Abstract
Highly compacted DNA nanoparticles, composed of single molecules of plasmid DNA compacted with block copolymers of polyethylene glycol and poly-L-lysine (PEG-CK30), have shown considerable promise in human gene therapy clinical trials in the nares, but may be less capable of transfecting cells that lack surface nucleolin. To address this potential shortcoming, we formulated pH-responsive DNA nanoparticles that mediate gene transfer via a nucleolin-independent pathway. Poly-L-histidine was inserted between PEG and poly-L-lysine to form a triblock copolymer system, PEG-CH12K18. Inclusion of poly-L-histidine increased the buffering capacity of PEG-CH12K18 to levels comparable with branched polyethyleneimine. PEG-CH12K18 compacted DNA into rod-shaped DNA nanoparticles with similar morphology and colloidal stability as PEG-CK30 DNA nanoparticles. PEG-CH12K18 DNA nanoparticles entered human bronchial epithelial cells (BEAS-2B) that lack surface nucleolin by a clathrin-dependent endocytic mechanism followed by endo-lysosomal processing. Despite trafficking through the degradative endo-lysosomal pathway, PEG-CH12K18 DNA nanoparticles improved the in vitro gene transfer by ~ 20-fold over PEG-CK30 DNA nanoparticles, and in vivo gene transfer to lung airways in BALB/c mice by ~ 3-fold, while maintaining a favorable toxicity profile. These results represent an important step toward the rational development of an efficient gene delivery platform for the lungs based on highly compacted DNA nanoparticles.
1. Introduction
A number of cellular barriers limit efficient delivery of therapeutic genes to the nucleus of lung epithelial cells [1–2], including poor cellular uptake across the apical membrane [3], unproductive intracellular trafficking [4–5], and inefficient nuclear import [6]. A particularly promising platform that overcomes many of these barriers is the highly compacted DNA nanoparticles, composed of single molecules of plasmid DNA compacted with block copolymers of polyethylene glycol and poly-L-lysine linked by a cysteine residue (PEG-CK30) [6–7]. Preclinical studies in mice demonstrated that these DNA nanoparticles mediate effective gene delivery to the brain, eyes and lungs with minimal toxicity and immunogenicity [7–11]. Liu et al. reported that only PEG-CK30 DNA nanoparticles having a minor diameter less than 25 nm, the approximate maximum inner diameter of the nuclear pore complex (NPC), mediated gene transfer following intra-cytoplasmic injection [6]. Recently, Chen et al. reported that nucleolin is a potential cell surface receptor for PEG-CK30 DNA nanoparticles, and appears to be involved in their intracellular trafficking within a non-degradative pathway that leads to nuclear import via the NPC [12–13]. However, nucleolin is not ubiquitously expressed on cell surfaces. In the mouse airways, the level of immunohistochemical staining for surface nucleolin varied among individual cells with a pattern resembling the patchy transgene expression by PEG-CK30 DNA nanoparticles [7, 13]. In addition, DNA nanoparticles generally fail to transfect in vitro cell lines that lack significant cell surface nucleolin (Copernicus Therapeutics, Inc., unpublished data). Thus, depending on the cell type and/or target tissue, highly compacted PEG-CK30 DNA nanoparticles will likely enter cells via other pathways, such as clathrin-mediated endocytosis (CME), which subjects the DNA nanoparticles to endo-lysosomal trafficking. Endo-lysosomal trafficking represents a major obstacle to non-viral gene therapy [14], where the cargo DNA may be degraded in the acidic and enzyme-rich late endosomes and lysosomes before reaching the nucleus. Furthermore, gene carriers sequestered in late endosomes and lysosomes are unable to enter the nucleus via the NPC.
A limitation of poly-L-lysine based gene carrier systems, including PEG-CK30 DNA nanoparticles, is that they lack an efficient mechanism for endosomal escape [15]. A number of endosome escape mechanisms have been incorporated into non-viral gene carrier systems to enhance gene transfer. A common strategy is to incorporate functional groups with buffering capacity between pH 7.4 – 5.1, which presumably mediates endo-lysosomal escape [16–17] via the proton sponge effect [18]. For example, due to the presence of secondary and tertiary amines, PEI- or PAMAM-based gene carriers have been shown to facilitate endosome escape by the proton sponge effect [19]. However, their clinical use is limited by toxicity induced by their highly cationic surface charge.
We hypothesized that incorporating pH-responsive poly-L-histidine into highly compacted PEG-CK30 based DNA nanoparticles would enhance their gene transfer efficiency, while retaining their unique physicochemical properties. To study the effect of pH-buffering on DNA nanoparticle mediated gene transfer, we inserted a poly-L-histidine segment between PEG and poly-L-lysine to form a triblock copolymer system. The triblock copolymer design ensures that a neutral surface charge is maintained by the terminal PEG chains, and that poly-L-histidine is localized in an intermediate layer, where it is available to buffer pH without interfering with DNA compaction by poly-L-lysine. Based on previous findings that polymers containing 40 – 50 % imidazole groups, the side chain of histidine, showed the highest in vitro transfection efficiencies [20–21], we synthesized CH12K18 by substituting histidine for 12 of the N-terminal lysines in CK30 (40% substitution). This degree of substitution ensured that a balance between DNA binding and pH buffering capacity was maintained, as a minimum of 15 lysines (K15) are required to ensure efficient compaction of DNA (Copernicus Therapeutics, Inc., data not shown). Here, we report the formulation and characterization of PEG-CH12K18 DNA nanoparticles containing a pH-responsive poly-L-histidine segment, including DNA nanoparticle morphology, colloidal stability, cellular uptake, intracellular trafficking, and in vitro and in vivo gene transfer efficiencies.
2. Materials & Methods
2.1. Reagents
All chemicals were purchased from Sigma-Aldrich (St. Louis, MO) and used as received, unless stated otherwise. The peptides CH12K18 and CK30 were synthesized by Fmoc-mediated solid-phase peptide synthesis using an automated peptide synthesizer (Symphony Quartet, Protein Technologies, Tucson, AZ). CH12K18 and CK30 were purified by HPLC and identity confirmed by mass spectrometry. Thiol reactive PEG, Methoxy-PEG-maleimide, MW 5 kDa, was purchased from Rapp Polymere (Tübingen, Germany). Lab-Tek glass-bottom tissue culture plates were from ThermoFisher Scientific (Rochester, NY). Alexa Fluor 555-labeled Cholera Toxin subunit B (CTB), tetramethylrhodamine-labeled Transferrin (TR-Transferrin), Alexa Fluor 488 sulfodichlorophenol (5-SDP) ester, Hoechst 34580, LysoTracker Red, Organelle Lights Endosome-RFP and Organelle Lights Lysosome-RFP were purchased from Invitrogen (Carlsbad, CA). Cy5 Label-IT nucleic acid labeling kits were purchased from Mirus Bio (Madison, WI). Tissue lysis and luciferase assay reagents were obtained from Promega (Madison, WI). Protein concentration assays were performed using the BCA Protein Assay Kit from Pierce (Rockford, IL). Antibodies against nucleolin (MS-3), ICAM-1 (15.2) and isotype control, normal mouse IgG1 were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA).
2.2. Reporter genes and plasmid preparation
Two plasmids encoding luciferase were used: pd1GL3-RL (7.9 kbp) and pBAL (5.1 kbp). The plasmid pd1GL3-RL containing two CMV promoted Luc genes, firefly (GL3) and Renilla (RL), the SV40 late polyadenylation signal, and kanamycin resistance was propagated in Escherichia coli DH5α (Kan/Neo selection). Plasmid DNA was collected and purified using Qiagen EndoFree Plasmid Giga kits (Qiagen Inc., Valencia, CA) per manufacturer's protocol. The plasmid pBAL containing the human beta-actin promoter, the firefly Luciferase gene, the SV40 late polyadenylation signal, and zeocin resistance was produced at Copernicus Therapeutics. Endotoxin levels were < 5 EU/mg plasmid. Compared to the transient transgene expression mediated by the CMV promoter, the beta-actin promoter is capable of sustained transgene expression.
2.3. Preparation of condensing peptides
A block co-polymer of polyethylene glycol and poly-L-lysine, PEG-CK30, was prepared as previously described [7], with the exception that the MW of PEG used was 5 kDa. Similarly, the triblock co-polymer poly(ethylene glycol)-block-poly(L-histidine)-block-poly(L-lysine), PEG-CH12K18, was prepared by substituting CH12K18 for CK30.
2.4. Preparation of fluorescently labeled condensing peptides and DNA
For co-localization and drug inhibition of uptake studies, PEG-CH12K18 and PEG-CK30 polymers were fluorescently labeled by conjugating the amine reactive probe Alexa Fluor 488 sulfodichlorophenol (5-SDP) ester (AF488) to the epsilon amines of lysine following the manufacturer's protocol (reaction stoichiometry: 1 AF488 per PEG-CK30). Unreacted AF488 was removed by fractionating the reaction mixture on a Sephadex G25 column (GE Healthcare, Piscataway, NJ) pre-equilibrated with 50 mM ammonium acetate (AA). For flow cytometric analysis of cellular uptake and co-localization studies, DNA was labeled with Cy5 using a Label-IT nucleic acid labeling kit according to the manufacturer's protocol.
2.5. Formulation of compacted DNA
DNA and polymer solutions were adjusted to pH 5.5 or 7.5 prior to DNA compaction. Compacted DNA nanoparticles were manufactured by the drop-wise addition of 9 volumes of DNA solution (0.222 mg/ml in water) to 1 volume of copolymer solution at a rate of 1 ml/min while vortexing at room temperature. For DNA nanoparticles prepared at pH 5.5, the final N:P ratio of positively charged amines (N) contributed by poly-L-histidine and/or poly-L-lysine to negatively charged DNA phosphates (P) was 2:1, whereas only the positively charged primary amines (N) of poly-L-lysine contributed to the final N:P ratio of 2:1 for DNA nanoparticles prepared at pH 7.5. After incubating at room temperature for 30 minutes, aggregates were removed by syringe filtration (0.2 μm). To exchange water for physiologic saline, DNA nanoparticles were diluted 10-fold with 0.9% NaCl and re-concentrated to 0.2 mg/ml using a Vivaspin 6 ultrafiltration device (100,000 MWCO, Vivaproducts, Inc., Littleton, MA) two times. For in vivo experiments, DNA nanoparticles were concentrated to 1.6 or 2 mg/ml and administered the same day. For long-term stability studies, DNA nanoparticles were stored at 4°C.
2.6. Formulation of PEI DNA nanoparticles
A 10 mg/ml aqueous solution of branched PEI (25 kDa) was prepared in sterile water at pH 7.4. PEI DNA nanoparticles were manufactured by the drop-wise addition of 9 volumes of DNA solution (0.222 mg/ml in water) to 1 volume of aqueous PEI solution at a rate of 1 ml/min while vortexing at room temperature. The final N:P ratio of PEI nitrogen amines (N) to DNA phosphates (P) was 10:1. After incubating at room temperature for 30 minutes, PEI DNA nanoparticles were diluted 10-fold with sterile water and re-concentrated to 0.2 mg/ml using a Vivaspin 6 centrifugal concentrator (100,000 MWCO, Vivaproducts, Inc., Littleton, MA) two times. For in vivo experiments, PEI DNA nanoparticles were concentrated to 2 mg/ml and administered the same day.
2.7. Physicochemical characterization of DNA nanoparticles
Compacted DNA nanoparticles were tested for quality assurance by transmission electron microscopy, gel electrophoresis, turbidity, and sedimentation, as described previously [7]. Size and ξ-potential (surface charge) were measured by dynamic light scattering and laser Doppler anemometry, respectively, using a Zetasizer Nano ZS90 (Malvern Instruments, Southborough, MA). Samples were diluted to 0.1 mg/ml in 10 mM NaCl pH 7.1 (viscosity 0.8894 cP and Refractive index 1.33 @ 25°C). Size measurements were performed at 25°C at a scattering angle of 90°. The zeta-potential values were calculated using the Smoluchowski equation.
2.8. Acid-base titration
The buffering capacities of CH12K18, CK30 and branched PEI (25 kDa) were determined by acid-base titration. First, 8 mg of peptide/polymer was dissolved in 10 ml of distilled water. Acid-base titrations were performed using a Titralab 856 automatic titrator (Radiometer Analytical, Lyon, France) as previously described [20]. The buffering capacity was calculated as the percentage of (protonatable) amine groups becoming protonated from pH 7.4 to 5.1 as previously described [22].
2.9. Cell culture
Human bronchial epithelial (BEAS-2B) cells (ATCC, Manassas, VA) were cultured at 37°C and 5 % CO2 in DMEM/F12 (Invitrogen Corp., Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS, Invitrogen Corp.) and 1% penicillin/streptomycin (Invitrogen Corp.). In select experiments, BEAS-2B cells were pre-transfected with Organelle Lights Endosome-RFP or Organelle Lights Lysosome-RFP kits according to the manufacturer's protocol, which contains a gene sequence that encodes for Rab5a-RFP (early endosome marker) or Lamp1 (lysosome marker), respectively. For live-cell microscopy, cells were seeded between 2.0 to 2.5 × 103 cells per plate onto Lab-Tek glass-bottom culture plates and incubated overnight at 37°C. After overnight incubation, culture medium was replaced with fresh media before particles were added.
2.10. Cellular uptake of DNA nanoparticles
BEAS-2B cells were seeded onto 6-well plates at an initial density of 3.0 × 105 cells/well and incubated at 37°C. After 24 hrs, cells were incubated with Cy5 labeled DNA nanoparticles (1 μg DNA/well) in media for 3 h at 37°C. Subsequently, the cells were washed with 1× PBS and extracellular fluorescence was quenched with 0.4% (w/v) trypan blue. Cells were then washed with 1× PBS and harvested with 0.05% Trypsin/EDTA and resuspended in 1× PBS. Mean fluorescence intensity was analyzed using FACSCalibur flow cytometer (Becton Dickinson, Franklin Lake, NJ). Data from 10,000 events were gated using forward and side scatter parameters to exclude cell debris.
2.11. In vitro transfection
BEAS-2B cells were seeded onto 12-well plates at an initial density of 5.0 × 104 cells/well and incubated at 37°C. After 24 hrs, cells were incubated with DNA nanoparticles (2 μg DNA/well) in media for 6 hrs at 37°C. Subsequently, nanoparticles and culture medium were replaced with fresh media. After additional 42 hrs incubation at 37°C, media was removed and 0.25 ml of 1× Reporter Lysis Buffer was added and then subjected to a freeze-and-thaw cycle. Cells were detached and collected using a cell scraper, and supernatants were obtained by centrifugation. Luciferase activity in the supernatants was analyzed using a standard luciferase assay kit (Promega, Madison, WI) and a 20/20n luminometer (Turner Biosystems, Sunnyvale, CA). Each sample was read in duplicate. Data are shown as relative light unit (RLU)/mg protein.
2.12. In vitro toxicity
BEAS-2B cells were seeded onto 96-well plates at an initial density of 1.0 × 104 cells/well and incubated at 37°C. After 24 hrs, cells were incubated with various doses of DNA nanoparticles (0.2, 2, 5, or 10 μg DNA/well) in media for 48 hrs at 37°C. After 48 hrs, cells were treated with 0.5 mg/ml MTT (3-(4,5-dimethyl-thiazol-2-yl)-2,5-diphenyl tetrazonlium bromide) solution for 4 hrs, and subsequently the solution was removed and replaced with DMSO. The metabolic activity was measured spectrophotometrically at 570 nm using a plate reader.
2.13. Confocal microscopy
LSM 510 Meta confocal microscope (Carl Zeiss Inc., Thornwood, NY) was used to capture images and time-lapse movies. For multi-color microscopy, samples were excited with 405, 488, 543 and 633nm laser lines, and images were captured by multi-tracking to avoid bleed-through between fluorophores. All samples were maintained at 37°C and 5% CO2 and observed under a 63× Plan-Apo/1.4 NA oil-immersion lens.
2.14. Cellular uptake and distribution in cytoplasm
Prior to imaging, BEAS-2B cells were treated for 10 min with Hoechst 33258 (5 μg/ml) to stain the nucleus. After staining, cells were washed with 1× PBS and incubated in Opti-MEM (Invitrogen Corp.) for imaging using confocal microscope. To evaluate the intracellular spatial distribution of gene carriers, the cytoplasm of each cell was divided into four radially equal quadrants (Q1 through Q4), where Q1 is closest to the nucleus and Q4 the furthest. DNA nanoparticle distributions in each quadrant were then quantified using MetaMorph software (Universal Imaging Corp., Downingtown, PA) on a per-pixel basis.
2.15. Co-localization with endocytic markers
In order to identify the endocytic pathway involved for PEG-CH12K18 DNA nanoparticles, co-localization studies with endocytic markers were performed in live BEAS-2B cells. In select experiments, cells were treated with TR-Transferrin (10 μg/ml) for 1 hr, Alexa Fluor 555-CTB (3 μg/ml) for 30 min, or FITC-Dextran (1 mg/ml) for 30 min. In another experiment, cells were treated with LysoTracker (100 nM) for 30 min. The cells were subsequently washed with 1× PBS and incubated in Opti-MEM for confocal imaging. Next, nanoparticles were added and then imaged under confocal microscope. Image acquisition was limited to 200 ms per frame or less to maintain accurate co-localization information and then analyzed with MetaMorph software.
2.16. Drug inhibition of uptake pathways
BEAS-2B cells were seeded onto 6-well plates at an initial density of 3.0 × 105 cells/well. After 24 hrs, BEAS-2B cells were treated with chlorpromazine (10 μg/ml), genistein (200 uM), amiloride (10 uM), or methyl-β-cyclodextrin (10 mM) with lovastatin (1 μg/ml) in media for 1 hr at 37°C. Subsequently, labeled DNA nanoparticles were added and incubated for another 2 hrs, after which cells were washed with 1× PBS and extracellular fluorescence was quenched with 0.4% (w/v) trypan blue. 4°C samples were pre-chilled for 30 min prior to addition of DNA nanoparticles and maintained at 4°C during the incubation period. Cells were then washed with 1× PBS and harvested with 0.05% Trypsin/EDTA and resuspended in 1× PBS. Mean fluorescence intensity was analyzed using FACSCalibur flow cytometer (Becton Dickinson). Data from 10,000 events were gated using forward and side scatter parameters to exclude cell debris.
2.17. Animals and nanoparticle administration
All procedures performed with mice were approved by the Johns Hopkins University Animal Care and Use Committee. BALB/c mice (female, 6–8 weeks old) were anesthetized by intraperitoneal (IP) injection of tribromoethanol (Avertin) at a dose of 250 mg/kg. Pulmonary administration was performed by oropharyngeal aspiration, as previously described [23]. Briefly, anesthetized mice were suspended at a 45° angle by their incisors on a plexiglass stand. The tongue was gently pulled aside and maintained in full extension by forceps. A solution (50 μl) was pipetted at the base of the tongue and tongue restraint was continued until 2 deep breaths were completed.
2.18. In vivo safety profile
DNA nanoparticles or naked DNA in saline, or PEI DNA nanoparticles in water were administered by oropharyngeal aspiration at a dose of 100 μg per mouse (n = 7 per group). Control mice were treated with saline or water (n = 7 per group). After 48 hrs, animals were sacrificed and lungs were harvested for either bronchoalveolar lavage fluid (BALF) collection (n = 5) or tissue histology (n = 2). BALF was collected by sequentially perfusing the lungs three times with 1 ml PBS, the recovered fluids were pooled for each animal. Cells were collected by centrifugation and supernatant was stored at −80°C until assayed for cytokines. Red blood cells were lysed with ACK lysing buffer (Quality Biological, Inc., Gaithersburg, MD). Total cell counts were determined using a Vi-cell XR automated cell viability analyzer (Beckman Coulter). Cytospin preparations were prepared using a Shandon Cytospin 3 centrifuge (Shandon Scientific, Cheshire, England). Differential cell counts were performed on cytospin preparations after staining with hematoxylin and eosin (H&E). Cytokines (TNF-α and IL-6) were assayed using Quantikine ELISA kits from R&D Systems, Inc. (Minneapolis, MN). Cytokine concentrations in BALF were adjusted for dilution by the urea method [24] and expressed as pg/ml epithelial lining fluid (ELF). To prepare lungs for tissue sectioning, lungs were excised completely from the chest, inflated with 1 ml of 10% formalin, and then immersed in 10% formalin for 24 hrs before embedding in paraffin. Sections (5 μm) were cut from each lung and stained with H&E.
2.19. In vivo airway gene transfer
DNA nanoparticles or naked DNA in saline were administered by oropharyngeal aspiration at a dose of 80 μg per mouse (n = 11 per group). Control mice were treated with saline (n = 11). To examine airway gene transfer, we measured the luciferase activity of lung tissue homogenates. Mice were euthanized at 2 days (n = 4) or 14 days (n = 7) after administration. Hearts were perfused with 3 ml PBS and lung tissues were harvested and homogenized in 1 ml of reporter lysis buffer (Promega, Madison, WI) using the TissueLyser LT (Qiagen, Valencia, CA). The homogenates were subjected to a freeze-thaw cycle and supernatants were obtained by centrifugation. Luciferase activity in the supernatant was measured using a standard luciferase assay kit (Promega, Madison, WI) and a 20/20n luminometer (Turner Biosystems, Sunnyvale, CA). Each sample was read in duplicate. Data are shown as relative light unit (RLU)/mg protein.
2.20. Statistical analysis
Statistical analysis of data was performed by one-way analysis of variance (ANOVA) followed by Tukey HSD or Games-Howell tests using SPSS 18.0 software (SPSS Inc., Chicago, IL). Differences were considered to be statistically significant at a level of P < 0.05.
3. Results
3.1. Physicochemical characterization of compacted DNA nanoparticles
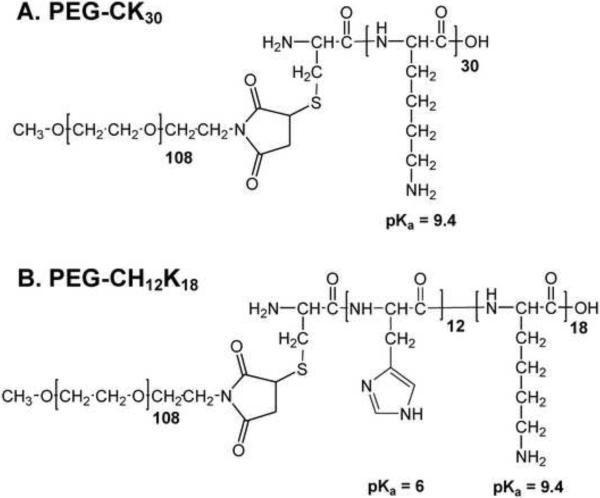
The conjugation of methoxy-PEG-mal to the condensing peptides, CK30 or CH12K18, was confirmed by 4,4'-dithiodipyridine release assay, indicating that all free thiol groups had fully reacted to yield either PEG-CK30 or PEG-CH12K18, respectively (Figure 1a,b). To determine the DNA binding strengths of the condensing peptides and the stability of resultant polyplexes, we performed an ethidium bromide (EtBr) fluorescence-quenching and recovery assay [20]. At pH 5.5, the interaction of PEG-CH12K18 with DNA was comparable to that observed with PEG-CK30, as indicated by a 60% quench of EtBr fluorescence (binding strength = ~ 0.6) (Figure S1a). At pH 7.5, the DNA binding strength for PEG-CH12K18 decreased (P < 0.001), likely due to the deprotonization of poly-L-histidine's imidazole groups (pKa = ~ 6.0). Regardless of formulation pH, polyplexes formed with PEG-CK30 or PEG-CH12K18 were resistant to dissociation at heparin:DNA charge ratios of 1:100 and 1:10, as indicated by the lack of EtBr fluorescence recovery (polyplex stability = ~ 1) (Figure S1b,c), whereas both types of polyplexes fully dissociated at a heparin:DNA charge ratio of 1:1 (polyplex stability = ~ 0) (Figure S1d). The exclusion of EtBr from DNA by PEG-CK30 or PEG-CH12K18, and the stability of resultant polyplexes, suggests the formation of compacted DNA.
Figure 1.
Chemical structures of (A) PEG-CK30 and (B) PEG-CH12K18.
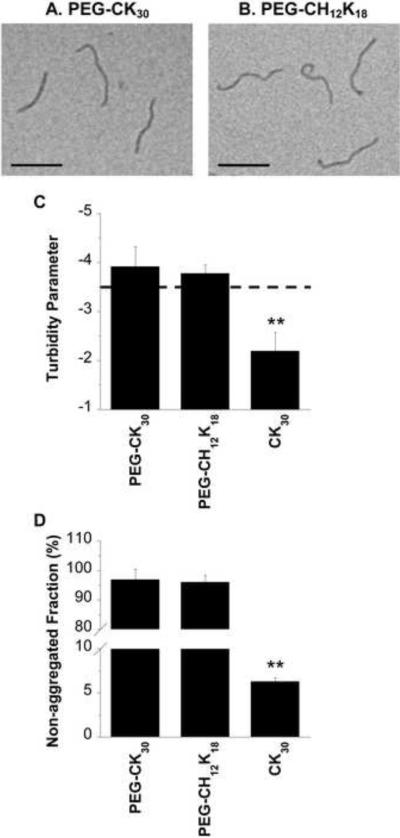
The formation of compacted DNA at pH 7.5 was confirmed by morphological examination via transmission electron microscopy (TEM), which revealed that both PEG-CK30 and PEG-CH12K18 produced flexible rod like nano-structures (Figure 2a,b), in agreement with previous reports for PEG-CK30 DNA nanoparticles [7, 25]. The average dimensions (length × width) for PEG-CH12K18 DNA nanoparticles and PEG-CK30 DNA nanoparticles formulated at pH 7.5 were ~ 325 × 13 nm and ~ 300 × 13 nm, respectively (Table 1). PEG-CH12K18 DNA nanoparticles formulated at pH 7.5 exhibited a slightly positive ζ-potential, as compared to PEG-CK30 DNA nanoparticles (P < 0.01). In contrast, PEG-CH12K18 DNA nanoparticles formulated at pH 5.5 exhibited a slightly negative ζ-potential that was comparable to PEG-CK30 DNA nanoparticles (Table S1). We monitored the colloidal stability in saline after storage at 4°C for 6 months using turbidity and sedimentation assays, dynamic light scattering, and laser Doppler anemometry. PEG-CH12K18 and PEG-CK30 DNA nanoparticles both exhibited acceptable turbidity parameters (Figure 2c; [25]) indicative of negligible aggregation, in good agreement with sedimentation assays that showed < 5 % of compacted DNA aggregated and sedimented during this period (Figure 2d). The size, particle size distribution (PDI), and ζ-potential of PEG-CH12K18 and PEG-CK30 DNA nanoparticles did not change significantly after storage at 4°C for 6 months (Table S2).
Figure 2.
Physicochemical characterization of DNA nanoparticles formulated at pH 7.5. Transmission electron microscopy of compacted DNA nanoparticles formulated with (A) PEG-CK30 and (B) PEG-CH12K18. Scale bar represents 200nm. Colloidal stability of compacted DNA nanoparticles after storage at 4°C for 6 months. (C) Turbidity parameters, an indicator of colloidal stability, for various DNA nanoparticle formulations. A turbidity parameter less than −3.5 (above the dashed line) indicates DNA nanoparticles that are compacted and non-aggregated, whereas values greater than −3.5 (below the dashed line) indicate significant aggregation of DNA nanoparticles. (D) Non-aggregated fraction of compacted DNA nanoparticles as determined by sedimentation. Data represents the mean +/− SD. * denotes statistical significance (** P < 0.01, Tukey HSD).
Table 1.
Physicochemical properties of DNA nanoparticles
| Formulationa | Length, nmb | Width, nmb | ζ-potential, mVc | Buffering Capacity, %d |
|---|---|---|---|---|
| PEG-CK30 | 300 ± 11 | 13 ± 0.2 | −1.4 ± 0.6 | 12 ± 5.0 |
| PEG-CH12K18 | 325 ± 11 | 13 ± 0.6 | 2.0 ± 0.3** | 24 ± 2.7* |
| PEI | 40 ± 3.2 | 40 ± 2.6 | 39 ± 0.6** | 23 ± 1.1* |
DNA compacted with PEG-CK30 or PEG-CH12K18 were formulated at pH 7.5 and at a final lysine to phosphate ratio (N:P) of 2:1. PEI DNA nanoparticles were formulated at pH 7.4 and at a final N:P ratio of 10:1.
DNA nanoparticle length and width as measured from TEM images, using ImageJ software package. Data represents the average of 3 independent experiments +/− SD.
Measured at pH 7.1. Data represents the average of 3 independent experiments +/− SD. * denotes statistical significance (** P < 0.001, Tukey HSD) as compared to all other groups.
P < 0.001
Buffering capacity, as measured by acid-base titration, represents the percentage of (protonable) amine groups becoming protonated from pH 7.4 to 5.1 [22]. Data represents the average of 3 independent experiments +/− SD. * denotes statistical significance (* P < 0.05, Games-Howell) as compared to PEG-CK30.
P < 0.05
3.2. Proton-Buffering capacity of DNA nanoparticles
The buffering capacity of the peptides PEG-CH12K18 and PEG-CK30, and branched polyethyleneimine (PEI, 25 kDa) was determined by acid-base titration. From the titration curves (data not shown), the buffering capacity was calculated as the percentage of (protonatable) amine groups becoming protonated in the endosomal pH range (pH 7.4 to 5.1) [22]. PEG-CK30 exhibited a relatively low buffering capacity, as only 12% of amine groups became protonated between pH 7.4 to 5.1 (Table 1). In comparison, PEG-CH12K18 exhibited a significantly higher buffering capacity (24%) that was comparable to the buffering capacity of PEI (23%).
3.3. Flow cytometric analysis of cell surface nucleolin on BEAS-2B cells
To evaluate the presence of cell surface nucleolin, flow cytometric analysis of BEAS-2B cells was performed using AF488 conjugated antibodies. BEAS-2B incubated with the monoclonal antibody against nucleolin (MS-3) showed only a slight shift of the fluorescence peak relative to cells treated with control IgG (Figure S2a). In contrast, cells incubated with a monoclonal antibody against intracellular adhesion molecule-1 (ICAM-1, 15.2), which is known to be constitutively expressed on BEAS-2B cells [26], resulted in a considerably greater shift of the fluorescence peak (Figure S2b). These results suggest that nucleolin is not strongly expressed on the surface of BEAS-2B cells.
3.4. In vitro and in vivo gene transfer
The in vitro gene transfer efficiency of highly compacted PEG-CH12K18 and PEG-CK30 DNA nanoparticles was evaluated in human bronchial epithelial cells (BEAS-2B) by using the pd1GL3-RL plasmid encoding firefly luciferase as the reporter gene. The transfection efficiency of PEG-CK30 DNA nanoparticles, regardless of formulation pH (5.5 or 7.5), was comparable to that obtained with naked DNA (Figure 3a), in good agreement with the nucleolin-dependent nature of PEG-CK30 DNA nanoparticle mediated gene transfer [12–13]. Similar transfection efficiencies were observed for PEG-CH12K18 DNA nanoparticles formulated at pH 5.5. In contrast, PEG-CH12K18 DNA nanoparticles formulated at pH 7.5 increased the transfection efficiency by 20-fold (P < 0.001).
Figure 3.
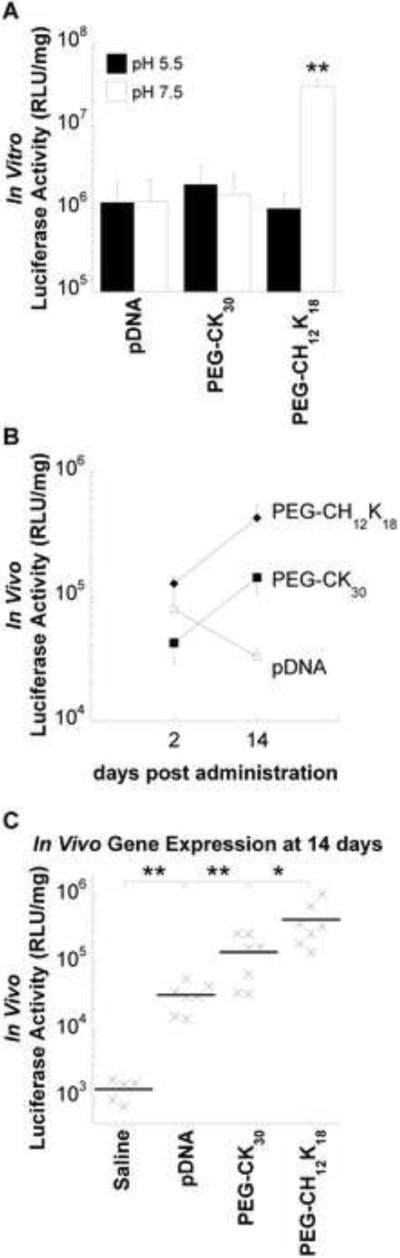
Gene transfer efficiency. (A) In vitro transfection of luciferase gene to BEAS-2B cells by DNA nanoparticles formulated at pH 5.5 or 7.5. BEAS-2B cells were incubated with DNA nanoparticles in medium containing 10% serum for 6 h, followed by an additional 42 h incubation without DNA nanoparticles. Data represents the mean +/− SD. * denotes statistical significance (** P < 0.001, Tukey HSD) as compared to all other groups. (B) Time course of in vivo gene expression. BALB/c mice received 80 μg compacted (formulated at pH 7.5) or naked pBAL DNA in saline by oropharyngeal aspiration. Luciferase activity in lung homogenates was measured at 2 days (n = 4) or 14 days (n = 7) after pulmonary administration. Data represents the mean +/− SEM. (C) In vivo gene expression at 14 days after pulmonary administration. Control mice (n=5) received saline alone. * denotes statistical significance (* P < 0.05, ** P < 0.001, Tukey HSD). pDNA, plasmid DNA; RLU, relative light unit.
To determine in vivo gene transfer efficiency, we administered PEG-CH12K18 DNA nanoparticles or PEG-CK30 DNA nanoparticles (formulated at pH 7.5) containing 80 μg of pBAL plasmid encoding firefly luciferase via oropharyngeal aspiration to the lungs of BALB/c mice. Control mice received either 80 μg of naked pBAL or saline. The luciferase activity of lung homogenates was measured at 2 days (n = 4) or 14 days (n = 7) after administration. At 2 days post administration, there was no significant difference in the luciferase activity for mice treated with PEG-CH12K18 DNA nanoparticles, PEG-CK30 DNA nanoparticles or naked DNA (Figure 3b). At 14 days post administration, the level of luciferase activity increased for mice that received DNA nanoparticles, whereas it decreased for mice that received naked DNA. Mice receiving highly compacted PEG-CH12K18 DNA nanoparticles exhibited the highest overall expression (Figure 3c), 13-fold higher than naked DNA (P < 0.001) and 3-fold higher than PEG-CK30 DNA nanoparticles (P < 0.05).
3.5. In vitro cellular uptake and intracellular trafficking
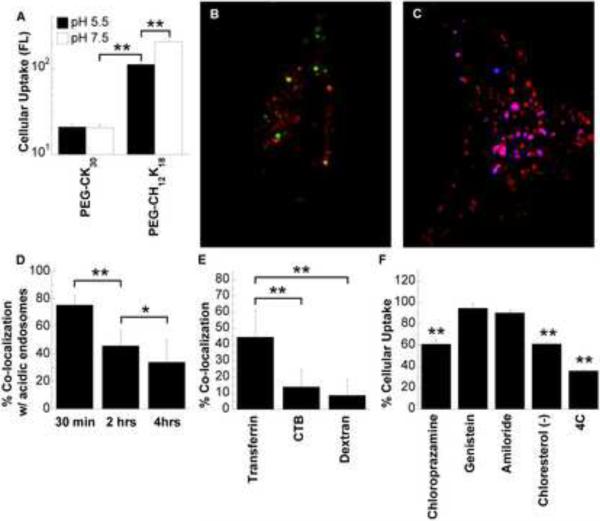
To investigate the mechanism of PEG-CH12K18 DNA nanoparticle mediated gene transfer, we studied cellular uptake and intracellular trafficking. Flow cytometry was used to evaluate the uptake of PEG-CH12K18 or PEG-CK30 DNA nanoparticles as a function of formulation pH. DNA plasmids were fluorescently labeled with Cy5, which did not affect the formation of DNA nanoparticles as confirmed by TEM (Figure S3), ζ-potential and sedimentation (data not shown). The cellular uptake of PEG-CH12K18 DNA nanoparticles formulated at pH 5.5 or 7.5 was 5- and 10-fold higher than PEG-CK30 DNA nanoparticles, respectively (P < 0.001) (Figure 4a). Formulation pH had no effect on the cellular uptake of PEG-CK30 DNA nanoparticles, whereas the cellular uptake of PEG-CH12K18 DNA nanoparticles was higher when nanoparticles were formulated at pH 7.5 (P < 0.001). Interestingly, although PEG-CH12K18 nanoparticles formulated at pH 5.5 had increased cell uptake compared to PEG-CK30 nanoparticles, they did not generate more luciferase activity (Figure 3a), suggesting a relatively inefficient cytoplasmic to nuclear trafficking pathway. In contrast, the improved uptake with PEG-CH12K18 nanoparticles formulated at pH 7.5 was associated with improved luciferase activity, suggesting enhanced nuclear uptake.
Figure 4.
In vitro cellular uptake and intracellular trafficking. (A) Cellular uptake of Cy5 labeled DNA nanoparticles by BEAS-2B cells. BEAS-2B cells were incubated with DNA nanoparticles in medium containing 10% serum for 2 hrs prior to flow cytometric analysis. (B) BEAS-2B cells pre-transfected with RFP-fused Rab5a protein (early endosome marker, red) were incubated with AF488 labeled PEG-CH12K18 DNA nanoparticles (green, formulated at pH 7.5) in Opti-MEM for 30 min prior to confocal microscopy. Yellow indicates co-localization. (C) BEAS-2B cells pretransfected with RFP-fused Lamp-1 protein (lysosomal marker, red) were incubated with Cy5 labeled PEG-CH12K18 DNA nanoparticles (blue, formulated at pH 7.5) in Opti-MEM for 30 min prior to confocal microscopy. Violet indicates co-localization. (D) Quantitative co-localization analysis between AF488 labeled PEG-CH12K18 DNA nanoparticles and LysoTracker at various time points. BEAS-2B cells were treated with LysoTracker (100 nM) for 30 min, washed with PBS and incubated with PEG-CH12K18 DNA nanoparticles in Opti-MEM prior to confocal microscopy. (E) Quantitative co-localization analysis between AF488 labeled PEG-CH12K18 DNA nanoparticles (formulated at pH 7.5) and endocytic markers. BEAS-2B cells were treated with Transferrin (10 μg/ml) for 1 hr, CTB (30 μg/ml) for 30 min, or FITC-Dextran (1 mg/ml) for 30 min, washed with PBS and incubated with PEG-CH12K18 DNA nanoparticles in Opti- MEM for 30 min prior to confocal microscopy. (F) Flow cytometry analysis for drug inhibition of uptake pathways. BEAS-2B cells were treated with the clathrin-mediated endocytosis inhibitor chloropromazine (10 μg/ml), the caveolae-mediated endocytosis inhibitor genistein (200 μM), the macropinocytosis inhibitor amiloride (10 μM), or cholesterol depleting agents methyl-β-cyclodextrin (10mM) and lovastatin (1 μg/ml) in media for 1 hr at 37°C, subsequently PEG-CH12K18 DNA nanoparticles (formulated at pH 7.5) were added and incubated for an additional 2 hrs at 37°C prior to harvesting cells for analysis by flow cytometry. 4°C samples were pre-chilled for 30 min prior to addition of PEG-CH12K18 DNA nanoparticles and maintained at 4°C during the incubation period. Data represents the mean +/− SD. * indicates statistically significant (* P < 0.05, ** P < 0.001, Tukey HSD). FL, fluorescence; CTB, Cholera Toxin subunit B; AF488, Alexa Fluor 488; RFP, Red Fluorescent Protein.
Recently we discovered that PEG-CK30 DNA nanoparticles enter BEAS-2B cells via caveolae-mediated endocytosis (CvME) and rapidly accumulate in the perinuclear region of cells via a non-endo-lysosomal pathway[27]. To determine whether PEG-CH12K18 DNA nanoparticles traffic within the same pathway as PEG-CK30 DNA nanoparticles, we investigated their intracellular trafficking in live BEAS-2B cells using confocal microscopy. We labeled the PEG-CH12K18 polymer with Alexa Fluor 488, which did not affect the formation of DNA nanoparticles as confirmed by TEM (Figure S3), ζ-potential and sedimentation (data not shown). PEG-CH12K18 DNA nanoparticles formulated at pH 7.5 were taken up by BEAS-2B cells within 2 hrs, and rapidly accumulated in the perinuclear region. On average, 37% of PEG-CH12K18 DNA nanoparticles were found in the perinuclear region (Q1) within 2 hrs post-incubation (Figure S4). To determine whether PEG-CH12K18 DNA nanoparticles formulated at pH 7.5 are trafficked via the endo-lysosomal pathway, we performed co-localization studies in BEAS-2B cells using Rab5a-RFP and Lamp-1-RFP, well-characterized markers for early endosomes and lysosomes, respectively. We found that PEG-CH12K18 DNA nanoparticles were highly co-localized with Rab5a-RFP and Lamp-1-RFP within 30 min (Figure 4b,c). PEG-CH12K18 DNA nanoparticles were also highly co-localized with LysoTracker, an acid sensitive probe that labels late endosomes and lysosomes, confirming that PEG-CH12K18 DNA nanoparticles are largely sequestered in acidic vesicles of the endo-lysosomal pathway; however the level of co-localization significantly decreased over time (Figure 4d), suggesting that PEG-CH12K18 DNA nanoparticles escaped from the endo-lysosomal pathway.
Previous studies have demonstrated that the endocytic mechanism influences the intracellular trafficking of nanoparticles [12, 28–29]. That PEG-CH12K18 DNA nanoparticles undergo trafficking within the endo-lysosomal pathway of BEAS-2B cells, whereas PEG-CK30 DNA nanoparticles do not [12–13, 27], suggests that different endocytic mechanisms may be involved. To determine the mechanism of cellular uptake, we co-incubated BEAS-2B cells with fluorescently labeled PEG-CH12K18 DNA nanoparticles and various endocytic markers: Transferrin (clathrin-mediated), Cholera Toxin subunit B (CTB, caveolae-mediated), and dextran (macropinocytosis), and measured the degree of co-localization of the distinct fluorophores using confocal microscopy. We found a substantial fraction of PEG-CH12K18 DNA nanoparticles were co-localized with Transferrin within BEAS-2B cells (Figure 4e), significantly higher (P < 0.001) than with either CTB or Dextran, suggesting they primarily enter BEAS-2B cells via CME.
To confirm our confocal microscopy observations with endocytic markers, we measured the uptake of DNA nanoparticles in BEAS-2B cells treated with different endocytic inhibitors using flow cytometry. The uptake of PEG-CH12K18 DNA nanoparticles formulated at pH 7.5 was significantly reduced (P < 0.001) by chlorpromazine (Figure 4f), which blocks CME by causing clathrin to accumulate in late endosomes. PEG-CH12K18 DNA nanoparticles uptake was unaffected by genistein or amiloride, inhibitors of caveolae-mediated uptake and macropinocytosis, respectively. Cellular uptake of PEG-CH12K18 DNA nanoparticles was also significantly reduced (P < 0.001) when BEAS-2B cells were incubated at 4°C, suggesting that DNA nanoparticles enter cells via an energy-dependent endocytic mechanism. The cellular uptake of PEG-CH12K18 DNA nanoparticles was significantly reduced (P < 0.001) by cholesterol depletion, using methyl-β-cyclodextrin (extracts cholesterol from plasma membrane) and lovastatin (inhibitor of de novo cholesterol synthesis), in good agreement with cholesterol-dependent nature of CME [30–32]. Together, these findings suggest PEG-CH12K18 DNA nanoparticles enter BEAS-2B cells primarily via CME.
3.6. In vitro and in vivo safety profile
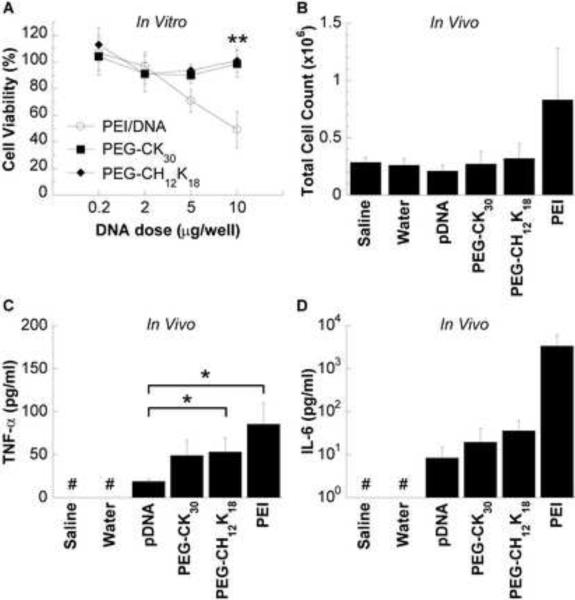
We tested the in vitro cytotoxicity of DNA nanoparticles made with PEI, PEG-CH12K18, or PEG-CK30, at various DNA doses, using the MTT assay. Results from BEAS-2B cells show that PEI DNA nanoparticles induced the greatest cytotoxicity, with a 50% reduction in metabolic activity at the highest dose (P < 0.01) (Figure 5a). In contrast, cells incubated with either PEG-CH12K18 DNA nanoparticles or PEG-CK30 DNA nanoparticles retained greater than 90% of their metabolic activity across all DNA doses tested.
Figure 5.
Safety profile of DNA nanoparticles formulated at pH 7.5. (A) In vitro metabolic activity of BEAS-2B cells as a function of DNA nanoparticle dose. * indicates statistically significant (** P < 0.01, Tukey HSD) as compared to PEI DNA nanoparticles. Bronchoalveolar lavage fluid (BALF) analysis of mice (n=5) treated with 100 μg of compacted or naked DNA by oropharyngeal aspiration and sacrificed 48 h after administration. (B) Total inflammatory cell counts. (C) Tumor necrosis factor alpha (TNF-α) concentration. # denotes values below the limit of detection. * denotes statistically significant (* P < 0.05, Games-Howell) as compared to pDNA. (D) Interleukin 6 (IL-6) concentration. # denotes values below the limit of detection. Data represents the mean +/− SD. Concentrations in BALF are adjusted for dilution by the urea method.
To determine the potential in vivo toxicity of DNA nanoparticles, we administered either PEG-CK30 DNA nanoparticles or PEG-CH12K18 DNA nanoparticles suspended in normal saline at a dose of 100 μg DNA per mouse (n = 7), by oropharyngeal aspiration. Groups of control mice (n = 7 per group) received either 100 μg DNA compacted with PEI suspended in water, 100 μg naked DNA suspended in saline, saline or water. At 48 hrs post administration, mice were sacrificed for histological examination of the lung (n = 2) or BAL fluid collection (n = 5), which was analyzed for total/differential cell counts and pro-inflammatory cytokines (TNF-α and IL-6). There was no significant difference in the total cell counts (Figure 5b) or differential cell counts for neutrophils, macrophages, lymphocytes, and eosinophils across all treatment groups (data not shown). All mice treated with saline or water had TNF-α and IL-6 levels below the limit of detection (5.0 pg/ml and 1.6 pg/ml, respectively). As compared to mice treated with naked DNA, there was a slight increase in TNF-α levels for mice treated with PEG-CK30 DNA nanoparticles (P = 0.06), PEG-CH12K18 DNA nanoparticles (P < 0.05), or PEI DNA nanoparticles (P < 0.05) (Figure 5c). There were detectable levels of IL-6 (3/5 mice receiving naked DNA, 4/5 mice receiving PEG-CK30 DNA nanoparticles, 4/5 mice receiving PEG-CH12K18 DNA nanoparticles, and 5/5 mice receiving PEI DNA nanoparticles), with PEI DNA nanoparticle treated mice exhibiting the highest values, ~ 100-fold greater than that elicited by PEG-CH12K18 DNA nanoparticles or PEG-CK30 DNA nanoparticles, although the differences were not significant (Figure 5d).
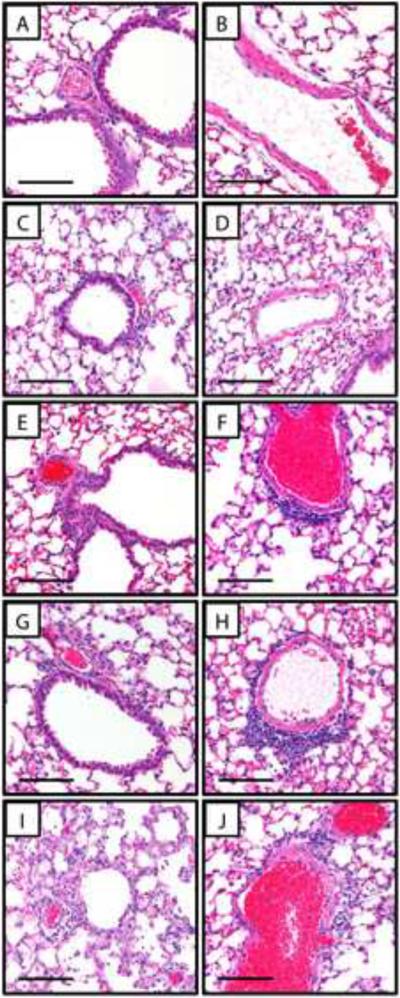
In the lungs, three mice (one saline treated, one treated with PEG-CK30 DNA nanoparticles, and one treated with PEG-CH12K18 DNA nanoparticles) had rare, small foci of supportive inflammation centered around plant material (not shown), likely due to inadvertent aspiration of food particles during pulmonary administration. The lungs of the remaining mice treated with saline or naked DNA were normal and did not show any significant peribronchial, alveolar, or perivascular infiltrates (Figure 6a–d). The lungs of mice treated with PEG-CH12K18 DNA nanoparticles or PEG-CK30 DNA nanoparticles did not show any significant peribronchial or alveolar infiltrates not associated with aspirated food particles, but did show trace to mild perivascular infiltrates of mononuclear inflammatory cells (lymphocytes, plasma cells, and macrophages) (Figure 6e–h). The lungs of mice treated with PEI DNA nanoparticles showed widespread epithelial necrosis and infiltration of neutrophils (Figure 6i,j); whereas the lungs of the mice treated with water were normal and did not show any significant peribronchial, alveolar, or perivascular infiltrates (not shown).
Figure 6.
Sample lung histology 48 hrs post-administration showing representative airways and pulmonary vessels from BALB/c mice treated with (A, B) saline, (C, D) 100 μg naked DNA, (E, F) 100 μg DNA compacted with PEG-CH12K18 at pH 7.5, (G, H) 100 μg DNA compacted with PEG-CK30 at pH 7.5, or (I, J) 100 μg DNA compacted with PEI at pH 7.4. All images were photographed at 20x magnification. Scale bar represents 100 μm.
4. Discussion
Efficient gene transfer by highly compacted PEG-CK30 DNA nanoparticles is thought to be mediated by cell surface nucleolin, which appears to be involved in DNA nanoparticle uptake and trafficking to the nucleus [12–13]. However, the utility of nucleolin-mediated gene delivery is limited to the population of cells that have nucleolin on their surface. In the airways of mice, immunohistochemical staining for cell surface nucleolin shows a patchy expression pattern [13] that resembles the pattern of transfection by PEG-CK30 DNA nanoparticles [7]. Uniform gene expression is required to effectively treat certain diseases, and in the case of cystic fibrosis, delivery to at least 25% of airway epithelial cells may be required [33]. To potentially improve the distribution of gene expression in the airways, we sought to formulate DNA nanoparticles that efficiently transfect cells by utilizing CME, which is constitutively active in all mammalian cells. Thus, we incorporated pH-responsive poly-L-histidine into PEG-CK30 based DNA nanoparticles to overcome the endo-lysosomal barrier that plagues many synthetic gene carriers (although not the PEG-CK30 DNA nanoparticles that enter cell nuclei via binding to cell surface nucleolin). Unexpectedly, we report that PEG-CH12K18 nanoparticles (formulated at pH 7.5) enter cells by CME, in contrast to PEG-CK30 DNA nanoparticles of similar size and morphology. Although the mechanism for CME uptake of PEG-CH12K18 nanoparticles is not yet defined, this cytoplasmic trafficking pathway presents new opportunities for gene transfer in tissues lacking significant cell surface nucleolin expression.
We report that DNA nanoparticles formulated with a pH-responsive poly-L-histidine segment, PEG-CH12K18, are colloidally stable over time and mediate enhanced gene transfer. Inclusion of poly-L-histidine into the polymer backbone increased the buffering capacity to levels comparable to PEI, a cationic polymer well known for its high buffering capacity [19], without eliciting any associated toxicity. Unlike PEG-CK30 based DNA nanoparticles, which rely on nucleolin for cellular uptake and nuclear import [12–13], PEG-CH12K18 DNA nanoparticles mediated enhanced gene transfer to human bronchial epithelial cells (BEAS-2B) that lack cell surface nucleolin. PEG-CH12K18 DNA nanoparticles also mediated enhanced gene transfer to the lungs of BALB/c mice, ~ 3-fold improvement over PEG-CK30 DNA nanoparticles at 14 days post administration (P < 0.05). A similar increase was observed at 2 days post administration but the difference was not significant possibly due to the limited number of animals at this time point.
The therapeutic potential of PEG-CH12K18 DNA nanoparticles is highlighted by the promising results from a Phase I/IIa study in CF subjects, in which the level of gene transfer to human nasal mucosa mediated by PEG-CK30 DNA nanoparticles, as measured by PCR analysis of vector DNA, was comparable to the highest levels observed in an AAV intrasinus trial [34]. Furthermore, PEG-CH12K18 DNA nanoparticles have an excellent safety profile that is comparable to PEG-CK30 based DNA nanoparticles, which were previously shown to be non-toxic, non-inflammatory, and non-immunogenic in the mouse lung and human nares [10, 34]. Together, these results suggest that PEG-CH12K18 DNA nanoparticles represent a promising platform for gene delivery applications.
PEG-CH12K18 DNA nanoparticles formulated at pH 7.5 mediated the highest levels of expression in vitro. Since the imidazole group has a pKa = 6, poly-L-histidine remains relatively unprotonated (i.e. uncharged) at pH 7.5 and does not participate in DNA compaction [21], whereas at pH 5.5, poly-L-histidine is protonated (i.e. positively charged) and can interact electrostatically with DNA. The finding that poly-L-histidine enhanced DNA nanoparticle mediated gene transfer only when histidine was not bound electrostatically with DNA suggests that the enhanced gene transfer is mediated by the side-chains of poly-L-histidine. Differences in the relative DNA binding strengths for PEG-CH12K18 and PEG-CK30 may influence the kinetics of DNA release and, consequently, gene transfer efficiency. For example, Wong et al. recently reported that among a combinatorial library of cationic polymers, those cofunctionalized with up to 50% imidazole and at least 50% primary amines resulted in the highest transfection efficiencies in vitro [20]. Imidazole's transfection-enhancing effect was attributed to non-electrostatic interactions with DNA to produce stable polyplexes that were more resistant to premature dissociation. However, this is unlikely the case for the highly compacted DNA nanoparticles studied here, since both PEG-CH12K18 and PEG-CK30 formulations have similar stabilities in the presence of competing polyanions.
Alternatively, the enhanced gene transfer by PEG-CH12K18 DNA nanoparticles is more likely attributed to increased cellular uptake and altered intracellular trafficking (Figure 4). Since the PEG coating could not completely block access of DNase I to cargo DNA [25, 35], the coating presumably would also allow access of cell surface glycoproteins to the peptide/DNA core, as may be the case for the direct binding of nucleolin to PEG-CK30 DNA nanoparticles [12]. The more positive surface charge on PEG-CH12K18 DNA nanoparticles formulated at pH 7.5, as compared to PEG-CH12K18 DNA nanoparticles and PEG-CK30 DNA nanoparticles formulated at pH 5.5 and 7.5, respectively, further suggests that poly-L-histidine is accessible on the nanoparticle surface. Thus, it is possible that interactions between cell surface glycoproteins and poly-L-histidine result in the differential sorting of PEG-CH12K18 DNA nanoparticles to clathrin coated pits and subsequent cellular uptake by CME, which has more rapid internalization kinetics than CvME [15, 25, 33], the predominant endocytic pathway for PEG-CK30 DNA nanoparticles uptake by BEAS-2B cells [27]. While the identity of the membrane glycoproteins responsible for the uptake of PEG-CH12K18 DNA nanoparticles is not yet known, the effects of nanoparticle surface chemistry on the kinetics and route of cellular uptake have been widely reported [36–40]. Harush-Frenkel et al. reported that positively charged nanoparticles are rapidly internalized into HeLa cells via CME, whereas negatively charged nanoparticles are internalized at a slower rate via a clathrin-independent pathway [37]. Others have also reported increased cellular uptake of positively charged gold [38] and iron oxide [39] nanoparticles as compared to negatively or neutrally charged particles.
Increased cellular uptake is likely not the sole reason for greater gene transfer, since the in vitro gene transfer efficiencies of PEG-CH12K18 DNA nanoparticles and PEG-CK30 DNA nanoparticles formulated at pH 5.5 were comparable (Figure 3a) despite a 5-fold higher level of cellular uptake of PEG-CH12K18 DNA nanoparticles (Figure 4a). The enhanced gene transfer efficiency may also be attributed to the buffering capacity of poly-L-histidine that, according to the proton sponge hypothesis, facilitates endosomal rupture and escape from lysosomal degradation [18]. Our results are consistent with this hypothesis, since the decrease in gene transfer efficiency of PEG-CH12K18 DNA nanoparticles formulated at pH 5.5 corresponds to the condition in which poly-L-histidine is bound electrostatically with DNA and is thus no longer available to buffer pH. Furthermore, the co-localization between PEG-CH12K18 DNA nanoparticles formulated at pH 7.5 and late endosomes/lysosomes labeled with lysotracker decreased over time, suggesting that nanoparticles successfully escaped from endosomes. However, the possibility that the AF488 dye has been cleaved from the condensing peptide and is able to diffuse out of the lysosome cannot be completely ruled out.
To further assess the potential of PEG-CH12K18 DNA nanoparticles for clinical development, we tested their safety and efficacy in BALB/c mice. BALB/c mice treated with PEG-CH12K18 DNA nanoparticles were indistinguishable from mice treated with PEG-CK30 DNA nanoparticles with regard to histological examination of the lung and BAL fluid markers for inflammation (cell counts and cytokines) after 48 hrs. Our results are consistent with a previous report that mice treated with clinically-tested PEG-CK30 DNA nanoparticles displayed a mild and transient response in the lung [10]. In comparison, mice treated with PEI DNA nanoparticles showed the highest levels of BAL fluid makers for inflammation, widespread epithelial necrosis and infiltration of neutrophils, consistent with previous reports [35–37]. Membrane destabilization by PEI has been associated with the induction of oxidative stress and pro-apoptotic signaling cascades [41–42]. In addition, both free PEI and PEI DNA nanoparticles are known to activate the complement system, which provokes an inflammatory response [43–44]. In comparison, short chain poly-L-lysine and poly-L-lysine DNA complexes are less potent activators of complement, and surface modification with PEG further reduces complement activation [44].
Optimization of plasmid DNA expression vectors is crucial for maximizing in vivo gene transfer performance. Plasmid DNA expression vectors under the control of viral promoters, such as that from cytomegalovirus (CMV), may result in transient transgene expression that peaks within the first 48 hrs and precipitously drops to baseline values within two weeks [45–47]. Such transient expression profiles are not ideal for the treatment of chronic lung disease, which requires sustained transgene expression at therapeutic levels. Alternatively, a number of eukaryotic promoters are capable of sustained pulmonary transgene expression, including the ubiquitin C [45, 47] and ubiquitin B [46, 48] promoters. In the present study, the human beta-actin promoter resulted in sustained luciferase activity that increased over time (up to two weeks) in mice treated with PEG-CH12K18 DNA nanoparticles or PEG-CK30 DNA nanoparticles by oropharyngeal aspiration. The sustained luciferase activity mediated by DNA nanoparticles is consistent with a recent report by Ziady et al. in which PEG-CK30 DNA nanoparticles containing a luciferase plasmid driven by the ubiquitin B promoter resulted in sustained pulmonary expression in C57BL/6 mice that peaked within the first 14 days [46]. The authors also reported negligible luciferase activity in mice that received naked DNA, suggesting that compaction was necessary for efficient uptake and expression of DNA. Similarly, a hundred-fold increase in luciferase activity was observed at day 2 in the lungs of C57BL/6j mice dosed with PEG-CK30 compacted DNA compared to naked DNA [7]. In comparison, we observed pulmonary luciferase activity in BALB/c mice treated with naked DNA at levels comparable to mice that received DNA nanoparticles at day 2 post administration; the reason for this difference is unclear but may be due to strain differences or other health factors. Interestingly, in our study the activity of naked DNA diminished after day 2 whereas luciferase activity increased for compacted DNA at day 14. The different time course of expression by DNA nanoparticles and naked DNA may be a result of differences in the kinetics of cellular uptake/processing and/or increased susceptibility of naked DNA to nucleases. These results suggest that DNA compaction may facilitate sustained pulmonary transgene expression.
5. Conclusion
In summary, we report that DNA nanoparticles formulated with a new pH-responsive polymer, PEG-CH12K18, mediate significant gene transfer in vitro and in vivo. Unlike PEG-CK30 DNA nanoparticles which enter cells and traffic to the nucleus via a nucleolin-mediated pathway, PEG-CH12K18 DNA nanoparticles formulated at pH 7.5 enter cells via clathrin-coated pits, where the poly-L-histidine moieties appear to have a proton sponge effect and result in escape from lysosomes. That PEG-CH12K18 DNA nanoparticles were more active than PEG-CK30 DNA nanoparticles in a surface nucleolin-negative cell line was expected, although the improvement in a pre-clinical lung model is noteworthy. We appreciate that the complexity of pathways utilized in nanoparticle uptake, as documented here, imply that the dynamics of cell surface nucleolin expression and other processes controlling CME will contribute significantly to the optimal nanoparticle design for safe and efficient gene transfer. Our data suggest that PEG-CH12K18 DNA nanoparticles may have a significant role in gene therapeutics and potential for clinical development. As uptake pathways for nanoparticles in various tissues and disease states are characterized, PEG-CH12K18 DNA nanoparticles may find use for the local delivery of therapeutic DNA to the lung and perhaps other organs.
Supplementary Material
Acknowledgements
Funding was provided by the National Institutes of Health (NIH R01EB003558 and P01HL51811) and a Wilmer Microscopy Core Facility Grant (EY001765). We are grateful to Professor Alexander Klibanov (Department of Chemistry, the Massachusetts Institute of Technology) for providing the plasmid pd1GL3-RL. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Biomedical Imaging and Bioengineering, the National Heart, Lung, and Blood Institute, or the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Griesenbach U, Alton EW. Gene transfer to the lung: lessons learned from more than 2 decades of CF gene therapy. Adv Drug Deliv Rev. 2009;61(2):128–39. doi: 10.1016/j.addr.2008.09.010. [DOI] [PubMed] [Google Scholar]
- [2].Ziady AG, Davis PB, Konstan MW. Non-viral gene transfer therapy for cystic fibrosis. Expert Opin Biol Ther. 2003;3(3):449–58. doi: 10.1517/14712598.3.3.449. [DOI] [PubMed] [Google Scholar]
- [3].Fasbender A, Zabner J, Zeiher BG, Welsh MJ. A low rate of cell proliferation and reduced DNA uptake limit cationic lipid-mediated gene transfer to primary cultures of ciliated human airway epithelia. Gene Ther. 1997;4(11):1173–80. doi: 10.1038/sj.gt.3300524. [DOI] [PubMed] [Google Scholar]
- [4].Suh J, Wirtz D, Hanes J. Efficient active transport of gene nanocarriers to the cell nucleus. Proc Natl Acad Sci U S A. 2003;100(7):3878–82. doi: 10.1073/pnas.0636277100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Schaffer DV, Fidelman NA, Dan N, Lauffenburger DA. Vector unpacking as a potential barrier for receptor-mediated polyplex gene delivery. Biotechnol Bioeng. 2000;67(5):598–606. doi: 10.1002/(sici)1097-0290(20000305)67:5<598::aid-bit10>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- [6].Liu G, Li D, Pasumarthy MK, Kowalczyk TH, Gedeon CR, Hyatt SL, et al. Nanoparticles of compacted DNA transfect postmitotic cells. J Biol Chem. 2003;278(35):32578–86. doi: 10.1074/jbc.M305776200. [DOI] [PubMed] [Google Scholar]
- [7].Ziady AG, Gedeon CR, Miller T, Quan W, Payne JM, Hyatt SL, et al. Transfection of airway epithelium by stable PEGylated poly-L-lysine DNA nanoparticles in vivo. Mol Ther. 2003;8(6):936–47. doi: 10.1016/j.ymthe.2003.07.007. [DOI] [PubMed] [Google Scholar]
- [8].Yurek DM, Fletcher AM, Smith GM, Seroogy KB, Ziady AG, Molter J, et al. Long-term transgene expression in the central nervous system using DNA nanoparticles. Mol Ther. 2009;17(4):641–50. doi: 10.1038/mt.2009.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Farjo R, Skaggs J, Quiambao AB, Cooper MJ, Naash MI. Efficient non-viral ocular gene transfer with compacted DNA nanoparticles. PLoS ONE. 2006;1:e38. doi: 10.1371/journal.pone.0000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ziady AG, Gedeon CR, Muhammad O, Stillwell V, Oette SM, Fink TL, et al. Minimal toxicity of stabilized compacted DNA nanoparticles in the murine lung. Mol Ther. 2003;8(6):948–56. doi: 10.1016/j.ymthe.2003.09.002. [DOI] [PubMed] [Google Scholar]
- [11].Ding XQ, Quiambao AB, Fitzgerald JB, Cooper MJ, Conley SM, Naash MI. Ocular delivery of compacted DNA-nanoparticles does not elicit toxicity in the mouse retina. PLoS ONE. 2009;4(10):e7410. doi: 10.1371/journal.pone.0007410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Chen X, Kube DM, Cooper MJ, Davis PB. Cell surface nucleolin serves as receptor for DNA nanoparticles composed of pegylated polylysine and DNA. Mol Ther. 2007;16(2):333–42. doi: 10.1038/sj.mt.6300365. [DOI] [PubMed] [Google Scholar]
- [13].Chen X, Shank S, Davis PB, Ziady AG. Nucleolin-mediated cellular trafficking of DNA nanoparticle is lipid raft and microtubule dependent and can be modulated by glucocorticoid. Mol Ther. 2011;19(1):93–102. doi: 10.1038/mt.2010.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Rejman J, Bragonzi A, Conese M. Role of clathrin- and caveolae-mediated endocytosis in gene transfer mediated by lipo- and polyplexes. Mol Ther. 2005;12(3):468–74. doi: 10.1016/j.ymthe.2005.03.038. [DOI] [PubMed] [Google Scholar]
- [15].Nguyen DN, Green JJ, Chan JM, Langer R, Anderson DG. Polymeric materials for gene delivery and DNA vaccination. Adv Mater. 2009;21(8):847–67. doi: 10.1002/adma.200801478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Pack DW, Hoffman AS, Pun S, Stayton PS. Design and development of polymers for gene delivery. Nat Rev Drug Discov. 2005;4(7):581–93. doi: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]
- [17].Wong SY, Pelet JM, Putnam D. Polymer systems for gene delivery--past, present, and future. Prog Polym Sci. 2007;32(8–9):799–837. [Google Scholar]
- [18].Behr J-P. The proton sponge: a trick to enter cells the viruses did not exploit. Chimia. 1997;51:34–6. [Google Scholar]
- [19].Sonawane ND, Szoka FC, Verkman AS. Chloride accumulation and swelling in endosomes enhances DNA transfer by polyamine-DNA polyplexes. J Biol Chem. 2003;278(45):44826–31. doi: 10.1074/jbc.M308643200. [DOI] [PubMed] [Google Scholar]
- [20].Wong SY, Sood N, Putnam D. Combinatorial evaluation of cations, pH-sensitive and hydrophobic moieties for polymeric vector design. Mol Ther. 2009;17(3):480–90. doi: 10.1038/mt.2008.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Midoux P, Monsigny M. Efficient gene transfer by histidylated polylysine/pDNA complexes. Bioconj Chem. 1999;10(3):406–11. doi: 10.1021/bc9801070. [DOI] [PubMed] [Google Scholar]
- [22].Zhong Z, Feijen J, Lok MC, Hennink WE, Christensen LV, Yockman JW, et al. Low molecular weight linear polyethylenimine-b-poly(ethylene glycol)-b-polyethylenimine triblock copolymers: synthesis, characterization, and in vitro gene transfer properties. Biomacromolecules. 2005;6(6):3440–8. doi: 10.1021/bm050505n. [DOI] [PubMed] [Google Scholar]
- [23].Rao GV, Tinkle S, Weissman DN, Antonini JM, Kashon ML, Salmen R, et al. Efficacy of a technique for exposing the mouse lung to particles aspirated from the pharynx. J Toxicol Environ Health A. 2003;66(15):1441–52. doi: 10.1080/15287390306417. [DOI] [PubMed] [Google Scholar]
- [24].Rennard SI, Basset G, Lecossier D, O'Donnell KM, Pinkston P, Martin PG, et al. Estimation of volume of epithelial lining fluid recovered by lavage using urea as marker of dilution. J Appl Physiol. 1986;60(2):532–8. doi: 10.1152/jappl.1986.60.2.532. [DOI] [PubMed] [Google Scholar]
- [25].Boylan NJ, Suk JS, Lai SK, Jelinek R, Boyle MP, Cooper MJ, et al. Highly compacted DNA nanoparticles with low MW PEG coatings: in vitro, ex vivo and in vivo evaluation. J Controlled Release. 2011 doi: 10.1016/j.jconrel.2011.08.031. doi:10.1016/j.jconrel.2011.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Atsuta J, Sterbinsky SA, Plitt J, Schwiebert LM, Bochner BS, Schleimer RP. Phenotyping and cytokine regulation of the BEAS-2B human bronchial epithelial cell: demonstration of inducible expression of the adhesion molecules VCAM-1 and ICAM-1. Am J Respir Cell Mol Biol. 1997;17(5):571–82. doi: 10.1165/ajrcmb.17.5.2685. [DOI] [PubMed] [Google Scholar]
- [27].Kim AJ, Boylan NJ, Suk JS, Lai SK, Hanes J. Non-degradative intracellular trafficking of highly compacted polymeric DNA nanoparticles. J Controlled Release. 2011 doi: 10.1016/j.jconrel.2011.10.031. doi:10.1016/j.jconrel.2011.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Lai SK, Hida K, Man ST, Chen C, Machamer C, Schroer TA, et al. Privileged delivery of polymer nanoparticles to the perinuclear region of live cells via a non-clathrin, non-degradative pathway. Biomaterials. 2007;28(18):2876–84. doi: 10.1016/j.biomaterials.2007.02.021. [DOI] [PubMed] [Google Scholar]
- [29].Rejman J, Oberle V, Zuhorn IS, Hoekstra D. Size-dependent internalization of particles via the pathways of clathrin- and caveolae-mediated endocytosis. Biochem J. 2004;377(Pt 1):159–69. doi: 10.1042/BJ20031253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Zuhorn IS, Kalicharan R, Hoekstra D. Lipoplex-mediated transfection of mammalian cells occurs through the cholesterol-dependent clathrin-mediated pathway of endocytosis. J Biol Chem. 2002;277(20):18021–8. doi: 10.1074/jbc.M111257200. [DOI] [PubMed] [Google Scholar]
- [31].Subtil A, Gaidarov I, Kobylarz K, Lampson MA, Keen JH, McGraw TE. Acute cholesterol depletion inhibits clathrin-coated pit budding. Proc Natl Acad Sci U S A. 1999;96(12):6775–80. doi: 10.1073/pnas.96.12.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Rodal SK, Skretting G, Garred Ø , Vilhardt F, van Deurs B, Sandvig K. Extraction of cholesterol with methyl-β-cyclodextrin perturbs formation of clathrin-coated endocytic vesicles. Mol Biol Cell. 1999;10(4):961–74. doi: 10.1091/mbc.10.4.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zhang L, Button B, Gabriel SE, Burkett S, Yan Y, Skiadopoulos MH, et al. CFTR delivery to 25% of surface epithelial cells restores normal rates of mucus transport to human cystic fibrosis airway epithelium. PLoS Biol. 2009;7(7):e1000155. doi: 10.1371/journal.pbio.1000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Konstan MW, Davis PB, Wagener JS, Hilliard KA, Stern RC, Milgram LJ, et al. Compacted DNA nanoparticles administered to the nasal mucosa of cystic fibrosis subjects are safe and demonstrate partial to complete cystic fibrosis transmembrane regulator reconstitution. Hum Gene Ther. 2004;15(12):1255–69. doi: 10.1089/hum.2004.15.1255. [DOI] [PubMed] [Google Scholar]
- [35].Suk JS, Boylan NJ, Trehan K, Tang BC, Schneider CS, Lin J-MG, et al. N-acetylcysteine enhances cystic fibrosis sputum penetration and airway gene transfer by highly compacted DNA nanoparticles. Mol Ther. 2011 doi: 10.1038/mt.2011.160. doi:10.1038/mt.2011.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Gratton S, Napier M, Ropp P, Tian S, DeSimone J. Microfabricated particles for engineered drug therapies: elucidation into the mechanisms of cellular internalization of PRINT particles. Pharm Res. 2008;25(12):2845–52. doi: 10.1007/s11095-008-9654-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Harush-Frenkel O, Debotton N, Benita S, Altschuler Y. Targeting of nanoparticles to the clathrin-mediated endocytic pathway. Biochem Biophys Res Commun. 2007;353(1):26–32. doi: 10.1016/j.bbrc.2006.11.135. [DOI] [PubMed] [Google Scholar]
- 38.Cho EC, Xie J, Wurm PA, Xia Y. Understanding the role of surface charges in cellular adsorption versus internalization by selectively removing gold nanoparticles on the cell surface with a I2/KI etchant. Nano Lett. 2009;9(3):1080–4. doi: 10.1021/nl803487r. [DOI] [PubMed] [Google Scholar]
- [39].Villanueva A, et al. The influence of surface functionalization on the enhanced internalization of magnetic nanoparticles in cancer cells. Nanotechnology. 2009;20(11):115103. doi: 10.1088/0957-4484/20/11/115103. [DOI] [PubMed] [Google Scholar]
- [40].Verma A, Stellacci F. Effect of surface properties on nanoparticle-cell interactions. Small. 2010;6(1):12–21. doi: 10.1002/smll.200901158. [DOI] [PubMed] [Google Scholar]
- [41].Moghimi SM, Symonds P, Murray JC, Hunter AC, Debska G, Szewczyk A. A two-stage poly(ethylenimine)-mediated cytotoxicity: implications for gene transfer/therapy. Mol Ther. 2005;11(6):990–5. doi: 10.1016/j.ymthe.2005.02.010. [DOI] [PubMed] [Google Scholar]
- [42].Beyerle A, Irmler M, Beckers J, Kissel T, Stoeger T. Toxicity pathway focused gene expression profiling of PEI-based polymers for pulmonary applications. Mol Pharm. 2010;7(3):727–37. doi: 10.1021/mp900278x. [DOI] [PubMed] [Google Scholar]
- [43].Merkel OM, Urbanics R, Bedocs P, Rozsnyay Z, Rosivall L, Toth M, et al. In vitro and in vivo complement activation and related anaphylactic effects associated with polyethylenimine and polyethylenimine-graft-poly(ethylene glycol) block copolymers. Biomaterials. 2011;32(21):4936–42. doi: 10.1016/j.biomaterials.2011.03.035. [DOI] [PubMed] [Google Scholar]
- [44].Plank C, Mechtler K, Szoka FC, Wagner E. Activation of the complement system by synthetic DNA complexes: a potential barrier for intravenous gene delivery. Hum Gene Ther. 1996;7(12):1437–46. doi: 10.1089/hum.1996.7.12-1437. [DOI] [PubMed] [Google Scholar]
- [45].Hyde SC, Pringle IA, Abdullah S, Lawton AE, Davies LA, Varathalingam A, et al. CpG-free plasmids confer reduced inflammation and sustained pulmonary gene expression. Nat Biotechnol. 2008;26(5):549–51. doi: 10.1038/nbt1399. [DOI] [PubMed] [Google Scholar]
- [46].Ziady AG, Kotlarchyk M, Bryant L, McShane M, Lee Z. Bioluminescent imaging of reporter gene expression in the lungs of wildtype and model mice following the administration of PEG-stabilized DNA nanoparticles. Microsc Res Tech. 2010;73(9):918–28. doi: 10.1002/jemt.20855. [DOI] [PubMed] [Google Scholar]
- [47].Gill DR, Smyth SE, Goddard CA, Pringle IA, Higgins CF, Colledge WH, et al. Increased persistence of lung gene expression using plasmids containing the ubiquitin C or elongation factor 1alpha promoter. Gene Ther. 2001;8(20):1539–46. doi: 10.1038/sj.gt.3301561. [DOI] [PubMed] [Google Scholar]
- [48].Yew NS, Przybylska M, Ziegler RJ, Liu D, Cheng SH. High and sustained transgene expression in vivo from plasmid vectors containing a hybrid ubiquitin promoter. Mol Ther. 2001;4(1):75–82. doi: 10.1006/mthe.2001.0415. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.