Abstract
Using water soluble, fluorescent, flexible polymers, we have devised a novel methodology for identification and differentiation of prostate cancer cells. By using a step-wise linear discriminant analysis we demonstrate that the differential modulations of the polymer emission intensities in the presence of conditioned cell culture media can be used to distinguish between prostate cancer subtypes and between cancerous and non-cancer cells. The differences in the compositions of the conditioned cell culture media are likely contributing to different fluorescence spectral patterns of the polymers. This in vitro approach may provide a novel platform for the development of an alternative prostate cancer diagnostic and subtyping technique.
Keywords: fluorescent polymers, prostate cancer, linear discriminant analysis
Fluorescent polymers have been successfully used to distinguish different proteins,1 isozymes,2 and for differentiating cancerous, non-cancerous and malignant cell lines.3 Unfortunately, translation of polymer technology has not yet evolved into any clinical applications. For clinical diagnosis of prostate cancer, analysis of blood, urine, or tumor markers such as prostate-specific antigen (PSA)4 are used. However, the PSA analysis may provide ambiguous results, leading to over diagnosis of prostate cancer.5,6 Fluorescent,7 luminescent,8 or other dye-based imaging techniques9 can also be used to detect different prostate cancer biomarkers. However, these diagnostic techniques do not differentiate the cancer cells into subtypes.
Cellular subtyping technologies come primarily from the field of microbiology. One classical example is the oligonucleotide sequencing employed in DNA microarrays and southern blots.10 A more recent development is microRNA sequencing for identification of cancerous biomarkers.11 However, the employment of both of these techniques is dependent upon phenotypically expressed DNA. The development of epigenetic studies (e.g., histone acetylation and methylation, DNA methylation, etc.) demonstrates that phenotype expression can be varied independent of DNA sequences.12,13 Thus, to obtain accurate results, we need to monitor at post-translational levels (i.e., expressed proteins) for subtyping cancer cells.
To develop an in vitro approach for subtyping prostate cancer cells, we choose to explore the use of water soluble, fluorescent polymers. We have recently demonstrated that the water-soluble, fluorescent polymers can be prepared for selective interactions with the isozyme matrix metalloproteianse-9 (MMP-9) compared to MMP-7 and −10.2 MMP-9 is a Zn2+ containing metalloenzyme overexpressed and secreted at different concentrations by different cancer cells.14,15 The enzyme contributes to the growth and metastasis of a large number of cancers.16 Besides MMP-9, various other extracellular (e.g., MMP-7, urokinase plasminogen activator etc.) and membrane-bound enzymes (a disintegrin and a metalloproteinase, ADAMs) are also overexpressed by metastatic cancer cells, albeit different amounts.17-19 We reasoned that the differential expression levels of various extracellular enzymes by the cancer cells will lead to differential modulations of fluorescence emission intensity from the water soluble polymers in the presence of conditioned cell culture media. Herein, we demonstrate that this strategy can be used for distinguishing prostate cancer cells from non-cancerous cells and for subtyping different prostate cancer cells. Human prostate and other cancer cells have been detected employing monoclonal antibodies as the recognition elements.20-22 However, preparation and production of monoclonal antibodies in large scale (> 1 g) can be really challenging. Proper storage and handling conditions must be followed to ensure that the monoclonal antibodies are not denatured and retains the selective binding property. In contrast, the polymers reported here are easy to prepare on a large scale and no special storage and handling procedures are needed.
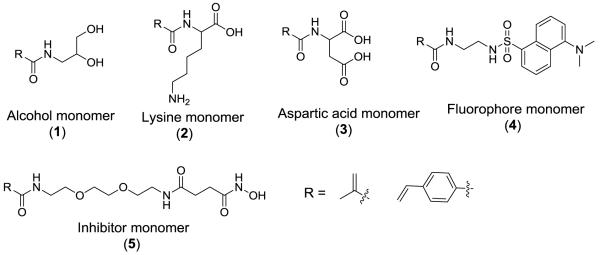
We used the monomers 1 – 5 (Scheme 1) to prepare the water-soluble, random polymers P1 and P2 (Table 1) employing AIBN as the free-radical initiator. We have previously observed that these two polymers were optimal for distinguishing recombinant human MMP-9 from MMP-7 and −10.2 Polymer P1 was prepared using the monomers with methacrylamide as the polymerizable group (starting with 50 mol% of monomer 1, 10 mol% of monomer 2, 10 mol% of monomer 3, 10 mol% of monomer 4, and 20 mol% monomer 5); P2 was prepared with the monomers containing 4-vinylbenzamide as the polymerizable group (starting with 45 mol% of monomer 1, 9 mol% of monomer 2, 9 mol% of monomer 3, 18 mol% of monomer 4, and 19 mol% of monomer 5). These polymers were then characterized by gel permeation chromatography (Table 1).
Scheme 1.
The structures of the monomers used in the preparation of the water-soluble, fluorescent polymers.
Table 1.
Molecular weights of the polymers P1 and P2 determined by gel permeation chromatography.
| Polymer P1 | Polymer P2 | |
|---|---|---|
| Mw | 117,191 | 114,428 |
| Mn | 64,577 | 78,161 |
| P.I. | 1.81 | 1.46 |
| Concentration used | 31 nM | 27 nM |
Polymers P1 and P2 are expected to interact with MMP-9 using a variety of non-covalent interactions. In contrast to the reported polymers for differential interactions with cells,3 our polymers contain attached MMP inhibitors (from the inhibitor monomer 5). The hydroxamic acid moiety will interact with the Zn2+ ion in active site pocket.23 We have previously demonstrated that the polymers P1 and P2 (100 nM solution) effectively inhibit recombinant MMP-9.2 This interaction could serve as the initial anchoring site for the enzymes to the polymer and facilitate the formation of the additional surface binding interactions to the MMP-9 enzyme. For example, the polyamide backbones of the polymers can form hydrogen bonds with the enzyme surface. Polymer P2 contains the benzamide groups and has the potential to interact with surface amino acids that containing conjugation.24 Lysine (positive charge) and aspartic acid (negative charge) groups on the polymers can interact with complementary charges on the enzyme’s surface. Hydrogen bonding interactions with the enzyme are also possible from the polymerized alcohol monomer 1.
For the fluorescence experiments, we used the dye-free conditioned cell culture media from the prostate cancer cell lines (22Rv1 and PC-3), pancreatic cancer (PANC1), and non-cancerous cell line (HEK-293). The experiments were conducted in 30 mM phosphate buffer (pH = 7.4). Amongst the selected prostate cancer cells, 22Rv1 is androgen-dependent and PC-3 is androgen-independent (more malignant).25,26 These cells were grown in dye-free media in a humidified atmosphere with 5% CO2 at 37°C to a state of confluence. Upon reaching a confluent state, the cell’s conditioned media were then harvested for fluorescence experiments. Further details regarding cell culture studies and fluorescence experiments can be found in the Supporting Information.
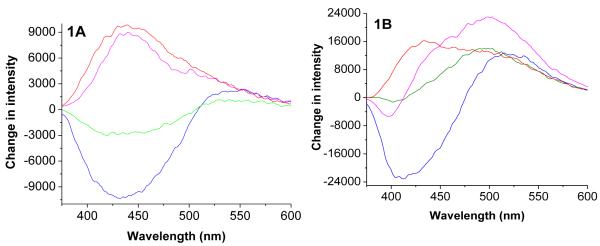
The experiments revealed variations in emission spectra of the polymers P1 and P2 when exposed to the conditioned cell culture media from different cells. The emission spectra of the polymers in the presence of media before cell culture were used as the controls for these experiments. The control emission spectra of the polymers were subtracted from the emission spectra in presence of the conditioned media to generate the corresponding difference spectra (Fig. 1). The emission spectra of the polymers P1 and P2 were found to be blue-shifted in the presence of the conditioned cell culture media, indicating the naphthalenesulfonamide fluorophores in the polymer are experiencing more hydrophobic microenvironment.27 We observed that the emission intensity of P2 increased in the presence of the conditioned media from the cells 22Rv1 and HEK-293 and decreased in the presence of the conditioned media from PANC1 and PC3 (Figure 1A). This trend was different for the polymer P2 (Figure 1B).
Figure 1.
The difference emission spectra for the polymer P1 (1A) and P2 (1B) in the presence of conditioned cell culture media from the cancer cells PANC-1 (blue trace), PC3 (green trace), 22Rv1 (pink trace) and HEK-293 (red trace). The emission spectra in the presence of unconditioned media were subtracted from the emission spectra in the presence of conditioned media to generate these plots.
In order to determine if these intensity changes correlate with the levels of MMP-9 secreted by these cells, we determined the total concentration (i.e., active and inactive) of MMP-9 in the conditioned cell culture media employing a commercially available ELISA kit. We observed that the PANC1 cells secreted the highest amount of MMP-9 in the conditioned media (2 ng/mL) and the amounts of MMP-9 secreted by the other cells were similar (740 – 780 pg/mL). Clearly, the changes in the emission intensity of the polymers do not correlate with the concentration of secreted MMP-9. Other proteins in the conditioned media are interacting with the polymers, causing the observed intensity changes in the emission spectra. While we do not have an explanation yet for the observed pattern of emission intensity changes, we reasoned that these differential modulations can be used to distinguish the cancer cells. In this endeavor, we calculated the ratios of emission intensity (410, 510 and 541 nm for P1; 420, 520 and 541 nm for P2) in the presence of conditioned and unconditioned culture media (Table S-2, Supporting Information). These ratios were subsequently subjected to linear discriminant analysis (LDA).
LDA was applied to evaluate which of these three ratios provided the maximum discrimination (i.e. best predictor) between the cell lines.1,3,28,29 We applied LDA in a stepwise fashion. First, we applied LDA to each polymer separately, where we evaluated each potential peak value based on its ability to discriminate between (or predict) the four cell lines. Each of these analyses was conducted using 32 observations (4 cell lines × 8 replications) and 4 variables (the cell line indicator and the three emission intensity wavelength variables). This LDA analyses step identifies the optimal intensity of 410 nm for the P2 polymer and 420 nm for the P1 polymer.
Using this optimal wavelength, LDA was applied to each polymer to determine the polymer’s ability to effectively discriminate across cell lines. Standard F-tests (Table 2) indicate significant (joint) differences in emission intensities across the four cell lines. Two eigenvalues were characterized by a canonical discriminant function and a canonical correlation. The first eigenvalue explains 99.9% of the variation in the data, while the second explains the remaining 0.1%. Chi-square tests indicate that only the first of these is significant at the 5% level.
Table 2.
The emission intensity ratio for polymer P1 and P2 in the presence of conditioned and unconditioned cell culture media.
| Cell Line | P1 | P2 |
|---|---|---|
| PANC1 | 0.768 | 0.948 |
| PC-3 | 0.850 | 0.975 |
| 22Rv1 | 1.462 | 1.204 |
| HEK-293 | 1.805 | 1.444 |
| Wilks’ Lambda | 0.02 | 0.457 |
| F-Statistic | 466.379 | 11.809 |
| P-Value | < 0.001 | < 0.001 |
As chemically expected, our structure matrix does suggest that the P1 and P2 polymers do contribute to the LDA analysis differently, likely due to the variation in the hydrophilic/hydrophobic properties of the polymers. We observed that the hydrophilic polymer (P1) displays a much greater contribution to the separation of the matrix. Usually hydrophilic amino acid residues are exposed on the surface of globular proteins.30 It is likely that these charged residues on the surface of MMP-9 and other proteins secreted by the cancer cells are contributing to stronger interactions with the hydrophilic polymer P1 as compared to polymer P2.
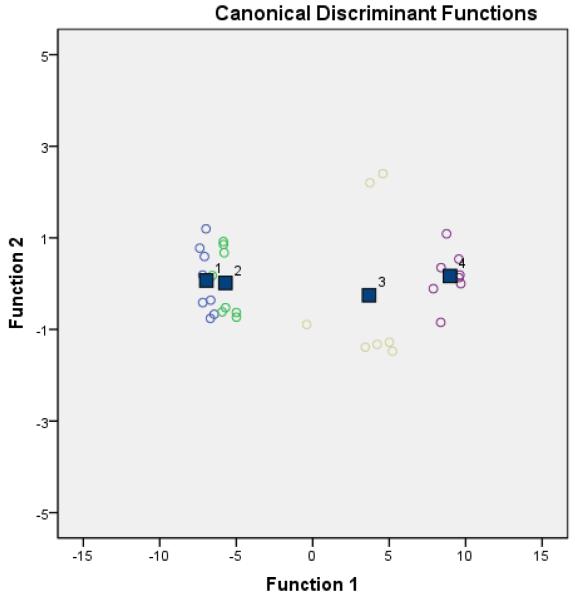
Figure 2 (Figures S-1, S-2 and S-3, Supporting Information) is a plot of the two canonical discriminant functions. This LDA model distinguishs between the two subtypes of prostate cancer (22Rv1 and PC-3) and non-cancer cells (HEK-293). But suprisingly it did not form a noticible differentiation between PC-3 and PANC-1 (pancreatic cancer) cells. Traditional and cross-validated discriminant functions each correctly predicted 93.8% and 87.5% of the cell lines, respectively, indicating a reasonable degree of interval validity.
Figure 2.
This figure shows the canonical discriminant functions plots of polymers P1. This clearly shows the separation between the cell lines PANC1 (blue #1), PC3 (green #2), 22Rv1 (yellow #3), and HEK-293 (purple #4). The boxes represent the four group’s centroid.
In conclusion, we have prepared two water-soluble, fluorescent polymers incorporating an inhibitor for the Zn2+ containing metalloenzyme MMP-9. These polymers showed differential modulations in emission spectra in the presence of conditioned cell culture media from cancer cells. It is likely that besides MMP-9, other proteins in the conditioned media are non-specifically interacting with our polymers. Despite these non-specific interactions, we can distinguish between the two prostate cancer cell lines and non-cancer cell line using this in vitro polymer-based approach. We noted that the polymers P1 and P2 do not have good ability to discriminate the PC3 cells from the PANC1 cells. Due to the complex nature of the conditioned cell culture media, we do not have an explanation yet for this lack of discrimination between these two cell lines. We are currently investigating the effects of incorporating selective MMP-9 inhibitors in the polymer in improving the ability to differentiate a wide variety of cancer cell subtypes.
Supplementary Material
ACKNOWLEDGMENT
This research was supported by NIH grants 1R01 CA 113746, 1R01 CA 132034 and NSF grants DMR 1005011 and CBET 0959422 to SM.
Footnotes
SUPPORTING INFORMATION AVAILABLE: Synthetic details and detailed discussion of the statistical analysis. this material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- (1).Miranda OR, Creran B, Rotello VM. Curr Opin Chem Biol. 2010;14:728. doi: 10.1016/j.cbpa.2010.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Dutta R, Scott MD, Haldar MK, Ganguly B, Srivastava DK, Friesner DL, Mallik S. Bioorg Med Chem Lett. 2011;21:2007. doi: 10.1016/j.bmcl.2011.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Bajaj A, Miranda OR, Phillips R, Kim IB, Jerry DJ, Bunz UH, Rotello VM. J Am Chem Soc. 2010;132:1018. doi: 10.1021/ja9061272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Oesterling JE. J Urol. 1991;145:907. doi: 10.1016/s0022-5347(17)38491-4. [DOI] [PubMed] [Google Scholar]
- (5).Mokete M, Shackley DC, Betts CD, O’Flynn KJ, Clarke NW. BJU Int. 2006;97:266. doi: 10.1111/j.1464-410X.2005.06011.x. [DOI] [PubMed] [Google Scholar]
- (6).Cooperberg MR, Lubeck DP, Meng MV, Mehta SS, Carroll PR. J Clin Oncol. 2004;22:2141. doi: 10.1200/JCO.2004.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Zhang H, Uselman RR, Yee D. Expert Opin Med Diagn. 2011;5:241. doi: 10.1517/17530059.2011.566858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Maldiney T, Byk G, Wattier N, Seguin J, Khandadash R, Bessodes M, Richard C, Scherman D. Int J Pharm. 2011 doi: 10.1016/j.ijpharm.2011.06.048. [DOI] [PubMed] [Google Scholar]
- (9).Gao X, Cui Y, Levenson RM, Chung LW, Nie S. Nat Biotechnol. 2004;22:969. doi: 10.1038/nbt994. [DOI] [PubMed] [Google Scholar]
- (10).Hsing AW, Gao YT, Wu G, Wang X, Deng J, Chen YL, Sesterhenn IA, Mostofi FK, Benichou J, Chang C. Cancer Res. 2000;60:5111. [PubMed] [Google Scholar]
- (11).Porkka KP, Pfeiffer MJ, Waltering KK, Vessella RL, Tammela TL, Visakorpi T. Cancer Res. 2007;67:6130. doi: 10.1158/0008-5472.CAN-07-0533. [DOI] [PubMed] [Google Scholar]
- (12).Minucci S, Pelicci PG. Nat Rev Cancer. 2006;6:38. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- (13).Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, Zhang LL, Scherer B, Sinclair DA. Nature. 2003;425:191. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- (14).Banerjee J, Hanson AJ, Gadam B, Elegbede AI, Tobwala S, Ganguly B, Wagh AV, Muhonen WW, Law B, Shabb JB, Srivastava DK, Mallik S. Bioconjug Chem. 2009;20:1332. doi: 10.1021/bc9000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Banerjee J, Hanson AJ, Nyren-Erickson EK, Ganguli B, Wagh A, Muhonen WW, Law B, Shabb JB, Srivastava DK, Mallik S. Chem Commun (Camb) 2010;46:3209. doi: 10.1039/b926554f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Roy R, Yang J, Moses MA. J Clin Oncol. 2009;27:5287. doi: 10.1200/JCO.2009.23.5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Koskensalo S, Mrena J, Wiksten JP, Nordling S, Kokkola A, Hagstrom J, Haglund C. Tumour Biol. 2010;31:149. doi: 10.1007/s13277-010-0020-1. [DOI] [PubMed] [Google Scholar]
- (18).Duffy MJ, McKiernan E, O’Donovan N, McGowan PM. Clin Chim Acta. 2009;403:31. doi: 10.1016/j.cca.2009.01.007. [DOI] [PubMed] [Google Scholar]
- (19).Almasi CE, Brasso K, Iversen P, Pappot H, Hoyer-Hansen G, Dano K, Christensen IJ. Prostate. 2010 [Google Scholar]
- (20).Taylor RM, Huber DL, Monson TC, Ali AM, Bisoffi M, Sillerud LO. J Nanopart Res. 2011;13:4717. doi: 10.1007/s11051-011-0439-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Li Q, Qi H, Zhou HX, Deng CY, Zhu H, Li JF, Wang XL, Li FR. Int J Nanomedicine. 2011;6:2175. doi: 10.2147/IJN.S24731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Ziegler VG, Knaup J, Stahl D, Krammer B, Plaetzer K. Lasers Surg Med. 2011;43:548. doi: 10.1002/lsm.21089. [DOI] [PubMed] [Google Scholar]
- (23).Zucker S, Cao J. Cancer Biol Ther. 2009;8:2371. doi: 10.4161/cbt.8.24.10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Samanta U, Pal D, Chakrabarti P. Acta Crystallogr D Biol Crystallogr. 1999;55:1421. doi: 10.1107/s090744499900726x. [DOI] [PubMed] [Google Scholar]
- (25).Sramkoski RM, Pretlow TG, 2nd, Giaconia JM, Pretlow TP, Schwartz S, Sy MS, Marengo SR, Rhim JS, Zhang D, Jacobberger JW. In Vitro Cell Dev Biol Anim. 1999;35:403. doi: 10.1007/s11626-999-0115-4. [DOI] [PubMed] [Google Scholar]
- (26).Mazor M, Kawano Y, Zhu H, Waxman J, Kypta RM. Oncogene. 2004;23:7882. doi: 10.1038/sj.onc.1208068. [DOI] [PubMed] [Google Scholar]
- (27).Tanaka T, Hidaka H. J Biol Chem. 1980;255:11078. [PubMed] [Google Scholar]
- (28).Nyren-Erickson EK, Haldar MK, Gu Y, Qian SY, Friesner DL, Mallik S. Anal Chem. 2011;83:5989. doi: 10.1021/ac2009993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Bunz UH, Rotello VM. Angew Chem Int Ed Engl. 2010;49:3268. doi: 10.1002/anie.200906928. [DOI] [PubMed] [Google Scholar]
- (30).Voet D, Voet JG. Biochemistry. 3rd ed. J. Wiley & Sons; New York: 2004. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.