Abstract
Background
Whether Folic acid is a potential drug that may prevent the progression of colorectal carcinoma and when to use are important healthy issues we focus on. Our study is to examine the effect of folic acid on the development of the CRC and the optimal time folic acid should be provided in a mouse-ICR model induced by 1, 2-Dimethylhydrazine. Also, we investigated the gene expression profile of this model related to folic acid.
Method
Female ICR mouse (n = 130) were divided into 7 groups either with the treatment of 1, 2-Dimethylhydrazine (20 mg/kg bodyweight) weekly or folic acid (8 mg/kg bodyweight) twice a week for 12 or 24 weeks. Using a 4 × 44 K Agilent whole genome oligo microarray assay, different gene expression among groups (NS, DMH, FA2, FA3) were identified and selected genes were validated by real-time polymerase chain reaction.
Results
Animals with a supplementary of folic acid showed a significant decrease in the incidence, the maximum diameter and multiplicity of adenocarcinomas (P < 0.05). Furthermore, there were fewer adenomas or adenocarcinomas developed in the group of folic acid supplementation in pre-adenoma stage compared to group of post-adenoma stage. Meanwhile, about 1070 genes that were changed by 1, 2-Dimethylhydrazine can be reversed by folic acid and 172 differentially genes were identified between the groups of pre- and post- adenoma stage using microarray gene expression analysis.
Conclusion
Our study demonstrated that folic acid supplementary was significantly associated with the decrease risk of CRC. And the subgroup of providing folic acid without precancerous lesions was more effective than that with precancerous lesions.
Introduction
It is known that colorectal cancer (CRC) is one of the most common cancers especially in western countries, referred to a multiple process, multiple factors with high recurrence and high mortality [1]. Chemoprevention methods for CRC have obtained increasing attention as surgery and chemotherapy strategies perform little function once diagnosed to be tumor that invades the muscularis propria. Also, the Non-steroidal anti-inflammatory drugs (NSAIDs), such as COX-2 inhibitors, are not always successful, and may have some harmful side-effects [2]. Generally, clinical trials require at least 3-5 years follow up and a large number of patients are difficult to control their lifestyles such as smoking and wine intake which may affect the incidence of cancer [3,4]. Therefore, we choose animal model induced by chemistry drugs 1, 2-dimethylhydrazine (DMH) to simulate the formation of CRC. As azoxymethane (AOM) or 1, 2-dimethylhydrazine (DMH)-induced colon carcinogenesis in mice or rat have been identified as a useful tool [5-9]. In the previous study, we have successfully induced CRC in this model using ICR mice [9].
Folic Acid (FA) is one kind of water-solubility vitamin, which has been believed to be chemo-preventive agent that can provide methy-group to DNA thus impact DNA synthesis and DNA methylation [10]. Abbreviations in DNA synthesis often lead to DNA mutation, DNA strand break and the impairment of DNA repair, which finally result in cancer formation [11].
However, there are many conflicting data about whether FA can inhibit or promote colorectal adenoma (CRA) from clinical or preclinical studies. Epidemiologic study shows that folic acid is significant associated with lower risk and not related to the increased risk of colorectal cancer, supporting folic acid as a protective role for colon mucosa [12,13], including several large prospective studies in 99,523 participants in the American Cancer Prevention Study II (CPS-II) Nutrition Cohort [12] and in double-blind, randomized clinical trial (RCT) conducted by 9 clinical centers incorporating 1091 participants for 3 years follow up [13]. However, the Aspirin/Folate Polyp Prevention Trial demonstrated that about 67% increased risk of advanced lesions with high malignant potential, and an increased risk of having multiple adenomas among the folic acid supplementation group by providing folic acid for 6 years at 1 mg/d [14]. While other researches have reported that there is no relation or positive association between folic acid supplementation and the risk of colon adenoma [15]. Therefore, a systematic description from RCTs investigating the relation between folic acid supplementation and the risk of colorectal cancer was conducted by many groups. One recent Meta-analysis data revealed that folic acid supplementary for 3 years had no effect on the adenoma recurrence while had an increased risk of adenoma lesion for those who received folic acid over 3 years [16]. Another Meta-analysis divided the RCTs into different groups including populations with a history of adenoma and with an-average risk populations. They concluded that the evidence that folic acid was effective in the chemoprevention of colorectal cancer was not enough in both populations [17].
Further, many researchers consider that the role of folic acid might be two-sided, that is to prevent in early phage of adenoma formation and to promote in late stage depending on the time of folic acid administration. Preclinical studies have suggested that folic acid may only protect against the development of CRC in normal colon-rectum rather in mucosa with an Aberrant Crypt Foci (ACF) status [18], which is the earliest pre-neoplastic lesion that can be recognized based on the morphology and pathology features [19,20], and the results were consistent with an AOM induced rat model of CRC [21]. These experiments demonstrated that folic acid had dual effects on the development of CRC depending on the timing and dose of the intervention of folic acid [11] However, the function that folic acid may perform to the exiting adenomas in chemicals induced mouse model and the possible mechanism is still un-established now.
In this study, we use ICR mice with 1, 2-Dimethylhydrazine (DMH) interfered models to analyze the impact of folic acid on different timing courses during the processes of CRC. We have previously demonstrated that 4 weeks old ICR mice given high dosage (8 mg/ml) folic acid for 20 weeks have much more apparent effects to prevent CRC incidence than low folic acid dosage (4 mg/kg bodyweight) group using DMH-induced mice model [9]. Therefore, to investigate the role of folic acid in the process of adenoma formation, we use the dose of 8 mg/kg bodyweight. Meanwhile, we inferred that the occurrence of adenoma may take place at the course of 12 weeks based on the performance of mice in previously study, so we designed the 12th week as the division of the prophase or advanced stage of CRC. The study is expected to guide the clinical application of folic acid and to identify the mechanism of folic acid in a microarray gene expression profile.
Materials and methods
Ethics Statement
Our study had been approved by Animal Care and Use Committee of Shanghai Jiao-Tong University School of Medicine Ren-Ji Hospital, Shanghai, China (approval ID: 2007-036. All animal procedures were performed according to guidelines developed by the China Council on Animal Care and protocol approved by Shanghai Jiao-Tong University School of Medicine Ren-Ji Hospital, Shanghai, China.
Chemicals
1, 2-Dimethylhydrazine (DMH) and Folic acid (FA, F8758) were obtained from Sigma Chemical Co. (St. Louis, MO, USA). The PH value of DMH is adjusted with NaHCO3 to 6.5-7.0. DMH was dissolved with Normal saline and Folic Acid with drinking water.
Experimental animals
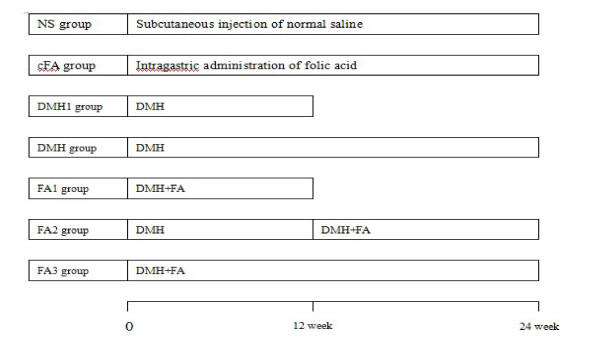
130 females, 4 weeks old ICR mice (weight, 18-20 g; grade, specific pathogen-free (SPF)) were bought from the Chinese Academy of Sciences (Shanghai, China). The mice were raised at constant temperature of 22°C with a relative humidity of 60% and 12-hour light/dark cycles; they were supplied a standard laboratory diet and drinking water. These 130 mice were randomly divided into 7 groups (Figure 1): NS group = 20 (Subcutaneous injection of physiological saline); DMH1 group = 20(Subcutaneous injection of DMH for 12 weeks); DMH group = 20 (Subcutaneous injection of DMH for 24 weeks); Cfa (control Folic Acid) = 10 (only intragastric administration of folic acid without DMH injection; FA1 = 20 (intragastric administration of folic acid with DMH injection for early 12 weeks); FA2 = 20(intragastric administration of folic acid with DMH injection for later 12 weeks); FA3 = 20 (intragastric administration of folic acid with DMH injection for 24 weeks). DMH was given subcutaneous injection once a week at the dosage of 20 mg/kg and folic acid was given by intragastric administration twice a week. All mice were weighted once a week. At the 12th weeks after DMH injection, 10 of NS and groups of DMH1, FA1 were killed and the conditions of organs were recorded. The mass number and size were assessed using a micrometer. Some fresh colon and rectal tissues were maintained immediately in liquid nitrogen, and others include liver or gastric tissues were fixed in formalin solution and embedded in paraffin blocks for pathological analysis. At the end of 24th weeks, all remaining mice were killed using the same methods.
Figure 1.
Groups of this study. NS group = Subcutaneous injection of physiological saline; DMH1 group = Subcutaneous injection of DMH for 12 weeks; DMH group = Subcutaneous injection of DMH for 24 weeks; cFA (control Folic Acid) = only intragastric administration of folic acid without DMH injection; FA1 = intragastric administration of folic acid with DMH injection for early 12 weeks; FA2 = intragastric administration of folic acid with DMH injection for later 12 weeks; FA3 = intragastric administration of folic acid with DMH injection for 24 weeks.
Histological Analysis
For pathology analysis, 4-μm thick sections of formalin-fixed, paraffin-embedded tissues were prepared. After hematoxylin and eosin staining, the sections of each tumor were examined under a light microscope (Olympus, Japan).
RNA extraction and Real-time polymerase chain reaction labeling, hybridization, and analysis
Total RNAs from normal colonic mucosa of all groups were got using TRIzol (Invitrogen, USA) according to manufacturer's instruction. RNA content and purity were measured using Nanodrop ND-1000, and denaturing gel electrophoresis was performed. Next, Reverse transcription and quantification of gene expression was performed according to the manufacture's introduction (Takara). We used 18s as an internal control in Real- time PCR. Next, 3 samples of non-tumor colon of the group of NS, DMH, FA2, FA3 were amplified and labeled with the Agilent Quick Amp labeling kit and hybridized using Agilent whole genome oligo microarray (Agilent Technologies, Palo Alto, CA, USA) by using Agilent SureHyb Hybridization Chambers. Then, the processed slides were scanned with the Agilent DNA microarray scanner according to the settings provided by Agilent Technologies.
The microarray data sets were normalized by Agilent GeneSpring GX software (version 11.0) using the Agilent FE one-color scenario (mainly median normalization). Differentially expressed genes were identified via the fold-change (FC) and p values of the t-test. Differentially expressed genes are identified to have an FC of ≥ 1.5 and a p value of ≤ 0.05 between two groups. Functional differences of the differentially expressed genes was analyzed using the Gene Ontology (GO; http://www.geneontology.gov/).
Statistical analysis
The results of the animal experiments and real-time PCR were analyzed using SAS 9.2 software (SAS Institute Inc. USA) with data presented in the forms of means ± SD. Student's t-test was used to compare values between two independent groups. Differences were considered to be significance when p < 0.05.
Results
Results of Animal Experiment
In the 12th week, 2 of 20 mice in DMH group were discovered average 2 × 3 mm adenoma, while there is none in FA1 and NS groups. Thus, the 12th week after DMH treatment might be considered to be the pre-stage that adenomas formed in DMH-induced model.
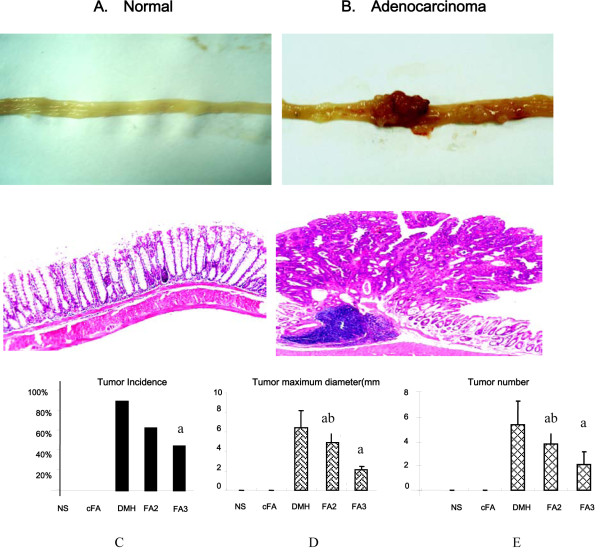
We have successfully induced CRC in the animal model with injection DMH for 24 weeks, which were identified as adenocarcinoma by histology analysis (Figure 2A, B). Figure 1 shows mainly results of the experiment. We can see that the incidence of DMH-induced group is 90%, much higher than any other groups such as FA2, FA3, which are 63%, 45% respectively (Figure 2C). There is significant difference between groups of FA3 and DMH but not between FA2 and DMH groups. However, the multiplicity and the size of the maximum masses in FA2 and FA3 groups are much smaller compared to the DMH group (5.39 ± 1.97, 6.44 ± 1.72 mm), indicating that folic acid may prevent the growth of adenomas.
Figure 2.
Main results of the animal experiment after sacrificed at the 24 weeks. A. The morphology of normal colon in macroscopic observation (Upper) and microscopy (HE stained) (Lower). Neither signs of injury nor tumor were found in NS group and cFA group. B. The morphology of colon adenocacinoma in macroscopic observation (Upper) and microscopy (HE stained) (Lower). C. The incidences of DMH-induced colorectal tumor in different groups. DMH group is 90%, which is much higher than any other groups such as FA2, FA3 which are 63%, 45% respectively. Meanwhile, there is none in NS and cFA group. D. Maximum diameter of tumor among the 5 groups (NS, cFA, DMH, FA1 and FA2). E. Tumor number in mice among the above 5 groups. (a: P < 0.05, FA3 and FA2 compared to DMH group; b: P < 0.05, FA2 compared to FA3 group)
Although the incidence in FA2 is higher than FA3, no significant difference was seen between them (63% vs 45%). However, the number and the maximum diameter of the masses in FA3 group (2.11 ± 1.05, 2.11 ± 0.60 mm) showed a significant smaller than FA2 group (3.83 ± 1.11, 4.92 ± 1.24 mm), P < 0.05 (Figure 2D and Figure 2E).
There is no tumor shaped and weight loss in Folic Acid control group and the mice behavior normal, so we conclude that folic acid is safe to normal colon. Meanwhile, there was no significant difference in the growth and development of mice among DMH and FA2, FA3 groups but groups between NS and DMH. Also, a macroscopic and microscopic examination of their kidneys, stomachs, lungs, liver, and spleen showed no obvious abnormalities (data not shown).
FA-mediated differential gene expression profile in mouse colorectal carcinogenesis model induced by DMH
With the quality control step, all twelve colonic tissues were analyzed as described in the Methods section. The microarray analysis was conducted in the NS group (3 samples), DMH group (3 samples), FA2 group (3 samples) and FA3 group (3 samples). Then we compared the gene expression levels between the samples of group NS and DMH, FA3 and DMH, FA2 and FA3.
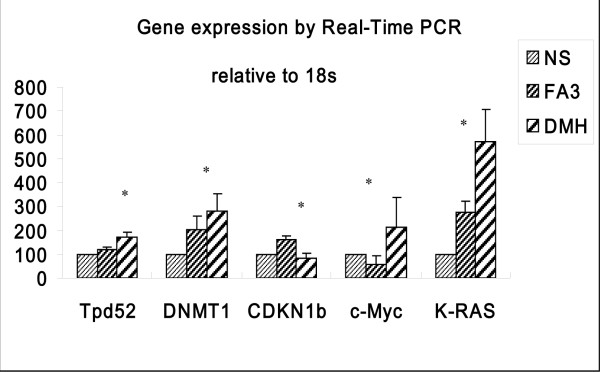
A homogenous expression profile among the samples of each group was shown after the hierarchical clustering analysis. And when the Fold Change (FC) is set > 1.5 and the p value at ≤ 0.05, we found that the expression of 12395 genes was significantly altered in the DMH group compared to those in the NS group (see additional file 1). Together with the result of FA3 vs DMH (see additional file 2), we found that 642 genes down-regulated and 428 genes up-regulated in FA3 group compared to DMH, which may indicate that folic acid can reverse the gene expression that changed by DMH (see additional file 3). Most of these genes are metabolic-related enzymes and regulators which may perform cellular binding and enzymatic activity, involved in the biological regulation and developmental process. Other genes which are differentially expressed are closely to carcinogenesis such as cell cycle, cell invasion and apoptosis. In table 1, the most changed genes comparing FA3 group and DMH group are listed, among which are some oncogenes, for example, Oil (oncoprotein induced transcript 1), Tnfrsf11b (tumor necrosis factor receptor superfamily, member 11b), Hmgn5 (high-mobility group nucleosome binding domain 5) are down-regulated while tumor suppressors such as Hnf4a (hepatic nuclear factor 4, alpha), Cdhr2 (cadherin-related family member 2), Muc2 (mucin 2) are up-regulated. From the results of the microarray analysis, we selected 5 genes i.e., K-ras, c-MYC, DNMT1, Tpd52, CDKN1b for PCR confirmation because they are already considered as tumor-related genes. The primers for these genes are shown in Table 2.
Table 1.
List of the most differentially expressed genes whose changes due to DMH treatment could be reversed by folic acid
| Accession number | Gene symbol | Gene Description | Fold change | P value |
|---|---|---|---|---|
| Downregulated genes | ||||
| NM_207634 | Rps24 | ribosomal protein S24 (Rps24), transcript variant 2 | 0.002356454 | 2.05154E-06 |
| NM_012052 | Rps3 | ribosomal protein S3 (Rps3) | 0.00933479 | 6.38113E-06 |
| NM_033073 | Krt7 | keratin 7 | 0.024674534 | 0.001286211 |
| NM_024478 | Grpel1 | GrpE-like 1, mitochondrial (Grpel1) | 0.029123617 | 3.65271E-05 |
| NM_024243 | Fuca1 | fucosidase, alpha-L- 1 | 0.031740456 | 0.000162318 |
| NM_146050 | Oit1 | oncoprotein induced transcript 1 | 0.032247549 | 0.001799574 |
| NM_013614 | Odc1 | ornithine decarboxylase, structural 1 | 0.032361 | 4.48641E-05 |
| NM_025431 | Llph | LLP homolog, long-term synaptic facilitation (Aplysia) | 0.036784284 | 1.18163E-06 |
| NM_008764 | Tnfrsf11b | tumor necrosis factor receptor superfamily, member 11b | 0.041187965 | 7.03729E-05 |
| NM_009402 | Pglyrp1 | peptidoglycan recognition protein 1 | 0.041272749 | 0.009299333 |
| NM_010106 | Eef1a1 | eukaryotic translation elongation factor 1 alpha 1 | 0.041438052 | 7.22246E-06 |
| NM_001008700 | Il4ra | interleukin 4 receptor, alpha | 0.043141894 | 0.000223171 |
| NM_182930 | Plekha6 | pleckstrin homology domain containing, family A member 6 | 0.04544609 | 0.001545018 |
| NM_011463 | Spink4 | serine peptidase inhibitor, Kazal type 4 | 0.045587012 | 0.000688366 |
| NM_016710 | Hmgn5 | high-mobility group nucleosome binding domain 5 | 0.046928235 | 0.000333311 |
| NM_016981 | Slc9a1 | solute carrier family 9 (sodium/hydrogen exchanger), member 1 | 0.052191789 | 5.29847E-05 |
| NM_145533 | Smox | spermine oxidase (Smox), transcript variant 2 | 0.053274908 | 6.23127E-05 |
| NM_008305 | Hspg2 | perlecan (heparan sulfate proteoglycan 2) | 0.056450624 | 0.001205571 |
| NM_172051 | Tmcc3 | transmembrane and coiled coil domains 3 | 0.058793481 | 0.001122075 |
| NM_009768 | Bsg | basigin (Bsg), transcript variant 1 | 0.061259044 | 0.000407939 |
| Upregulted genes | ||||
| NM_009946 | Cplx2 | complexin 2 | 1109.786672 | 0.000155322 |
| NM_001039493 | Plekhm3 | pleckstrin homology domain containing, family M, member 3 | 56.2494337 | 0.000450001 |
| NM_024272 | Ssbp2 | single-stranded DNA binding protein 2 (Ssbp2), transcript variant 2 | 54.215495 | 2.06403E-05 |
| NM_175013 | Pgm5 | phosphoglucomutase 5 | 47.38198278 | 1.84156E-05 |
| NM_008222 | Hccs | holocytochrome c synthetase | 39.34022581 | 0.000130923 |
| NM_001033364 | Cdhr2 | cadherin-related family member 2 | 38.97741927 | 0.000749154 |
| NM_023566 | Muc2 | mucin 2 | 30.63268666 | 0.02159023 |
| NM_010418 | Herc2 | hect domain and RCC1 (CHC1)-like domain (RLD) 2 | 29.34751955 | 0.003432199 |
| NM_008261 | Hnf4a | hepatic nuclear factor 4, alpha | 28.66993377 | 0.000234502 |
| NM_176850 | Bptf | bromodomain PHD finger transcription factor | 26.66298996 | 0.000156324 |
Fold change and P values are the results comparing FA3 group and DMH group.
Table 2.
Primer sequence for real-time pcr
| Gene name | Forward sequence | Reverse sequence | Product |
|---|---|---|---|
| length | |||
| Tpd52 | tctaaagtaggaggagccaagc | gctctctgtcatctgttctgga | 117 |
| DNMT1 | caagaagaaaggcaaggtcaac | cctggatgctctcaagtaggtc | 212 |
| c-Myc | atttctatcaccagcaacagcag | aacataggatggagagcagagc | 137 |
| K-RAS | tggtcctggtagggaataagtg | cccatctttgctcatcttttct | 191 |
| CDKN1b | cttgcccgagttctactacagg | agagtttgcctgagacccaat | 127 |
| Tnfrsf12a | cgaccacacagcgacttct | ccaaaaccaggaccagactaag | 106 |
| VDR | tgaaggagttcatcctcacaga | gataatgtgctgttgctcctca' | 128 |
| 18S rRNA | cggacaggattgacagattgatagc | tgccagagtctcgttcgttatcg | 150 |
However, from the analysis of microarray there are only 172 differentially genes expressed between FA2 group and FA3 group (see additional file 4). Consistent with the animal experiment that FA2 group have increase number and diameter of multiple masses, there are some tumor suppressors down-regulated in FA2 group, such as VDR (vitamin D receptor, FC = 0.30101), CDX2(FC = 0.24596), and oncogenes up-regulated, i.e, FN1 (fibronectin 1, FC = 3.859909), TNFRSF12A (tumor necrosis factor receptor superfamily, member12a, FC = 2.515130), NPM1(nucleophosmin1, FC = 1.557789) that have been functional in the process of cell proliferation, cell adhesion, cell differentiation and apoptosis(see table 3). It is the first study that different genes are identified caused by the time that folic acid is provided either in the pre- or post- carcinoma stage.
Table 3.
Partial list of the differentially expressed genes between FA2 and FA3
| Accession number | Gene symbol | Gene Description | Fold change | P value |
|---|---|---|---|---|
| Upregulated genes | ||||
| NM_009758 | BMPR1A | bone morphogenetic protein receptor, type 1A | 2.044809816 | 0.015778782 |
| NM_008722 | Npm1 | nucleophosmin 1 | 1.557789177 | 0.019815969 |
| NM_022563 | Ddr2 | discoidin domain receptor family, member 2 | 3.237694059 | 0.036468073 |
| NM_026653 | Rpa1 | replication protein A1 | 1.568298305 | 0.049492698 |
| NM_010730 | ANXA1 | annexin A1 | 3.666236872 | 0.034499347 |
| NM_009242 | SPARC | secreted acidic cysteine rich glycoprotein | 2.576417983 | 0.004456278 |
| NM_025866 | Cdca7 | cell division cycle associated 7 | 2.483199204 | 0.032125313 |
| NM_013749 | TNFRSF12A | tumor necrosis factor receptor superfamily, member 12a | 2.515130632 | 0.001750863 |
| NM_026148 | LIMS1 | LIM and senescent cell antigen-like domains 1 | 1.897061785 | 0.022103283 |
| NM_010233 | Fn1 | fibronectin 1 | 3.859908549 | 0.036063689 |
| NM_133918 | EMILIN1 | elastin microfibril interfacer 1 | 2.165900048 | 0.018411074 |
| NM_133721 | ITGA9 | integrin alpha 9 | 2.471522431 | 0.019449109 |
| NM_022563 | DDR2 | discoidin domain receptor family, member 2 | 3.237694059 | 0.036468073 |
| NM_178665 | LPP | LIM domain containing preferred translocation partner in lipoma | 4.202943318 | 0.034835063 |
| NM_026361 | PKP4 | plakophilin 4 | 1.685566251 | 0.028039843 |
| NM_010480 | HSP90AA1 | heat shock protein 90, alpha (cytosolic), class A member 1 | 1.656494408 | 0.029335434 |
| NM_010135 | ENAH | enabled homolog (Drosophila) (Enah), transcript variant 1 | 2.96541359 | 0.030677412 |
| NM_013885 | CLIC4 | chloride intracellular channel 4 | 1.737725253 | 0.044653582 |
| NM_010663 | KRT17 | keratin 17 | 3.435610932 | 0.02165621 |
| NM_001081185 | Flnc | filamin C, gamma | 4.041058771 | 0.02814183 |
| Downregulated genes | ||||
| NM_007673 | Cdx2 | caudal type homeobox 2 | 0.24596643 | 0.030973362 |
| NM_145953 | CTH | cystathionase | 0.31273227 | 0.002366272 |
| NM_008885 | PMP22 | peripheral myelin protein 22 | 0.576303226 | 0.031915491 |
| NM_011146 | Pparg | peroxisome proliferator activated receptor gamma | 0.483425898 | 0.035947091 |
| NM_138942 | Dbh | dopamine beta hydroxylase | 0.411709887 | 0.018408936 |
| NM_020257 | CLEC2I | C-type lectin domain family 2, member i | 0.572216631 | 0.009695318 |
| NM_010708 | LGALS9 | lectin, galactose binding, soluble 9 | 0.610346325 | 0.033584593 |
| NM_011146 | PPARG | peroxisome proliferator activated receptor gamma | 0.483425898 | 0.035947091 |
| NM_009504 | VDR | vitamin D receptor | 0.30101348 | 0.021805069 |
| NM_015789 | DKKL1 | dickkopf-like 1 | 0.628957018 | 0.004386895 |
Fold change and P values are the results comparing FA2 group and FA3 group.
Using the GO and KEGG software, we analyzed our microarray dataset (on the basis of the results shown in additional file 3) to identify whether specific biological pathways or functional gene groups were differentially affected by the supplementary of folic acid (see additional file 5). We found that there are 63 signaling pathways including some tumor-related pathways such as Mismatch repair, focal adhesion, cell cycle and mTOR signaling pathway et al. (see additional file 6). Importantly, there are some key enzymes of metabolism pathways including fatty acid metabolism, oxidative phosphorylation decreased in FA3 group compared with DMH group, which may indicate that the decrease of the ability of the metabolism is unfavorable to tumor growth. And the most enriched pathways are shown in table 4.
Table 4.
The most enrichment pathways related to tumorgegesis by KEGG
| Pathway ID | Pathway name | Selection Count | Count | Enrichment |
|---|---|---|---|---|
| mmu05219 | Bladder cancer - Mus musculus (mouse) | 22 | 44 | 3.709033 |
| mmu05216 | Thyroid cancer - Mus musculus (mouse) | 17 | 31 | 3.597993 |
| mmu03430 | Mismatch repair - Mus musculus (mouse) | 13 | 23 | 3.030142 |
| mmu05211 | Renal cell carcinoma - Mus musculus (mouse) | 30 | 77 | 2.524291 |
| mmu04520 | Adherens junction - Mus musculus (mouse) | 29 | 79 | 2.035831 |
| mmu04912 | GnRH signaling pathway - Mus musculus (mouse) | 36 | 104 | 1.939698 |
| mmu05214 | Glioma - Mus musculus (mouse) | 27 | 74 | 1.892937 |
| mmu04110 | Cell cycle - Mus musculus (mouse) | 46 | 140 | 1.872654 |
| mmu05215 | Prostate cancer - Mus musculus (mouse) | 31 | 94 | 1.446692 |
| mmu04150 | mTOR signaling pathway - Mus musculus (mouse) | 20 | 56 | 1.429803 |
| mmu05200 | Pathways in cancer - Mus musculus (mouse) | 98 | 345 | 1.369825 |
| mmu05221 | Acute myeloid leukemia - Mus musculus (mouse) | 21 | 61 | 1.309804 |
"SelectionCounts" stands for the Count of the DE genes' entities directly associated with the listed PathwayID;
"Count" stands for the count of the chosen background population genes' entities associated with the listed PathwayID;
Discussion
In this analysis with a DMH-induced CRC model, we concluded that the supplementation of folic acid can decrease the risk of CRC and the subgroup of providing folic acid without precancerous lesions was more effective than that with precancerous lesions. Significantly, there was a reduction in the tumor mass diameter and multiplicity in folate supplementation group. Moreover, the study is consistent with many other studies either in rodent models or clinical medical researches. Recently, a study that investigated 2299 incidents and 5655 CRA in Nurses' Health Study and Health Professionals Follow-Up Study showed that folic acid intake 12-16 y before diagnosis was inversely associated with CRC and identified the latency that folic acid should be provided. However, the study didn't analyze the results that folic acid was provided after diagnosis [22]. With the same kind of chemical in a rat model of CRC, folate deficiency was found to enhance the development of neoplasia compared to the diets containing 8 mg/kg folic acid [21] but the study had no related mechanisms. However, some studies observed the opposite results. Le Leu [23] believed that folate deficiency can decrease the development of the intestinal tumors in AOM-induced SD-rat model. To this point, we think that the animal strain, experimental condition, experiment skills, folic acid manufactories, folic acid intervention time et al may contribute to these differences in varies studies. Also, there is a possibility that excessive intake of folic acid could have promoted the growth of pre-neoplastic lesions so that our study support that enteroscope should be conducted for the cases in clinical studies before incorporated.
On the other hand, there are still no significant differences in the incidence of cancers between group FA2 and FA3 even though the maximum diameter and the number of the tumor mass are significantly decreased in FA3 group. It may be due to too small number of mice or too much difference among individuals. In another respect, not all the mice had adenomas in the 12th week as the incidence was only 10% among DMH1 group. So, further study should extend the number of samples to get more objective results.
Next, we use microarray gene expression profile analysis to study the mechanism of folic acid-mediated prevention of colon tumors and the difference in folic acid intervention time. To our knowledge, this is the first investigation to use microarray technology to study the role of folic acid in the prevention of CRC and the difference of folic acid intervention times.
Firstly, when the FC was set to ≥ 1.5, 642 genes that changed with the treatment of DMH could be reversed with folic acid supplementary. We selected 5 known tumor-related genes i.e., K-ras, c-MYC, DNMT1, Tpd52, CDKN1b for PCR confirmation [Figure 3]. It is known that genetic alterations may contribute substantially to the pathogenesis of colon cancer. Point mutation of K-ras (occurring in 40% of sporadic CRCs) is an established predictor of absence of response to epidermal growth factor receptor (EGFR) -targeted agents [24,25]. Hutchins [26] reported that KRAS mutant tumors were more evenly distributed: 40% right colon, 28% left colon, and 36% rectal tumors compared to BRAF mutant tumors. Meanwhile, the relationship between Folic acid and KRAS has been studied. Some suggested that the effect of folate on rectal cancer risk is different to men and women which may depend on the status of K-ras mutation of tumors. They believed that folate intake was related to a decreased risk of G > A transitions (RR-0.08, 95% CI = 0.01-0.53) while an inversely risk of G > T and G > C transversions in tumors (RR = 2.69, 95% CI = 1.43-5.09)[27].
Figure 3.
Differentially expressed genes validated by real-time polymerase chain reaction (q-PCR). We used 18s rRNA as an internal control. Relative mRNA expression was calculated according to the 2-ΔΔT method. Data are expressed as the mean ± SD of 10 samples. The significance of the varieties between the average values of groups DMH and FA3 was analyzed through student's t-t test. (*: P < 0.05, between FA3 and DMH group)
CDKN1b (cyclin-dependent kinase inhibitor 1B, FC = 7.992979) which is also known as p27 encodes a protein which belongs to the Cip/Kip family of cyclin dependent kinase (Cdk) inhibitor proteins [28] It is often considered as a cell cycle inhibitor protein because its major function is to control the cell cycle progression at G1 phase so that can prevent the development of cancer. Reduced p27 levels were found in different cancerous stages in hepatocelluar carcinomas [29]. Some studies demonstrated that loss of p27 expression is associated with a higher response rate to CRC chemo-therapy [30]. The p27KIP1 null (-/-) mouse shows a significant increase in cell proliferation, resulting in approximately 30% increase in mass size, multiple organ hyperplasia [31]. Together, these researches supported p27 as an important tumor suppressor and suggest that events leading to p27 upregulation may inhibit the tumor progression.
The methylation of genomic DNA in malignant cells is catalyzed by DNA methytransferases(DNMT)which include maintenance DNA methyltransferase (Dnmt1), DNMT1, de novo DNA methyltransferases (Dnmt3a and 3b), 3a/3b. DNA methylation is an important form of epigenetic that can regulate some gene expression such as c-Myc, CDKN2a, CDH1 and VDR et al [32-34]. We have seen that the expression of DNMT1 was increased in FA3 compared to DMH, which is consistent with the research that the folate - and methyl-deficient diet alters components of the DNA methylation via both transcriptional and posttranscriptional mechanisms in in livers of F344 rats [35]. Meanwhile, some methylation-related genes that are functional in carcinogenesis can also be regulated by folic acid in terms of DNA methylation [36].
Tumor necrosis factor receptor superfamily, member 12a (Tnfrsf12a), also known as fn14 or TWEAK-R have been implicated in a variety of pathological processes including chronic inflammation and cancers [37]. And fn14 expression is at a relative lower level in normal tissues while much higher in cancer cells or tissues [38]. Kawashima [39] reported that IL-13 may damage the mucosa of colon via the function of TWEAK and Fn14 pathway and Fn14 could aggravate intestinal inflammation in patients with UC. So the relation between fn14 and diseases might suggest fn14 and TWEAK are targets for cancer therapy [37]. In our study, Tnfrsf12a's expression is 2.5 fold changes higher in FA2 group than FA3, which may be explained that the degree of colon mucosal damage in FA2 was much worse and was prone to develop to cancers compared to FA3. In this aspect, the high expression of fn14 may contribute to the growth of masses in FA2 group.
Vitamin D Receptor gene (VDR) is involved in the progress of cancers or chronic diseases [40]. Some argued that the polymorphism of VDR and CDX2 was not associated with increased risk of CRCs [41]. While others suggested that significant associations with VDR polymorphisms was found not in colorectal cancers but much stronger in cancers of breast, prostate and renal cell carcinomas [42]. And the association between VDR polymorphisms and folic acid has not been reported yet. In another respect, VDR is considered to be an epithelial marker in the process of Epithelial to mesenchymal transition (EMT) and thus might have a suppressive function of invasion [43]. Therefore, the expression of many tumor suppressors such as VDR was much lower (FC = 0.3010) compared with group FA2 and FA3, which was opposite to oncogenes.
However, there are some limitations of our study should be mentioned. First, we ignored the usage of the B Vitamins in the animal experiment, which is important in the process of Folic acid' transport and storage in liver. Therefore, Folic acid supplements may sometimes include vitamin B12 supplements with simultaneous administration of vitamin B12 [22]. However, some studies do not think there are any influences exiting with or without vitamin B12 [44]. Others even found that treatment with folic acid plus vitamin B(12) was associated with increased cancer outcomes [45]. Thus, consideration should be given to the potential value of providing with or without vitamin B12 in addition to the current mandatory folic acid supplementation.
Second, since folic acid is important in many processes of metabolism and might help to protect against the cardiovascular, mental diseases, cancer and birth defects [46]. However, we have no indicators to find other adverse effects but to observe the injury of organs in this study. Even though there are no abnormalities discovered in other organs except colon and rectum, the function of folic acid is needed to be further studied in terms of being effective to therapy.
Finally, although some similarities do exist between chemical rodent models of colon cancer and human natural CRCs, several respects of differs may also exist indeed. For example, the dose and duration of folic acid supplementation used in our study may be different from human studies. So, considering the safety of chemoprevention in clinical application, the optimal researches should be established in humans based on these findings with an initial colonoscopy before incorporated.
In summary, for the first time, our data suggest that folic acid supplementary in pre-cancerous era is much more protective than that in post-cancerous stage in a DMH induced mouse model and identify differential genes that folic acid can reversed and that between groups of pre or post-adenoma induced by folic acid using microarray gene expression profile. Not only to the reason that floate supplementation facilitates the progression of (pre)neoplastic lesions though providing nucleotide precursors to the rapidly replicating transformed cells, thus accelerating proliferation [11]. We also clarified that in gene expression profile, certain oncogenes that promote tumor growth, cell cycle, cell invasion such as TNFRSF12A, fibronectin 1, Cdca7 are high expressed in FA2 group compared to FA3 group while tumor suppressors are down-regulated such as VDR, CDX2, which may partly explain the result. However, the mechanism why folic acid provided in different phages can change these genes' expression remains to be studied.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
YL, JW, LR, JH carried out the molecular genetic studies, participated in the sequence alignment. YL, JW, HC, YZ participated in animal experiment. YL, JW, RL, JH, JF conceived of the study and participated in its design and coordination. YL, JW performed in the statistical analysis and drafted the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Table S1. Complete list of differentially expressed genes in the DMH group compared with the Control group. the file contains all different genes identified by micro-array between DMH group and Control group.
Table S2. Complete list of differentially expressed genes in the FA3 group compared with the DMH group. the file contains all different genes identified by micro-array between FA3 group and DMH group.
Table S3. Complete list of genes whose changes due to DMH treatment could be reversed by folic acid. the file contains all genes that could be reserved by folic acid when treated with DMH
Table S4. Complete list of differentially expressed genes in FA2 group and FA3 group. the file contains complete differential genes between FA3 group and FA2 group by the micro-array
Table S5. Complete list of the GO terms based on the genes whose changes due to DMH treatment could be reversed by folic acid. the file contains GO terms based on the differential genes between FA3 group and DMH group by the micro-array
Table S6. Complete list of pathways based on the genes whose changes due to DMH treatment could be reversed by folic acid. the file contains complete pathways that could be affected by folic acid when treated with DMH
Contributor Information
Yan-Wei Lin, Email: yanweilin2008@yahoo.cn.
Ji-Lin Wang, Email: wangjilin811123@163.com.
Hui-Min Chen, Email: huimin.chen@yahoo.com.cn.
Yan-Jie Zhang, Email: zhangyanjie2007@126.com.
Rong-Lu, Email: rong_lu@urmc.rochester.edu.
Lin-Lin Ren, Email: successrll@163.com.
Jie Hong, Email: jiehong97@gmail.com.
Jing-Yuan Fang, Email: jingyuanfang2007@126.com.
Acknowledgements
We thank Chen X, Peng Y, Cui Y, Gu W and Zhu H, who made a significant contribution to the performance and successful completion of the study. We also thank KangChen Bio-tech Inc (Shanghai, China) for the excellent microarray services.
This work was supported by a grant from the grants from the National Science Found of China (30830055) and the Ministry of Public Health, China (No. 200802094).
References
- Centers for Disease Control and Prevention (CDC) Vital signs: Colorectal cancer screening, incidence, and mortality--United States, 2002-2010. MMWR Morb Mortal Wkly Rep. 2011;60:884–9. [PubMed] [Google Scholar]
- Holt K. Common side effects and interactions of colorectal cancer therapeutic agents. J Pract Nurs. 2011;61:7–20. [PubMed] [Google Scholar]
- Kohne CH, Bruce C, Folprecht GA, udisio R. Role of new agents in the treatment of colorectal cancer. Surg Oncol. 2004;13:75–81. doi: 10.1016/j.suronc.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Buchanan DD, Sweet K, Drini M, Jenkins MA, Win AK, English DR, Walsh MD, Clendenning M, McKeone DM, Walters RJ, Roberts A, Pearson SA, Pavluk E, Hopper JL, Gattas MR, Goldblatt J, George J, Suthers GK, Phillips KD, Woodall S, Arnold J, Tucker K, Muir A, Field M, Greening S, Gallinger S, Perrier R, Baron JA, Potter JD, Haile R, Frankel W, de la Chapelle A, Macrae F, Rosty C, Walker NI, Parry S, Young JP. Risk factors for colorectal cancer in patients with multiple serrated polyps: a cross-sectional case series from genetics clinics. PLoS One. 2010;5:e11636. doi: 10.1371/journal.pone.0011636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Femia AP, Luceri C, Toti S, Giannini A, Dolara P, Caderni G. Gene expression profile and genomic alterations in colonic tumours induced by 1,2-dimethylhydrazine (DMH) in rats. BMC Cancer. 2010;10:194. doi: 10.1186/1471-2407-10-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perse M, Cerar A. Morphological and molecular alterations in 1,2 dimethylhydrazine and azoxymethane induced colon carcinogenesis in rats. J Biomed Biotechnol. 2011;2011:473964. doi: 10.1155/2011/473964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery ML, Wolff RK, Herrick JS, Curtin K, Caan BJ, Samowitz W. Alcohol consumption and rectal tumor mutations and epigenetic changes. Dis Colon Rectum. 2010;53:1182–9. doi: 10.1007/DCR.0b013e3181d325db. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Femia AP, Paulsen JE, Dolara P, Alexander J, Caderni G. Correspondence between flat aberrant crypt foci and mucin-depleted foci in rodent colon carcinogenesis. Anticancer Res. 2008;28:3771–5. [PubMed] [Google Scholar]
- Lu R, Wang X, Sun DF, Tian XQ, Zhao SL, Chen YX, Fang JY. Folic acid and sodium butyrate prevent tumorigenesis in a mouse model of colorectal cancer. Epigenetics. 2008;3:330–5. doi: 10.4161/epi.3.6.7125. [DOI] [PubMed] [Google Scholar]
- Choi SW, Mason JB. Folate status: Effects on pathwaysof colorectal carcinogenesis. J Nutr. 2001;132:2413S–2418S. doi: 10.1093/jn/132.8.2413S. [DOI] [PubMed] [Google Scholar]
- Kim YI. Folate and colorectal cancer: an evidence-based critical review. Mol Nutr Food Res. 2007;51:267–92. doi: 10.1002/mnfr.200600191. [DOI] [PubMed] [Google Scholar]
- Stevens VL, McCullough ML, Sun J, Jacobs EJ, Campbell PT, Gapstur SM. High Levels of Folate From Supplements and Fortification Are Not Associated With Increased Risk of Colorectal Cancer. Gastroenterology. 2011;141:98–105. doi: 10.1053/j.gastro.2011.04.004. [DOI] [PubMed] [Google Scholar]
- Cole BF, Baron JA, Sandler RS, Haile RW, Ahnen DJ, Bresalier RS, McKeown-Eyssen G, Summers RW, Rothstein RI, Burke CA, Snover DC, Church TR, Allen JI, Robertson DJ, Beck GJ, Bond JH, Byers T, Mandel JS, Mott LA, Pearson LH, Barry EL, Rees JR, Marcon N, Saibil F, Ueland PM, Greenberg ER. Polyp Prevention Study Group. Folic acid for the prevention of colorectal adenomas: a randomized clinical trial. JAMA. 2007;297:2351–9. doi: 10.1001/jama.297.21.2351. [DOI] [PubMed] [Google Scholar]
- Sie KK, Medline A, van Weel J, Sohn KJ, Choi SW, Croxford R, Kim YI. Effect of maternal and postweaning folic acid supplementation on colorectal cancer risk in the offspring. Gut. 2011;60:1687–94. doi: 10.1136/gut.2011.238782. [DOI] [PubMed] [Google Scholar]
- Lonn E, Yusuf S, Arnold MJ, Sheridan P, Pogue J, Micks M, McQueen MJ, Probstfield J, Fodor G, Held C, Genest J Jr. Heart Outcomes Prevention Evaluation (HOPE) 2 Investigators Homo-cysteine lowering with folic acid and B vitamins in vasculardisease. N Engl J Med. 2006;354:1567–1577. doi: 10.1056/NEJMoa060900. [DOI] [PubMed] [Google Scholar]
- Fife J, Raniga S, Hider PN, Frizelle FA. Folic acid supplementation and colorectal cancer risk: a meta-analysis. Colorectal Dis. 2011;13:132–7. doi: 10.1111/j.1463-1318.2009.02089.x. [DOI] [PubMed] [Google Scholar]
- Carroll C, Cooper K, Papaioannou D, Hind D, Tappenden P, Pilgrim H, Booth A. Meta-analysis: folic acid in the chemoprevention of colorectal adenomas and colorectal cancer. Aliment Pharmacol Ther. 2010;31:708. doi: 10.1111/j.1365-2036.2010.04238.x. [DOI] [PubMed] [Google Scholar]
- Kim YI. Folic acid supplementation and cancer risk: point. CancerEpidemiol Biomarkers Prev. 2008;17:2220–2225. doi: 10.1158/1055-9965.EPI-07-2557. [DOI] [PubMed] [Google Scholar]
- Bird RP. Role of aberrant crypt foci in understanding the pathogenesis of colon cancer. Cancer Lett. 1995;93:55–71. doi: 10.1016/0304-3835(95)03788-X. [DOI] [PubMed] [Google Scholar]
- Pretlow TP, O'Riordan MA, Pretlow TG, Stellato TA. Aberrant crypts in human colonic mucosa: putative preneoplastic lesions. J Cell Biochem Suppl. 1992;16G:55–62. doi: 10.1002/jcb.240501111. [DOI] [PubMed] [Google Scholar]
- Lindzon GM, Medline A, Sohn KJ, Depeint F, Croxford R, Kim YI. Effect of folic acid supplementation on the progression of colorectal aberrant crypt foci. Carcinogenesis. 2009;30:1536–43. doi: 10.1093/carcin/bgp152. [DOI] [PubMed] [Google Scholar]
- Lee JE, Willett WC, Fuchs CS, Smith-Warner SA, Wu K, Ma J, Giovannucci E. Folate intake and risk of colorectal cancer and adenoma: modification by time. Am J Clin Nutr. 2011;93:817–25. doi: 10.3945/ajcn.110.007781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Leu RK, Young GP, McIntosh GH. Folate deficiency reduces the development of colorectal cancer in rats. Carcinogenesis. 2002;21:2261–5. doi: 10.1093/carcin/21.12.2261. [DOI] [PubMed] [Google Scholar]
- Dempke WC, Heinemann V. Kas mutational status is a biomarker for resistance to EGFR inhibitors in colorectal carcinoma. Anticancer Res. 2010;30:4673–7. [PubMed] [Google Scholar]
- Heinemann V, Stintzing S, Kirchner T, Boeck S, Jung A. Clinical relevance of EGFR- and KRAS-status in colorectal cancer patients treated with monoclonal antibodies directed against the EGFR. Cancer Treat Rev. 2009;35:262–271. doi: 10.1016/j.ctrv.2008.11.005. [DOI] [PubMed] [Google Scholar]
- Hutchins G, Southward K, Handley K, Magill L, Beaumont C, Stahlschmidt J, Richman S, Chambers P, Seymour M, Kerr D, Gray R, Quirke P. Value of mismatch repair, KRAS, and BRAF mutations in predicting recurrence and benefits from chemotherapy in colorectal cancer. J Clin Oncol. 2011;29:1261–70. doi: 10.1200/JCO.2010.30.1366. [DOI] [PubMed] [Google Scholar]
- Brink M, Weijenberg MP, de Goeij AF, Roemen GM, Lentjes MH, de Bruïne AP, van Engeland M, Goldbohm RA, van den Brandt PA. Dietary folate intake and k-ras mutations in sporadic colon and rectal cancer in The Netherlands Cohort Study. Int J Cancer. 2005;114:824–30. doi: 10.1002/ijc.20775. [DOI] [PubMed] [Google Scholar]
- Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–12. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- Tannapfel A, Grund D, Katalinic A, Uhlmann D, Köckerling F, Haugwitz U, Wasner M, Hauss J, Engeland K. Wittekind CDecreased expression of p27 protein is associated with advanced tumor stage in hepatocellular carcinoma. Int J Cancer. 2000;89:350–5. doi: 10.1002/1097-0215(20000720)89:4<350::AID-IJC6>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Ogino S, Meyerhardt JA, Cantor M, Brahmandam M, Clark JW, Namgyal C, Kawasaki T, Kinsella K, Michelini AL, Enzinger PC, Kulke MH, Ryan DP, Loda M, Fuchs CS. Molecular alterations in tumors and response to combination chemotherapy with gefitinib for advanced colorectal cancer. Clin Cancer Res. 2005;11:6650–6. doi: 10.1158/1078-0432.CCR-05-0738. [DOI] [PubMed] [Google Scholar]
- Hikosaka A, Ogawa K, Sugiura S, Asamoto M, Takeshita F, Sato SY, Nakanishi M, Kohri K, Shirai T. Susceptibility of p27 kip1 knockout mice to urinary bladder carcinogenesis induced by N-butyl-N-(4-hydroxybutyl)nitrosamine may not simply be due to enhanced proliferation. Int J Cancer. 2008;122:1222–8. doi: 10.1002/ijc.23249. [DOI] [PubMed] [Google Scholar]
- Du YP, Peng JS, Sun A, Tang ZH, Ling WH, Zhu HL. Assessment of the effect of betaine on p16 and c-myc DNA methylation and mRNA expression in a chemical induced rat liver cancer model. BMC Cancer. 2009;9:261. doi: 10.1186/1471-2407-9-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler JM, Kim HC, Efstathiou JA, Ilyas M, Mortensen NJ, Bodmer WF. Hypermethylation of the promoter region of the E-cadherin gene (CDH1) in sporadic and ulcerative colitis associated colorectal cancer. Gut. 2001;48:367–71. doi: 10.1136/gut.48.3.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnoff P, Liel Y, Gnainsky J, Shany S, Schwartz B. The protective effect of estrogen against chemically induced murine colon carcinogenesis is associated with decreased CpG island methylation and increased mRNA and protein expression of the colonic vitamin D receptor. Oncol Res. 1999;11:255–64. [PubMed] [Google Scholar]
- Ghoshal K, Li X, Datta J, Bai S, Pogribny I, Pogribny M, Huang Y, Young D, Jacob STA. folate- and methyl-deficient diet alters the expression of DNA methyltransferases and methyl CpG binding proteins involved in epigenetic gene silencing in livers of F344 rats. J Nutr. 2006;136:1522–7. doi: 10.1093/jn/136.6.1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cravo ML, Mason JB, Dayal Y, Hutchinson M, Smith D, Selhub J, Rosenberg IH. Folate deficiency enhances the development of colonic neoplasia in dimethylhydrazine-treated rats. Cancer Res. 1992;52:5002–6. [PubMed] [Google Scholar]
- Winkles JA, Tran NL, Berens ME. TWEAK and Fn14: new molecular targets for cancer therapy? Cancer Lett. 2006;235:11–7. doi: 10.1016/j.canlet.2005.03.048. [DOI] [PubMed] [Google Scholar]
- Feng SL, Guo Y, Factor VM, Thorgeirsson SS, Bell DW, Testa JR, Peifley KA, Winkles JA. The Fn14 immediate-early response gene is induced during liver regeneration and highly expressed in both human and murine hepatocellular carcinomas. Am J Pathol. 2000;156:1253–61. doi: 10.1016/S0002-9440(10)64996-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima R, Kawamura YI, Oshio T, Son A, Yamazaki M, Hagiwara T, Okada T, Inagaki-Ohara K, Wu P, Szak S, Kawamura YJ, Konishi F, Miyake O, Yano H, Saito Y, Burkly LC, Dohi T. Interleukin-13 Damages Intestinal Mucosa via TWEAK and Fn14 in Mice-a Pathway Associated with Ulcerative Colitis. Gastroenterology. 2011;141:2119–2129. doi: 10.1053/j.gastro.2011.08.040. e8. [DOI] [PubMed] [Google Scholar]
- Norman AW. Minireview: vitamin D receptor: new assignments for an already busy receptor. Endocrinology. 2006;147:5542–8. doi: 10.1210/en.2006-0946. [DOI] [PubMed] [Google Scholar]
- Slattery ML, Herrick J, Wolff RK, Caan BJ, Potter JD, Sweeney C. CDX2 VDR polymorphism and colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2007;16:2752–5. doi: 10.1158/1055-9965.EPI-07-2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köstner K, Denzer N, Müller CS, Klein R, Tilgen W, Reichrath J. The relevance of vitamin D receptor (VDR) gene polymorphisms for cancer: a review of the literature. Anticancer Res. 2009;29:3511–36. [PubMed] [Google Scholar]
- Peña C, García JM, Larriba MJ, Barderas R, Gómez I, Herrera M, García V, Silva J, Domínguez G, Rodríguez R, Cuevas J, de Herreros AG, Casal JI, Muñoz A, Bonilla F. SNAI1 expression in colon cancer related with CDH1 and VDR downregulation in normal adjacent tissue EMT genes in normal tissue adjacent to tumor. Oncogene. 2009;28:4375–4385. doi: 10.1038/onc.2009.285. [DOI] [PubMed] [Google Scholar]
- Malouf R, Grimley Evans J. Folic acid with or without vitamin B12 for the prevention and treatment of healthy elderly and demented people. Cochrane Database Syst Rev. 2008;8:CD004514. doi: 10.1002/14651858.CD004514.pub2. [DOI] [PubMed] [Google Scholar]
- Ebbing M, Bønaa KH, Nygård O, Arnesen E, Ueland PM, Nordrehaug JE, Rasmussen K, Njølstad I, Refsum H, Nilsen DW, Tverdal A, Meyer K, Vollset SE. Cancer incidence and mortality after treatment with folic acid and vitamin B12. JAMA. 2009;302:2119–26. doi: 10.1001/jama.2009.1622. [DOI] [PubMed] [Google Scholar]
- Hoey L, McNulty H, Askin N, Dunne A, Ward M, Pentieva K, Strain J, Molloy AM, Flynn CA, Scott JM. Effect of a voluntary food fortification policy on folate, related B vitamin status, and homocysteine in healthy adults. Am J Clin Nutr. 2007;86:1405–13. doi: 10.1093/ajcn/86.5.1405. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Complete list of differentially expressed genes in the DMH group compared with the Control group. the file contains all different genes identified by micro-array between DMH group and Control group.
Table S2. Complete list of differentially expressed genes in the FA3 group compared with the DMH group. the file contains all different genes identified by micro-array between FA3 group and DMH group.
Table S3. Complete list of genes whose changes due to DMH treatment could be reversed by folic acid. the file contains all genes that could be reserved by folic acid when treated with DMH
Table S4. Complete list of differentially expressed genes in FA2 group and FA3 group. the file contains complete differential genes between FA3 group and FA2 group by the micro-array
Table S5. Complete list of the GO terms based on the genes whose changes due to DMH treatment could be reversed by folic acid. the file contains GO terms based on the differential genes between FA3 group and DMH group by the micro-array
Table S6. Complete list of pathways based on the genes whose changes due to DMH treatment could be reversed by folic acid. the file contains complete pathways that could be affected by folic acid when treated with DMH