Abstract
Noroviruses are the major cause of food- or water-borne gastroenteritis outbreaks in humans. The norovirus protease that cleaves a large viral polyprotein to nonstructural proteins is essential for virus replication and an attractive target for antiviral drug development. Noroviruses show high genetic diversity with at least five genogroups, GI–GV, of which GI and GII are responsible for the majority of norovirus infections in humans. We cloned and expressed proteases of Norwalk virus (GI) and MD145 virus (GII) and characterized the enzymatic activities with fluorescence resonance energy transfer substrates. We demonstrated that the GI and GII proteases cleaved the substrates derived from the naturally occurring cleavage site in the open reading frame (ORF) 1 of G1 norovirus with similar efficiency, and that enzymatic activity of both proteases was inhibited by commercial protease inhibitors including chymostatin. The interaction of chymostatin to Norwalk virus protease was validated by nuclear magnetic resonance (NMR) spectroscopy.
Keywords: Antiviral, Viral proteases, Fluorescence resonance energy transfer, Norwalk virus, MD145 virus, Nuclear magnetic resonance spectroscopy, Chymostatin
Introduction
Noroviruses are responsible for more than 60% of all food- or water-borne gastroenteritis outbreaks in humans with an estimated 23 million annual cases in the U.S. alone, followed by Salmonella infection (C.D.C., 2010, Fankhauser et al., 1998). The symptoms of noroviral gastroenteritis such as nausea, vomiting, diarrhea, headaches, fever, chills, myalgias and sore throat usually last for 24 to 48 h (Green et al., 2001). Although noroviruses generally cause mild to moderate gastroenteritis, it can incapacitate affected individuals in military troops on ships or in war zones (Green et al., 2001), and can be severe to life-threatening in the young, elderly and immunocompromised patients (Atmar and Estes, 2006, Dolin, 2007). Recent studies have shown that noroviral diarrhea can persist for up to 4 weeks (Rockx et al., 2002, Sakai et al., 2001) and the viruses can be excreted for up to 3 weeks (Rockx et al., 2002). Furthermore, it has been reported that norovirus diarrhea and shedding lasted longer than 2 years in an immunocompromised patient (Nilsson et al., 2003).
The Norwalk virus (NV) is the first enteric calicivirus discovered in 1972 (Kapikian et al., 1972). Since the discovery of the first norovirus, at least five genogroups have been established in the genus Norovirus. Among them GI, GII, and rarely GIV viruses infect humans. The GI and GII noroviruses are further subdivided into genotypes GI/1–7 and GII/1–15 (Green, 2007). NV is the most studied prototype virus and is classified as GI/1 strain. In recent years, GII/4 noroviruses became predominantly associated with norovirus outbreaks and sporadic cases worldwide (Siebenga et al., 2010, Zheng et al., 2010). Overall, norovirus strains belonging to the GII are found in 75–100% of sporadic cases of norovirus infections (Patel et al., 2009), and GII/4 strains account for 60–70% of all reported norovirus outbreaks globally (Kroneman et al., 2008, Siebenga et al., 2009).
However, no vaccine or antiviral drug is currently available for norovirus infections, which is largely due to the absence of cell culture systems and animal models for human noroviruses. Noroviruses show high diversity, and immunity to one strain does not necessarily provide protection from infection with another strain. In addition, inadequate long-term immunity against noroviruses is indicated by repeated infections in adults (Glass et al., 2009, Green, 2007). Although noroviruses do not multiply in food or water, they can cause large outbreaks because as few as 10–100 virions are sufficient to cause illness in a healthy adult (Green, 2007). Noroviruses are classified as NIAID category B priority pathogens (NIAID) due to their highly contagious nature and a potential to cause a serious public health challenge. Therefore, development of antiviral drugs is highly desirable for preventing and treating norovirus infections.
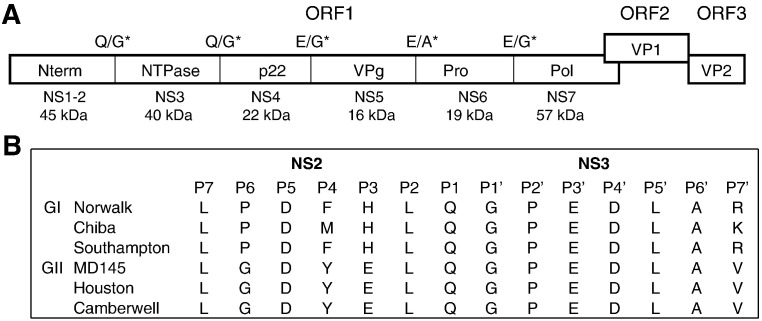
Noroviruses are single-stranded RNA viruses and encode three open reading frames (ORFs) for a nonstructural polyprotein and minor and major structural proteins. The gene organization of the norovirus nonstructural polyproteins encoded by ORF1 is N-terminal protein (Nterm, NS1-2), NTPase (NS3), p22 (3A-like protein, NS4), VPg (NS5), protease (Pro, NS6), and RNA-dependent RNA polymerase (Pol, NS7) (Green et al., 2001) (Fig. 1 ). Norovirus protease specifically recognizes and cleaves LQ/GP (Nterm/NTPase), LQ/GP (NTPase/p20), PE/GK (p20/VPg), FE/AP (VPg/Pro), and LE/GG (Pro/Pol) junctions to produce the mature proteins during viral replication (Belliot et al., 2003, Hardy et al., 2002, Liu et al., 1999, Sosnovtsev et al., 2006). Norovirus protease is classified as 3C-like cysteine protease due to its similarity to picornavirus 3C protease, which has a catalytic triad of amino acids composed of C, H, and E or D (Green, 2007, Nakamura et al., 2005). Since norovirus protease is essential for viral replication, viral protease represents an attractive target for antiviral drug development. The design and screening of antiviral agents targeting viral protease can be greatly facilitated by the availability of an assay that is suitable for large scale screening of potential novel drugs targeting viral protease.
Fig. 1.
Norovirus genome organization and proteolytic map. A. The cleavages at NS2/3 (between NS1–2 and NS3 proteins) and NS3/4 sites in ORF1 occur more efficiently than other cleavage sites in ORF1 of GI and GII noroviruses. *Cleavage dipeptide. B. Cleavage dipeptide and surrounding residues (P7–P7′) at NS2/3 site in ORF1 of GI and GII noroviruses. The designation of substrate residues for P1 and P1′ starts at the scissile bond and counts toward the N- or C-terminus, respectively, as suggested by Schechter and Berger (1967).
The fluorescence resonance energy transfer (FRET) protease assay has been developed to provide a rapid and specific identification of protease inhibitors for various cellular and viral proteases including foot-and-mouth virus and severe acute respiratory syndrome (SARS) coronavirus (Blanchard et al., 2004, Chen et al., 2005, Jaulent et al., 2007). In this assay system, substrates have a fluorescence donor and a quencher on each end, and the donor fluorescence signal in the uncleaved substrate is inhibited by the interaction of the fluorescence donor and quencher. Once substrates are cleaved by a protease, the donor fluorescence is no longer quenched, yielding an increase in fluorescence intensity. Addition of protease inhibitors to the assay inhibits the cleavage of the substrates, which leads to reduced fluorescence intensity, enabling screening of potential protease inhibitors. The FRET protease assay system is also a useful tool for measuring the activity and substrate specificity of a protease. So far, most of information on the proteolytic processing of the norovirus proteases has been obtained using in vitro transcription–translation assay systems (Belliot et al., 2003, Hardy et al., 2002, Scheffler et al., 2007, Seah et al., 1999, Someya and Takeda, 2009). Limited studies are reported on the kinetics of GI norovirus proteases using fluorogenic (Someya et al., 2008) or chromogenic substrates (Hussey et al., 2011). However studies on those of GII norovirus protease are not available. Furthermore, a FRET-based protease assay suitable as a high-throughput screening of protease inhibitors has not yet been reported for norovirus proteases of GI or GII noroviruses (Zeitler et al., 2006).
In this study, we describe the development and application of a high-throughput FRET assay using norovirus proteases from NV (NVpro) and MD145 virus (MD145pro) that belong to genogroups I (GI/1) and II (GII/4), respectively. Based on the assay, similar kinetic parameters of the fluorogenic substrates and inhibition by a selection of standard protease inhibitors including chymostatin were observed for NVpro and MD145pro. The binding of chymostatin and NVpro was also studied to validate the interactions using nuclear magnetic resonance (NMR) spectroscopy.
Results
MD145pro efficiently cleaved substrates originated from the cleavage site of GI norovirus
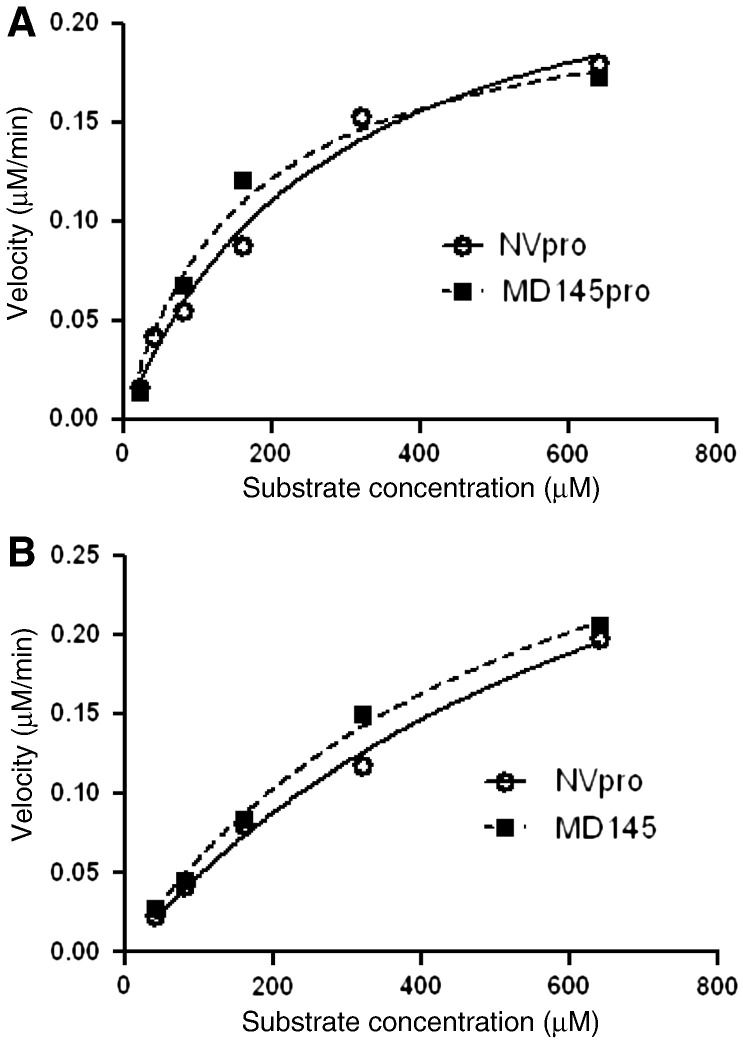
The NVpro and MD145pro exhibited enzymatic activity with similar cleavage efficiency on each substrate (kcat/km) (Fig. 2 , Table 1 ), suggesting that MD145pro is able to efficiently recognize and cleave the substrates derived from a cleavage site of GI norovirus. Both proteases showed higher cleavage efficiency towards the truncated substrate (DFHLQGP) as indicated by higher kcat/km value, compared to the substrate of 14 residues (EPDFHLQGPEDLAK) (Table 1). The mutant NV protease (C139A) did not increase fluorescence signal on addition of substrates, as expected (data not shown). The edans-DFHLQGP-dabcyl was used as a substrate for further experiments.
Fig. 2.
The enzyme kinetics of NVpro and MD145pro with fluorogenic substrates EPDFHLQGPEDLAK (A) and DFHLQGP (B). The reaction velocity as a function of the substrate concentration was plotted for each protease. NVpro and MD145pro are denoted by the solid line with open circles and the dotted line with filled rectangles, respectively.
Table 1.
Kinetic parameters for cleavage of the substrates by NVpro and MD145pro.
| Substrate | Protease | Kcat (min− 1) | Km (μM) | Kcat/Km (M− 1 min− 1) |
|---|---|---|---|---|
| EPDFHLQGPEDLAK | NV | 0.4376 ± 0.0655 | 790 ± 184.9 | 0.554 × 103 |
| MD145 | 0.3857 ± 0.03416 | 544.8 ± 84.52 | 0.708 × 103 | |
| DFHLQGP | NV | 0.2626 ± 0.02562 | 272.3 ± 58.19 | 1.111 × 103 |
| MD145 | 0.2201 ± 0.02443 | 159.8 ± 46.55 | 1.873 × 103 |
It is to note that the kinetic values are based on the concentration of the product (substrate) converted from the corrected RFU as described in the Materials and Methods section.
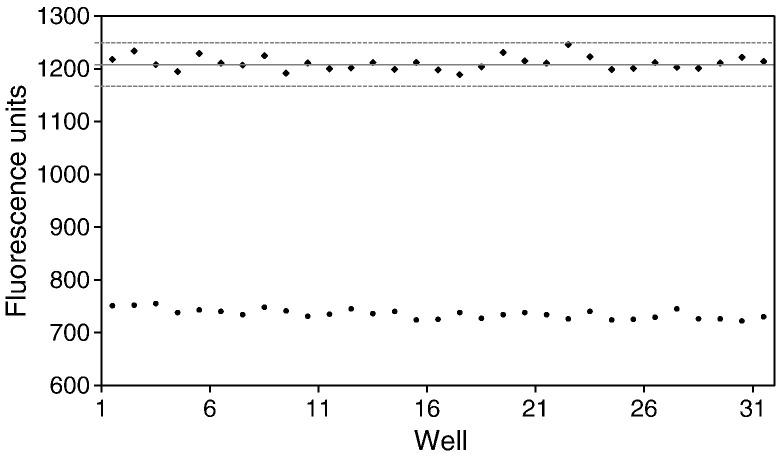
Z factor analysis
The Z factor is a measure of the distance between the standard deviation for the positive (fluorescence signal) and negative (background) controls of the assay, and is used to evaluate the overall quality of a high-throughput screening assay (Inglese et al., 2007, Zhang et al., 1999). A Z factor of 0.5 or greater is considered robust and reliable indicator for an assay system for screening purpose (Blanchard et al., 2004, Zhang et al., 1999). Fig. 3 is the scatter plot of fluorescence units from positive or negative control wells, showing assay range and the data variation. The mean Z factor for our FRET protease assay using NVpro and edans-DFHLQGP-dabcyl as a substrate was calculated as 0.86, demonstrating excellent signal-to-background ratio, and robustness and sensitivity of the assay.
Fig. 3.
Z factor analysis. The scatter plot represents positive control data (filled diamond data points) and negative control data (filled round data point). Solid lines represent mean fluorescence, and dashed lines represent three standard deviations above and below each set of positive control data points. Experiments are repeated more than three times with consistent results.
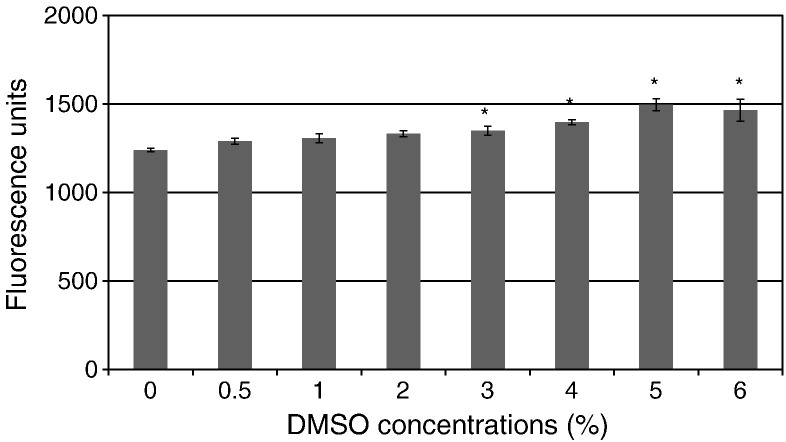
DMSO tolerance
The effect of DMSO on the FRET-assay with NVpro was determined and shown in Fig. 4 . DMSO concentrations of 0.5–2% (final concentration, v/v) did not significantly affect enzymatic activity, indicating that this assay is tolerant of DMSO up to 2%. All the experiments in this study contained less than 1.5% DMSO.
Fig. 4.
DMSO tolerance. Substrate was incubated with NVpro in the presence of 0–6% DMSO (v/v) in assay buffer at 37 °C. Fluorescence signals were measured at 1 h incubation using a plate reader. *Statistically significant (p < 0.05) compared to DMSO 0% by Student's t-test.
NVpro and MD145pro showed a similar inhibitor profile
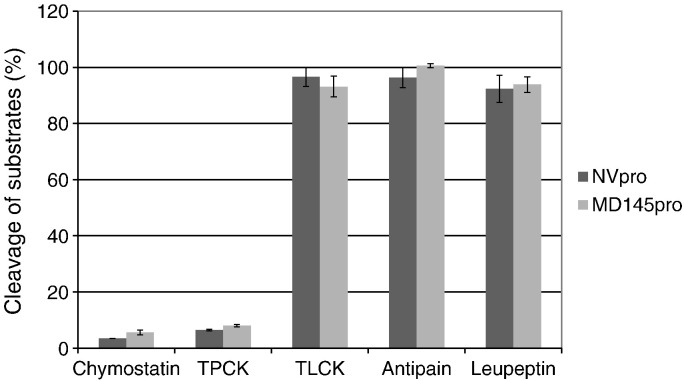
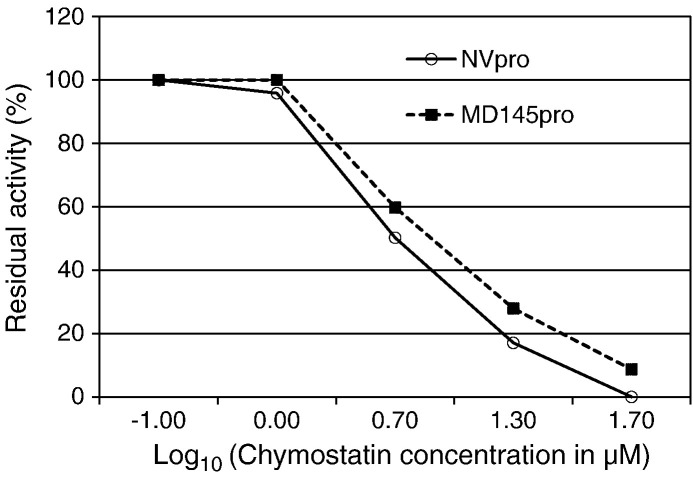
Four commercially available serine protease inhibitors (chymostatin, leupeptin, TPCK, and TLCK) and a papain-like cysteine protease inhibitor (antipain) were used to probe the inhibitor specificity of each protease. Chymostatin and TPCK at 50 μM markedly decreased fluorescence intensity from inhibition of substrate cleavage by NVpro and MD145pro (Fig. 5 ). However, TLCK, antipain and leupeptin failed to show significant inhibition of NVpro or MD145pro under the same condition. Then, chymostatin was used to determine the IC50 and inhibition constants Ki against both proteases. The dose response curves for chymostatin on NVpro and MD145pro are shown in Fig. 6A. Chymostatin inhibited NVpro and MD145pro with a similar efficiency indicated by comparable IC50 and inhibition constant Ki values (Table 2 ). These results indicate that proteases from two predominant norovirus genogroups share a similar inhibitor profile.
Fig. 5.
Inhibitor profiles. A range of commercially available protease inhibitors were tested for their activity against NVpro and MD145pro. Each bar represents the mean percentage of substrates (± SEM) cleaved by each protease in the absence (control) or presence of each inhibitor at 50 μM. NVpro and MD145pro are denoted by the dark and light bars, respectively.
Fig. 6.
Dose–responsive curves of chymostatin on NVpro and MD145pro. The dot represents percentage of substrates cleaved by NVpro in the presence of chymostatin at different concentrations, compared with control. NVpro and MD145pro are denoted by the solid line with open circles and the dotted line with filled rectangles, respectively.
Table 2.
IC50 and Ki of chymostatin for the inhibition of NVpro and MD145pro.
| Protease | Substrate | IC50 (μM) | Ki (M) |
|---|---|---|---|
| NV | DFHLQGP | 5.489 | 4.063 ± 0.6687 × 10− 6 |
| MD145 | DFHLQGP | 9.861 | 6.169 ± 0.7115 × 10− 6 |
Chemical shift mapping to study the interaction of NVpro and chymostatin
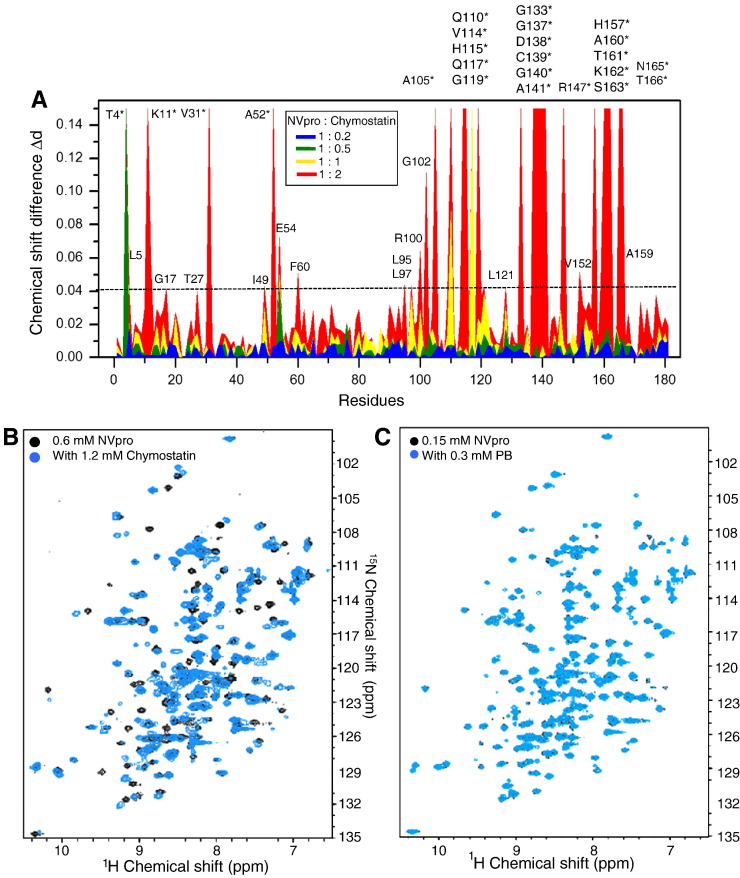
Since chymostatin significantly inhibited norovirus proteases in the FRET assay, the binding of chymostatin and NVpro was validated using NMR spectroscopy. A series of 1H–15N HSQC spectra of NVpro with increasing concentrations of chymostatin were analyzed to identify the residues of NVpro involved in the interaction with chymostatin. NMR chemical shift perturbations of backbone amide (1H and 15N) resonance in the 1H–15N HSQC spectra are sensitive probes for changes in chemical environments surrounding amino acid residues and/or in the relative populations of different conformations in protein structural ensemble (Shuker et al., 1996). The weighted chemical shift differences of 1H–15N resonances for NVpro in the presence of varying concentrations of chymostatin were compared to those observed in the absence of chymostatin (Fig. 7A). Significant chemical shift changes were observed during chymostatin titration in a concentration-dependent manner (Figs. 7A and B), indicating the specific interaction between NVpro and chymostatin. On the other hand, PB compound added as an unrelated control caused no significant chemical shift perturbation for NVpro (Fig. 7C). In the presence of 2-fold molar excess of chymostatin, the residues of NVpro that showed significantly shifts (Δd ≥ 0.04) were T4*, K11*, G17, T27, V31*, I49, A52*, E54, G60, L97, R100, G102, A105*, Q110*,V114*, H115*, Q117*, G119*, A141*, R147*, G133*, G137*, D138*, C139*, G140*, A141*, R147*, V152, H157*, A159, A160*, T161*, K162*, S163*, N165*, and T166*. The residues that could not be traced out due to the peak disappearance and/or line broadening are marked with asterisks and the largest positive values (Δd ≥ 0.15) in Fig. 7A. The line-broadening effects are likely indicative of chemical exchange on the slow time scale (μs–ms) due to the conformational dynamics of NVpro interacting with chymostatin.
Fig. 7.
Chymostatin specifically interacts with NVpro. A. Weighted chemical shift differences of the 1H and 15N resonances for NVpro when titrated with increasing amounts of chymostatin. Largest values (0.15) and asterisks are used to indicate the residues that cannot be assigned after adding 2-fold molar excess of chymostatin due to the peak disappearance or broadness. Weighted chemical shift difference is calculated by an equation, Δd = [1/2(dH2 + 1/25dN2)]1/2. B. Overlay of 1H–15N HSQC spectra of NVpro in the absence (black) and in the presence of 2-fold excess of chymostatin (blue). C. Comparison of 1H–15N HSQC spectra of NVpro in the absence and presence of 0.3 mM (2-fold molar excess) PB compound shows that PB titrated as a control causes generally no chemical shift perturbation on the spectra.
Docking model of NVpro with chymostatin
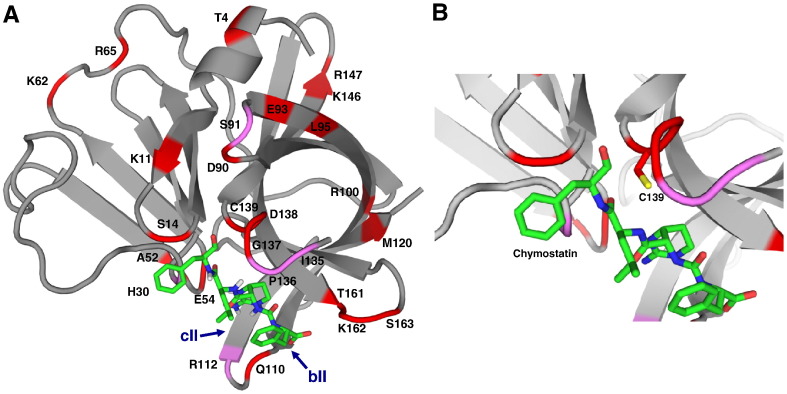
To corroborate the result obtained for NVpro-chymostatin interaction using NMR spectroscopy, we constructed a docking model for NVpro complexed with chymostatin using HADDOCK program (de Vries et al, Dominguez et al., 2003). During the docking, experimental information on the interface between NVpro and chymostatin derived from NMR chemical shift perturbation was required as ambiguous interaction restraints (AIRs) to drive the docking of two molecules. Based on the NVpro chemical shift perturbation observed in the presence of chymostatin and higher solvent accessibility, T4, K11, S14, A52, K62, R65, D90, E93, L95, R100, Q110, M120, K146, R147, T161, K162, and S163 are designated as active residues in AIRs. In addition, catalytically important residues E54, G137, and C139, which displayed significant perturbation but lower solvent accessibility were also selected as active residues. H30, S91, R112, I135, and P136 adjacent to those active residues were defined as passive residues. The positions of the active and passive residues for NVpro are indicated on the structure (Figs. 8A and B). As a result of the docking calculation, several different clusters with similar HADDOCK score were obtained. For all those clusters, it is indicated that chymostatin is positioned near the active and substrate binding sites of NVpro. The structural ensemble in the lowest energy cluster has RMSD value of 0.8 ± 0.2 Å over all backbone atoms. The representative structure of the lower energy cluster is presented in Fig. 8. The structure has favorable energy values, van der Waals energy of − 27.1 kcal mol− 1 and electrostatic energy of − 145.3 kcal mol− 1, but higher restraint violation energy of 169.9 kcal mol− 1. This higher violation is mostly due to our unbiased selection of active residues showing chemical shift perturbation. As clearly seen in Fig. 8, several residues do not have any direct interactions with chymostatin (T4, K11, K62, R65, S91, E93, L95, R100, M120, K146, and R147). It is highly probable that those residues distant from the binding site were indirectly affected and experienced a certain degree of conformational changes upon binding of chymostatin. The docking model depicted in Fig. 8 demonstrates that chymostatin bound to NVpro in a canonical substrate binding mode as also suggested by NMR data; NVpro-chymostatin interactions occur mainly in C-terminal domain and an arch formed by two β-sheets, βII and cII, covers the chymostatin.
Fig. 8.
HADDOCK docking model of NVpro complexed with chymostatin. The structure belonging to the lower energy cluster is presented. Chymostatin is shown as stick model (green). Active (red) and passive residues (pink) are labeled. (B) Close up view of chymostatin binding site.
Discussion
Noroviruses are the leading cause of food- or water-borne gastroenteritis outbreaks worldwide, affecting millions of people in the U.S. alone. Norovirus infection causes debilitating illness in people of all age groups at military bases, as well as in cruise ships, schools, and hospitals. Noroviruses require only a few viral particles to initiate infection and are highly contagious. Moreover, long-lasting immunity following natural infection rarely develops in people. Considering these challenges, antiviral drugs for controlling norovirus infections are in urgent need. Norovirus protease is known to be responsible for critical virus-mediated post-translational cleavages of the polypeptide encoding the viral proteins. Therefore, inhibition of the viral protease has the capacity to prevent viral replication. There are several viral protease inhibitors on the market or under development for pathogenic viruses such as human immunodeficiency virus, hepatitis C virus, and SARS coronavirus (Ghosh et al., 2007, Kazmierski et al., 2006, Tsantrizos, 2008, Walker et al., 2003). Robust and reliable assays for high throughput screening of potential norovirus protease inhibitors can greatly facilitate the development of antiviral drugs against the viral protease.
The genogroup I and II noroviruses are associated with majority of norovirus infections in humans. The amino acid homology of proteases among GI noroviruses is 90–94%, while the homology between GI and GII norovirus proteases is about 65–69%. However, the amino acid sequence analysis showed that the key residues of the catalytic site and residues forming the S1 subsite are conserved in GI and GII noroviruses (Ng and Parra, 2010). A co-transcription and translation assay study using MD145 ORF1 on the enzymatic activity of MD145 (GII) protease suggested that substrate specificity requirements are similar to those for NV (GI) based on western blots and radioactive microsequencing mapping (Belliot et al., 2003). However, there is little information on the comparative kinetic properties of proteases from GI and GII noroviruses. Therefore, we developed a FRET protease assay with high-throughput capability to use for screening of norovirus protease inhibitors, and study the substrate and inhibitor specificities of norovirus proteases of GI and GII noroviruses.
The conserved amino acid sequences of P7-P6′ at the N2/3 cleavage site of NV and MD145 noroviruses differ in P3 (H/E), P4 (F/Y) and P6 (P/G) residues, of which P4 position is reported to be important for maximal activity by earlier mapping studies of the cleavage sites recognized by calicivirus proteases (Hardy et al., 2002). In our study, MD145pro cleaved the substrates spanning P7–P7′ or P5–P2′ residues from the NS2/3 cleavage site of NV with an efficiency similar to NVpro, indicating that MD145pro recognized and cleaved the substrates with H and F in P3 and P4 positions, respectively, as efficiently as NVpro. In addition, NVpro and MD145pro cleaved the substrate with P5–P2′ residues more efficiently than substrate with P7–P7′ residues, suggesting that P3′–P7′ and P6–P7 residues are not necessary for substrate binding and cleavage by both proteases. These results are in line with an earlier report that p-nitroaniline peptide with P5–P1 residues was most efficiently cleaved by GI protease (Southampton strain) compared to those containing P6–P1 or P4–P1 residues, and the substrate of P3–P1 residues was not cleaved (Hussey et al., 2011). In addition, Chiba virus protease (GI)-substrate complex modeling based on crystal structure suggested that the substrate binding is stabilized by interactions of substrate residues P5–P2 to the corresponding binding sites of the protease (Nakamura et al., 2005).
We then evaluated the inhibitor specificities of NVpro and MD145pro using a set of commercially available standard protease inhibitors. Norovirus protease is structurally similar to the chymotrypsin superfamily based on X-ray crystallography (Nakamura et al., 2005, Zeitler et al., 2006). The standard protease inhibitors share a common mechanism of action, although they do not share the primary sequence for the three-dimensional structures (Bode and Huber, 1992, Laskowski and Kato, 1980). The screening of protease inhibitors in our assay revealed that the activities of both proteases were significantly inhibited by the presence of chymostatin and TPCK. Detailed kinetic studies of the inhibition of NVpro and MD145pro by chymostatin confirmed that both proteases have similar inhibition profiles, suggesting that protease inhibitors may be developed against multiple norovirus genogroups. In addition, the versatility of the assay allows incorporation of proteases from different genogroups so that screening of compounds against various norovirus genogroups can be performed. The quality and suitability of the FRET assay for high throughput screening were demonstrated by high Z factor of 0.86.
The interaction of chymostatin and NVpro was validated by the NMR studies. The analysis of NMR chemical shift mapping data revealed that the resonances of residues that are involved in substrate binding sites or active sites are greatly affected. Chymostatin that exhibited strong inhibitory effects against both proteases contains G–L–F moiety. Previous structural studies on the interaction of chymotrypsin and serine standard protease inhibitors showed that side chain of the P1 residue (F) of chymostatin fits into the specificity subsite of chymotrypsin, and the carbonyl oxygen extends toward the oxyanion hole formed by the G and S residues located in the active site of the enzyme. The P1–P3 residues of chymostatin also interact with the enzyme by forming a short, anti-parallel β-sheet interaction (Coombs et al., 1999). These interactions represent a substrate-like binding mode for enzymes. Our docking results using HADDOCK based on NMR chemical shift perturbation study indicate that chymostatin specifically interacts with NVpro in a canonical binding mode that can be seen in the protease-substrate/substrate-directed inhibitor complexes. At this stage we are not able to give a clear answer for whether chymostatin covalently modifies the nucleophilic thiol group and forms hemithioacetal adduct or forms non-covalent enzyme-inhibitor complex. However our results indicate that critical residues for the catalysis are targeted by chymostatin.
In conclusion, we demonstrate a development and application of a FRET norovirus protease (GI and GII) assay with high-throughput capability for screening potential antiviral compounds and monitoring enzymatic reactions, and the interaction of chymostatin and norovirus protease by NMR spectroscopy. These results provide valuable information for the study of antivirals and enzyme activities.
Materials and methods
The expression and purification of proteases from NV and MD145 viruses
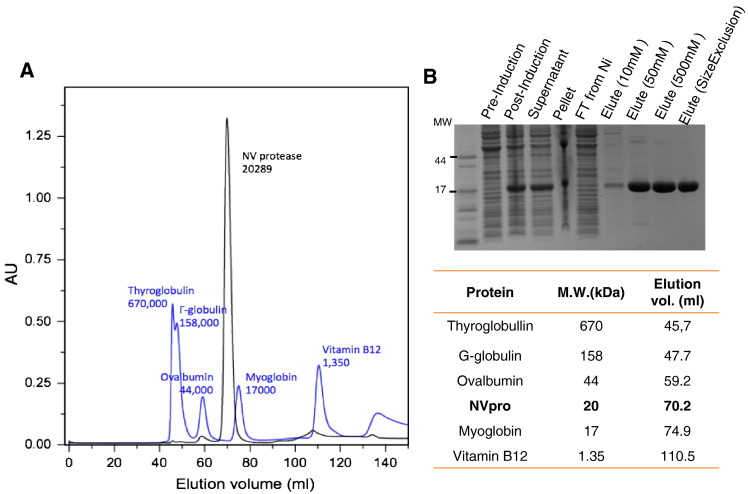
Full-lengths codon-optimized cDNAs corresponding to the complete amino acid sequence of NVpro and MD145pro were synthesized and cloned into the pET28a vector (GenScript, Piscataway, NJ). The synthesized cDNA sequences include start and stop codons as well as sequences encoding N-terminal six H for Ni column purification. The plasmid encoding NVpro or MD145pro was transformed into E. coli BL21 cells. Each protein was expressed in a regular Luria-Bertani broth by induction with 1 mM isopropyl β-d-thiogalactopyranoside (IPTG) for 4 h at 37 C° in a shaking incubator. The harvested cells were sonicated and ultracentifuged. Both proteases were soluble and the supernatants were applied to a Ni-NTA affinity column (QIAGEN, Valencia, CA) for purification. The mutant NVpro with C (nucleophile) to A substitution at position 139 (C139A) was generated by site-directed mutagenesis. All proteins were monomers with a size of approximately 20 kDa as determined by size exclusion chromatography (Fig. 9 ), which is in contrast to previous reports that norovirus proteases stably formed a dimer (NVpro) or were predominantly dimeric in solution (Southampton virus protease) (Hussey et al., 2011, Zeitler et al., 2006). Interestingly, our preliminary x-ray crystallography revealed NVpro as dimers, suggesting that the His-tag at the N-terminus does not interfere with dimerization (data not shown). These results indicate that the concentration and/or condition of the prepared protease may influence the monomer-dimer equilibrium of NVpro.
Fig. 9.
Purification of NVpro. A. His-tagged NVpro was purified by size exclusion chromatography. B. SDS-PAGE analysis of purified NVpro. The untagged version of the protease has a predicted molecular weight of 19 kDa. The (His) 6-tagged proteases are shifted to a slightly higher molecular weight.
For NMR studies, uniformly 15N-labeled NVpro was expressed in the E. coli BL21 cells grown in M9 minimal media supplemented with 1 g/L 15NH4Cl (Cambridge Isotope Laboratories, Andover, MA) and purified as previously described (Takahashi et al., 2011). Briefly, the cells containing the expression plasmid encoding NVpro were grown in a starter culture consisting of 50 ml Luria Bertani media at 37 °C for 6–8 h. The cells were then grown in M9 minimal media to an OD600 value of 1.0 and induced with 1.0 mM IPTG for 5 h. The cells were harvested by centrifugation, subsequently resuspended in buffer containing 50 mM sodium phosphate (pH 8.0), 300 mM sodium chloride, and 10 mM imidazole, and sonicated at 4 °C. After centrifugation of the cell lysates, NVpro was purified from the cell lysate supernatant using a Ni-NTA affinity column (Qiagen, Valencia, CA). Size exclusion chromatography on a Superdex 75 prep grade (GE healthcare, Amersham, UK) was applied as a final purification step. Gel-filtration standard (Biorad, Hercules, CA) containing thyroglobulin (670 kDa), γ-globulin (158 kDa), ovalbumin (43 kDa), and myoglobin (17 kDa) was used to analyze the molecular weight of the eluted proteins. NMR samples contained uniformly 15N-labeled NVpro (0.1–0.6 mM), 50 mM sodium phosphate (pH 6.5), 100 mM NaCl, 5 mM DTT, and 3 mM NaN3 in 90% H2O/10% D2O unless otherwise noted.
FRET assay with proteases from Norwalk and MD145 noroviruses
Substrates
Two fluorogenic substrates, edans-EPDFHLQGPEDLAK-dabcyl (Zeitler et al., 2006) and edans-DFHLQGP-dabcyl derived from the P7–P7′ and P5-P2′ residues on the NS2/3 cleavage site in ORF1 of NV, respectively, were synthesized (GenScript, Piscataway, NJ). The designation of substrate residues for P1 and P1′ starts at the scissile bond and counts toward the N- or C-terminus, respectively, as suggested by Schechter and Berger (1967).
FRET protease assays
Stock solutions (10 mM) of the substrates were prepared in DMSO, and diluted in assay buffer (50 mM HEPES buffer [pH 8.0] containing 50 mM NaCl, 0.4 mM EDTA, 60% Glycerol, and 6 mM DTT). The 25 μl of protease (0.5 μM, final concentration) was mixed with 25 μl of serially diluted substrates in assay buffer in a 96-well black plate (Nalge Nunc International, Rochester, NY). The fluorescence signals were detected using an excitation wavelength of 360 nm and an emission wavelength of 460 nm on a fluorescence microplate reader (FLx800, Biotek, Winooski, VT) every 5 min for 30 min at 37 °C.
Correction factor for the inner filter effects
The background (substrate control well without protease)-subtracted relative fluorescence units (RFU) were corrected for inner filter effects. The inner filter effects are produced at higher substrate concentrations due to the absorption of fluorescence by the quencher on the adjacent substrates, and cause attenuation of fluorescence intensity, abolishing the linear correlation between the cleaved substrate concentration and the fluorescence intensity (Cuerrier et al., 2005, Jaulent et al., 2007). Briefly, serially diluted edans reference standards (free edans) were added to wells in a 96-well plate with or without equal concentration of substrate, and fluorescence readings were obtained on a microplate reader. The correction factors to compensate for the inner filter effects were determined empirically as described previously (Cuerrier et al., 2005, Jaulent et al., 2007).
Determination of kcat/km
The corrected RFU was converted to the concentration of the product of protease reaction by plotting edans fluorescence reference standard as RFU versus concentration. The initial velocities (Vo), rate of enzyme reaction, were calculated using the first 30 min of the progress curve, and plotted against substrate concentrations (Michaelis–Menten plot) using GraphPad Prism program (GraphPad Software, San Diego, CA) to determine the ratio of kcat/km, which is the measure of efficiency of the catalytic reaction (specificity constant).
Determination of Z factor
The substrate edans-DFHLQGP-dabcyl (16 μM, final concentration) was added to wells containing NVpro at 0.5 μM of final concentration (positive control wells) or assay buffer (negative control wells) in a 96-well black plate. Since Z factor is dependent on means and standard deviations, each combination had more than 30 wells. The plate was incubated at 37 °C for up to 1 h and the fluorescence readings were obtained. The means and standard deviations of fluorescence were calculated for negative and positive wells to determine Z factor as previously described (Blanchard et al., 2004, Inglese et al., 2007).
Where, σp = standard deviation of positive controls, σn = standard deviation of negative controls, μp = means of positive controls, μn = means of negative controls.
DMSO tolerance
To investigate the tolerance of the assay toward DMSO, a solvent typically used to dissolve test compounds, 50 μl reactions containing 0.5 μM NVpro and 16 μM fluorogenic substrate (edans-DFHLQGP-dabcyl) in assay buffer were supplemented with DMSO. The DMSO concentrations varied between 0% and 6% (v/v) at otherwise constant conditions. Fluorescence was measured following incubation at 37 °C for 60 min. All reactions were performed in triplicate to determine the effect of DMSO on enzyme activity.
FRET protease assay with protease inhibitors
Pilot screening of protease inhibitors
Commercially available standard protease inhibitors including serine protease inhibitors (chymostatin, leupeptin, N-tosyl-l-phenylalanine chloromethyl ketone [TPCK], and tosyl-l-lysine chloromethyl ketone [TLCK]) and a papain-like cysteine protease inhibitor (antipain) were obtained from Sigma-Aldrich (St Louis, MO). Stock solutions (10 mM) of inhibitors were prepared in DMSO, and further diluted in assay buffer. The final concentrations of DMSO in the assay did not exceed 1.5% (vol/vol). NVpro and MD145pro at a final concentration of 0.5 μM and the substrate (edans-DFHLQGP-dabcyl) at 16 μM that give a strong signal and minimal inner filter effects were used for the studies. Inhibitors at a final concentration of 50 μM were pre-incubated with NVpro or MD145pro in 25 μl for 30 min at 37 °C, and the same volume of substrate was added to a 96-well black plate, followed by incubation. The mixtures were incubated at 37 °C, and fluorescence readings were obtained on a microplate reader after 60 min of incubation. The substrate concentration used for pilot screening was well below the km, but substrate depletion was not observed in the control well (without an inhibitor) during 60 min incubation time. The reduction of cleaved products by each preparation was calculated by the comparison to that of control without any inhibitor.
Determination of IC50 and Ki of chymostatin
Continuous monitoring of activity kinetics of NVpro and MD145pro was performed to evaluate the detailed efficacy of chymostatin. Proteases (0.5 μM, final concentration) were mixed with serially diluted chymostatin (0–50 μM) in 25 μl of assay buffer and incubated at 37 °C for 30 min, followed by the addition of various concentrations of substrates (30–100 μM). The mixtures were incubated at 37 °C, and fluorescence readings were obtained on a microplate reader at every 5 min for up to 30 min. The RFUs were corrected for inner filter effects and converted to the concentration of the product of protease reaction. The dissociation constant for inhibitor binding (Ki) values was determined by nonlinear regression analysis using GraphPad Prism software. The dose-dependent FRET inhibition curves were fitted with variable slope (four parameters) using GraphPad Prism software in order to determine the concentration of chymostatin that reduces enzyme velocity by half (IC50). The IC50 values were determined in the presence of substrate at 30 μM at 60 min.
Solution NMR spectroscopy of NVpro
All NMR experiments were performed at 25 °C on a Varian VNMR 500 MHz spectrometer equipped with a 5 mm triple resonance inverse detection cryogenic NMR probe. All NMR spectra were processed using NMRPipe (Delaglio et al., 1995), and analyzed with Sparky (Goddard and Kneller, 2006) and CARA software (http://www.nmr.ch) (Keller, 2004). Inhibitor titration experiments were carried out by direct addition of 10 mM stock solution of chymostatin dissolved in DMSO-d6 (Cambridge Isotope Laboratories, Andover, MA) into uniformly 15N-labeled NVpro solution. 2D 1H–15N HSQC spectra of NVpro were monitored during addition of 0.2, 0.5, 1 and 2 M equivalents of chymostatin. Complex points 1024 × 128 and spectral widths of 14 × 38 ppm were used to record all the spectra. Recently we have reported a total of 94% of the backbone 1H and 15N resonance assignments of free NVpro (Takahashi et al., 2011). The assignment of the HSQC spectra in the presence of chymostatin was used to investigate the interaction with the inhibitor by following the peak shifts during titration. Since the use of DMSO-d6 as a co-solvent caused a slight but traceable resonance peak shifts in dose-dependent manner, the titrations were repeated with DMSO-d6 alone. Each spectrum with varying amount of chymostatin was compared with that of DMSO-d6 at a corresponding titration point. As a control, similar experiments were repeated using an unrelated compound dissolved in DMSO-d6 that does not inhibit NVpro activity (PB compound). Chemical shift changes were characterized using a weighted chemical shift difference (Δδ − [1/2{δH 2 + 1/25δN 2}]1/2) (Grzesiek et al., 1996).
Docking calculations
The docking models of NVpro in complex with chymostatin were built using the HADDOCK (high-ambiguity driven biomolecular docking) program (de Vries et al, Dominguez et al., 2003), in which the experimental information on residues involved in intermolecular interaction can be used as ambiguous interaction restraints (AIRs) to drive the docking. Protein residues directly involved in intermolecular interaction are defined as “active residues”, while neighboring residues of active residues as “passive residues”. HADDOCK docking protocols consist of three-stage procedure, a rigid-body energy minimization, a semi-flexible refinement in torsion angle space and a final refinement in explicit solvent. Crystal structure of NVpro (PDB code: 2FYQ) (Zeitler et al., 2006) was used as the starting structure. NVpro residues showing peak disappearance or chemical shift perturbation greater than the average shift value, 0.019 ppm, in 1H–15N HSQC spectrum of NVpro with chymostatin and possessing higher solvent accessibility were selected as active residues. Solvent accessibility was calculated using GETAREA (Robert Fraczkiewicz, 1998), where residues are defined as solvent exposed when the ratio of side-chain surface area to its random coil value exceeds 50%. All solvent accessible surface neighbors of active residues were then defined as passive residues in the docking calculation. Whole residues of the small ligand, chymostatin, were selected as active residues. Initially, a total of 1000 structures were calculated at the rigid body minimization stage. From this calculation, the 200 lowest-energy structures were then selected for the subsequent semi-flexible simulated annealing and the refinement in explicit solvent. The final structures were clustered using the backbone root-mean-square deviation (RMSD) with a cutoff of 7.5 Å. All the structures were drawn with the PyMol (DeLano).
Statistics
The student t-test was used to compare the significance of the unpaired sample means. P values < 0.05 were considered significant.
Acknowledgment
We would like to thank David George and Samira Najm for technical assistant. This work was supported by NIH grant, U01 AI081891, 1S10RR025441, RR17686, RR17708 and Johnson Center for Basic Cancer Research, and Targeted Excellence Programs of Kansas State University.
References
- Atmar R.L., Estes M.K. The epidemiologic and clinical importance of norovirus infection. Gastroenterol. Clin. North Am. 2006;35(2):275–290. doi: 10.1016/j.gtc.2006.03.001. (viii) [DOI] [PubMed] [Google Scholar]
- Belliot G., Sosnovtsev S.V., Mitra T., Hammer C., Garfield M., Green K.Y. In vitro proteolytic processing of the MD145 norovirus ORF1 nonstructural polyprotein yields stable precursors and products similar to those detected in calicivirus-infected cells. J. Virol. 2003;77(20):10957–10974. doi: 10.1128/JVI.77.20.10957-10974.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard J.E., Elowe N.H., Huitema C., Fortin P.D., Cechetto J.D., Eltis L.D., Brown E.D. High-throughput screening identifies inhibitors of the SARS coronavirus main proteinase. Chem. Biol. 2004;11(10):1445–1453. doi: 10.1016/j.chembiol.2004.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode W., Huber R. Natural protein proteinase inhibitors and their interaction with proteinases. Eur. J. Biochem. 1992;204(2):433–451. doi: 10.1111/j.1432-1033.1992.tb16654.x. [DOI] [PubMed] [Google Scholar]
- C.D.C. 2010. Morbidity and Mortality Weekly Report (MMWR): Surveillance for Foodborne Disease Outbreaks — United States, 2007. [PubMed] [Google Scholar]
- Chen S., Chen L.L., Luo H.B., Sun T., Chen J., Ye F., Cai J.H., Shen J.K., Shen X., Jiang H.L. Enzymatic activity characterization of SARS coronavirus 3C-like protease by fluorescence resonance energy transfer technique. Acta Pharmacol. Sin. 2005;26(1):99–106. doi: 10.1111/j.1745-7254.2005.00010.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombs G.S., Rao M.S., Olson A.J., Dawson P.E., Madison E.L. Revisiting catalysis by chymotrypsin family serine proteases using peptide substrates and inhibitors with unnatural main chains. J. Biol. Chem. 1999;274(34):24074–24079. doi: 10.1074/jbc.274.34.24074. [DOI] [PubMed] [Google Scholar]
- Cuerrier D., Moldoveanu T., Davies P.L. Determination of peptide substrate specificity for mu-calpain by a peptide library-based approach: the importance of primed side interactions. J. Biol. Chem. 2005;280(49):40632–40641. doi: 10.1074/jbc.M506870200. [DOI] [PubMed] [Google Scholar]
- de Vries, S.J., van Dijk, M., Bonvin, A.M., 2010. The HADDOCK web server for data-driven biomolecular docking. Nat. Protoc. 5 (5), 883–897. [DOI] [PubMed]
- Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6(3):277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- DeLano, W.L., 2010. The PyMOL Molecular Graphics System. DeLano Scientific LLC, San Carlos, CA.
- Dolin R. Noroviruses—challenges to control. N. Engl. J. Med. 2007;357(11):1072–1073. doi: 10.1056/NEJMp078050. [DOI] [PubMed] [Google Scholar]
- Dominguez C., Boelens R., Bonvin A.M. HADDOCK: a protein–protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 2003;125(7):1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- Fankhauser R.L., Noel J.S., Monroe S.S., Ando T., Glass R.I. Molecular epidemiology of “Norwalk-like viruses” in outbreaks of gastroenteritis in the United States. J. Infect. Dis. 1998;178(6):1571–1578. doi: 10.1086/314525. [DOI] [PubMed] [Google Scholar]
- Ghosh A.K., Xi K., Grum-Tokars V., Xu X., Ratia K., Fu W., Houser K.V., Baker S.C., Johnson M.E., Mesecar A.D. Structure-based design, synthesis, and biological evaluation of peptidomimetic SARS-CoV 3CLpro inhibitors. Bioorg. Med. Chem. Lett. 2007;17(21):5876–5880. doi: 10.1016/j.bmcl.2007.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass R.I., Parashar U.D., Estes M.K. Norovirus gastroenteritis. N. Engl. J. Med. 2009;361(18):1776–1785. doi: 10.1056/NEJMra0804575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard T.D., Kneller K.D. University of California; San Francisco: 2006. SPARKY 3.113. [Google Scholar]
- Green K.Y. In: Fields Virology. 5th ed. Knipe D.M., Howley P.M., editors. Lippincott Williams & Wilkins; Philadelphia: 2007. Caliciviruses: the noroviruses; p. 1. [Google Scholar]
- Green K.Y., Chanock R.M., Kapikian A.Z. In: 4 ed. Knipe D.M., Howley P.M., editors. Vol. 1. Lippincott Williams & Wilkins; Philadelphia: 2001. Human caliciviruses; pp. 841–874. (Fields Virology). [Google Scholar]
- Grzesiek S., Bax A., Clore G.M., Gronenborn A.M., Hu J.S., Kaufman J., Palmer I., Stahl S.J., Wingfield P.T. The solution structure of HIV-1 Nef reveals an unexpected fold and permits delineation of the binding surface for the SH3 domain of Hck tyrosine protein kinase. Nat. Struct. Biol. 1996;3(4):340–345. doi: 10.1038/nsb0496-340. [DOI] [PubMed] [Google Scholar]
- Hardy M.E., Crone T.J., Brower J.E., Ettayebi K. Substrate specificity of the Norwalk virus 3C-like proteinase. Virus Res. 2002;89(1):29–39. doi: 10.1016/s0168-1702(02)00114-4. [DOI] [PubMed] [Google Scholar]
- Hussey R.J., Coates L., Gill R.S., Erskine P.T., Coker S.F., Mitchell E., Cooper J.B., Wood S., Broadbridge R., Clarke I.N., Lambden P.R., Shoolingin-Jordan P.M. A structural study of norovirus 3C protease specificity: binding of a designed active site-directed peptide inhibitor. Biochemistry. 2011;50:240–249. doi: 10.1021/bi1008497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglese J., Johnson R.L., Simeonov A., Xia M., Zheng W., Austin C.P., Auld D.S. High-throughput screening assays for the identification of chemical probes. Nat. Chem. Biol. 2007;3(8):466–479. doi: 10.1038/nchembio.2007.17. [DOI] [PubMed] [Google Scholar]
- Jaulent A.M., Fahy A.S., Knox S.R., Birtley J.R., Roque-Rosell N., Curry S., Leatherbarrow R.J. A continuous assay for foot-and-mouth disease virus 3C protease activity. Anal. Biochem. 2007;368(2):130–137. doi: 10.1016/j.ab.2007.05.026. [DOI] [PubMed] [Google Scholar]
- Ng K., Parra F. In: Caliciviruses: Molecular and Cellular Virology. Hansman G.S., Jiang X.J., Green K.Y., editors. Caister Academic Press; Norfolk: 2010. Calicivirus protein structures; pp. 95–110. [Google Scholar]
- Kapikian A.Z., Wyatt R.G., Dolin R., Thornhill T.S., Kalica A.R., Chanock R.M. Visualization by immune electron microscopy of a 27-nm particle associated with acute infectious nonbacterial gastroenteritis. J. Virol. 1972;10(5):1075–1081. doi: 10.1128/jvi.10.5.1075-1081.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazmierski W.M., Kenakin T.P., Gudmundsson K.S. Peptide, peptidomimetic and small-molecule drug discovery targeting HIV-1 host-cell attachment and entry through gp120, gp41, CCR5 and CXCR4. Chem. Biol. Drug Des. 2006;67(1):13–26. doi: 10.1111/j.1747-0285.2005.00319.x. [DOI] [PubMed] [Google Scholar]
- Keller R. Verlag Goldau; Switzerland: 2004. The Computer Aided Resonance Assignment Tutorial. [Google Scholar]
- Kroneman A., Harris J., Vennema H., Duizer E., van Duynhoven Y., Gray J., Iturriza M., Bottiger B., Falkenhorst G., Johnsen C., von Bonsdorff C.H., Maunula L., Kuusi M., Pothier P., Gallay A., Schreier E., Koch J., Szucs G., Reuter G., Krisztalovics K., Lynch M., McKeown P., Foley B., Coughlan S., Ruggeri F.M., Di Bartolo I., Vainio K., Isakbaeva E., Poljsak-Prijatelj M., Grom A.H., Bosch A., Buesa J., Fauquier A.S., Hernandez-Pezzi G., Hedlund K.O., Koopmans M. Data quality of 5 years of central norovirus outbreak reporting in the European Network for food-borne viruses. J. Public Health (Oxf) 2008;30(1):82–90. doi: 10.1093/pubmed/fdm080. [DOI] [PubMed] [Google Scholar]
- Laskowski M., Jr., Kato I. Protein inhibitors of proteinases. Annu. Rev. Biochem. 1980;49:593–626. doi: 10.1146/annurev.bi.49.070180.003113. [DOI] [PubMed] [Google Scholar]
- Liu B.L., Viljoen G.J., Clarke I.N., Lambden P.R. Identification of further proteolytic cleavage sites in the Southampton calicivirus polyprotein by expression of the viral protease in E. coli. J. Gen. Virol. 1999;80(Pt. 2):291–296. doi: 10.1099/0022-1317-80-2-291. [DOI] [PubMed] [Google Scholar]
- Nakamura K., Someya Y., Kumasaka T., Ueno G., Yamamoto M., Sato T., Takeda N., Miyamura T., Tanaka N. A norovirus protease structure provides insights into active and substrate binding site integrity. J. Virol. 2005;79(21):13685–13693. doi: 10.1128/JVI.79.21.13685-13693.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIAID Category A, B, and C Priority Pathogens. http://www.niaid.nih.gov/topics/biodefenserelated/biodefense/research/pages/cata.aspx In.
- Nilsson M., Hedlund K.O., Thorhagen M., Larson G., Johansen K., Ekspong A., Svensson L. Evolution of human calicivirus RNA in vivo: accumulation of mutations in the protruding P2 domain of the capsid leads to structural changes and possibly a new phenotype. J. Virol. 2003;77(24):13117–13124. doi: 10.1128/JVI.77.24.13117-13124.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel M.M., Hall A.J., Vinje J., Parashara U.D. Noroviruses: a comprehensive review. J. Clin. Virol. 2009;44(1):1–8. doi: 10.1016/j.jcv.2008.10.009. [DOI] [PubMed] [Google Scholar]
- Robert Fraczkiewicz W.B. Exact and efficient analytical calculation of the accessible surface areas and their gradients for macromolecules. J. Comput. Chem. 1998;19(3):319–333. [Google Scholar]
- Rockx B., De Wit M., Vennema H., Vinje J., De Bruin E., Van Duynhoven Y., Koopmans M. Natural history of human calicivirus infection: a prospective cohort study. Clin. Infect. Dis. 2002;35(3):246–253. doi: 10.1086/341408. [DOI] [PubMed] [Google Scholar]
- Sakai Y., Nakata S., Honma S., Tatsumi M., Numata-Kinoshita K., Chiba S. Clinical severity of Norwalk virus and Sapporo virus gastroenteritis in children in Hokkaido, Japan. Pediatr. Infect. Dis. J. 2001;20(9):849–853. doi: 10.1097/00006454-200109000-00005. [DOI] [PubMed] [Google Scholar]
- Schechter I., Berger A. On the size of the active site in proteases. I. Papain. Biochem. Biophys. Res. Commun. 1967;27(2):157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- Scheffler U., Rudolph W., Gebhardt J., Rohayem J. Differential cleavage of the norovirus polyprotein precursor by two active forms of the viral protease. J. Gen. Virol. 2007;88(Pt 7):2013–2018. doi: 10.1099/vir.0.82797-0. [DOI] [PubMed] [Google Scholar]
- Seah E.L., Marshall J.A., Wright P.J. Open reading frame 1 of the Norwalk-like virus Camberwell: completion of sequence and expression in mammalian cells. J. Virol. 1999;73(12):10531–10535. doi: 10.1128/jvi.73.12.10531-10535.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuker S.B., Hajduk P.J., Meadows R.P., Fesik S.W. Discovering high-affinity ligands for proteins: SAR by NMR. Science. 1996;274(5292):1531–1534. doi: 10.1126/science.274.5292.1531. [DOI] [PubMed] [Google Scholar]
- Siebenga J.J., Vennema H., Zheng D.P., Vinje J., Lee B.E., Pang X.L., Ho E.C., Lim W., Choudekar A., Broor S., Halperin T., Rasool N.B., Hewitt J., Greening G.E., Jin M., Duan Z.J., Lucero Y., O'Ryan M., Hoehne M., Schreier E., Ratcliff R.M., White P.A., Iritani N., Reuter G., Koopmans M. Norovirus illness is a global problem: emergence and spread of norovirus GII.4 variants, 2001–2007. J. Infect. Dis. 2009;200(5):802–812. doi: 10.1086/605127. [DOI] [PubMed] [Google Scholar]
- Siebenga J.J., Duizer E., Koopmans M.P.G. In: Caliciviruses. Hansman G.S., Jians X.J., Green K.Y., editors. Caister Academic Press; Norfolk: 2010. Norovirus epidemiology; pp. 1–24. [Google Scholar]
- Someya Y., Takeda N. Insights into the enzyme–substrate interaction in the norovirus 3C-like protease. J. Biochem. 2009;146(4):509–521. doi: 10.1093/jb/mvp094. [DOI] [PubMed] [Google Scholar]
- Someya Y., Takeda N., Wakita T. Saturation mutagenesis reveals that GLU54 of norovirus 3C-like protease is not essential for the proteolytic activity. J. Biochem. 2008;144(6):771–780. doi: 10.1093/jb/mvn130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosnovtsev S.V., Belliot G., Chang K.O., Prikhodko V.G., Thackray L.B., Wobus C.E., Karst S.M., Virgin H.W., Green K.Y. Cleavage map and proteolytic processing of the murine norovirus nonstructural polyprotein in infected cells. J. Virol. 2006;80(16):7816–7831. doi: 10.1128/JVI.00532-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi D., Kim Y., Chang K., Anbanandam A., Prakash O. Backbone and side-chain assignments of Norwalk virus protease. Biomol. NMR Assign. 2011 doi: 10.1007/s12104-011-9316-3. Jun 8. doi: 10.1007/s12104-011-9316-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsantrizos Y.S. Peptidomimetic therapeutic agents targeting the protease enzyme of the human immunodeficiency virus and hepatitis C virus. Acc. Chem. Res. 2008;41(10):1252–1263. doi: 10.1021/ar8000519. [DOI] [PubMed] [Google Scholar]
- Walker M.P., Appleby T.C., Zhong W., Lau J.Y., Hong Z. Hepatitis C virus therapies: current treatments, targets and future perspectives. Antivir. Chem. Chemother. 2003;14(1):1–21. doi: 10.1177/095632020301400101. [DOI] [PubMed] [Google Scholar]
- Zeitler C.E., Estes M.K., Venkataram Prasad B.V. X-ray crystallographic structure of the Norwalk virus protease at 1.5-A resolution. J. Virol. 2006;80(10):5050–5058. doi: 10.1128/JVI.80.10.5050-5058.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.H., Chung T.D., Oldenburg K.R. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 1999;4(2):67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- Zheng D.P., Widdowson M.A., Glass R.I., Vinje J. Molecular epidemiology of genogroup II-genotype 4 noroviruses in the United States between 1994 and 2006. J. Clin. Microbiol. 2010;48(1):168–177. doi: 10.1128/JCM.01622-09. [DOI] [PMC free article] [PubMed] [Google Scholar]