX. The availability of the later can transform MEIS1 action from proliferative into pro-apoptotic [28].
In the cell lines studied, we observed that an apoptotic stimulus induces MEIS1 up- and down-regulation (Jurkat and K562, respectively). A strategy of tumor cells for survival could be down-regulation of MEIS1. In this respect, through lowering its proliferation rate, tumor cells avoid DNA damage, which can induce apoptosis.
Regarding MEIS2 expression, this gene has been found in immature neuronal precursor cells, lens proliferative cells, ovarian cancer, and other tumor cell types, which underlies its possible role in sustaining proliferation [30]. We observed strong expression in leukemia-derived cell lines compared with control cells, which is in agreement with the findings of Smith et al. [31]; however, when we analyzed its expression in patients, we found no variation in the expression of this gene (Figure 3).
To a greater extent, we observed that all studied cell lines express PREP1, but not PREP2. PREP1 has been described to be ubiquitously expressed in adult tissues [32] and PREP2 is depicted as possessing more restricted expression, being negative in peripheral blood leukocytes [2]. After apoptosis induction by etoposide, CEM cells greatly increase PREP1 gene expression, PREP1 has been directly involved in the regulation of apoptosis: it has been described that BCLXL, an intrinsic apoptotic-pathway regulator, is a direct target of PREP1 [22]. PREP proteins interact with PBX members to achieve their functions [33]. Interaction of PREP with PBX1 and PBX2 increases the stability of PBX proteins and additionally increases the affinity of PREP for DNA binding [34,35]; the expression of BCLXL and p53 has been reported to be regulated by PREP1 in cooperation with PBX1b [22,36]. In etoposide-treated CEM cells, it was observed that expression of PBX2 and PBX4 increases (Additional file 1); PBX2 has been reported as a negative apoptosis modulator through negative regulation of BCL2 [37]. Up-regulation of PREP1 together with down-regulation of PBX genes could account for the etoposide-resistant phenotype of CEM cells; this resistance could be mediated by BCLXL and p53. High levels of p53 have been associated with apoptosis but, in the presence of BCLXL-mediated survival signals, p53 can induce senescence instead of apoptosis [38].
Conclusions
In conclusion, our study shows that MEIS1 and PREP1 mRNA levels are significantly up-regulated in patients with ALL in comparison with healthy controls and inversely, that PBX4 is down-regulated in patients with ALL. Importantly, utilizing silencing assays, we confirmed that down-modulation of MEIS1 produces a lower leukemic-cell proliferation rate, an effect that was most notorious in the K562 myeloblastic cell line. Etoposide- induced apoptosis leads to changes in the expression of PREP1 and MEIS1; up-regulation of PREP1 and down-regulation of MEIS1 were independently related with resistance to apoptosis. Taken together, these results support the important role that TALE genes play in leukemic cell proliferation and survival, in addition to their probable involvement during leukemia development. Therefore, it could be important to evaluate MEIS1 and PREP1 expression in patients with leukemia prior to and after chemotherapeutic treatment and to correlate these findings with the clinical response.
Methods
Cells and cell culture
We used five commercially available human leukemia-derived cell lines: MOLT-4; Jurkat and CEM cells derived from lymphoid leukemia; HL-60 derived from promyelocytic leukemia, and K562 from erythroleukemia. Cells were grown in RPMI-1640 medium supplemented with 10% Fetal bovine serum (FBS), penicillin (100 U/mL,) and streptomycin (100 μg/mL); all products mentioned previously were obtained from GIBCO™ (Invitrogen Corp., Carlsbad, CA, USA). Cultures were maintained at 37°C in a humidified atmosphere with 5% CO2.
Patients and sample collection
Peripheral blood samples were collected from 14 patients with Acute lymphoblastic leukemia (ALL) according to World Health Organization (WHO) classification criteria at the Centro Médico Nacional de Occidente of the Mexican Social Security Institute (CIBO-IMSS) and the Hospital Civil de Guadalajara Fray Antonio Alcalde. Additionally, blood samples from 19 healthy donors were also collected from the IMSS Blood Bank. Letters of informed consent and protocols were approved by the CLIS-1305 Ethical Board of CIBO-IMSS.
Drugs and in vitro cell treatments
Etoposide was obtained from Lemery Laboratorios, México. The drug was stored at 4°C for <4 days and adjusted to the desirable concentration with DMEM culture medium immediately prior to utilization. The concentration employed was 170 μM etoposide.
RNA extraction and cDNA synthesis
Total RNA was isolated by using the PureLink™ Micro-to-Midi Total RNA Purification System (Invitrogen Corp.) from 5 × 106 cultured cells as described by the manufacturer. Peripheral blood samples from patients with leukemia and from healthy donors were collected with EDTA as anticoagulant and mixed immediately after collection with 45 mL of RNA/DNA Stabilization Reagent for Blood/Bone Marrow (Roche Applied Science, Germany) and stored at ‒80°C for preservation. The stabilized samples were utilized for mRNA isolation via a two-step procedure by means of magnetic separation employing the mRNA Isolation kit for blood/bone marrow (Roche Applied Science). mRNA was finally eluted from the magnetic pearls in 20 μL of water and stored at ‒80°C until use.
cDNA synthesis was performed from 5 μg of total RNA or 12 uL mRNA employing the Transcriptor First Strand cDNA Synthesis kit primed with oligo(dT) (cat. no. 04897030001, Roche Applied Science). The protocol was conducted as recommended by the manufacturer. cDNA were stored at ‒20°C and aliquots were utilized as templates for PCR and RT-PCR reactions.
PCR and RT-PCR
PCR reactions were carried out utilizing the set of primers presented in Table 1; the primers were designed using Oligo v6.0 software from sequences obtained from the NCBI-website GenBank Nucleotide database. PCR was performed using Taq DNA Polymerase (cat. no. 11146173001, Roche Applied Science) and Deoxynucleoside triphosphates (cat. no. 1969064, Roche Applied Science) in a PX2 Thermal Cycler (Thermo Electron Corp.). All reactions were conducted in 20 μL at the specified Tm (see Table 1). PCR products were resolved in 2% agarose gels containing 0.1 μg/mL ethidium bromide (Sigma Aldrich, Germany), visualized under Ultraviolet (UV) light, and documented with a DigiDoc-It System, (UVP, UK). RT-PCR analysis was achieved by employing the LightCycler-FastStart DNA MasterPLUS SYBR Green I kit (cat. no. 03515885001, Roche Applied Science) in the LightCycler 1.5 System (Roche Diagnostics GmbH, Mannheim, Germany). Data were normalized to the expression of the reference genes RPL32 (L32 Ribosomal Protein) and ACTB (β-actin).
ΔCP analysis
To normalize target gene expression, we employed two different reference genes. We calculated the Crossing point (CP) for target and reference genes in each sample and subsequently calculated the ΔCP value of each sample, i.e., the target gene CP minus the reference gene CP. This facilitated analysis by taking only the intrinsic values of each sample. CPs from ACTB, and RLP32 were employed for this analysis. It is extremely noteworthy that ΔCP is inversely proportional to the expression of the target gene.
Lentivirus production and infection
Oligonucleotides used to construct shRNAs were: forward 5'- GAT CCG CGG GAC TCA CCA TCC TTC AAG TGA ATT CAA GAG ATT CAC TTG AAG GAT GGT GAG TCC CGT TTT TTG-3' and reverse 5'-AAT TCA AAA AAC GGG ACT CAC CAT CCT TCA AGT GAA TCT CTT GAA TTC ACT TGA AGG ATG GTG AGT CCC GCG-3' directed to MEIS1 exon 9 (E9); forward 5'-GAT CCG CCG TGT GTT TAG AAG CCT AAT TCA AGA GAT TAG GCT TCT AAA CAC ACG GCT TTT TTA CGC GTG-3 and reverse 5'- AAT TCA CGC GTA AAA AAG CCG TGT GTT TAG AAG CCT AAT CTC TTG AAT TAG GCT TCT AAA CAC ACG GCG-3' directed to MEIS1 exon 13 (E13); this latter sequence has been previously used [24]. Complementary primers were annealed and cloned into the vector pLVX-shRNA1 (Clontech Laboratories, USA) using the restriction sites BamHI and EcoRI (NEB-Biolabs, USA). To produce infectious viral particles, Lenti-X 293T cells were transient-transfected by Lentiphos HT/Lenti-X HT Packaging Systems with lentiviral vectors pLVX-Puro or pLVX-shRNA1-E9 or pLVX-shRNA1-E13 as described by the manufacturer (Clontech Laboratories, USA). After 48 h, supernatants were checked with Lenti-X GoStix (Clontech Laboratories, USA) to determine whether sufficient viral particles were produced before transducing target cells. Supernatants were filtered through a 0.22-μm PES filter to eliminate detached cells, were aliquoted, and subsequently stored at ‒80°C until use. Jurkat and K562 cells (2.5 × 105) were transduced with approximately 4.5 × 105 IFU/mL of supernatants. RNA extractions were obtained after at least 2 weeks of puromycin selection (1 μg/mL).
Cell survival determination
Cell survival was determined by cleavage of tetrazolium salt WST-1 to formazan by cellular mitochondrial dehydrogenase enzymes. After different treatment periods, cells were incubated with 10 μL/well of WST-1/ECS solution (BioVision Research, Mountain View, CA, USA) for 3 h. Absorbance (450 nm) of treated and untreated samples was determined on a microtiter plate reader (Synergy™ HT Multi-Mode Microplate Reader; Biotek, Winooski, VT, USA). Data are reported as percentage of cell survival taking untreated control cells as 100% of cell survival.
Apoptosis detection
Cell death was measured by flow cytometry using propidium iodide (cat. no. P4864, Sigma-Aldrich) and Annexin-V-FlUOS (cat. no. 1828681, Roche Applied Science) as recommended by these manufacturers. Cells were seeded in 6-well plates at a density of 3 × 105 cells per well in 1 mL RPMI medium containing or not etoposide (170 μM). After 5, 15, and 25 h, each sample was analyzed in a FACS Aria cytometer (BD Biosciences).
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
JAR-A, JT-F, and AA-L carried out the PCR experiments but were also involved in all of the experimental work. GH-F, PCO-L, and JML-D made up the cell culture and devised drug treatment and flow cytometry for apoptosis detection. RdC determined cell survival. AB-C, CG-D, OG-R, and EB-C were involved in the recruitment of patients with leukemia and controls. AA-L and LFJ-S performed the statistical analysis, conceived of and designed the study, and wrote the manuscript. All authors helped to draft the manuscript and in reading and approving this final version.
Supplementary Material
Modulation of PBX1-4 expression after etoposide treatment. Jurkat and CEM cells were treated with 170 μM etoposide for 1 and 2 h; thereafter, total RNA was extracted and retrotranscribed. Real time-PCR assays were performed to determine the relative expression levels of PBX1-4. Expression analysis was carried out by normalizing with non-treated cells and employing RPL32 as reference gene. The bars represent means ± Standard deviations (SD) of two independent experiments.
Contributor Information
Judith A Rosales-Aviña, Email: judithabisag1@gmail.com.
Jorge Torres-Flores, Email: torresflores_md@yahoo.com.mx.
Adriana Aguilar-Lemarroy, Email: adry.aguilar.lemarroy@gmail.com.
Carmen Gurrola-Díaz, Email: carmenhpv@yahoo.de.
Georgina Hernández-Flores, Email: geodic1967@yahoo.com.mx.
Pablo C Ortiz-Lazareno, Email: paulcesar05@hotmail.com.
José M Lerma-Díaz, Email: drlermamed@gmail.com.
Ruth de Celis, Email: ruthdecelis@hotmail.com.
Óscar González-Ramella, Email: glezramella@hotmail.com.
Esperanza Barrera-Chaires, Email: vivibarrera2010@hotmail.com.
Alejandro Bravo-Cuellar, Email: abravocster@gmail.com.
Luis F Jave-Suárez, Email: lfjave@yahoo.com.
Acknowledgements
We thank our technicians María de Jesús Delgado-Ávila and Leticia Ramos-Zavala for their efficient support. This work was supported by grants CB-2005-25121/51502-M (CONACyT-México), FIS/2005/1/I/022, and FIS/2006/1A/I/051 (IMSS) to LFJ-S.
References
- Burglin TR. Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals. Nucleic Acids Res. 1997;25:4173–4180. doi: 10.1093/nar/25.21.4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fognani C, Kilstrup-Nielsen C, Berthelsen J, Ferretti E, Zappavigna V, Blasi F. Characterization of PREP2, a paralog of PREP1, which defines a novel sub-family of the MEINOX TALE homeodomain transcription factors. Nucleic Acids Res. 2002;30:2043–2051. doi: 10.1093/nar/30.9.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monica K, Galili N, Nourse J, Saltman D, Cleary ML. PBX2 and PBX3, new homeobox genes with extensive homology to the human proto-oncogene PBX1. Mol Cell Biol. 1991;11:6149–6157. doi: 10.1128/mcb.11.12.6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner K, Mincheva A, Korn B, Lichter P, Popperl H. Pbx4, a new Pbx family member on mouse chromosome 8, is expressed during spermatogenesis. Mech Dev. 2001;103:127–131. doi: 10.1016/S0925-4773(01)00349-5. [DOI] [PubMed] [Google Scholar]
- Di Giacomo G, Koss M, Capellini TD, Brendolan A, Popperl H, Selleri L. Spatio-temporal expression of Pbx3 during mouse organogenesis. Gene Expr Patterns. 2006;6:747–757. doi: 10.1016/j.modgep.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Selleri L, Depew MJ, Jacobs Y, Chanda SK, Tsang KY, Cheah KS, Rubenstein JL, O'Gorman S, Cleary ML. Requirement for Pbx1 in skeletal patterning and programming chondrocyte proliferation and differentiation. Development. 2001;128:3543–3557. doi: 10.1242/dev.128.18.3543. [DOI] [PubMed] [Google Scholar]
- Toresson H, Parmar M, Campbell K. Expression of Meis and Pbx genes and their protein products in the developing telencephalon: implications for regional differentiation. Mech Dev. 2000;94:183–187. doi: 10.1016/S0925-4773(00)00324-5. [DOI] [PubMed] [Google Scholar]
- DiMartino JF, Selleri L, Traver D, Firpo MT, Rhee J, Warnke R, O'Gorman S, Weissman IL, Cleary ML. The Hox cofactor and proto-oncogene Pbx1 is required for maintenance of definitive hematopoiesis in the fetal liver. Blood. 2001;98:618–626. doi: 10.1182/blood.V98.3.618. [DOI] [PubMed] [Google Scholar]
- Pillay LM, Forrester AM, Erickson T, Berman JN, Waskiewicz AJ. The Hox cofactors Meis1 and Pbx act upstream of gata1 to regulate primitive hematopoiesis. Dev Biol. 2010. [DOI] [PubMed]
- Hisa T, Spence SE, Rachel RA, Fujita M, Nakamura T, Ward JM, Devor-Henneman DE, Saiki Y, Kutsuna H, Tessarollo L. et al. Hematopoietic, angiogenic and eye defects in Meis1 mutant animals. EMBO J. 2004;23:450–459. doi: 10.1038/sj.emboj.7600038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskow JJ, Bullrich F, Huebner K, Daar IO, Buchberg AM. Meis1, a PBX1-related homeobox gene involved in myeloid leukemia in BXH-2 mice. Mol Cell Biol. 1995;15:5434–5443. doi: 10.1128/mcb.15.10.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Largaespada DA, Shaughnessy JD Jr, Jenkins NA, Copeland NG. Cooperative activation of Hoxa and Pbx1-related genes in murine myeloid leukaemias. Nat Genet. 1996;12:149–153. doi: 10.1038/ng0296-149. [DOI] [PubMed] [Google Scholar]
- Calvo KR, Knoepfler PS, Sykes DB, Pasillas MP, Kamps MP. Meis1a suppresses differentiation by G-CSF and promotes proliferation by SCF: potential mechanisms of cooperativity with Hoxa9 in myeloid leukemia. Proc Natl Acad Sci USA. 2001;98:13120–13125. doi: 10.1073/pnas.231115398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Blanco E, Bruns I, Neumann F, Fischer JC, Graef T, Rosskopf M, Brors B, Pechtel S, Bork S, Koch A. et al. Molecular signature of CD34(+) hematopoietic stem and progenitor cells of patients with CML in chronic phase. Leukemia. 2007;21:494–504. doi: 10.1038/sj.leu.2404549. [DOI] [PubMed] [Google Scholar]
- Mann RS, Affolter M. Hox proteins meet more partners. Curr Opin Genet Dev. 1998;8:423–429. doi: 10.1016/S0959-437X(98)80113-5. [DOI] [PubMed] [Google Scholar]
- Moens CB, Selleri L. Hox cofactors in vertebrate development. Dev Biol. 2006;291:193–206. doi: 10.1016/j.ydbio.2005.10.032. [DOI] [PubMed] [Google Scholar]
- Chung EY, Liu J, Homma Y, Zhang Y, Brendolan A, Saggese M, Han J, Silverstein R, Selleri L, Ma X. Interleukin-10 expression in macrophages during phagocytosis of apoptotic cells is mediated by homeodomain proteins Pbx1 and Prep-1. Immunity. 2007;27:952–964. doi: 10.1016/j.immuni.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deflorian G, Tiso N, Ferretti E, Meyer D, Blasi F, Bortolussi M, Argenton F. Prep1.1 has essential genetic functions in hindbrain development and cranial neural crest cell differentiation. Development. 2004;131:613–627. doi: 10.1242/dev.00948. [DOI] [PubMed] [Google Scholar]
- Penkov D, Di Rosa P, Fernandez Diaz L, Basso V, Ferretti E, Grassi F, Mondino A, Blasi F. Involvement of Prep1 in the alphabeta T-cell receptor T-lymphocytic potential of hematopoietic precursors. Mol Cell Biol. 2005;25:10768–10781. doi: 10.1128/MCB.25.24.10768-10781.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti E, Villaescusa JC, Di Rosa P, Fernandez-Diaz LC, Longobardi E, Mazzieri R, Miccio A, Micali N, Selleri L, Ferrari G, Blasi F. Hypomorphic mutation of the TALE gene Prep1 (pKnox1) causes a major reduction of Pbx and Meis proteins and a pleiotropic embryonic phenotype. Mol Cell Biol. 2006;26:5650–5662. doi: 10.1128/MCB.00313-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roschke AV, Tonon G, Gehlhaus KS, McTyre N, Bussey KJ, Lababidi S, Scudiero DA, Weinstein JN, Kirsch IR. Karyotypic complexity of the NCI-60 drug-screening panel. Cancer Res. 2003;63:8634–8647. [PubMed] [Google Scholar]
- Micali N, Ferrai C, Fernandez-Diaz LC, Blasi F, Crippa MP. Prep1 directly regulates the intrinsic apoptotic pathway by controlling Bcl-XL levels. Mol Cell Biol. 2009;29:1143–1151. doi: 10.1128/MCB.01273-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen TD, Zhu YX, Hawley TS, Hawley RG. TALE homeoproteins as HOX11-interacting partners in T-cell leukemia. Leuk Lymphoma. 2000;39:241–256. doi: 10.3109/10428190009065824. [DOI] [PubMed] [Google Scholar]
- Kumar AR, Li Q, Hudson WA, Chen W, Sam T, Yao Q, Lund EA, Wu B, Kowal BJ, Kersey JH. A role for MEIS1 in MLL-fusion gene leukemia. Blood. 2009;113:1756–1758. doi: 10.1182/blood-2008-06-163287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geerts D, Schilderink N, Jorritsma G, Versteeg R. The role of the MEIS homeobox genes in neuroblastoma. Cancer Lett. 2003;197:87–92. doi: 10.1016/S0304-3835(03)00087-9. [DOI] [PubMed] [Google Scholar]
- Kawagoe H, Humphries RK, Blair A, Sutherland HJ, Hogge DE. Expression of HOX genes, HOX cofactors, and MLL in phenotypically and functionally defined subpopulations of leukemic and normal human hematopoietic cells. Leukemia. 1999;13:687–698. doi: 10.1038/sj.leu.2401410. [DOI] [PubMed] [Google Scholar]
- Argiropoulos B, Yung E, Humphries RK. Unraveling the crucial roles of Meis1 in leukemogenesis and normal hematopoiesis. Genes Dev. 2007;21:2845–2849. doi: 10.1101/gad.1619407. [DOI] [PubMed] [Google Scholar]
- Wermuth PJ, Buchberg AM. Meis1-mediated apoptosis is caspase dependent and can be suppressed by coexpression of HoxA9 in murine and human cell lines. Blood. 2005;105:1222–1230. doi: 10.1182/blood-2004-03-0802. [DOI] [PubMed] [Google Scholar]
- Fujino T, Yamazaki Y, Largaespada DA, Jenkins NA, Copeland NG, Hirokawa K, Nakamura T. Inhibition of myeloid differentiation by Hoxa9, Hoxb8, and Meis homeobox genes. Exp Hematol. 2001;29:856–863. doi: 10.1016/S0301-472X(01)00655-5. [DOI] [PubMed] [Google Scholar]
- Pennartz S, Belvindrah R, Tomiuk S, Zimmer C, Hofmann K, Conradt M, Bosio A, Cremer H. Purification of neuronal precursors from the adult mouse brain: comprehensive gene expression analysis provides new insights into the control of cell migration, differentiation, and homeostasis. Mol Cell Neurosci. 2004;25:692–706. doi: 10.1016/j.mcn.2003.12.011. [DOI] [PubMed] [Google Scholar]
- Smith JE, Afonja O, Yee HT, Inghirami G, Takeshita K. Chromosomal mapping to 15q14 and expression analysis of the human MEIS2 homeobox gene. Mamm Genome. 1997;8:951–952. doi: 10.1007/s003359900621. [DOI] [PubMed] [Google Scholar]
- Ferretti E, Schulz H, Talarico D, Blasi F, Berthelsen J. The PBX-regulating protein PREP1 is present in different PBX-complexed forms in mouse. Mech Dev. 1999;83:53–64. doi: 10.1016/S0925-4773(99)00031-3. [DOI] [PubMed] [Google Scholar]
- Diaz VM, Bachi A, Blasi F. Purification of the Prep1 interactome identifies novel pathways regulated by Prep1. Proteomics. 2007;7:2617–2623. doi: 10.1002/pmic.200700197. [DOI] [PubMed] [Google Scholar]
- Berthelsen J, Zappavigna V, Mavilio F, Blasi F. Prep1, a novel functional partner of Pbx proteins. EMBO J. 1998;17:1423–1433. doi: 10.1093/emboj/17.5.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longobardi E, Blasi F. Overexpression of PREP-1 in F9 teratocarcinoma cells leads to a functionally relevant increase of PBX-2 by preventing its degradation. J Biol Chem. 2003;278:39235–39241. doi: 10.1074/jbc.M304704200. [DOI] [PubMed] [Google Scholar]
- Micali N, Longobardi E, Iotti G, Ferrai C, Castagnaro L, Ricciardi M, Blasi F, Crippa MP. Down syndrome fibroblasts and mouse Prep1-overexpressing cells display increased sensitivity to genotoxic stress. Nucleic Acids Res. 2010. [DOI] [PMC free article] [PubMed]
- Qiu Y, Song B, Zhao G, Deng B, Makino T, Tomita Y, Wang J, Luo W, Doki Y, Aozasa K, Morii E. Expression level of Pre B cell leukemia homeobox 2 correlates with poor prognosis of gastric adenocarcinoma and esophageal squamous cell carcinoma. Int J Oncol. 2010;36:651–663. doi: 10.3892/ijo_00000541. [DOI] [PubMed] [Google Scholar]
- Zuckerman V, Wolyniec K, Sionov RV, Haupt S, Haupt Y. Tumour suppression by p53: the importance of apoptosis and cellular senescence. J Pathol. 2009;219:3–15. doi: 10.1002/path.2584. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Modulation of PBX1-4 expression after etoposide treatment. Jurkat and CEM cells were treated with 170 μM etoposide for 1 and 2 h; thereafter, total RNA was extracted and retrotranscribed. Real time-PCR assays were performed to determine the relative expression levels of PBX1-4. Expression analysis was carried out by normalizing with non-treated cells and employing RPL32 as reference gene. The bars represent means ± Standard deviations (SD) of two independent experiments.