Abstract
G protein-coupled receptors (GPCRs) comprise the most “prolific” family of cell membrane proteins, which share a common mechanism of signal transduction, but greatly vary in ligand recognition and function. Crystal structures are now available for rhodopsin, adrenergic, and adenosine receptors in both inactive and activated forms, as well as for chemokine, dopamine, and histamine receptors in inactive conformations. Here we review common structural features, outline the scope of structural diversity of GPCRs at different levels of homology and briefly discuss impact of the structures on drug discovery. Given the current set of GPCR crystal structures, a distinct modularity is now being observed between the extracellular (ligand-binding) and intracellular (signaling) regions. The rapidly expanding repertoire of GPCR structures provides a solid framework for experimental and molecular modeling studies, and helps to chart a roadmap for comprehensive structural coverage of the whole superfamily and an understanding of GPCR biological and therapeutic mechanisms.
Keywords: adrenergic, adenosine, chemokine, dopamine, histamine, rhodopsin, G protein-coupled receptor
Breakthroughs in GPCR crystallography
The G protein-coupled receptor (GPCR) superfamily comprises more than 800 distinct human proteins that share a common seven transmembrane α-helical (7TM) fold. Tracing their origins to the first eukaryotes [1], GPCRs have diverged in vertebrates into five major classes and numerous subfamilies (Figure 1) [2, 3], and continue to play a key dynamic role in mammalian evolution [4]. As highly versatile membrane sensors, GPCRs respond to a great variety of extracellular signals, ranging from photons, ions and sensory stimuli, to lipids, neurotransmitters and hormones [5], converting them into cellular responses via G-proteins, β-arrestins and other downstream effectors [6]. Signaling through GPCRs controls major biological and pathological processes in neural, cardiovascular, immune and endocrine systems, as well as in cancer [7], making GPCRs the most prominent therapeutic target family that mediates action of more than 40% of clinically approved drugs[8].
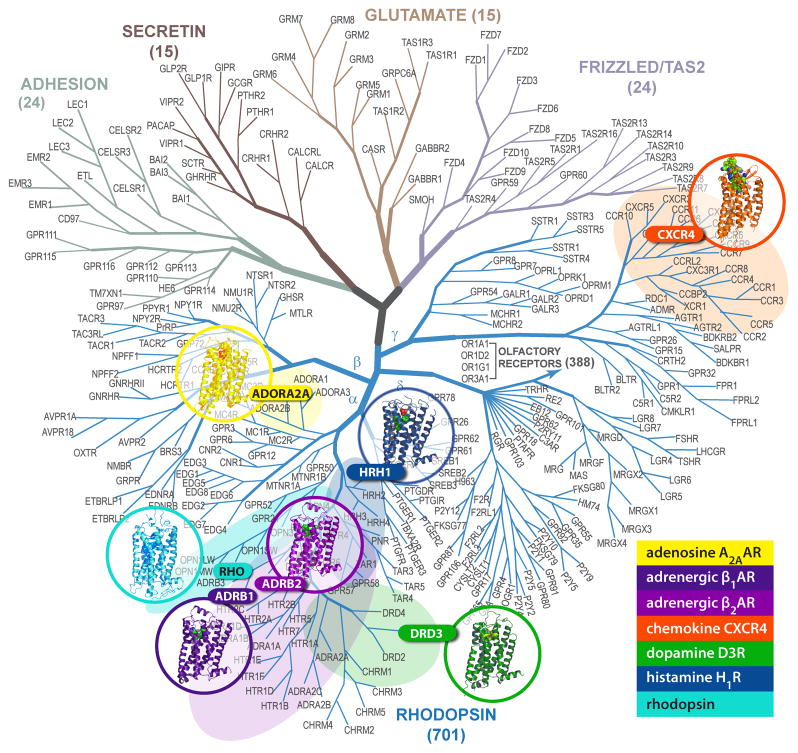
Fig. 1.
Current status of GPCR family structural coverage. Published high-resolution structures of GPCRs are shown on the sequence homology tree (modified from [2]). Highlighted areas show close homologs of the crystal structures with better than 35% sequence identity in the TM helices, which are likely to be amenable for accurate comparative modeling.
Due to challenges in expression and crystallization, the GPCR family has remained the largest “terra incognita” of structural biology for decades, which hampered molecular interpretation of biophysical and biochemical findings, and rational drug discovery applications. The bovine rhodopsin structure was first solved in 2000 [9], 15 years after the first membrane protein crystal structure [10], and another seven years of extensive research and technology developments were needed to obtain the high-resolution structure of the human β2-adrenergic receptor (β2AR) – the first example of a GPCR with a diffusible ligand [11, 12] [13]. That structure was followed by other Class A (rhodopsin-like) GPCRs, including β1AR [14], A2A adenosine (A2AAR) [15], chemokine CXCR4 (CXCR4) [16], dopamine D3 (D3R) [17], and most recently histamine H1 (H1R) [18] receptors (see Table I). While corroborating the common 7TM GPCR fold, the structures provide the first insights into the scope of structural diversity in GPCRs at various levels of homology. Thus, the structures represent closely related GPCR subtypes (β1AR and β2AR with 65% sequence identity), different subfamilies within the aminergic family (β2AR, D3R and H1R with ~35% identity), different sub-branches within the same major α-branch (β2AR and A2AAR with ~30% identity), as well as different major α- and γ-branches of Class A GPCRs (β2AR and CXCR4 with 25% identity) (Figure 1). In this review, we highlight both common and diverse structural features of GPCRs and discuss how these crystallographic insights and initial comparative modeling efforts help to outline a path to comprehensive coverage of the structure and biology of the GPCR family. We will focus on inactive conformations of GPCRs in complex with antagonists and inverse agonists, while the impact of structure-function efforts on understanding GPCR activation mechanisms is discussed in several recent and upcoming reviews (e.g. see [19]).
Table I.
Crystal structures of GPCRs.
| Receptor | Engineereda | Ligand Typeb | Ligand | Year | Ref | PDB ID (resolution, Å) |
|---|---|---|---|---|---|---|
| A2A Adenosine A2AAR (HUMAN) | ICL3 fusion | IAG | ZM241385 | 2008 | [15] | 3EML (2.6) |
| AGO* | UK-432097 | 2011 | [47] | 3QAK (2.71) | ||
| Point mutations | AGO* | Adenosine | 2011 | [29] | 2YDO (3.0) | |
| NECA | 2YDV (2.6) | |||||
| ANT | ZM241385 | 2011 | [77] | 3PWH (3.30) | ||
| caffeine | 2011 | 3REY (3.31) | ||||
| IAG | ZM241385 | 2011 | 3RFM (3.60) | |||
| β1 Adrenergic β1AR (TURKEY) | Point mutations, partial ICL3 deletion | IAG | Cyanopindolol | 2008 | [14] | 2VT4 (2.7) |
| PAG | Dobutamine | 2011 | [78] | 2Y00 (2.5), 2Y01 (2.6) | ||
| AGO | Carmoterol | 2Y02 (2.6) | ||||
| AGO | Isoprenaline | 2Y03 (2.85) | ||||
| PAG | Salbutamol | 2Y04 (3.05) | ||||
| IAG | Carazolol | 2011 | [61] | 2YCW (3.0) | ||
| Cyanopindolol | 2YCX (3.25), 2YCY (3.15), 2YCZ (3.65) | |||||
| β2 Adrenergic β2AR (HUMAN) | ICL3 fusion | IAG | Not Resolved + Fabd | 2007 | [13] | 2R4R (3.4), 2R4S (3.4) |
| IAG | Carazolol | 2007 | [11, 12] | 2RH1 (2.4) | ||
| IAG | Timolol | 2008 | [23] | 3D4S (2.8) | ||
| IAG | ICI118551 | 2010 | [24] | 3NY8 (2.84) | ||
| IAG | Compd #1[44] | 3NY9 (2.84) | ||||
| ANT | Alprenolol | 3NYA (3.16) | ||||
| IAG | Not Resolved + Fabd | 2010 | [79] | 3KJ6 (3.4) | ||
| AGO* | BI-167107 +Nanobodye | 2011 | [26] | 3P0G (3.5) | ||
| AGO | FAUC50c | 2011 | [80] | 3PDS (3.5) | ||
| N-term fusion | AGO* | BI-167107 + Gαβγ + nanobody | 2011 | [25] | 3SN6 (3.2) | |
| Chemokine CXCR4 (HUMAN) | ICL3 fusion | ANT | IT1t | 2010 | [16] | 3ODU (2.5), 3OE6 (3.2), 3OE8 (3.1), 3OE9 (3.1) |
| ANT | CVX15 peptide | 3OE0 (2.9) | ||||
| Dopamine D3R (HUMAN) | ICL3 fusion | ANT | Eticlopride | 2010 | [17] | 3PBL (2.89) |
| Histamine H1R (HUMAN) | ICL3 fusion | IAG | Doxepin | 2011 | [18] | 3RZE (3.1) |
| Rhodopsin (BOVINE) | IAG | 11-cis retinalc | 2000 2004 |
[9]f [81] |
1F88 (2.8) 1U19 (2.2) 1GZM (2.65), L9H (2.6), 1HZX (2.8), 2I35(3.8), 2I36(4.), 2I37 (4.), 2J4Y(3.4), 3C9L(2.65), 3C9M (3.4), 3OAX (2.6) |
|
| 9-cis retinalc | 2007 | [82] | 2PED (2.95) | |||
| AGO | All-trans retinalc | 2006 | [83] | 2G87 (2.6), 2HPY (2.8) | ||
| E113Q | AGO* | All-trans retinalc +GαPeptidee | 2011 | [48] | 2X72 (3.0) | |
| 2011 | [49] | 3PQR (2.85), 3PXO (3.0) | ||||
| Opsin (BOVINE) | APO* | 2008 | [50] | 3CAP (2.9) | ||
| APO* | GαPeptidee | 2008 | [51] | 3DQB (3.2) | ||
| Rhodopsin (SQUID) | IAG | 11-cis retinalc | 2008 | [84] | 2ZIY (3.7) | |
| 2008 | [85] | 2Z73 (2.5) |
All non-rhodopsin GPCRs have truncated/modified N and C-termini
Co-crystallized ligand types: IAG inverse agonist, , ANT-antagonist, AGO – agonist, PAG – partial agonist, APO – no ligand.
Covalently bound ligand
Monoclonal antibody assisted crystallization
G-protein mimic, bound on the intracellular side
For the dark state bRho, only the references for the first and the highest resolution structures are shown.
Activated form of receptor
Structural and functional modularity of GPCRs
GPCRs are comprised of a bundle of seven transmembrane (7TM) α-helices, connected by three extracellular loops (ECL1-3) and three intracellular loops (ICL1-3) (Figure 2). The extracellular (EC) part, responsible for ligand binding, also includes an N-terminus, which can range from relatively short, often unstructured sequences in some of the rhodopsin-like and bitter taste receptors to large globular EC domains in other GPCR classes [5]. The intracellular (IC) part interacts with G proteins, arrestins and other downstream effectors, and, in addition to ICLs, usually includes helix VIII and a C-terminus sequence that may carry palmitoylation and other signal sites [20, 21].
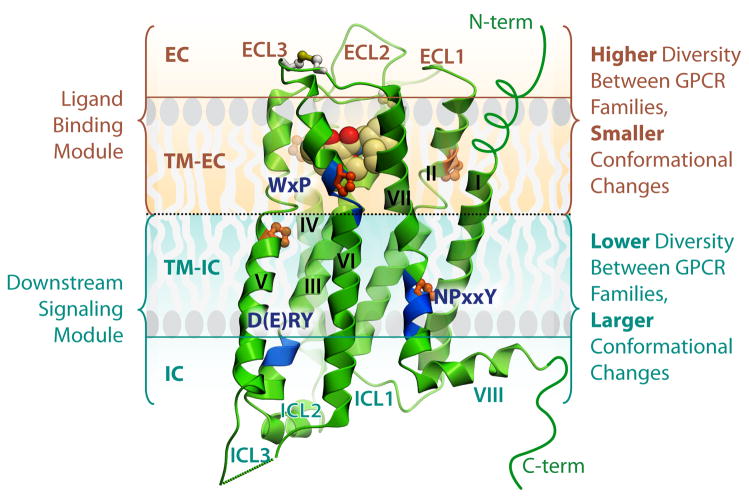
Fig. 2.
General architecture and modularity of GPCRs. Major regions and structural features of GPCRs are shown on example of the D3R crystal structure (PDB ID 3PBL). The EC region includes three ECLs and the N-terminus, which may be represented by a short unstructured peptide (as in most class A GPCRs), or a longer globular-like domain. The 7TM helical bundle contains a number of proline-dependent kinks (prolines shown in orange), that approximately divide the receptor into two modules. The EC module (EC and TM-EC regions) is responsible for binding diverse ligands and has much higher structural diversity. In contrast, the IC module (IC and IC-TM regions), involved in binding downstream effectors including G proteins and arrestins, is more conserved between GPCRs, but undergoes larger conformational changes upon receptor activation. Blue ribbon patches highlight highly conserved, functionally relevant motifs in the TM helices of Class A GPCRs. The C-terminus in most GPCRs is comprised of helix VIII, and in many receptors also has a palmitoylation site anchoring helix VIII to the membrane (not shown).
The 7TM helical bundle has been long recognized as the most conserved component of GPCRs. It shows characteristic hydrophobic patterns and harbors several functionally important signature motifs, such as the D[E]RY motif in helix III (part of the so-called “ionic lock”), the WxP motif in helix VI and the NPxxY motif in helix VII [22]. Although the available crystal structures of rhodopsin-like GPCRs (see Table 1) confirm the overall structural conservation of the 7TM fold, they also reveal a remarkable structural diversity, not only in the loop regions, but also in the helical bundle itself. These variations are especially pronounced on the EC side of receptors, reflecting a distinct evolutionary and functional modularity between EC (ligand-binding) and IC (downstream signaling) modules of GPCRs (Figure 2).
Conformational stability and structural diversity of the extracellular loops
One of the most striking features in the EC region revealed by the high-resolution GPCR structures involves the highly ordered conformations of their ECLs, stabilized by various secondary structure elements, disulfide bonds and interactions with the 7TM bundle. Thus, several structures of β2AR [11, 23, 24] reveal virtually the same conformation in each of their ECLs, despite being crystallized by different approaches, in different crystal packing orientations, and with different antagonists and inverse agonists bound; even in the activated agonist-bound structures[25, 26], the changes are rather limited. The ECL conformations are also very similar between two molecules within the asymmetric unit of the D3R crystal structure [17], between different complexes and different crystal forms of the CXCR4 structures [16], as well as between the ECL backbone structures of very closely related β2AR and β1AR subtypes that share 65% sequence identity. Importantly, the high conformational stability of ECLs in β2AR was recently validated by hydrogen-deuterium exchange (HDX) data, where the measured deuterium exchange ratios were consistent with the crystal structures [27, 28]. The only regions of potential high instability in ECLs that have been identified so far are in small distal portions of ECL2 in A2AAR and H1R, which were not resolved in the corresponding crystal structures [15, 18]. Although a more recent structure of a highly thermostabilized A2AAR observed about a 2-turn α-helix in this region[29], its stability and physiological relevance need further evaluation, as this short α-helix in ECL2 is sandwiched between two α-helices from C-termini of other crystallographic units in the particular crystal form.
One common structural feature of ECL2 is a conserved disulfide bridge connecting the loop with the tip of helix III. This bridge divides ECL2 into two regions –ECL2a and ECL2b, the latter serving as a covalent “linker” between helices III and V. The distinct structural elements especially in the ECLa region show remarkable diversity between distinct GPCR subfamilies (Figure 3). Unlike in rhodopsin structures, in which the β-hairpin of ECL2 “seals” the retinal binding pocket, the ECL2s in other GPCR structures keep the pocket open and readily accessible for ligands. The structural diversity in ECL2 is evident even between related GPCRs within the aminergic subfamily; for example, the ECLa in β2AR contains 2.5-turns of an α-helix, whereas the shorter ECL2a in D3R and H1R lack any secondary structure. In contrast, A2AAR contains both a 1-turn α-helix in ECLb and a short β-strand in ECL2a interacting with a β-strand in ECL1. Finally, in the more distant (by sequence homology) CXCR4, ECL2 forms a β-hairpin, which is positioned very differently than the β-hairpin in rhodopsin, and is critical for CXCR4 binding of either the small molecule IT1t, or the peptide antagonist CVX15 that mimics the V3 loop of the HIV spike protein gp120.
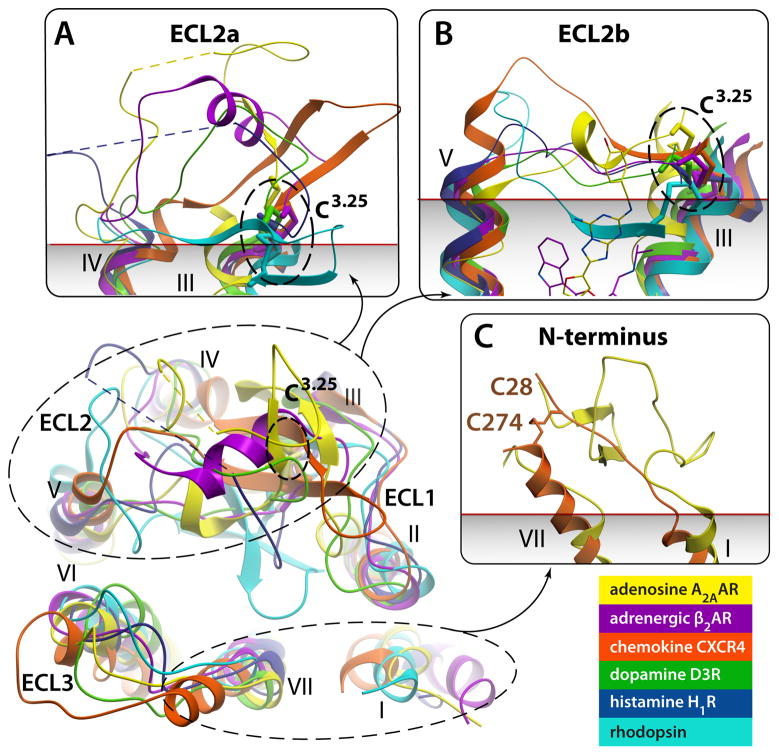
Fig. 3.
Remarkable diversity in the EC region. Top view of ECLs from superimposed GPCR structures with close-up side views in panels (A)–(C). Note different secondary structure elements (or lack thereof) in ECL2, as well as large deviations in positions of extracellular tips of the TM helices. N-termini of Rhodopsin and CXCR4 are removed for clarity. (A) Close up of ECL2a, a part of ECL2 from helix IV to the conserved disulfide bond connecting ECL2 to helix III. The ECL2a is highly variable in length and secondary structure elements, showing distinct structure between structures of all GPCRs solved; the only exception being the practically identical ECL2a conformations ((RMSD(Cα)=0.5 A)) between very closely related β2AR (PDB ID 2RH1) and β1AR (PDB ID 2VT4, not shown). Short unresolved portions of ECL2a in A2AAR and H1R are shown by dashed lines. (B) Close up side view of ECL2b – the linker part connecting helices V and III through the disulfide bond to Cys3.25. Note that in some GPCRs the ECL2b element is as short as 4–5 amino acids and fully extended, while longer sequence in others can comprise small secondary structure elements. Relative positions of Carazolol and ZM241385 interacting with ECL2b residues are shown by thin lines. (C) N-terminus is fully resolved in Rhodopsin (PDB ID 1GZM; residues 1:34) and partially in CXCR4 (PDB ID 3ODU; residues 27:34) structures; in other known structures N-termini are likely disordered. Approximate membrane boundary is shown in panels (A)–(C), as predicted by OPM database [86].
The ECL2b linker is tightly integrated with the 7TM bundle and, in many GPCRs, participates in binding ligands. Crystal structures show important consequences of the ECL2b length variations even in closely related GPCRs. Thus, although in β2AR, ECL2b has five residues and an almost fully extended backbone conformation, in A2AAR, it is seven residues long and comprises a 1-turn α-helix that accommodates the extra residues. By contrast, in D3R and other D2-like dopamine receptors, ECL2b has only four residues, which apparently pull the EC tips of helices III and V about 3.5 Å closer together than in β2AR [17]. Intriguingly, the D1 and D5 dopamine receptors are more homologous to β2AR than to D2-like dopamine receptors, and, like β2AR also have five residues in their ECL2b, suggesting that variations in the ECL2b between dopamine receptor subtypes can play a role in their different response profile to dopamine [30].
Although the structural diversity is much less pronounced in ECL1s and ECL3s, which are 5–6 and 6–8 amino acids long, respectively, in most solved GPCR structures, they still show several distinct features. For example, in A2AAR, ECL1 is dramatically shifted inward as compared to other known GPCR structures and forms a unique short β-strand with ECL2, shaping the entrance to the ligand binding pocket. In A2AAR, D3R and H1R, ECL3 is further stabilized by a disulfide bond, and in A2AAR and β2AR, the side chains of ECL3 also form salt bridges to ECL2, which apparently impact ligand binding. In CXCR4, the distinct shape of ECL3 is defined by helix VII, which is elongated by an additional 2.5 turns, and a disulfide bond that bridges ECL3 with the N-terminus; this disulfide crosslink is important for shaping the site of interaction with peptide antagonists, and potentially with its native SDF-1 ligand.
Apparently, this is only the tip of the GPCR family-wide repertoire of structural arrangements, secondary structure elements, and disulfide bonding patterns in the ECL region. Note, for example, that even the ECL2-helix III disulfide bond – the only common feature observed in the ECL2 of all GPCR crystal structures published to date – is not conserved in many Class A receptors that lack a cysteine in position 3.25 (i.e. S1P receptor family). Additional structural features can also be expected for the N-terminus (Figure 3C), which so far has only been resolved in rhodopsin and partially in CXCR4. In CXCR4, the extended conformation of residues 28 to 38 of the N-terminus is stabilized by a disulfide bond to ECL3; the rest of this N-terminus is probably unstructured in the absence of its chemokine binding partner [16].
Precise 3D structural knowledge of the EC regions is of great value in GPCR drug discovery, as the N-terminus and/or ECLs play major roles in subtype selectivity for both peptide and small molecule ligands. This 3D knowledge is even more critical for understanding the binding and mechanisms of action for allosteric modulators and “bitopic” ligands of GPCRs [6, 31–33], as these promising new classes of therapeutic compounds often specifically target the loop regions of receptors.
Structural diversity in the extracellular portion of the 7TM helical bundle
As expected from the characteristic 7TM sequence pattern and early 3D modeling, crystal structures confirm the overall structural conservation of the 7TM helical bundle, with Cα root mean squared deviations (RMSD) of <3 Å in TM-helices between any pairs of GPCRs. At the same time, the structures reveal dramatic local variations between different GPCRs even within the 7TM helices; in particular, at the sites involving bulges, kinks and other deviations from canonical α-helices that are usually associated with prolines (Figure 4).
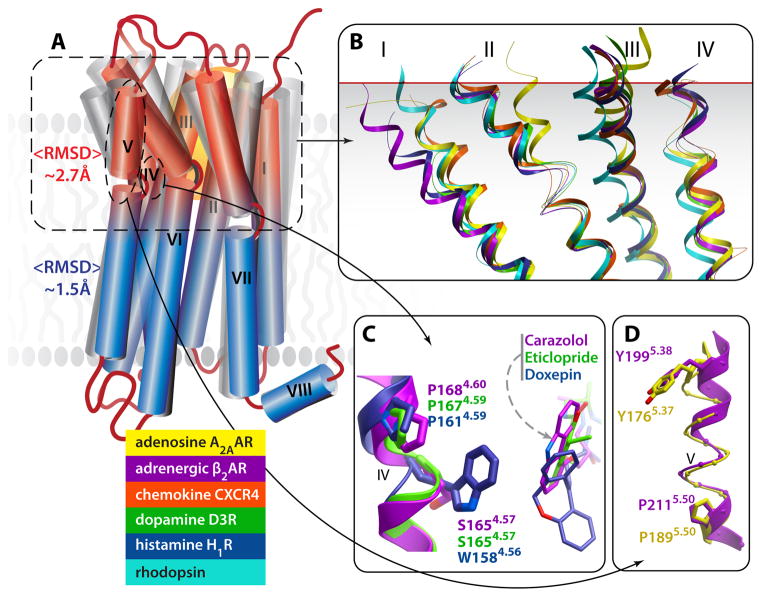
Fig. 4.
Structural diversity and conformational plasticity in GPCRs. (A) A cartoon illustrating 7TM helical bundles of known GPCR structures. In general, structural deviations between receptors are larger in TM-EC module (red) than in TM-IC module (blue) (average RMSDs are ~2.7 A vs. 1.5 A). (B) Structural superimposition of TM helical bundle structures of different GPCR shows especially high deviations in the extracellular half of the bundle, with strong deviation observed in each of the helices (only helices I to IV are shown for clarity). (C) An insertion/deletion in structural alignment, shown in an example of a conserved proline kink in helix IV. Note that Trp in position 4.56 of H1R structurally aligns with Ser in position 4.57 of β2AR and D3R, while the Pro in position 4.60 of β2AR structurally aligns with Pro in position 4.59 in D3R and H1R. (D) Observed π-helical structure in the extracellular portion of A2AAR helix V. The π-helix (i+5) has more residues per helical turn than standard α-helical (i+4) structure, resulting in “phase shift” and structural alignment of Tyr in position 5.37 of A2AAR with Tyr in position 5.38 of β2AR.
Some less obvious variations, such as those including helix IV of H1R (Figure 4C) [18] and helix II of CXCR4 [16], can be represented as an insertion or deletion of one residue in the backbone structure, accompanied by some local adjustments of a few neighboring residues. The corresponding sequence alignment should, therefore, incorporate such one-residue insertions/deletions. Comparative analysis of GPCR structures, however, has traditionally relied on gapless alignments of TM helices and specialized residue numbering, e.g. Ballesteros-Weinstein [34], which, in many cases, may not reflect a real structural alignment of the residues. Special care should be used to identify and predict such local deviations often dramatically affecting functional sites in the ligand-binding pocket. Moreover, some other deviations, like π-helical (i+5) structure of the helix V in A2AAR (Figure 5D), or sharp bends of the EC tips of helices II and III in A2AAR, cannot be adequately described by sequence alignment. All these non-canonical features represent a significant challenge for molecular modeling [35, 36], emphasizing the role of high-resolution crystallography as an indispensible source of accurate 3D knowledge for GPCRs.
Fig. 5.
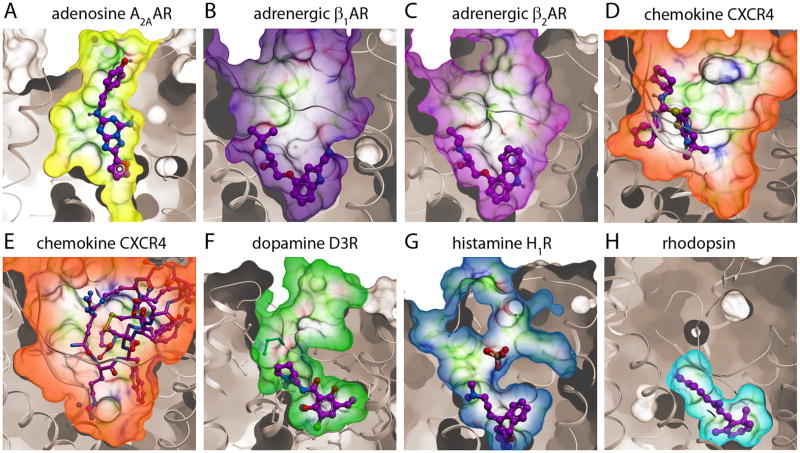
Diversity of the ligand binding pocket shape and properties in GPCR crystal structures. Structural diversity, including large backbone deviations of ECLs and TM helices, results in dramatic variations in size, shape and binding properties of GPCR pockets: (A) In A2AAR (PDB ID 3EML), the pocket forms an accessible channel with the ligand (antagonist ZM241385) positioned vertically towards the EC region. (B) (C) The β1AR (PDB ID 2VT4) and β2AR (PDB ID 2RH1) have almost identical highly accessible pockets that share the same contact residues for antagonists cyanopindolol and carazolol, respectively. (D) (E) The chemokine CXCR4 receptor has a large and open pocket, which can accommodate not only small molecules such as IT1t (D) (PDB ID 3ODU), but also molecules as large as the 16-residue cyclic peptide CVX15 (E) (PDB ID 3OE0). (F) The D3R pocket (PDB ID 3PBL) has distinct EC and “core” sub-pockets, the latter one occupied by the antagonist eticlopride (see also Fig 6B). (G) The H1R pocket (PDB ID 3RZE) reaches deeper into the TM than in other receptors, while the PO43− ion modulates ligand access to the pocket. (H) The retinal pocket in Rhodopsin (PDB ID 1GZM) is small, hydrophobic and completely enclosed. All pockets are shown in the same orientation.
It is important to note, in the structures solved to date, that structural variations are especially pronounced in the EC-TM half of the helical bundle-module, as reflected in up to 7Å shifts in the EC helical tips. On average, the protein backbone RMSD in the EC-TM region between different GPCR pairs is almost two times larger than the RMSD value for the IC-TM module (Figure 4A,B). Such a contrast between ligand binding and downstream signaling modules in their structural variability is also evident from a comparison on the individual residue level. For example, only 6% of residues are exactly conserved in the EC-TM region across all published GPCR structures, as compared to 26% of residues in the IC-TM region.
Diversity in the ligand binding pocket
A highly diverse repertoire of structural features in the ligand-binding pockets of different GPCR subfamilies apparently reflects evolutionary pressure to selectively recognize ligands that vary greatly in shapes, sizes and electrostatic properties. Even within the preserved overall 7TM bundle architecture, there is a remarkable diversity of the binding pocket shapes and features between GPCRs subfamilies (Figure 5), manifested in both side-chain diversity and variations in backbone conformations of TM helices and ECLs. Thus, unlike rhodopsin that contains a tightly enclosed hydrophobic pocket, GPCRs that bind diffusible ligands have more open pockets, accessible from the EC side. In the β2AR and other aminergic receptors, the core binding site sits rather deep in the 7TM bundle, and many high affinity ligands do not make significant contacts with the ECL residues. In contrast, the A2AAR binding site is located much closer to the loops and the high affinity binding requires ligand interaction with at least one ECL2 side chain, Phe168. In the chemokine-binding CXCR4, the ligand pocket is much larger and shallower than in other solved GPCR structures, and binding of its peptide antagonists involves extensive interactions with ECL2, which probably mimic interactions with a native SDF-1 ligand and the V3 loop of the HIV gp120 protein.
In contrast, GPCR subtypes that bind the same endogenous ligand are expected to have rather high levels of binding pocket conservation. Among them, the crystal structures of β2AR and β1AR represent extreme cases, with 100% conserved contact residues and RMSD <1.0 Å in these side chains (compared to heavy-atom RMSD ~0.9 Å in the same side chains between β2AR crystal structures with different ligands). More typically, about 50–60% of residues are identical between pockets in GPCR subtypes binding the same ligand. Conformational modeling studies supported by ligand binding data, for example for adenosine receptor subtypes [37, 38], suggest a high level of structural conservation between such subtypes, allowing accurate prediction of the pockets from one or more representative structures in the subfamily. As crystallization of each GPCR subtype may be impractical, such “close-range” modeling can be useful to fill the remaining gaps in structural knowledge to better understand ligand subtype selectivity, which is essential for rational design of tool compounds and safer drug candidates for GPCR targets.
Another important feature of GPCRs revealed by analysis of multiple crystal structures is the relative rigidity of their binding pockets. Despite the overall conformational flexibility of GPCRs, crystal structures show that conformational rearrangements (ligand induced fit) in the binding pockets are usually rather limited within their inactive states. For example, in several crystal structures of β2AR with different antagonists and inverse agonists [11, 23, 24], RMSD of the common binding pocket’s side chains does not exceed 1.0 Å. Of course, binding of some bulky conformationally selective ligands, e.g. the 16-residue cyclic peptide antagonist CVX15 [16], can induce some substantial deviations in the side chains and helices (compare Figure 5D and E), however such deviations are usually minor for small drug-like compounds. This result has a major implication for the applicability of GPCR crystal structures to rational drug discovery [39, 40], suggesting that one (or very few) conformations of the receptor can effectively explain binding of majority of drug-like compounds. Applicability of the high-resolution GPCR structures to drug discovery has been further corroborated by retrospective Virtual Ligand Screening (VLS) (e.g. [41, 42]) and exceptionally high hit rates obtained in prospective VLS-based studies that have already identified a number of novel high-affinity antagonists for β2AR [43, 44] and A2AAR [45, 46].
Structural characterization of agonist-bound active-state GPCRs is much more challenging due to their reduced stability, though both crystallography [26, 29, 47–51] and conformational modeling [52, 53] are starting to provide the first insights into binding with this class of GPCR ligands, as reviewed in [19, 40, 54].
Structural conservation and conformational flexibility in the IC region
In contrast to the high structural diversity observed in the EC module, crystal structures of inactive GPCRs reveal high structural conservation (RMSD ~1.5 Å) in the IC module, which is involved in binding to G proteins and arrestins and downstream signal transduction through mechanisms that are believed to be similar across GPCRs [55, 56]. The IC-TM region also harbors several functionally important features conserved in the Class A subfamily, including the D[E]RY motif in helix III and Glu6.30, which form the so-called “ionic lock” in some inactive GPCR structures [57], as well as the NPxxY motif in helix VII [58]. GPCR structures reveal that a high level of structural conservation also extends to the short ICL1 and helix VIII. The 6-residue ICL1 has a similar backbone conformation and a conserved leucine side chain inserted back into the TM bundle in all known GPCR structures. Helix VIII is a 3–4 turn α-helix that runs parallel to the membrane and is characterized by a common [F(RK)xx(FL)xxx(LF)] amphiphilic motif. In many GPCRs, helix VIII is also anchored to the membrane by palmitoylation and is essential for receptor expression and function [59]. Among the known GPCR structures, CXCR4 is the only exception in which helix VIII shows a disordered behavior, probably because one of the phenylalanines in the amphiphilic motif is missing, although this does not preclude the possibility of its formation in CXCR4 in cell membranes under certain conditions.
At the same time, the IC region undergoes large conformational changes as a part of the activation mechanism in each GPCR [26, 29, 47–51], therefore one can expect much higher conformational flexibility and/or instability of some of ICLs. Indeed, dramatic variations in ICL2 structure (9–12 residues long), have been observed within the same receptor, and even within the same crystal form. Thus, in the D3R structure, crystallized with two receptors per asymmetric unit, molecule A has a well-resolved 2.5-turn α-helix in ECL2 that runs parallel to the membrane, while in molecule B this loop is disordered and no electron density is observed. Similarly, both extended and α-helical conformations have been observed for ICL2s of β2AR [11, 12] and β1AR [14, 60], which have almost the same sequence. These crystallographic, as well as HDX observations [28], suggest that ICL2 can be structurally ambivalent and its conformational state may depend on the functional state of the receptor and its interactions with G proteins.
Unlike other IC regions, ICL3 has a highly variable length, ranging from as short as five residues in CXCR4, up to hundreds of residues in some other receptors. Variations in length and sequence motifs in ICL3 are high even in closely related subtypes, as ICL3 is believed to controls receptor selectivity to different G proteins. ICL3 regions in crystal structures are usually disordered or replaced by a stabilizing fusion protein. At the same time, there are indications that in some GPCRs the ICL3 has a propensity for forming α-helical structures, which elongate helices V and VI by at least 2–3 turns [50, 61]. In β1AR [61], two alternative conformations of this region have been recently resolved in a crystal structure of the inactive receptor: one of them includes a bent helix VI allowing formation of the ionic lock, whereas another with straight helix VI lacks the ionic lock interaction. By contrast, the high proteolytic susceptibility of ICL3, also supported by recent HDX data for β2AR [27, 28], suggests that such secondary structure elements in ICL3 are rather unstable and/or flexible, at least in the absence of a binding partner.
Further structural and biophysical studies, especially in GPCR complexes with G-proteins [25] and arrestins will be needed to understand the structure and dynamics of the IC region and nature of its multifaceted pattern of selectivity to downstream effector molecules. While is not directly involved in binding of orthosteric ligands or drugs, the IC region is critical for the functional selectivity of drug candidates and is also considered a potential alternative target for allosteric drugs [62].
Allosteric sites and dimerization
GPCR function involves specific interactions with numerous binding partners, which include not only signaling ligands and downstream effectors, such as G proteins and arrestins, but also other regulatory proteins [63, 64], lipids and sterols [59, 65] and ions [66]. Moreover, allosteric sites [67] for binding small molecule modulators have been identified in many GPCRs [68]. Such sites can be located in direct proximity of the ligand binding pocket [69], in the 7TM helical bundle for Class B and C GPCRs [70], or even in the IC part of the receptor [62, 71]. Molecules targeting regulatory and other allosteric sites are important for understanding GPCR biological pathways, and also may be highly beneficial in therapeutic applications as they can modulate specific parameters of the native ligand signaling without completely hijacking or blocking receptor activity [72].
Although the number of studies characterizing these types of interactions biochemically and functionally has been growing, understanding the molecular basis of allosteric binding in GPCRs remained limited until recently owing to the lack of corresponding structural information. Moreover, allosteric sites are usually located in less-conserved regions of the protein, as opposed to orthosteric sites, and, therefore, are less amenable for homology modeling. The recent availability of GPCR crystal structures has provided a detailed atomic structural framework that can greatly assist biochemical methods traditionally used for identification and analysis of allosteric sites [33]. Crystal structures can also directly suggest novel allosteric sites with unusual properties and selectivity (Figure. 6). For example, a specific cholesterol binding site has been repeatedly observed in β2AR [11, 23], with cholesterol modulating receptor thermostability and affinity to inverse agonists [23, 73]. Moreover, the allosteric effect of cholesterol and some of its close analogs, found to be critical for full activation of the oxytocin receptor, is likely mediated by the same and possibly other allosteric sites [74]. Binding of a phosphate ion in the EC subpocket of the H1R structure was also found to allosterically and specifically modulate binding of different zwitterion antihistamines [18]. Some other “druggable” non-orthosteric pockets or subpockets have been described in crystal structures, for example a selectivity subpocket in D3R [17], which can be further explored as a putative target for allosteric and/or bitopic ligands.
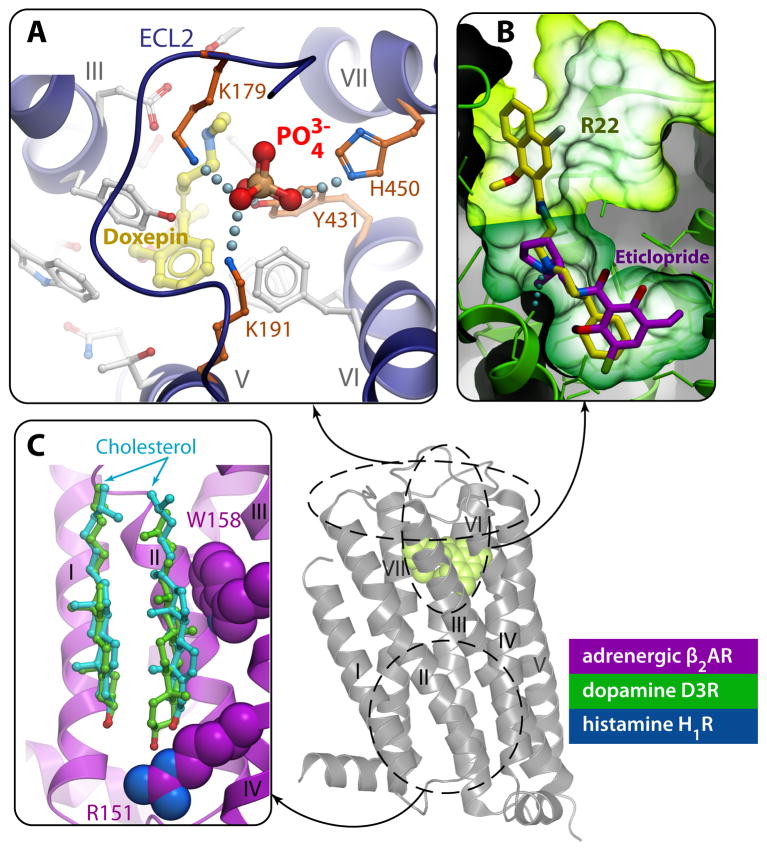
Fig. 6.
Examples of allosteric interaction sites identified in GPCR crystal structures. A) Binding of a phosphate ion in the ECLs of H1R (PDB ID 3RZE) is coordinated by four receptor side chains, and affects the accessibility of the ligand pocket. This site is key for H1 subtype selectivity to second-generation antihistamines [18]. B) The ligand-binding pocket in the D3R (PDB ID 3PBL) consists of a core site (dark green) with bound antagonist eticlopride and a secondary or allosteric site (lime). Docking the D3 subtype-selective ligand R22 into the D3R crystal structure reveals the bitopic nature of this compound occupying the core site and extending into the secondary site. Interactions between R22 and the secondary site define most of the subtype selectivity for this ligand [17]. C) The cholesterol binding site on the lipid-TM interface has been reproduced in different crystal forms of the β2AR, (shown here by cyan sticks for PDB IDs 2RH1 and by green sticks for 3D4S), and is likely conserved in many other GPCRs [23].
Another important type of interactions is homo- and hetero-dimerization of GPCRs. These interactions can modulate the signaling properties of the receptors and mediate crosstalk between GPCR pathways [75]. Even for the most studied dimers, however, unambiguous identification of the functionally relevant interfaces has proved to be very difficult, partially due to a transient mode of the interactions and technical challenges of separating specific and non-specific binding in a crowded membrane environment [76]. In crystallization studies, non-specific dimerization of a GPCR can be detrimental for obtaining crystals, as it introduces heterogeneity in samples, and for that reason it was largely avoided. Therefore, in most crystal structures so far, the GPCR molecules have been found to pack in non-functional (e.g. antiparallel) orientations. An important exception is represented by the recent crystal structures of CXCR4, which have revealed a consistent parallel dimer arrangement [16]. The dimer interface is virtually identical in five different crystal packing forms of CXCR4 complexes with both peptide and small molecule antagonists, suggesting its functional relevance. It is likely that future crystallization efforts, also specifically optimizing dimerization conditions, will bring about more structures of GPCR functional dimers, including heterodimers.
Towards comprehensive structural coverage of GPCR superfamily
Due to the rapid progress in GPCR crystallography, in a mere four years we have come from reliance on rhodopsin as the only representative structure for the GPCR superfamily to high resolution structural characterization of seven distinct GPCRs. Three of them [16–18] have been resolved just within the last year. The determined crystal structures represent two (α and γ) out of four major branches in Class A GPCRs [2] (Figure 1) and exemplify very different levels of sequence and structural homology between them. The structures allow major insights into the rich structural complexity of GPCRs, making it possible to let go of the natural simplifications employed for analysis in the long period of the “structure drought”.
At the same time, the new structures can be considered as the first steps on the way to comprehensive coverage of the superfamily, which defines the future strategy towards this major goal. Selection of targets for the first phase of the NIH NIGMS PSI:Biology center GPCR Network was based on the assumption that an optimum use of limited resources can be achieved by crystallization of one to three representative receptors in each GPCR subfamily, also taking into account the level of homology and the number of GPCRs in the subfamily. Although still limited, the initial sampling provided by the solved GPCR crystal structures supports this strategy. The structures show remarkable structural variations between GPCR subfamilies, especially in the ligand binding region, which cannot be reliably and accurately predicted by current modeling techniques[36]. On the other hand, genetically and functionally related subtypes within GPCR subfamilies are also close structurally. The crystal structures, therefore, may provide a solid structural framework for comparative analysis of some other subtypes with close structural homology, making it possible to fill (at least temporarily) the gaps in the structural knowledge and understanding of the 3D basis of ligand selectivity.
Another dimension in deciphering GPCR biology is provided by the structural characterization of representative receptors bound to different ligands. Multiple inactive structures with antagonists suggest a generally low level of induced fit in GPCRs, making possible accurate prediction of binding conformations for most ligands [24]. However, binding of certain conformationally selective compounds, including both antagonists and agonists can reveal a number of new interesting features including characterization of new functionally selective states. These states are of great interest to GPCR biology and drug discovery, and should be actively pursued by structural studies. Finally, although crystallography has recently achieved a first milestone in characterizing activated states of adrenergic and adenosine receptors, we are still at the very beginning of the quest for understanding the nature of GPCR function. Further structural, biophysical and computational studies will be required for deciphering ligand-dependent activation triggers in other receptors, are well as the structural basis of GPCR selective interactions with G-proteins, arrestins and other modulators.
Acknowledgments
This work was supported in part by the NIH Roadmap grant P50 GM073197 and NIH PSI:Biology grant U54 GM094618. We are very grateful to the accumulation of more than 20 years of structure determination efforts on GPCR structural biology and technology by a number of different groups worldwide, including our own lab. We are particularly thankful to our biochemistry/chemistry collaborators in the different receptor systems including Ad IJzerman and Ken Jacobsen (A2A adenosine), Brian Kobilka (β2 adrenergic), Tracy Handel and Alexei Brooun (chemokine CXCR4), Jonathan Javitch and Amy Newman (dopamine D3), and So Iwata and Tatsuro Shimamura (histamine H1). We thank K. Kadyshevskaya for assistance with figure preparation and A. Walker for assistance with manuscript preparation.
References
- 1.Nordstrom KJ, et al. Independent HHsearch, Needleman-Wunsch-based and motif analyses reveals the overall hierarchy for most of the G protein-coupled receptor families. Mol Biol Evol. 2011;28(9):2471–80. doi: 10.1093/molbev/msr061. [DOI] [PubMed] [Google Scholar]
- 2.Fredriksson R, et al. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol. 2003;63(6):1256–72. doi: 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- 3.Fredriksson R, Schioth HB. The repertoire of G-protein-coupled receptors in fully sequenced genomes. Mol Pharmacol. 2005;67(5):1414–25. doi: 10.1124/mol.104.009001. [DOI] [PubMed] [Google Scholar]
- 4.Strotmann R, et al. Evolution of GPCR: change and continuity. Mol Cell Endocrinol. 2011;331(2):170–8. doi: 10.1016/j.mce.2010.07.012. [DOI] [PubMed] [Google Scholar]
- 5.Lagerstrom MC, Schioth HB. Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat Rev Drug Discov. 2008;7(4):339–57. doi: 10.1038/nrd2518. [DOI] [PubMed] [Google Scholar]
- 6.Whalen EJ, Rajagopal S, Lefkowitz RJ. Therapeutic potential of beta-arrestin-and G protein-biased agonists. Trends Mol Med. 2011;17(3):126–39. doi: 10.1016/j.molmed.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lappano R, Maggiolini M. G protein-coupled receptors: novel targets for drug discovery in cancer. Nat Rev Drug Discov. 2011;10(1):47–60. doi: 10.1038/nrd3320. [DOI] [PubMed] [Google Scholar]
- 8.Wise A, Gearing K, Rees S. Target validation of G-protein coupled receptors. Drug Discov Today. 2002;7(4):235–46. doi: 10.1016/s1359-6446(01)02131-6. [DOI] [PubMed] [Google Scholar]
- 9.Palczewski K, et al. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289(5480):739–45. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 10.Deisenhofer J, et al. X-ray structure analysis of a membrane protein complex. Electron density map at 3 A resolution and a model of the chromophores of the photosynthetic reaction center from Rhodopseudomonas viridis. J Mol Biol. 1984;180(2):385–98. doi: 10.1016/s0022-2836(84)80011-x. [DOI] [PubMed] [Google Scholar]
- 11.Cherezov V, et al. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318(5854):1258–65. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosenbaum DM, et al. GPCR engineering yields high-resolution structural insights into beta2-adrenergic receptor function. Science. 2007;318(5854):1266–73. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- 13.Rasmussen SG, et al. Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature. 2007;450(7168):383–7. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 14.Warne T, et al. Structure of a beta(1)-adrenergic G-protein-coupled receptor. Nature. 2008;454(7203):486–91. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jaakola VP, et al. The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science. 2008;322(5905):1211–7. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu B, et al. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science. 2010;330(6007):1066–71. doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chien EY, et al. Structure of the human dopamine D3 receptor in complex with a D2/D3 selective antagonist. Science. 2010;330(6007):1091–5. doi: 10.1126/science.1197410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shimamura T, et al. Structure of the human histamine H1 receptor complex with doxepin. Nature. 2011;475(7354):65–70. doi: 10.1038/nature10236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deupi X, Standfuss J. Structural insights into agonist-induced activation of G-protein-coupled receptors. Curr Opin Struct Biol. 2011;21(4):541–51. doi: 10.1016/j.sbi.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 20.Maeda A, et al. Palmitoylation stabilizes unliganded rod opsin. Proc Natl Acad Sci U S A. 2010;107(18):8428–33. doi: 10.1073/pnas.1000640107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tobin AB, Butcher AJ, Kong KC. Location, location, location...site-specific GPCR phosphorylation offers a mechanism for cell-type-specific signalling. Trends Pharmacol Sci. 2008;29(8):413–20. doi: 10.1016/j.tips.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nygaard R, et al. Ligand binding and micro-switches in 7TM receptor structures. Trends Pharmacol Sci. 2009;30(5):249–59. doi: 10.1016/j.tips.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 23.Hanson MA, et al. A specific cholesterol binding site is established by the 2.8 A structure of the human beta2-adrenergic receptor. Structure. 2008;16(6):897–905. doi: 10.1016/j.str.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wacker D, et al. Conserved binding mode of human beta2 adrenergic receptor inverse agonists and antagonist revealed by X-ray crystallography. J Am Chem Soc. 2010;132(33):11443–5. doi: 10.1021/ja105108q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rasmussen SG, et al. Crystal structure of the beta(2) adrenergic receptor-Gs protein complex. Nature. 2011 doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rasmussen SG, et al. Structure of a nanobody-stabilized active state of the beta(2) adrenoceptor. Nature. 2011;469(7329):175–80. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang X, et al. Dynamics of the beta2-adrenergic G-protein coupled receptor revealed by hydrogen-deuterium exchange. Anal Chem. 2010;82(3):1100–8. doi: 10.1021/ac902484p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.West GM, et al. Ligand-dependent perturbation of the conformational ensemble of the GPCR beta2 adrenergic receptor revealed by HDX. Structure. 2011 doi: 10.1016/j.str.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lebon G, et al. Agonist-bound adenosine A2A receptor structures reveal common features of GPCR activation. Nature. 2011;474(7352):521–5. doi: 10.1038/nature10136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Civelli O, Bunzow JR, Grandy DK. Molecular diversity of the dopamine receptors. Annu Rev Pharmacol Toxicol. 1993;33:281–307. doi: 10.1146/annurev.pa.33.040193.001433. [DOI] [PubMed] [Google Scholar]
- 31.Gao ZG, Jacobson KA. Keynote review: allosterism in membrane receptors. Drug Discov Today. 2006;11(5–6):191–202. doi: 10.1016/S1359-6446(05)03689-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kenakin T, Miller LJ. Seven transmembrane receptors as shapeshifting proteins: the impact of allosteric modulation and functional selectivity on new drug discovery. Pharmacol Rev. 2010;62(2):265–304. doi: 10.1124/pr.108.000992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keov P, Sexton PM, Christopoulos A. Allosteric modulation of G protein-coupled receptors: a pharmacological perspective. Neuropharmacology. 2011;60(1):24–35. doi: 10.1016/j.neuropharm.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 34.Ballesteros JA, Weinstein H. Integrated methods for the construction of three dimensional models and computational probing of structure function–relations in G-protein coupled receptors. Methods Neurosci. 1995;25:366–428. [Google Scholar]
- 35.Michino M, et al. Community-wide assessment of GPCR structure modelling and ligand docking: GPCR Dock 2008. Nat Rev Drug Discov. 2009;8(6):455–63. doi: 10.1038/nrd2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kufareva I, et al. Status of GPCR Modeling and Docking as Reflected by Community-wide GPCR Dock 2010 Assessment. Structure. 2011;19(8):1108–26. doi: 10.1016/j.str.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Katritch V, Kufareva I, Abagyan R. Structure based prediction of subtype-selectivity for adenosine receptor antagonists. Neuropharmacology. 2011;60(1):108–15. doi: 10.1016/j.neuropharm.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jaakola VP, et al. Ligand binding and subtype selectivity of the human A(2A) adenosine receptor: identification and characterization of essential amino acid residues. J Biol Chem. 2010;285(17):13032–44. doi: 10.1074/jbc.M109.096974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reynolds K, Abagyan R, Katritch V. Structure and Modeling of GPCRs: Implications for Drug Discovery. In: Gilchrist A, editor. GPCR Molecular Pharmacology and Drug Targeting: Shifting Paradigms and New Directions. Wiley & Sons, Inc; Hoboken, NJ: 2010. pp. 385–433. [Google Scholar]
- 40.Congreve M, et al. Progress in structure based drug design for G protein-coupled receptors. J Med Chem. 2011;54(13):4283–311. doi: 10.1021/jm200371q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Topiol S, Sabio M. Use of the X-ray structure of the Beta2-adrenergic receptor for drug discovery. Bioorg Med Chem Lett. 2008;18(5):1598–602. doi: 10.1016/j.bmcl.2008.01.063. [DOI] [PubMed] [Google Scholar]
- 42.Reynolds KA, Katritch V, Abagyan R. Identifying conformational changes of the beta(2) adrenoceptor that enable accurate prediction of ligand/receptor interactions and screening for GPCR modulators. J Comput Aided Mol Des. 2009;23(5):273–88. doi: 10.1007/s10822-008-9257-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sabio M, Jones K, Topiol S. Use of the X-ray structure of the beta2-adrenergic receptor for drug discovery. Part 2: Identification of active compounds. Bioorg Med Chem Lett. 2008;18(20):5391–5. doi: 10.1016/j.bmcl.2008.09.046. [DOI] [PubMed] [Google Scholar]
- 44.Kolb P, et al. Structure-based discovery of beta2-adrenergic receptor ligands. Proc Natl Acad Sci U S A. 2009;106(16):6843–8. doi: 10.1073/pnas.0812657106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Katritch V, et al. Structure-based discovery of novel chemotypes for adenosine A(2A) receptor antagonists. J Med Chem. 2010;53(4):1799–809. doi: 10.1021/jm901647p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carlsson J, et al. Structure-based discovery of A2A adenosine receptor ligands. J Med Chem. 2010;53(9):3748–55. doi: 10.1021/jm100240h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu F, et al. Structure of an Agonist-Bound Human A2A Adenosine Receptor. Science. 2011;332(6027):322–327. doi: 10.1126/science.1202793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Standfuss J, et al. The structural basis of agonist-induced activation in constitutively active rhodopsin. Nature. 2011;471(7340):656–60. doi: 10.1038/nature09795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Choe HW, et al. Crystal structure of metarhodopsin II. Nature. 2011;471(7340):651–5. doi: 10.1038/nature09789. [DOI] [PubMed] [Google Scholar]
- 50.Park JH, et al. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature. 2008;454(7201):183–7. doi: 10.1038/nature07063. [DOI] [PubMed] [Google Scholar]
- 51.Scheerer P, et al. Crystal structure of opsin in its G-protein-interacting conformation. Nature. 2008;455(7212):497–502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- 52.Katritch V, et al. Analysis of full and partial agonists binding to beta2-adrenergic receptor suggests a role of transmembrane helix V in agonist-specific conformational changes. J Mol Recognit. 2009;22(4):307–18. doi: 10.1002/jmr.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vilar S, et al. In silico analysis of the binding of agonists and blockers to the beta2-adrenergic receptor. J Mol Graph Model. 2011;29(6):809–17. doi: 10.1016/j.jmgm.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Katritch V, Abagyan R. Trends in Pharm Sciences. GPCR agonist binding revealed by modeling and crystallography. Accepted, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Milligan G, Kostenis E. Heterotrimeric G-proteins: a short history. Br J Pharmacol. 2006;147(Suppl 1):S46–55. doi: 10.1038/sj.bjp.0706405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gurevich VV, Gurevich EV. The structural basis of arrestin-mediated regulation of G-protein-coupled receptors. Pharmacol Ther. 2006;110(3):465–502. doi: 10.1016/j.pharmthera.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rovati GE, Capra V, Neubig RR. The highly conserved DRY motif of class A G protein-coupled receptors: beyond the ground state. Mol Pharmacol. 2007;71(4):959–64. doi: 10.1124/mol.106.029470. [DOI] [PubMed] [Google Scholar]
- 58.Fritze O, et al. Role of the conserved NPxxY(x)5,6F motif in the rhodopsin ground state and during activation. Proc Natl Acad Sci U S A. 2003;100(5):2290–5. doi: 10.1073/pnas.0435715100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chini B, Parenti M. G-protein-coupled receptors, cholesterol and palmitoylation: facts about fats. J Mol Endocrinol. 2009;42(5):371–9. doi: 10.1677/JME-08-0114. [DOI] [PubMed] [Google Scholar]
- 60.Dror RO, et al. Identification of two distinct inactive conformations of the beta2-adrenergic receptor reconciles structural and biochemical observations. Proc Natl Acad Sci U S A. 2009;106(12):4689–94. doi: 10.1073/pnas.0811065106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moukhametzianov R, et al. Two distinct conformations of helix 6 observed in antagonist-bound structures of a {beta}1-adrenergic receptor. Proc Natl Acad Sci U S A. 2011;108(20):8228–32. doi: 10.1073/pnas.1100185108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dowal L, et al. Identification of an antithrombotic allosteric modulator that acts through helix 8 of PAR1. Proc Natl Acad Sci U S A. 2011;108(7):2951–6. doi: 10.1073/pnas.1014863108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thompson MD, Cole DE, Jose PA. Pharmacogenomics of G protein-coupled receptor signaling: insights from health and disease. Methods Mol Biol. 2008;448:77–107. doi: 10.1007/978-1-59745-205-2_6. [DOI] [PubMed] [Google Scholar]
- 64.Ritter SL, Hall RA. Fine-tuning of GPCR activity by receptor-interacting proteins. Nat Rev Mol Cell Biol. 2009;10(12):819–30. doi: 10.1038/nrm2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Escriba PV, et al. Lipid-protein interactions in GPCR-associated signaling. Biochim Biophys Acta. 2007;1768(4):836–52. doi: 10.1016/j.bbamem.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 66.Ericksen SS, et al. Ligand selectivity of D2 dopamine receptors is modulated by changes in local dynamics produced by sodium binding. J Pharmacol Exp Ther. 2009;328(1):40–54. doi: 10.1124/jpet.108.141531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Changeux JP, Edelstein SJ. Allosteric mechanisms of signal transduction. Science. 2005;308(5727):1424–8. doi: 10.1126/science.1108595. [DOI] [PubMed] [Google Scholar]
- 68.Smith NJ, Bennett KA, Milligan G. When simple agonism is not enough: emerging modalities of GPCR ligands. Mol Cell Endocrinol. 2011;331(2):241–7. doi: 10.1016/j.mce.2010.07.009. [DOI] [PubMed] [Google Scholar]
- 69.May LT, et al. Allosteric modulation of G protein-coupled receptors. Annu Rev Pharmacol Toxicol. 2007;47:1–51. doi: 10.1146/annurev.pharmtox.47.120505.105159. [DOI] [PubMed] [Google Scholar]
- 70.Conn PJ, Christopoulos A, Lindsley CW. Allosteric modulators of GPCRs: a novel approach for the treatment of CNS disorders. Nat Rev Drug Discov. 2009;8(1):41–54. doi: 10.1038/nrd2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Covic L, et al. Activation and inhibition of G protein-coupled receptors by cell-penetrating membrane-tethered peptides. Proc Natl Acad Sci U S A. 2002;99(2):643–8. doi: 10.1073/pnas.022460899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bridges TM, Lindsley CW. G-protein-coupled receptors: from classical modes of modulation to allosteric mechanisms. ACS Chem Biol. 2008;3(9):530–41. doi: 10.1021/cb800116f. [DOI] [PubMed] [Google Scholar]
- 73.Yao Z, Kobilka B. Using synthetic lipids to stabilize purified beta2 adrenoceptor in detergent micelles. Anal Biochem. 2005;343(2):344–6. doi: 10.1016/j.ab.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 74.Gimpl G, et al. Oxytocin receptors: ligand binding, signalling and cholesterol dependence. Prog Brain Res. 2008;170:193–204. doi: 10.1016/S0079-6123(08)00417-2. [DOI] [PubMed] [Google Scholar]
- 75.Milligan G, et al. The role of GPCR dimerisation/oligomerisation in receptor signalling. Ernst Schering Found Symp Proc. 2006;2:145–61. doi: 10.1007/2789_2006_007. [DOI] [PubMed] [Google Scholar]
- 76.James JR, et al. A rigorous experimental framework for detecting protein oligomerization using bioluminescence resonance energy transfer. Nat Methods. 2006;3(12):1001–6. doi: 10.1038/nmeth978. [DOI] [PubMed] [Google Scholar]
- 77.Dore AS, et al. Structure of the Adenosine A(2A) Receptor in Complex with ZM241385 and the Xanthines XAC and Caffeine. Structure. 2011;19(9):1283–93. doi: 10.1016/j.str.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Warne T, et al. The structural basis for agonist and partial agonist action on a beta(1)-adrenergic receptor. Nature. 2011;469(7329):241–4. doi: 10.1038/nature09746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bokoch MP, et al. Ligand-specific regulation of the extracellular surface of a G-protein-coupled receptor. Nature. 2010;463(7277):108–12. doi: 10.1038/nature08650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rosenbaum DM, et al. Structure and function of an irreversible agonist-beta(2) adrenoceptor complex. Nature. 2011;469(7329):236–40. doi: 10.1038/nature09665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Okada T, et al. The retinal conformation and its environment in rhodopsin in light of a new 2.2 A crystal structure. J Mol Biol. 2004;342(2):571–83. doi: 10.1016/j.jmb.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 82.Sekharan S, et al. Protein assistance in the photoisomerization of rhodopsin and 9-cis-rhodopsin--insights from experiment and theory. J Am Chem Soc. 2007;129(5):1052–4. doi: 10.1021/ja066970p. [DOI] [PubMed] [Google Scholar]
- 83.Nakamichi H, Okada T. Local peptide movement in the photoreaction intermediate of rhodopsin. Proc Natl Acad Sci U S A. 2006;103(34):12729–34. doi: 10.1073/pnas.0601765103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shimamura T, et al. Crystal structure of squid rhodopsin with intracellularly extended cytoplasmic region. J Biol Chem. 2008;283(26):17753–6. doi: 10.1074/jbc.C800040200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Murakami M, Kouyama T. Crystal structure of squid rhodopsin. Nature. 2008;453(7193):363–7. doi: 10.1038/nature06925. [DOI] [PubMed] [Google Scholar]
- 86.Lomize MA, et al. OPM: orientations of proteins in membranes database. Bioinformatics. 2006;22(5):623–5. doi: 10.1093/bioinformatics/btk023. [DOI] [PubMed] [Google Scholar]