Abstract
Background and objective
Gemcitabine is widely used to treat non-small cell lung cancer (NSCLC). To assess the pharmacogenomic effects of the entire gemcitabine metabolic pathway, we genotyped SNPs within the 17 pathway genes using DNA samples from NSCLC patients treated with gemcitabine to determine the effect of genetic variants within gemcitabine pathway genes on overall survival (OS) of NSCLC patients after treatment of gemcitabine.
Methods
Eight of the 17 pathway genes were resequenced with DNA samples from Coriell lymphoblastoid cell lines (LCLs) using Sanger sequencing for all exons, exon-intron junctions and 5′-, 3′-UTRs. A total of 107 tag SNPs were selected based on the resequencing data for the 8 genes and on HapMap data for the remaining 9 genes, followed by successful genotyping of 394 NSCLC patient DNA samples. Association of SNPs/haplotypes with OS was performed using the Cox regression model, followed by functional studies performed with LCLs and NSCLC cell lines.
Results
5 SNPs in 4 genes (CDA, NT5C2, RRM1, and SLC29A1) showed associations with OS of those NSCLC patients, as well as 9 haplotypes in 4 genes (RRM1, RRM2, SLC28A3, and SLC29A1) with P < 0.05. Genotype imputation using the LCLs was performed for a region of 200kb surrounding those SNPs, followed by association studies with gemcitabine cytotoxicity. Functional studies demonstrated that downregulation of SLC29A1, NT5C2, and RRM1 in NSCLC cell lines altered cell susceptibility to gemcitabine.
Conclusion
These studies help identify biomarkers to predict gemcitabine response in NSCLC, a step toward the individualized chemotherapy of lung cancer.
Keywords: gemcitabine, pharmacogenomics, metabolic pathway, lymphoblastoid cell lines, non-small cell lung cancer (NSCLC)
Introduction
Lung cancer is one of the most common malignancies and the leading cause of cancer-related mortality in the United States [1, 2]. Approximately 222,520 new cases and 157,300 deaths from lung cancer were estimated to occur in the United States in 2010 [3]. In spite of the use of newer agents in lung cancer treatment, chemotherapy is still the front line treatment, and cytotoxic chemotherapy is the standard care for patients with advanced NSCLC [4–6]. Standard therapeutic regimens are platinum-based doublets (platinum plus another agent). The second agent used with platinum includes microtubule-targeted agents (paclitaxel, docetaxel, or vinorelbine), cytidine analogues (gemcitabine) [7], and DNA-damaging agents (irinotecan) [6, 7]. The efficacy of each of these combinations has been shown to be similar by a series of trials, with response rates from 30% to 40% [8–10]. Therefore, the investigation of potential genetic variation that might contribute to the efficacy of these chemotherapy regimens might help make it possible to better individualize therapies for lung cancer.
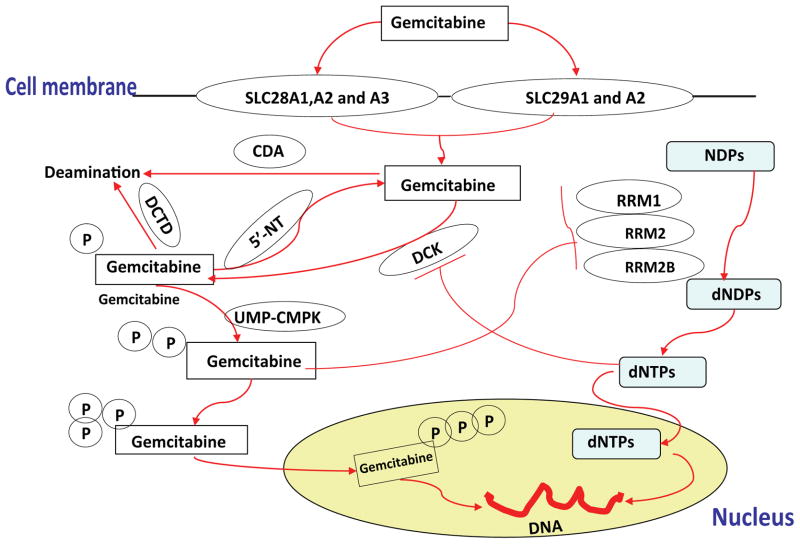
Gemcitabine, a cytidine analogue, shows significant therapeutic effect in a variety of cancers including NSCLC. It is approved by the FDA in combination with cisplatin for first-line treatment of patients with locally advanced (stage IIIA or stage IIIB) or metastatic (stage IV or cancer that has spread) NSCLC [7, 11]. Gemcitabine is a prodrug that must be transported into cells, activated by kinases to form active di- and triphosphorylated metabolites, and inactivated by dephosphorylation or deamination [12]. The triphosphorylated metabolite, gemcitabine TP, can be incorporated into DNA, terminating DNA synthesis, while gemcitabine DP can inhibit ribonucleotide reductases (RRs), enzymes that catalyze the conversion of ribonucleotides to deoxyribonucleotidase [13–15]. Seventeen genes are known to be involved in this cytidine analogue “pathway” (Fig. 1).
Figure 1. Schematic diagram of the gemcitabine drug metabolizing pathway.
Seventeen genes are known to be involved in the “gemcitabine metabolism pathway”. These include two different types of nucleoside transporters, human equilibrative nucleoside transporters (hENT) and human concentrative nucleoside transporters (hCNT). Gemcitabine has been shown to be a substrate for two hENT family members (SLC29A1 and 29A2) and three hCNT family members (SLC28A1, A2 and A3) [37, 52]. Once gemcitabine is inside the cell, it is phosphorylated by deoxycytidine kinase (DCK) to form a monophosphorylated metabolite, difluorodeoxycytidine monophosphate (gemcitabineMP) [13]. This metabolite is subsequently phosphorylated by nucleoside monophosphate (CMPK) and nucleoside diphosphate kinase to generate active forms of the drug, difluorodeoxycytidine diphosphate (gemcitabineDP) and difluorodeoxycytidine triphosphate (gemcitabineTP) [13]. These phosphorylated gemcitabine metabolites can be dephosphorylated by 5′-nucleotidase (5′-NT) family members [40], including 5 known 5′-nucleotidases: NT5C, NT5C1A, NT5C1B, NT5C2 and NT5C3. Gemcitabine is inactivated by deamination catalyzed by cytidine deaminase (CDA) and deoxycytidylate deaminase (DCTD) [23]. GemcitabineDP can inhibit ribonucleotide reductase (RRM1, RRM2 and RRM2B) enzymes that catalyze reactions which generate deoxynucleoside triphosphates required for DNA synthesis [41].
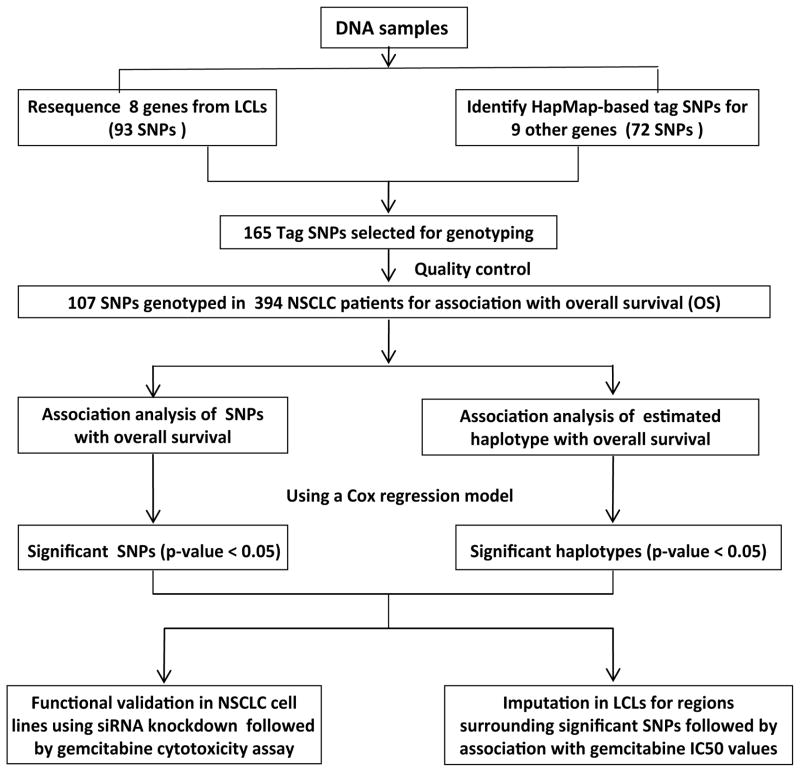
Although gemcitabine is commonly used in the treatment of NSCLC, response to this drug varies widely. Evidence has shown that sequence variation in genes involved in the gemcitabine transport, metabolism and bioactivation pathway contribute to variation in [16–21]. However, few comprehensive pharmacogenomic studies taking all of the pathway genes into account have been performed. Therefore, the objective of the present study is to test the hypothesis that SNPs in genes within gemcitabine pathway might be associated with treatment outcomes of NSCLC patients who were treated with gemcitabine (Fig. 2). To further validate the findings from the clinical study, we also performed functional studies by using siRNA knockdown of candidate genes in cultured cells. Taken together, the pathway-based approach with clinical patient samples, followed by functional studies could help us to identify and understand genetic biomarkers involved in the gemcitabine metabolism and activation pathway for the treatment of lung cancer.
Figure 2. Experimental strategy.
Tag SNPs were selected based on either our gene resequencing data or HapMap data, followed by genotyping of DNA samples from NSCLC patients. Top findings were validated by functional assays using siRNA knockdown in lung cancer cell lines and gemcitabine cytotoxicity in LCLs.
Materials and Methods
DNA samples and cell lines
DNA samples from 60 Caucasian-American (CA), 60 African-American (AA) and 60 Han Chinese-American (HCA) subjects (sample sets HD100CAU, HD100AA and HD100CHI) were obtained from the Coriell Cell Repository (Camden, NJ). These DNA samples have been widely used for human genetic studies [22–25]. Immortalized lymphoblastoid cell lines (LCLs) from the same subjects from whom the DNA was obtained are also available. We have already generated over 1.3 million SNPs, microarray expression data, and gemcitabine cytotoxicity data for these LCLs, as described previously [26, 27]. All of these DNA samples and cell lines were anonymized by the National Institute of General Medical Sciences prior to deposit, and all subjects had provided written consent for the use of their DNA and cells for experimental purposes. This study was reviewed and approved by Mayo Clinic Institutional Review Board. The human NSCLC A549 and H1437 cell lines were obtained from the American Type Culture Collection (ATCC, Manassas, VA), and were cultured in RPMI 1640 medium containing 10% FBS.
Lung cancer patient samples
Three hundred and ninety four lung cancer patients with gemcitabine-based treatment were enrolled and identified at Mayo Clinic (Rochester, MN) between 1997 and 2008. Details with regard to clinical characteristics of patients, patient enrollment, and data collection procedures were described previously [25, 28, 29]. Briefly, each case was identified through the Mayo Clinic pathologic diagnostic (Co-Path) system. After obtaining written informed consent, blood samples were collected from the patients. Patient characteristics of patients were abstracted from the medical record, including demographics, lung cancer pathology, anatomic site, and types and timing of treatment and chemotherapeutic agents. Clinical staging and recurrence or progression were determined based on the results of chest radiography, computerized tomography, bone scans, position emission tomography scans, and magnetic resonance imaging. All patients were actively followed during the initial six months after diagnosis, with subsequent annual follow-up by mailed questionnaires and annual verification of patients’ vital status. All research protocols were received and approved by the Mayo Clinic Institutional Review Board.
In addition to gemcitabine, many of the patients were also treated with radiation therapy and/or surgery, as well as 4 other classes of anticancer drugs: platinum agents, paclitaxel, etopside and EGFR inhibitors. Detailed patient demographic information is listed in Table 1. Overall survival (OS) time, was defined as the time from lung cancer diagnosis to either death or the last known date alive (i.e., censored at this date), was used as the primary endpoint.
Table 1.
Characteristics of 394 NSCLC patients who were treated with gemcitabine-based therapy
| Characteristics of diagnosis and treatment | Values and percentages |
|---|---|
| Age at Diagnosis | |
| Mean (SD) | 61.9 (10.94) |
| Median (range) | 63.0 (35.0 – 88.0) |
| Gender | |
| Female | 179 (45.4%) |
| Male | 215 (54.6%) |
| Cigarette smoking status | |
| Never | 100 (25.4%) |
| Former smokers | 194 (49.2%) |
| Current smokers | 96 (24.4%) |
| Some Smokers | 4 (1%) |
| NSCLC Stage | |
| Stage I and II | 69 (17.5%) |
| Stage III | 143 (36.3%) |
| Stage IV | 182 (46.2%) |
| Histologic Cell Type | |
| Adenocarcinoma/Bronchioloalveolar carcinoma | 253 (64.2%) |
| Squamous cell carcinoma | 60 (15.2%) |
| Large Cell carcinoma | 6 (1.5%) |
| Mixed and unspecified NSCLC | 70 (17.7%) |
| Others | 5 (1.3%) |
| Tumor Differentiation Grade | |
| Well differentiated | 44 (11.2%) |
| Moderately differerntiated | 167 (42.4%) |
| Poor/undifferentiated | 145 (36.8%) |
| Nongradable or unknown | 38 (9.6%) |
| Treatment Modality | |
| Chemotherapy | 132 (33.5%) |
| Surgery & Chemotherapy | 70 (17.8%) |
| Radiation & Chemotherapy | 126 (32%) |
| Surgery & Radiation & Chemotherapy | 66 (16.8%) |
|
| |
| Total | 394 (100%) |
Stage for NSCLC is defined on the basis of the tumor-node-metastasis classification.
Resequencing analysis
DNA samples from 3 different ethnic groups (60 CA, 60 HCA and 60 AA) were used to perform PCR amplifications to resequence 8 genes within the cytidine analogue metabolic pathway, including 5 that have been already published [22, 23, 30]. Resequencing for the other 3 genes was performed using the primers and PCR conditions described in Supplementary Table S1. Resequenced regions included all exons, intron-exon splice junctions, 1,000 bp of 5′-flanking regions for each of the noncoding exons, and the 3′-untranslated regions (UTR). Resequencing was performed using dye terminator sequencing chemistry. Amplicons were sequenced on both strands in the Mayo Molecular Biology Core Facility with an ABI 377 DNA sequencer. To exclude PCR-induced artifacts, independent amplifications were performed for samples in which a SNP was observed only once or for any sample with an ambiguous chromatogram. The chromatograms were analyzed with Mutation Surveyor (SoftGenetics, State College, PA).
Genotyping in lung cancer patients
To determine the effect of genetic variation in the gemcitabine pathway genes on response, tag SNPs in 17 pathway genes were selected on the basis of either our own in-depth resequencing data or HapMap Release 22 (Phase II) phased haplotype data by using Tagger, a tool for the selection and evaluation of tag SNPs [31, 32]. Tag SNPs were identified with a pair-wise linkage disequilibrium threshold of r2 ≥ 0.8, followed by genotyping 394 DNA samples from NSCLC patients, using the Illumina GoldenGate platform. Quality control tests for genotyping were performed to remove SNPs with call rate < 95%, minor allele frequencies (MAF) < 0.05, and Hardy-Weinberg equilibrium (HWE) P value < 0.001, as well as monomorphic SNPs. SNPs with Illumina scores < 0.4 were excluded, which resulted in 107 tag SNPs for further study. Concordance among three genomic control DNA samples present in duplicate was 100%.
Imputation analysis in LCLs and lung cancer samples
Imputation analysis was performed in a region 200kb up and downstream of candidate SNPs/or genes that were associated with OS of lung cancer patients. Imputation was performed with genome-wide SNP data obtained for LCLs with HapMap Release 22 (Phase II) phased haplotype data (http://hapmap.ncbi.nlm.nih.gov/) as the reference panel using MACH 1.0 software [33]. Specifically, SNP markers for AA subjects were imputed using both CEU and YRI data; SNPs for CA subjects were imputed based on CEU data; and SNP markers for HCA subjects were imputed using CHB and JPT data. For quality control purposes, imputed SNPs with score MACH Rsq values less than 0.3 were removed from the analysis.
Data analysis
Association analysis of SNPs/haplotypes with OS in NSCLC patients
Following genotyping of tag SNPs for each pathway gene, association analysis was performed with OS as the phenotype using the Cox proportional hazards model. Backward selection was performed to determine whether associations with SNPs should be adjusted for the clinical covariates of age at diagnosis, gender, smoking status, disease stage, and treatment. Disease stage was included in the final multivariate Cox-regression model since it was significantly associated with OS for the lung cancer patients. Disease stage was divided into stages I + II versus III versus IV. The association of SNPs with OS in NSCLC patients was assessed using a Cox regression model that included the effects of SNP genotype dosage (count of minor allele) and adjustment for stage. The association was repeated for the top 5 SNPs including both stage and treatment. Hazard ratios (HRs) and 95% confidence intervals (CI) were estimated for SNP alleles. HR values > 1 indicated that subjects who carried the minor allele had worse survival. We used 0.05 as a cutoff for P values (without adjustment for multiple testing) to select SNPs/genes for further functional validation.
Haplotypes have been shown to provide increased power to identify rare causal variants compared to single markers [34]. Therefore, we performed haplotype analysis for the subsequent association studies to supplement the single marker approach. To perform full haplotype analysis for every haplotype in each of the pathway gene, we used the haplo.stats software package (http://cran.r-project.org/web/packages/haplo.stats) to estimate haplotype frequencies and to obtain posterior probabilities of haplotype pairs for each subject conditional on the observed genotype data. We focused on those haplotypes with predicted count ≥ 5 for further association analysis. Using the haplo.stats framework, a Cox regression model was used with the haplotype design matrix, incorporating haplotype pair posterior probabilities and adjusted for tumor stage to test the effects of haplotypes across a gene, given the available tagging SNPs and stage [31].
Association analysis of SNPs with gemcitabine IC50 values and gene expression in LCLs
To help determine the possible function of SNPs found to be associated with OS in the NSCLC patients, we performed genotype-phenotype association studies with gemcitabine IC50 values in the LCLs, using the genotyped SNPs present on the Illumina 550K, Illumina 510s, and Affymetrix 6.0 SNP chips, plus imputed SNPs for the pathway genes, as described previously [35]. The associations of SNPs with mRNA expression levels of pathway genes in the same cell lines were assessed using Pearson correlation analysis, after normalization and covariate adjustment as described previously [35].
Additional statistical analyses
Gemcitabine cytotoxicity for lung cancer cell lines was determined by comparison of area under the dose-response curve (AUC) values between cells treated with negative control siRNA and gene-specific siRNA using a two-tailed unpaired t-test.
Transient transfection and cell proliferation assay
siRNA pools with a set of 4 specific siRNAs for candidate genes and negative non-target control siRNA pool were purchased from Dharmacon (Chicago, IL). Reverse transfection of siRNA was performed in 96-well plates with either A549 or H1437 cells, using 0.2 μL of lipofectamine RNAi-MAX reagent (Invitrogen, Carlsbad, CA), and siRNA pools at a final concentration of 50 nM for 24 hours. Cells were then treated with gemcitabine at final concentrations of 0, 001, 0.01, 0.1, 1, 10, 100, and 1000 μM. After 72 hours of incubation with gemcitabine, cytotoxicity assay were performed using the Cell Titer 96 AQueous Non-Radioactive Cell Proliferation Assay kit (Promega Corporation, Madison, WI), followed by absorbance measurement at 490 nm in a Safire2 microplate reader (Tecan AG, Switzerland).
Real-time quantitative reverse transcription-PCR (qRT-PCR)
Total RNA was isolated from cultured cells using the Quick-RNA™ MiniPrep kit (Zymo Research, Orange, CA), followed by qRT-PCR performed with the Power SYBR® Green RNA-to-CTTM 1-Step Kit (AB Foster CA). Specifically, primers purchased from QIAGEN were used to perform qRT-PCR using the Stratagene Mx3005P Real-Time PCR detection system (Agilent Technologies, Santa Clara, CA). All experiments were corrected by using beta-actin as an internal control.
Results
Gene resequencing and tag SNP selection
Among the 17 genes within the gemcitabine pathway (Figs. 1 and 2), we resequenced 8 genes, 5 genes (DCK, CMPK, DCTD, NT5C3, and CDA) published [22, 23, 30] as well as 3 additional genes encoding the ribonucleotide reductase subunits, RRM1, RRM2 and RRM2B. All of the resequencing data have been deposited in the PharmGKB NIH database (http://www.pharmgkb.org/). More than half of the SNPs that we obtained during resequencing were novel. For example, for the three unpublished genes, 24 of 72 SNPs in RRM1, 4 of 35 in RRM2, and 28 of 51 in RRM2B were in a publicly available database, while the remainder was not included in NCBI Build 36.3 (http://www.ncbi.nlm.nih.gov/nuccore).
Since this project was initiated over two years ago, and genotyping was performed prior to completion of the 1000 Genomes Project [36], we performed tag SNP selection based on our own resequencing data and HapMap Release 22 (Phase II) phased haplotype data. Specifically, 325 SNPs for those 8 resequenced genes were used for the subsequent tag SNP selection. One hundred and twelve of these 325 SNPs were recently identified by the 1000 Genomes Project. A total of 165 tag SNPs were selected, including 93 SNPs based on resequencing data for 8 genes and 82 based on HapMap data for the remaining 9 pathway genes (NT5C, NT5C1A, NT5C1B, NT5C2, SLC28A1, SLC28A2, SLC28A3, SLC29A1, and SLC29A) using Tagger [32]. After quality control as described in the Methods, 107 SNPs among these 17 pathway genes were genotyped with the Illumina GoldenGate platform using 394 DNA samples from NSCLC patients (Fig. 2 and Supplementary Table S2).
Survival analysis for gemcitabine pathway SNPs/haplotypes in NSCLC patients
To determine genetic variation in the gemcitabine pathway that might contribute to the outcome of gemcitabine treatment of NSCLC patients, we performed an association analysis of tag SNPs for those genes with OS (Fig. 2). The results showed that 5 SNPs located in introns of 4 genes, NT5C2, SLC29A1, RRM1, and CDA, were associated with OS (P < 0.05). However none of these associations achieved statistical significance after Bonferroni correction for multiple comparisons (Table 2A and Supplementary Table 2). We have adjusted for the stage during the analysis. However, since these were heterogeneous patients with regard to their tumor stage and treatment, we redid analysis with the top 5 SNPs, but including both stage and prior treatment as adjusting covariates, the P values remained <0.05. Haplotype association analysis was also performed to assess the effect of haplotypes for each gemcitabine pathway gene on OS. Nine haplotypes in 4 genes, SLC28A3, RRM1, RRM2, and SLC29A1, were associated with OS (P < 0.05) (Table 2B). Five haplotypes in SLC28A3, RRM1, and RRM2 showed association P values < 0.01 (Table 2B). None of these associations were significant after Bonferroni correction since we had conducted 135 tests with haplotypes. The most significant haplotype in SLC28A3, with an estimated frequency of 0.008, was associated with OS with an HR = 5.35 (P value = 0.0001).
Table 2.
Association analysis of SNPs/haplotypes with OS in NSCLC patients treated with gemcitabine
| (A). The top 5 SNPs associated with patient OS.
| ||||||||
|---|---|---|---|---|---|---|---|---|
| SNP | P_value | HR (95% CI) | Gene | Chromosome | Position | MAF_Caucasion | Location | Allele |
| rs2274341 | 0.011 | 1.256 (1.053 – 1.499) | NT5C2 | 10 | 105000000 | 0.229 | Intron | C/T |
| rs9394992 | 0.012 | 0.792 (0.660 – 0.950) | SLC29A1 | 6 | 44302323 | 0.317 | Intron | A/G |
| rs720106 | 0.016 | 1.899 (1.124 – 3.206) | RRM1 | 11 | 4096554 | 0.017 | Intron | A/G |
| rs747199 | 0.027 | 0.782 (0.630 – 0.972) | SLC29A1 | 6 | 44302323 | 0.192 | Intron | A/G |
| rs818194 | 0.039 | 1.209 (1.009 – 1.448) | CDA | 1 | 20804415 | 0.208 | Intron | C/T |
| (B). Haplotype analysis for OS in NSCLC patients.
| |||||
|---|---|---|---|---|---|
| Gene | Hap code | P_value | HR (95% CI) | Haplotype | Hap Freq |
| RRM1 | mhap.2 | 0.007 | 0.316 (0.137 – 0.730) | ACGA | 0.017 |
| mhap.4 | 0.008 | 0.458 (0.257 – 0.815) | CCAA | 0.097 | |
| mhap.5 | 0.033 | 0.565 (0.334 – 0.955) | CCGA | 0.590 | |
| mhap.1 | 0.046 | 0.574 (0.332 – 0.991) | ACAA | 0.242 | |
|
| |||||
| RRM2 | mhap.3 | 0.009 | 1.308 (1.070 – 1.599) | CA | 0.201 |
|
| |||||
| SLC28A3 | mhap.66 | 0.001 | 4.648 (1.890 – 11.433) | GAGCAGAAAGGGAACCGGAGACATTAAGA | 0.008 |
| mhap.143 | 0.022 | 0.523 (0.301 – 0.912) | GCACGAGCAAGGGACCGGAGACAATAAGA | 0.028 | |
| mhap.116 | 0.037 | 0.681 (0.474 – 0.977) | GAGCAGGCGGGGGGGCGGAGACATTAAGA | 0.087 | |
|
| |||||
| SLC29A1 | mhap.15 | 0.028 | 0.019 (0.001 – 0.649) | CGAGA | 0.007 |
HR, hazard ratio; HR > 1 indicates that subjects carrying the minor allele had a worse OS. The 95% confidence interval (CI) for the HR is listed with the P-value.
SNP association analysis with both gemcitabine IC50 values and pathway gene expression in LCLs
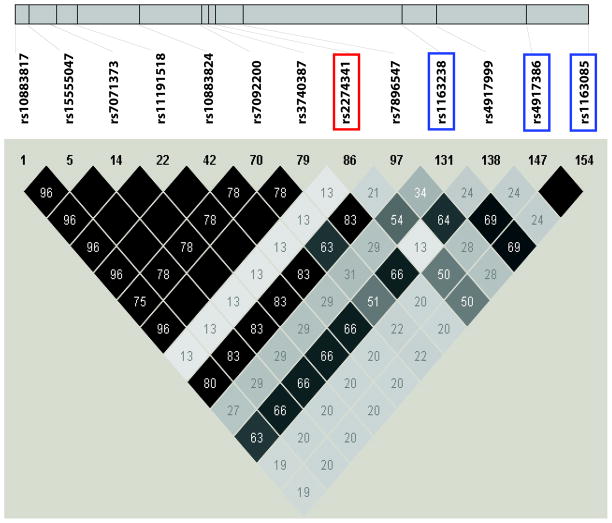
In an attempt to determine the possible functional implications of the top genetic variants observed in the NSCLC patient samples, we took advantage of LCLs for which we had obtained 1.3 million genome-wide SNPs, expression array data and gemcitabine IC50 values (Fig. 2) [27, 35]. Specifically, we examined a total of 1311 SNPs for the pathway genes that had been genotyped on our GWAS platforms and also imputed an additional 600 SNPs in regions extending 200kb up and downstream of the top SNPs in the LCLs within the 6 genes (NT5C2, CDA, SLC29A1, RRM1, RRM2, and SLC28A3) that were identified using the clinical DNAS samples, followed by association analysis of those SNPs with gemcitabine IC50 values to identify potential functional SNPs. As a result, the survival-associated SNP in NT5C2, rs2274341, was associated with gemcitabine IC50 values (P = 0.045), but so were 12 additional SNPs in this gene (P < 0.05) (Table 3 and Fig. 3). Among those 12 SNPs, the rs1163238 SNP was moderately linked to rs2274241 (r2 = 0.543, and D′ = 1) and had the smallest P value = 0.018 (Table 3 and Fig. 3). Furthermore, there were two clinical survival-associated SNPs in SLC29A1, rs9394992 and rs747199, which showed associations with gemcitabine IC50 values in LCLs, with P values of 0.026 and 0.155, respectively. In addition, 4 SNPs, rs9472236, rs4714772, rs1875324, and rs3757283 in SLC29A1, also showed associations with gemcitabine IC50 values with small P values (Psmallest = 0.013) (Table 3). Finally, one SNP (rs10868141) within the strongest clinical survival-associated haplotype in SLC28A3 was also associated with gemcitabine IC50 values (P = 0.048) (Table 3). Since the frequency of this haplotype in SLC28A3 was low, we did not have power to detect haplotype associations in LCLs.
Table 3.
Association of top SNPs with gemcitabine IC50 values in LCLs
| SNP | Gene | Chromosome | Position | SNP type | R-value | P-value | R2 | D′ |
|---|---|---|---|---|---|---|---|---|
| rs818194 | CDA | 1 | 20804415 | i | 0.034 | 0.653 | NA | NA |
|
| ||||||||
| rs1163238 | NT5C2 | 10 | 104933983 | i | 0.181 | 0.018 | 0.545 | 1.000 |
| rs4917999 | NT5C2 | 10 | 104953041 | o | 0.180 | 0.021 | 0.136 | 1.000 |
| rs10883817 | NT5C2 | 10 | 104745421 | o | 0.163 | 0.033 | 0.131 | 0.810 |
| rs4917386 | NT5C2 | 10 | 104997518 | o | −0.162 | 0.034 | 0.505 | 0.945 |
| rs1163085 | NT5C2 | 10 | 105029231 | o | −0.161 | 0.035 | 0.505 | 0.945 |
| rs7896547 | NT5C2 | 10 | 104856948 | i | 0.159 | 0.037 | 0.210 | 1.000 |
| rs7092200 | NT5C2 | 10 | 104834862 | o | 0.158 | 0.039 | 0.138 | 0.757 |
| rs3740387 | NT5C2 | 10 | 104839458 | o | 0.158 | 0.042 | 0.137 | 0.821 |
| rs12255047 | NT5C2 | 10 | 104750742 | i | 0.155 | 0.043 | 0.137 | 0.821 |
| rs7071373 | NT5C2 | 10 | 104761912 | i | 0.155 | 0.043 | 0.137 | 0.821 |
| rs11191518 | NT5C2 | 10 | 104772843 | i | 0.154 | 0.044 | 0.137 | 0.821 |
| rs2274341 | NT5C2 | 10 | 104843493 | i | 0.153 | 0.045 | 1.000 | 1.000 |
| rs10883824 | NT5C2 | 10 | 104802887 | o | 0.151 | 0.048 | 0.137 | 0.821 |
|
| ||||||||
| rs720106 | RRM1 | 11 | 4096554 | i | −0.026 | 0.731 | NA | NA |
|
| ||||||||
| rs4668664 | RRM2 | 2 | 10186249 | o | 0.049 | 0.521 | NA | NA |
| rs6741290 | RRM2 | 2 | 10182160 | o | −0.049 | 0.524 | NA | NA |
| rs6759180 | RRM2 | 2 | 10184014 | o | 0.024 | 0.758 | NA | NA |
| rs10868141 | SLC28A3 | 9 | 86111098 | i | 0.151 | 0.048 | NA | NA |
| rs4585823 | SLC28A3 | 9 | 86094005 | o | −0.143 | 0.061 | NA | NA |
| rs7035188 | SLC28A3 | 9 | 86123225 | o | −0.120 | 0.116 | NA | NA |
| rs4877832 | SLC28A3 | 9 | 86089767 | i | 0.119 | 0.119 | NA | NA |
| rs12004882 | SLC28A3 | 9 | 86127879 | i | −0.117 | 0.128 | NA | NA |
| rs11140523 | SLC28A3 | 9 | 86125404 | o | 0.110 | 0.152 | NA | NA |
| rs9792596 | SLC28A3 | 9 | 86115694 | i | −0.083 | 0.279 | NA | NA |
| rs10746739 | SLC28A3 | 9 | 86117750 | o | −0.083 | 0.279 | NA | NA |
| rs3812509 | SLC28A3 | 9 | 86144977 | o | 0.079 | 0.305 | NA | NA |
| rs17087148 | SLC28A3 | 9 | 86158872 | o | −0.068 | 0.376 | NA | NA |
| rs17343066 | SLC28A3 | 9 | 86135893 | o | 0.066 | 0.391 | NA | NA |
| rs11140507 | SLC28A3 | 9 | 86115568 | o | −0.061 | 0.432 | NA | NA |
| rs4877839 | SLC28A3 | 9 | 86118829 | o | −0.060 | 0.435 | NA | NA |
| rs11140525 | SLC28A3 | 9 | 86127135 | o | 0.054 | 0.483 | NA | NA |
| rs13283683 | SLC28A3 | 9 | 86131263 | o | 0.049 | 0.525 | NA | NA |
| rs4305983 | SLC28A3 | 9 | 86134271 | o | −0.047 | 0.544 | NA | NA |
| rs17087056 | SLC28A3 | 9 | 86106827 | o | 0.040 | 0.606 | NA | NA |
| rs4877831 | SLC28A3 | 9 | 86089704 | o | 0.038 | 0.618 | NA | NA |
| rs10435946 | SLC28A3 | 9 | 86145628 | o | 0.022 | 0.778 | NA | NA |
| rs11140544 | SLC28A3 | 9 | 86156579 | o | −0.010 | 0.892 | NA | NA |
| rs4877848 | SLC28A3 | 9 | 86140690 | o | 0.001 | 0.990 | NA | NA |
|
| ||||||||
| rs9472236 | SLC29A1 | 6 | 44314728 | o | 0.190 | 0.013 | 0.071 | 1.000 |
| rs4714772 | SLC29A1 | 6 | 44315095 | o | 0.190 | 0.013 | 0.076 | 1.000 |
| rs1875324 | SLC29A1 | 6 | 44330988 | o | 0.174 | 0.022 | 0.093 | 1.000 |
| rs3757283 | SLC29A1 | 6 | 44322395 | o | 0.174 | 0.022 | 0.098 | 1.000 |
| rs9394992 | SLC29A1 | 6 | 44303970 | o | −0.170 | 0.026 | 1.000 | 1.000 |
| rs747199 | SLC29A1 | 6 | 44302323 | o | −0.109 | 0.155 | 0.015 | 0.173 |
The SNPs in bold are the top SNPs associated with gemcitabine IC50 values in LCLs (P < 0.05). SNPs highlighted in gray are tag SNPs that were genotyped and showed SNP/or haplotype associations with OS in NSCLC patients (P < 0.05). R2 and D′ between the top survival-associated SNPs and linked SNPs identified based on the HapMap Phase II Release 22 are indicated. NA indicates R2 less than 0.1 and is considered no linkage between the two SNPs. “I” indicated SNPs imputed using HapMap data as the reference panel, and “o” indicated SNPs observed in LCLs on the basis of Illumina and Affymetrix SNP chip genotyping.
Figure 3. NT5C2 Haplotype structure.
The haplotype structure of NT5C2 was generated based on HapMap Phase II Release 22 data. Colors ranging from black to gray to white indicate the range of R2 values from high to low. The SNP that we identified in the survival analysis for NSCLC patients is in the red box, whereas SNPs in blue boxes are linked to this SNP with R2 > 0.5.
One possible mechanism by which these SNPs might have functional impact is through transcription regulation. Therefore, we also examined the relationship of these SNPs and haplotypes to gene expression in the LCLs through cis-regulation. We found that the survival-associated SNP in RRM1, rs720106, was associated with mRNA expression level of RRM1 in the LCLs (P = 9.9 × 103), suggesting that this SNP might have an effect on variation in gemcitabine response through cis-regulation of gene expression. However, none of the other SNPs showed a significant association with mRNA gene expression for the corresponding gene in the LCLs. These results indicated the existence of other potential mechanisms by which those SNPs might influence gemcitabine response if the associations that we observed can be replicated.
RNA inference functional validation of 6 pathway genes in NSCLC cell lines
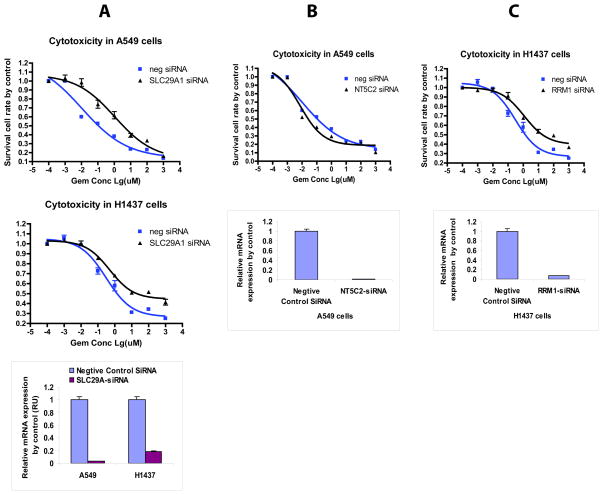
Functional studies of the 6 genes identified during our clinical study (NT5C2, RRM1, RRM2, CDA, SLC28A3, and SLC29A1) were performed using siRNA knockdown in human NSCLC A549 and H1437 cell lines (Fig. 2). Knockdown of SLC29A1 in both A549 and H1437 cells desensitized the cells to gemcitabine (Fig. 4A), which is consistent with the function of SLC29A1 since it is a transporter involved in uptake of gemcitabine into cells [37, 38]. Furthermore, knockdown of NT5C2 in A549 cells sensitized cells to gemcitabine, while downregulation of RRM1 in H1437 cells resulted in enhanced resistance, observations consistent with the biological functions of these genes (Fig. 4B and 4C) [39–41]. NT5C2 is a nucleotidase that dephosphorylates active phosphorylated gemcitabine metabolites to make the drug less active. Therefore, knockdown of NT5C2 would be expected to result in sensitization of cells to gemcitabine. RRM1 is a target for gemcitabine. Therefore, decreased expression of RRM1 would be anticipated to cause resistance to gemcitabine. In all of our experiments, knockdown efficiency was confirmed using real-time QRT-PCR (Fig. 4).
Figure 4. Functional validation in two human non-small cell lung cancer (NSCLC) cell lines.
Cells were treated with control siRNA or specific siRNA for (A) SLC29A1, (B) NT5C2 or (C) RRM1, followed by MTS assays. QRT-PCR was used to determine knockdown efficiency.
Discussion
Gemcitabine, a cytidine analogue, has been widely used to treat a variety of solid cancers [15, 42]. Seventeen genes are known to be involved in the so-called “gemcitabine metabolism pathway” [15] (Fig. 1). Therefore, pharmacogenomic studies of these pathway genes might help us to systematically understand the contribution of genetic variation to gemcitabine response in the treatment of cancer, including NSCLC. Our pathway-based pharmacogenomic study is complementary to other approaches such as genome-wide association studies (GWAS). Using GWAS approaches, we have previously identified variations in gene expression across the entire genome that appears to contribute to variation in gemcitabine response. One gene in the gemcitabine pathway, NT5C3, was identified using this GWAS and cell-based model system approach [26]. Furthermore, we also demonstrated that genetic variation in NT5C3 might affect protein function and potentially influence drug response based on gene resequencing and functional genomic studies [22]. Therefore, it is important to determine the contribution of genetic variation in pathway genes to variation in gemcitabine response during cancer therapy. In the present study, we selected tag-SNPs for pathway genes to genotype DNA samples obtained from NSCLC patients treated with gemcitabine-based chemotherapy in an attempt to understand the possible influence of genetic variation in these genes on gemcitabine response (Table 2).
Specifically, we genotyped 107 tag SNPs for these 17 gemcitabine pathway genes to determine their possible association with treatment outcome in NSCLC. We identified potential candidate SNPs within pathway genes that might contribute to variation in gemcitabine response and OS for NSCLC patients. Furthermore, several of these SNPs were functionally validated on the basis of association analysis of SNP genotype data with gemcitabine cytotoxicity in human LCLs. For example, the rs2274341SNP in NT5C2, associated with OS of NSCLC patients, was also associated with gemcitabine IC50 values in the LCLs (P = 0.04), as were 12 other linked NT5C2 SNPs, among which rs1163238 in moderate LD (r2 = 0.543) with rs2274341 might also be important for gemcitabine response since it had an even smaller P value, 0.018, in the LCLs (Table 3 and Fig. 3). In addition, the strongest association observed was for a haplotype in SLC28A3 (P = 0.0001, HR = 5.436), which contained one SNP (rs10868141) that was also associated with gemcitabine IC50 values (P = 0.048) (Table 3). Therefore, those SNPs in NT5C2 and SLC29A1, as well as the SLC28A3 haplotype, might be potential biomarkers for gemcitabine response, of course requiring further investigation and replication.
One of the possible mechanisms for which those genetic variants might have functional impact on drug response is through transcription regulation. Therefore, we also determined if SNPs and haplotypes might influence the expression of their own gene (cis-regulation) using LCL data. Few SNPs in the pathway genes were correlated with mRNA expression for their corresponding genes, suggesting that other mechanisms exist. Difference in tissue-specific transcription regulation between lung cancer tissue and LCLs is the most likely explanation. In addition, these SNPs might contribute to variation in gemcitabine response via trans-regulation of the expression of other genes, or additional variant sequence in the same LD region might be responsible for variation in gemcitabine response.
We also compared our findings with previous pharmacogenomic studies of gemcitabine. None of the SNPs identified in the current study have been previously reported for NSCLC patients treated with gemcitabine [19, 39, 42–44]. One study from Japan had genotyped germline DNA samples from 94 healthy Asian donors and 53 NSCLC patients receiving gemcitabine-based chemotherapy. They found that in NSCLC patients, a SNP in the coding region of CDA, rs1048977, was associated with a lower response rate (P = 0.026) and shorter time to progression (P = 0.016), while a SNP in the coding region of SLC28A1 (rs2242046) was associated with neutropenia (P = 0.030) and thrombocytopenia nadir (P = 0.037) [45]. However, we did not observe the same associations in our NSCLC samples, which might be due to differences in sample size and/or allele frequencies for the two variants among different populations. According to that report, the frequency of rs2242046 in SLC28A1 was significantly lower in healthy Chinese (12%), Malays (30%) and Indians (35%) compared with Caucasians (73%). The allele frequency of rs1048977 in CDA is 0.260 in CEPH, and 0.307 in CHB + JPT HapMap LCLs. In our previous study, we also found that the level of NT5C3 gene expression was significantly associated with gemcitabine sensitivity in pancreatic and breast cancer cell line [26]. However, none of the tag SNPs in NT5C3 associated with OS of NSCLC patients. One possibility is that rare variants in NT5C3 that were not genotyped or linked to the genotyped SNPs might contribute to the variation in gemcitabine response in NSCLC patients.
In the present study, we used LCL data to help us interpret association results for the patient samples. Obviously, there are limitations associated with the use of EBV transformed LCLs. Expression profiles might differ between lung cancer tissue and LCLs, and EBV transformation can influence both gene expression and sensitivity to chemotherapeutic agents. Other confounding factors such as ATP levels and cell growth rates might also influence drug cytotoxicity assays [46, 47]. However, we have used this same system to successfully interpret SNP signals for other clinical studies and have also used this system to help identify and understand pharmacogenomic candidates [48, 49]. In addition, our functional genomic studies performed with lung cancer cell lines validated several of the candidate SNPs (Fig. 4). Although we did not see an effect of NT5C2 on gemcitabine sensitivity in our previous study, this difference could be due to the different cell lines used in the two studies, where a pancreatic (SU86) and a breast cell line (MBA-MD-231) were used in our previous study, while NSCLC cell lines were used in the current study.
Clinical phenotypes for the patients included in this study have been well documented and some of the samples were used in previous pharmacogenomic studies to test different hypotheses [25, 50, 51]. However, our patient population is heterogeneous with regard to the treatment they received. Most patients were treated with multiple drugs besides gemcitabine, including platinum compounds, paclitaxel, etoposide and EGFR inhibitors, as well as with radiation therapy and/or surgery. To be more precise, we also repeated analyses for the 5 SNPs reported in Table 2, including both stage and prior treatment as adjusting covariates, and all 5 SNPs remained statistically significant (P value < 0.05). In addition, our studies with LCLs using gemcitabine cytotoxicity as a phenotype helped us to interpret results obtained by genotyping the clinical samples and our functional studies make it possible to directly assess the influence of SNPs on gemcitabine cytotoxicity and also help to validate our association study results.
In summary, results from these studies indicate that genetic variation in genes encoding proteins involved in the gemcitabine metabolism and activation pathway might contribute to variation in gemcitabine response in the treatment of lung cancer. Taken together, a pathway-based pharmacogenomic approach with clinical patient samples, followed by functional studies, can potentially help identify genetic biomarkers involved in the gemcitabine metabolism and activation pathway and understand its role in variation in response to the treatment of lung cancer.
Supplementary Material
Acknowledgments
We thank Luanne Wussow for her assistance with the preparation of this manuscript.
Sources of Support: This study was supported by U.S. National Institutes of Health research grants, R01 CA80127 (to PY), R01 CA84354 (to PY), Mayo Foundation funds (to PY), K22 CA130828 (to LW), R01 CA138461 (to LW), P50 CA102701 (to LW), U19 GM61388 (The Pharmacogenomics Research Network), ASPET-Astellas Award (to LW), a PhRMA Foundation “Center of Excellence in Clinical Pharmacology” Award (to LW), and the Gerstner Family Career Development Awards in Individualized Medicine (to LL).
Footnotes
Author disclosures: no conflicts of interest.
Authors contributions
Conceived and designed the experiments: LL and LW. Performed the experiments: LL, IM and BE. Analyzed the data: LL, DS, BLF, KK, GJ, AB, RA, LP, EW, ZS, PY and LW. Wrote the paper: LL, DS, BF and LW.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Peto R, Chen ZM, Boreham J. Tobacco--the growing epidemic. Nat Med. 1999;5:15–17. doi: 10.1038/4691. [DOI] [PubMed] [Google Scholar]
- 3.American Cancer Society. American Cancer Society: Cancer Facts and Figures 2010. Atlanta, GA: 2010. [Google Scholar]
- 4.Arriagada R, Bergman B, Dunant A, Le Chevalier T, Pignon JP, Vansteenkiste J. Cisplatin-based adjuvant chemotherapy in patients with completely resected non-small-cell lung cancer. N Engl J Med. 2004;350:351–360. doi: 10.1056/NEJMoa031644. [DOI] [PubMed] [Google Scholar]
- 5.Fossella F, Pereira JR, von Pawel J, Pluzanska A, Gorbounova V, Kaukel E, et al. Randomized, multinational, phase III study of docetaxel plus platinum combinations versus vinorelbine plus cisplatin for advanced non-small-cell lung cancer: the TAX 326 study group. J Clin Oncol. 2003;21:3016–3024. doi: 10.1200/JCO.2003.12.046. [DOI] [PubMed] [Google Scholar]
- 6.Wozniak AJ, Crowley JJ, Balcerzak SP, Weiss GR, Spiridonidis CH, Baker LH, et al. Randomized trial comparing cisplatin with cisplatin plus vinorelbine in the treatment of advanced non-small-cell lung cancer: a Southwest Oncology Group study. J Clin Oncol. 1998;16:2459–2465. doi: 10.1200/JCO.1998.16.7.2459. [DOI] [PubMed] [Google Scholar]
- 7.Sandler AB, Nemunaitis J, Denham C, von Pawel J, Cormier Y, Gatzemeier U, et al. Phase III trial of gemcitabine plus cisplatin versus cisplatin alone in patients with locally advanced or metastatic non-small-cell lung cancer. J Clin Oncol. 2000;18:122–130. doi: 10.1200/JCO.2000.18.1.122. [DOI] [PubMed] [Google Scholar]
- 8.Ohe Y, Ohashi Y, Kubota K, Tamura T, Nakagawa K, Negoro S, et al. Randomized phase III study of cisplatin plus irinotecan versus carboplatin plus paclitaxel, cisplatin plus gemcitabine, and cisplatin plus vinorelbine for advanced non-small-cell lung cancer: Four-Arm Cooperative Study in Japan. Ann Oncol. 2007;18:317–323. doi: 10.1093/annonc/mdl377. [DOI] [PubMed] [Google Scholar]
- 9.Scagliotti GV, De Marinis F, Rinaldi M, Crino L, Gridelli C, Ricci S, et al. Phase III randomized trial comparing three platinum-based doublets in advanced non-small-cell lung cancer. J Clin Oncol. 2002;20:4285–4291. doi: 10.1200/JCO.2002.02.068. [DOI] [PubMed] [Google Scholar]
- 10.Schiller JH, Harrington D, Belani CP, Langer C, Sandler A, Krook J, et al. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med. 2002;346:92–98. doi: 10.1056/NEJMoa011954. [DOI] [PubMed] [Google Scholar]
- 11.Cardenal F, Lopez-Cabrerizo MP, Anton A, Alberola V, Massuti B, Carrato A, et al. Randomized phase III study of gemcitabine-cisplatin versus etoposide-cisplatin in the treatment of locally advanced or metastatic non-small-cell lung cancer. J Clin Oncol. 1999;17:12–18. doi: 10.1200/JCO.1999.17.1.12. [DOI] [PubMed] [Google Scholar]
- 12.Heinemann V, Xu YZ, Chubb S, Sen A, Hertel LW, Grindey GB, et al. Cellular elimination of 2′,2′-difluorodeoxycytidine 5′-triphosphate: a mechanism of self-potentiation. Cancer Res. 1992;52:533–539. [PubMed] [Google Scholar]
- 13.Heinemann V, Hertel LW, Grindey GB, Plunkett W. Comparison of the cellular pharmacokinetics and toxicity of 2′,2′-difluorodeoxycytidine and 1-beta-D-arabinofuranosylcytosine. Cancer Res. 1988;48:4024–4031. [PubMed] [Google Scholar]
- 14.Mini E, Nobili S, Caciagli B, Landini I, Mazzei T. Cellular pharmacology of gemcitabine. Ann Oncol. 2006;17 (Suppl 5):v7–12. doi: 10.1093/annonc/mdj941. [DOI] [PubMed] [Google Scholar]
- 15.Plunkett W, Huang P, Searcy CE, Gandhi V. Gemcitabine: preclinical pharmacology and mechanisms of action. Semin Oncol. 1996;23:3–15. [PubMed] [Google Scholar]
- 16.Kim SR, Saito Y, Maekawa K, Sugiyama E, Kaniwa N, Ueno H, et al. Twenty novel genetic variations and haplotype structures of the DCK gene encoding human deoxycytidine kinase (dCK) Drug Metab Pharmacokinet. 2008;23:379–384. doi: 10.2133/dmpk.23.379. [DOI] [PubMed] [Google Scholar]
- 17.Kim SR, Saito Y, Maekawa K, Sugiyama E, Kaniwa N, Ueno H, et al. Thirty novel genetic variations in the SLC29A1 gene encoding human equilibrative nucleoside transporter 1 (hENT1) Drug Metab Pharmacokinet. 2006;21:248–256. doi: 10.2133/dmpk.21.248. [DOI] [PubMed] [Google Scholar]
- 18.Kwon WS, Rha SY, Choi YH, Lee JO, Park KH, Jung JJ, et al. Ribonucleotide reductase M1 (RRM1) 2464G>A polymorphism shows an association with gemcitabine chemosensitivity in cancer cell lines. Pharmacogenet Genomics. 2006;16:429–438. doi: 10.1097/01.fpc.0000204999.29924.da. [DOI] [PubMed] [Google Scholar]
- 19.Maring JG, Groen HJ, Wachters FM, Uges DR, de Vries EG. Genetic factors influencing pyrimidine-antagonist chemotherapy. Pharmacogenomics J. 2005;5:226–243. doi: 10.1038/sj.tpj.6500320. [DOI] [PubMed] [Google Scholar]
- 20.Sugiyama E, Kaniwa N, Kim SR, Kikura-Hanajiri R, Hasegawa R, Maekawa K, et al. Pharmacokinetics of gemcitabine in Japanese cancer patients: the impact of a cytidine deaminase polymorphism. J Clin Oncol. 2007;25:32–42. doi: 10.1200/JCO.2006.06.7405. [DOI] [PubMed] [Google Scholar]
- 21.Yonemori K, Ueno H, Okusaka T, Yamamoto N, Ikeda M, Saijo N, et al. Severe drug toxicity associated with a single-nucleotide polymorphism of the cytidine deaminase gene in a Japanese cancer patient treated with gemcitabine plus cisplatin. Clin Cancer Res. 2005;11:2620–2624. doi: 10.1158/1078-0432.CCR-04-1497. [DOI] [PubMed] [Google Scholar]
- 22.Aksoy P, Zhu MJ, Kalari KR, Moon I, Pelleymounter LL, Eckloff BW, et al. Cytosolic 5′-nucleotidase III (NT5C3): gene sequence variation and functional genomics. Pharmacogenet Genomics. 2009;19:567–576. doi: 10.1097/FPC.0b013e32832c14b8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilbert JA, Salavaggione OE, Ji Y, Pelleymounter LL, Eckloff BW, Wieben ED, et al. Gemcitabine pharmacogenomics: cytidine deaminase and deoxycytidylate deaminase gene resequencing and functional genomics. Clin Cancer Res. 2006;12:1794–1803. doi: 10.1158/1078-0432.CCR-05-1969. [DOI] [PubMed] [Google Scholar]
- 24.Ji Y, Olson J, Zhang J, Hildebrandt M, Wang L, Ingle J, et al. Breast cancer risk reduction and membrane-bound catechol O-methyltransferase genetic polymorphisms. Cancer Res. 2008;68:5997–6005. doi: 10.1158/0008-5472.CAN-08-0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moyer AM, Sun Z, Batzler AJ, Li L, Schaid DJ, Yang P, et al. Glutathione pathway genetic polymorphisms and lung cancer survival after platinum-based chemotherapy. Cancer Epidemiol Biomarkers Prev. 2010;19:811–821. doi: 10.1158/1055-9965.EPI-09-0871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li L, Fridley B, Kalari K, Jenkins G, Batzler A, Safgren S, et al. Gemcitabine and cytosine arabinoside cytotoxicity: association with lymphoblastoid cell expression. Cancer Res. 2008;68:7050–7058. doi: 10.1158/0008-5472.CAN-08-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niu N, Qin Y, Fridley BL, Hou J, Kalari KR, Zhu M, et al. Radiation pharmacogenomics: a genome-wide association approach to identify radiation response biomarkers using human lymphoblastoid cell lines. Genome Res. 2010;20:1482–1492. doi: 10.1101/gr.107672.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang P, Allen MS, Aubry MC, Wampfler JA, Marks RS, Edell ES, et al. Clinical features of 5,628 primary lung cancer patients: experience at Mayo Clinic from 1997 to 2003. Chest. 2005;128:452–462. doi: 10.1378/chest.128.1.452. [DOI] [PubMed] [Google Scholar]
- 29.Yang P, Sun Z, Krowka MJ, Aubry MC, Bamlet WR, Wampfler JA, et al. Alpha1-antitrypsin deficiency carriers, tobacco smoke, chronic obstructive pulmonary disease, and lung cancer risk. Arch Intern Med. 2008;168:1097–1103. doi: 10.1001/archinte.168.10.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kocabas NA, Aksoy P, Pelleymounter LL, Moon I, Ryu JS, Gilbert JA, et al. Gemcitabine pharmacogenomics: deoxycytidine kinase and cytidylate kinase gene resequencing and functional genomics. Drug Metab Dispos. 2008;36:1951–1959. doi: 10.1124/dmd.108.020925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002;70:425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Bakker PI, Yelensky R, Pe’er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 33.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clark AG. The role of haplotypes in candidate gene studies. Genet Epidemiol. 2004;27 :321–333. doi: 10.1002/gepi.20025. [DOI] [PubMed] [Google Scholar]
- 35.Li L, Fridley BL, Kalari K, Jenkins G, Batzler A, Weinshilboum RM, et al. Gemcitabine and arabinosylcytosin pharmacogenomics: genome-wide association and drug response biomarkers. PLoS One. 2009;4:e7765. doi: 10.1371/journal.pone.0007765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Durbin RM, Abecasis GR, Altshuler DL, Auton A, Brooks LD, Durbin RM, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baldwin SA, Beal PR, Yao SY, King AE, Cass CE, Young JD. The equilibrative nucleoside transporter family, SLC29. Pflugers Arch. 2004;447:735–743. doi: 10.1007/s00424-003-1103-2. [DOI] [PubMed] [Google Scholar]
- 38.Mackey JR, Mani RS, Selner M, Mowles D, Young JD, Belt JA, et al. Functional nucleoside transporters are required for gemcitabine influx and manifestation of toxicity in cancer cell lines. Cancer Res. 1998;58:4349–4357. [PubMed] [Google Scholar]
- 39.Ceppi P, Volante M, Novello S, Rapa I, Danenberg KD, Danenberg PV, et al. ERCC1 and RRM1 gene expressions but not EGFR are predictive of shorter survival in advanced non-small-cell lung cancer treated with cisplatin and gemcitabine. Ann Oncol. 2006;17 :1818–1825. doi: 10.1093/annonc/mdl300. [DOI] [PubMed] [Google Scholar]
- 40.Hunsucker SA, Mitchell BS, Spychala J. The 5′-nucleotidases as regulators of nucleotide and drug metabolism. Pharmacol Ther. 2005;107:1–30. doi: 10.1016/j.pharmthera.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 41.Nordlund P, Reichard P. Ribonucleotide reductases. Annu Rev Biochem. 2006;75:681–706. doi: 10.1146/annurev.biochem.75.103004.142443. [DOI] [PubMed] [Google Scholar]
- 42.Wong A, Soo RA, Yong WP, Innocenti F. Clinical pharmacology and pharmacogenetics of gemcitabine. Drug Metab Rev. 2009;41:77–88. doi: 10.1080/03602530902741828. [DOI] [PubMed] [Google Scholar]
- 43.Muller PJ, Dally H, Klappenecker CN, Edler L, Jager B, Gerst M, et al. Polymorphisms in ABCG2, ABCC3 and CNT1 genes and their possible impact on chemotherapy outcome of lung cancer patients. Int J Cancer. 2009;124:1669–1674. doi: 10.1002/ijc.23956. [DOI] [PubMed] [Google Scholar]
- 44.Gray JH, Mangravite LM, Owen RP, Urban TJ, Chan W, Carlson EJ, et al. Functional and genetic diversity in the concentrative nucleoside transporter, CNT1, in human populations. Mol Pharmacol. 2004;65:512–519. doi: 10.1124/mol.65.3.512. [DOI] [PubMed] [Google Scholar]
- 45.Soo RA, Wang LZ, Ng SS, Chong PY, Yong WP, Lee SC, et al. Distribution of gemcitabine pathway genotypes in ethnic Asians and their association with outcome in non-small cell lung cancer patients. Lung Cancer. 2009;63:121–127. doi: 10.1016/j.lungcan.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 46.Stark AL, Zhang W, Mi S, Duan S, O’Donnell PH, Huang RS, et al. Heritable and non- genetic factors as variables of pharmacologic phenotypes in lymphoblastoid cell lines. Pharmacogenomics J. 2010;10:505–512. doi: 10.1038/tpj.2010.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Choy E, Yelensky R, Bonakdar S, Plenge RM, Saxena R, De Jager PL, et al. Genetic analysis of human traits in vitro: drug response and gene expression in lymphoblastoid cell lines. PLoS Genet. 2008;4:e1000287. doi: 10.1371/journal.pgen.1000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ingle JN, Schaid DJ, Goss PE, Liu M, Mushiroda T, Chapman JA, et al. Genome-wide associations and functional genomic studies of musculoskeletal adverse events in women receiving aromatase inhibitors. J Clin Oncol. 2010;28:4674–4682. doi: 10.1200/JCO.2010.28.5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ingle JN, Liu M, Wickerham DL, Schaid DJ, Mushiroda T, Kubo M, et al. Genome-wide associations of breast events and functional genomics studies in high-risk women receiving tamoxifen or raloxifene on NSABP P1 and P2 prevention trials. A pharmacogenomics research network-RIKEN-NSABP collaboration. Cancer Res. 2010;70 :110s. [Google Scholar]
- 50.Li Y, Sun Z, Cunningham JM, Aubry MC, Wampfler JA, Croghan GA, et al. Genetic variations in multiple drug action pathways and survival in advanced stage non-small cell lung cancer treated with chemotherapy. Clin Cancer Res. 2011;17:3830–3840. doi: 10.1158/1078-0432.CCR-10-2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pankratz VS, Sun Z, Aakre J, Li Y, Johnson C, Garces YI, et al. Systematic evaluation of genetic variants in three biological pathways on patient survival in low-stage non-small cell lung cancer. J Thorac Oncol. 2011;6:1488–1495. doi: 10.1097/JTO.0b013e318223bf05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gray JH, Owen RP, Giacomini KM. The concentrative nucleoside transporter family, SLC28. Pflugers Arch. 2004;447:728–734. doi: 10.1007/s00424-003-1107-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.