Abstract
The assembly of ribonucleoprotein complexes occurs in a broad range of conditions, but the principles that promote assembly and allow function at high temperature are poorly understood. The ribosomal protein S8 from the hyperthemophilic bacterium Aquifex aeolicus (AS8) is unique in that there is a 41 residue insertion in the consensus S8 sequence. In addition, AS8 exhibits an unusually-high affinity for the 16S ribosomal RNA (rRNA), characterized by a picomolar dissociation constant that is approximately 26,000 fold tighter than the equivalent interaction from Escherichia coli. Deletion analysis demonstrated that binding to the minimal helix 21 occurred at the same nanomolar affinity found for other bacterial species. The additional affinity required the presence of a three-helix junction between helices 20, 21, and 22. The crystal structure of AS8 was solved, revealing the helix-loop-helix geometry of the unique AS8 insertion region, while the core of the molecule is conserved with known S8 structures. The AS8 structure was modeled onto the structure of the 30S ribosomal subunit from E. coli, suggesting the possibility that the unique subdomain provides additional backbone and side-chain contacts between the protein and an unpaired base within the three-way junction of helices 20, 21, and 22. Point mutations in the protein insertion subdomain resulted in a significantly reduced RNA binding affinity with respect to wild-type AS8. These results indicate that the AS8-specific subdomain provides additional interactions with the three-way junction that contribute to the extremely tight binding to rRNA.
Introduction
Understanding the adaptation of organisms to their environment often comes from studies of the relationships between protein thermostability and structure. A common and informative strategy is to compare the sequences, the three dimensional structures, and the thermodynamics of folding for homologous proteins from mesophilic and thermophilic organisms 1; 2; 3; 4; 5; 6. These types of comparisons can illuminate mechanisms of evolution and provide insights into the molecular forces that stabilize biological molecules.
Relatively little is known about the adaptations to high temperature of ribonucleoprotein complexes (RNPs). The prokaryotic ribosome provides a convenient source for investigating the general principles of RNP thermostability. Comparative studies of the thermodynamics of the interactions between ribosomal proteins (r-proteins) of the large subunit and the 23S ribosomal RNA (rRNA) have been conducted for L1 7, L10:L124 8, and L11 systems, while similar studies for the binding to the 16S rRNA have been conducted for the small subunit proteins S4 9; 10 and S8 11. The general trend is that binding is tighter in thermophilic systems with 10 to 1000 fold higher affinities being seen when compared to mesophilic systems.
Of all r-proteins, S8 is particularly well suited for the simultaneous comparison of sequence, structure, and thermodynamics because these data are available from multiple organisms including both mesophiles and thermophiles. A recent investigation into the assembly of the central domain of the 30S ribosomal subunit from Aquifex aeolicus reported a picomolar Kd between the A. aeolicus S8 (AS8) r-protein and the central domain of 16S rRNA at 65°C 12. This represents a 26,000 fold increase in binding in comparison to a similar interaction between S8 and 16S rRNA from Escherichia coli measured at 40°C 11; 13, but the structural and thermodynamic basis for this increase was unknown.
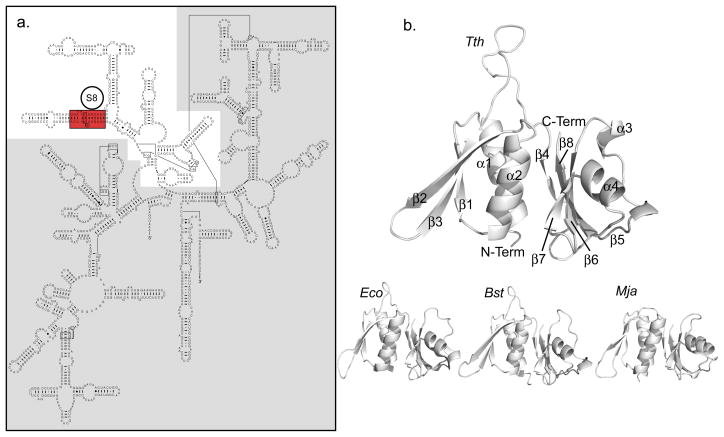
In E. coli, S8 is critical for the proper assembly of the 30S subunit and contributes to bacterial growth 14. S8 binds to the central domain of the 16S rRNA (Figure 1a) independently of other r-proteins 14; 15. In vitro assembly of the 30S subunit in the absence of S8 produces an incompletely formed RNP with highly reduced function 16, and mutations of the 16S rRNA that disrupt S8 binding correlate with greatly reduced growth rates 17; 18. Nuclease protection studies originally mapped the S8 binding site to helix 21 of the central domain 19; 20; 21; 22, and similar results where observed in both protein cross-linking 23 and chemical modification experiments 24; 25. Enzymatic and hydroxyl radical footprinting investigations identified additional interactions with helices 20, 22, 25, and 26a 10; 26. Also, the footprinting pattern of hydroxyl radicals generated from iron-dervatized S15 (Fe(II)-S15) in helices 24 and 26 depends on the presence of S8 27. Furthermore, protection from hydroxyl radicals generated from Fe(II)-S8 link its binding to progression through transition states of 16S rRNA folding with consequences throughout the entire subunit 28. Protein S8 is present in the crystal structures of the 30S subunits from both E. coli. (Protein Data Bank (PDB) code 2AVY) 29 (Figure 1b) and Thermus thermophilus (PDB code 1J5E) 30. The RNA contacts in these structures are in excellent agreement with the biochemical data and indicate major contacts along helix 21 and helix 25 31.
Figure 1.
a. Secondary structure of the E. coli 16S rRNA with the central domain highlighted in white. The minimal site for specific S8 binding is highlighted in red. b. Known structures of S8 proteins: Tth (T. thermophilus, PDB code 1AN7), Eco (E. coli, PDB code 1SO3), Bst (B. stearothermophilus, PDB code 1SEI), Mja (M. jannaschii, PDB code 1I6U). The conserved elements of S8 secondary structure are labeled on Tth. Tth and Bst are RNA-free structures, Mja and Eco were extracted from complexes with RNA.
Several thermodynamic studies have defined the interactions that provide the free energy of S8-rRNA binding. The original studies used filter-binding assays to monitor E. coli S8 binding to the 16S rRNA followed by studies using progressively smaller fragments of the 16S rRNA 13; 25; 32; 33. These studies most consistently identified interactions in the tenths of micromolar range and indicated that nearly all of the energy of binding derives from interactions with a short stretch of helix 21 within the region of the universally conserved A642 (Figure 1a). Nuclear magnetic resonance (NMR) studies of model oligomers based on the RNA target site in the absence and presence of E. coli S8 indicate little change in the RNA structure upon protein binding 34; 35. Furthermore, crystal structures of RNA-free S8 protein from T. Thermophilus 36 and Bacillus stearothermophilus 37 are available in addition to RNA-bound structures of S8 from several other species 29; 30; 38. Comparisons of these structures are consistent with a model of minimal conformational changes within the protein upon RNA binding. Most of the specific interactions are believed to originate from shape complementarities between S8 and the rRNA with organism specific modulation from interactions between bases and hydrophobic areas of protein side-chains 38.
The reported three-dimensional structures of S8 proteins from different organisms (Figure 1b) 36; 37; 38; 39 exhibit a conserved architecture. S8 folds into an N-terminal domain containing a three-stranded antiparallel β-sheet (strands β1, β2, and β3) packed against two α-helices on one side (α1 and α2) and a C-terminal domain composed of a four-stranded antiparallel β-sheet (β4, β6, β7 and β8) and two α-helices. An extended hydrophobic interface is formed between the two α–helices α1 and α2 of the N-terminal domain and the β-sheet of the C-terminal domain.
Interestingly AS8 contains a peptide insertion unique to Aquifex bacteria in the β-sheet region of the N-terminal domain. This peptide is 41 amino acids long (positions 51–92), and Blast searches (http://blast.ncbi.nlm.nih.gov/Blast.cgi) on the peptide sequence return only AS8 and the S8 protein of Aquifex pyrophilus. The position of the unique AS8 insertion led us to speculate that additional interactions between the rRNA of the central domain and the insertion sequence might occur, contributing to the extra affinity.
In this study we investigate the molecular mechanisms that result in the picomolar interaction between AS8 and the central domain of the 16S rRNA. A combination of fluorescence anisotropy and electrophoretic mobility shift assays (EMSA) were used to map the RNA contacts required for this uniquely tight binding. A three-dimensional structure of AS8 was determined by X-ray crystallography and homology modeling within the known structures of the 30S subunit of E. coli (PDB code 2AVY) and T. thermophilus (PDB code 1J5E) was performed to investigate the molecular rationale for the extremely tight interaction between AS8 and rRNA. In total, these experiments provide a structural framework for the conserved and AS8-specific interactions that contribute to the binding of the minimal rRNA site along helix 21 and unique interactions with the three-way junction of helices 20, 21, and 22 that contribute to the picomolar binding affinity of AS8 for the larger rRNA.
Results
Binding of AS8 and rRNA
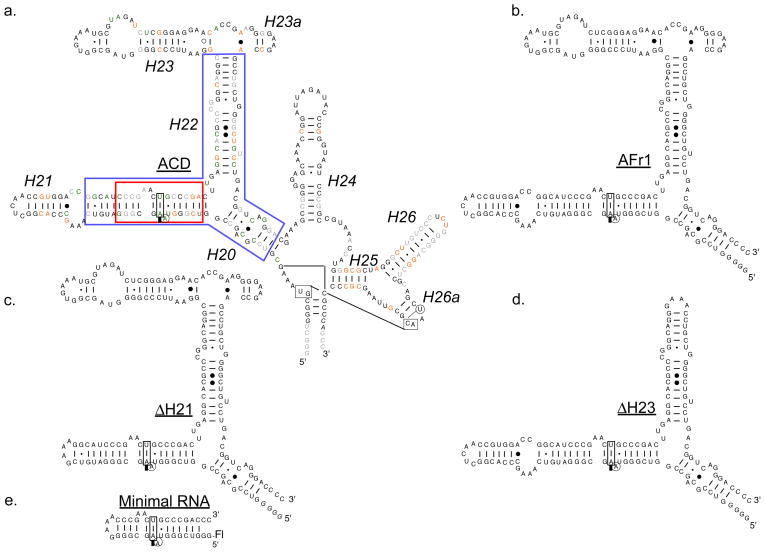
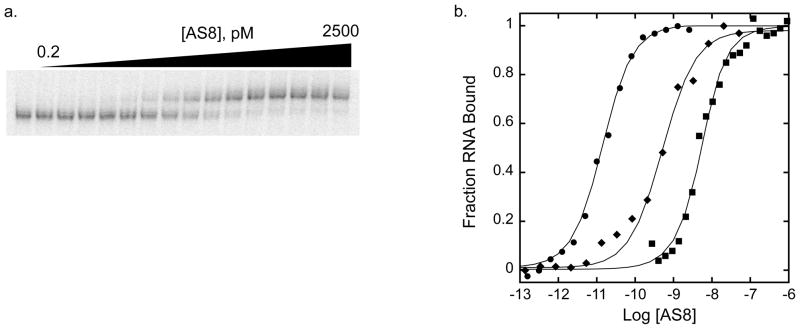
A series of RNA constructs were prepared based on the central domain of the A. aeolicus 16S rRNA (Figure 2a–e) to map the interaction between AS8 and the 16S rRNA by EMSA and fluorescence anisotropy assays (Figure 3a & b). AS8 binds to the AFr1 construct (Figure 2b) at 65°C with an apparent Kd of 21 pM (Figure 3). The affinity of AS8 for ΔH21 (Figure 2c) and ΔH23 (Figure 2d) RNA constructs were also measured by direct titration at 65°C (Table 1). The respective Kd values of 60 and 18 pM show only modest deviations from the value of AFr1 RNA and demonstrate that neither the stem loop region of helix 21 nor all of helix 23 are required for picomolar affinity. Both a larger construct containing the total central domain of A. aeolicus 16S rRNA (ACD in Figure 2a) and a helical construct containing only the A. aeolicus sequence homologous to the minimal S8-binding region of helix 21 identified in studies of E. coli (minimal RNA in Figure 2e) were investigated by competition assay. A small, two-fold deviation from the Kd observed with AFr1 RNA demonstrates that additional interactions with the central domain of 16S rRNA do not make a large contribution to the picomolar affinity. However, the interaction with the minimal site on helix 21 is dramatically reduced, with a Kd value of 1.5 nM.
Figure 2.
Primary and predicted secondary structures of RNA constructs used in this work. a. ACD, central domain of 16S rRNA from A. aeolicus. The ACD helices are labeled (italic) as the homologus secondary structure of E. coli 16S rRNA. Identical bases in the central domains of A. aeolicus, T. thermophilus, and E. coli are printed in black. A. aeolicus bases identical to E. coli, T. thermophilus, or neither are printed in green, orange, and gray respectively. The italic bases indicate non-ribosomal nucleotides included in the sequences for ease of transcription. The helical region corresponding to the minimal site for specific binding of E. coli S8 is boxed in red. The smallest sequence of RNA mapped to a picomolar Kd for AS8 is boxed in blue. b. AFr1 is the A. aeolicus 16S rRNA construct that was previously identified with a picomolar Kd for AS8. c and d. ΔH21 and ΔH23 were used in EMSA experiments to map the smallest RNA with a picomolar Kd for AS8 (blue box in Figure 2a). e. Minimal 16S rRNA construct required for specific binding to AS8. Protein binding to this site was monitored by both competition EMSA and fluorescence anisotropy assays. The position of the fluorescein label on the minimal RNA construct is labeled Fl.
Figure 3.
Thermodynamic mapping of AS8-rRNA interaction by EMSA and fluorescence anisotropy assays. a. Representative gel for EMSA of AS8 titrated into 32P AFr1 at 65°C. b. Comparison of titrations of AS8-AFr1 interaction (EMSA) (circles), the AS8 E85A, K88A, N91A triple mutant–AFr1 interaction (EMSA) (diamonds), and the AS8-minimal RNA interaction (fluorescence anisotropy)(squares). The titrations shown yield an apparent dissociation constant of about 14 pM (65°C), 0.5 nM (65°C), and 2nM (25°C) respectively.
Table 1.
Apparent Kd values for AS8 RNA Interactions
| RNA* | Kd (pM) |
|---|---|
| EMSA 65°C | |
| AFr1 | |
| (Wildtype AS8) | 21 ± 3 |
| (E83A, K86A, Q89A AS8) | 520 ± 80 |
| ACD** | 6.0 ± 0.5 |
| ΔH21 | 60 ± 7 |
| ΔH23 | 18 ± 4 |
| Minimal RNA** | >1500 |
| Fluorescence Anisotropy Assay 25°C | |
| Minimal RNA | |
| KCl (0.33M) | 2800 ± 500 |
| KCl (0.54M) | 2100 ± 400 |
| KCl (0.71M) | 7000 ± 2000 |
| KCl (0.91M) | 12000 ± 2000 |
Interaction is with wildtype AS8 except where indicated
EMSA of Competition with AFr1
The interaction of AS8 with the RNA corresponding to the minimal site on helix 21 was further monitored by fluorescence anisotropy. AS8 was titrated into fluorescein labeled rRNA at 25°C and the resulting interaction was measured by a change in anisotropy. The reaction was monitored in a range of salt conditions varying from 0.33 M to 0.90 M KCl. AS8 binds the minimal construct of helix 21 with a Kd of 2 nM at 25°C and 0.33 M KCl, which is consistent with both EMSA competition experiments conducted at 65°C and with past observations with other thermophilic S8 proteins. The reaction is largely independent of KCl concentration with only a five-fold increase in Kd measured in 0.90 M KCl, suggesting that S8-RNA interactions with this site are driven by shape complementarity with the protein backbone and modulated by hydrophobic interactions that are not affected by high ionic strength.
Taken in total, these results show that two regions of rRNA contribute significantly to the affinity of the interaction with AS8 (Figure 2a). One region consists of the minimal binding site surrounding the unpaired A of helix 21 and has a nanomolar affinity for AS8 (red box in Figure 2a). The other region contains the three-way junction of helices 20, 21, and 22 that contributes to picomolar affinity when combined with the minimal binding site (blue box Figure 2a).
Overall structure of AS8
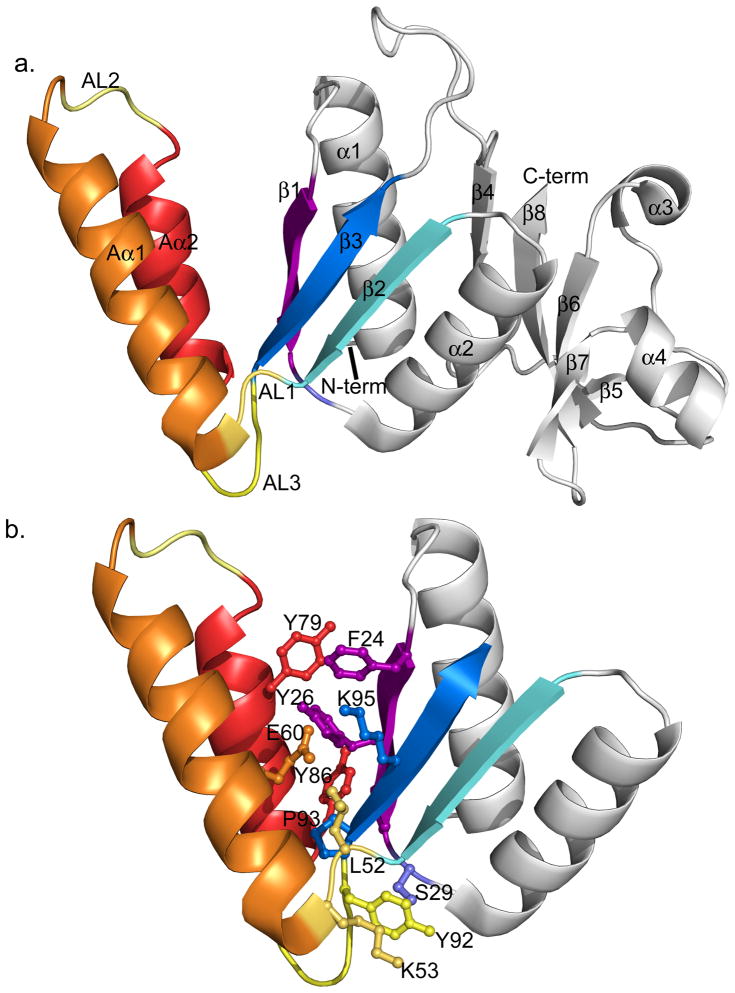
The crystal structure of AS8 was solved at 2.16 Å resolution. Data processing and refinement statistics are summarized in Table 2 and the three-dimensional structure is presented in Figure 4. AS8 maintains the core architecture of the N- and C-terminal domains found in other S8 proteins (Figure 1b) but also includes two additional α helices formed by the unique peptide insertion. The N-terminal domain is formed by helices α1 and α2 packed on one side against the three-stranded antiparallel β-sheet (formed by strands β1, β2 and β3) and flanked on the other side by two α helices formed by the AS8 peptide insertion (Aα1-Aα2). The insertion replaces the short loop that connects β2 and β3 in the known S8 structures. Aα1 is connected to β2 through an ordered loop (AL1), while Aα1 and Aα2 are connected by a loop (AL2) that is not well defined in the crystal structure. Aα2 is connected to β3 by a third loop (AL3). A cluster of positively charged residues is observed on the solvent exposed side of helix Aα2 (residues Lys75, Lys77, Arg78, Lys81). A loop of 13 amino acids connects the N- and C-terminal domains (loop between strands β3 and β4 in Figure 4a). This loop is shorter than the inter-domain loop in T. Thermophilus (PDB code 1AN7), but similar in length to B. Stearothermophilus S8 (PDB code 1SEI) and E.coli S8 (PDB code 2AVY). The C-terminal domain consists of a four-stranded antiparallel β-sheet (β4, β6, β7 and β8,) flanked by two short alpha helices (α3 and α4) on one side and packed against helices α1 and α2 of the N-terminal domain.
Table 2.
Data Collection And Refinement Statistics
| SeMeta | Nativeb | |||
|---|---|---|---|---|
| A. Diffraction data | ||||
| Peak | Inflection | Remote | ||
| Space group | P4 | P4 | P4 | P 41 21 2 |
| Cell dimensions | ||||
| a, b, c (Å) | a=b=96.711 | a=b=96.711 | a=b=96.736 | a=b=96.235 |
| c=37.970 | c=37.978 | c=37.978 | c=37.614 | |
| α, β, γ(°) | α=β=γ=90 | α=β=γ=90 | α=β=γ=90 | α=β=γ=90 |
| Wavelength (Å) | 0.97826 | 0.97862 | 0.91837 | 0.97945 |
| Resolution range (Å) | 50-2.60 | 50-2.60 | 50-2.60 | 50-2.16 |
| Total reflections | 208216 | 194405 | 2129366 | 121797 |
| Unique reflections | 11301 | 11285 | 11311 | 9985 |
| Rmerge (%) | 12.2 (36.5) | 11.0 (35.9) | 11.3 (36.8) | 6.1 (21.3) |
| I/σ(I) | 23.9 (5.8) | 25.6 (5.2) | 24.7 (5.4) | 38.5 (5.6) |
| Completeness (%) | 98.6 (90.3) | 98.2 (88.6) | 98.3 (89.6) | 99.4 (96.1) |
| Redundancy | 18.8 (14.6) | 9.3 (7.0) | 19 (15.5) | 6.8 (2.6) |
|
|
||||
| highest resolution shell=2.64-2.60 | highest resolution shell=2.24-2.16 | |||
| B. Refinement statistics | ||||
| Resolution range (Å) | 30.43-2.16 | |||
| Rwork | 0.223 | |||
| Rfree | 0.267 | |||
| Stereochemistry | ||||
| Ramachandran outlier residues (%) | 0.7 | |||
| Ramachandran favored residues (%) | 93.3 | |||
| Number of atoms | ||||
| Protein | 1333 | |||
| Water | 51 | |||
| Ligand | 25 | |||
| B factors (Å) | ||||
| Protein | 51.5 | |||
| Water | 47.0 | |||
| Ligand | 88.8 | |||
| RMSD from ideal values | ||||
| Bond lengths (Å) | 0.007 | |||
| Bond angles (°) | 0.990 | |||
| Residues not modeled | 1-3(MSA) | |||
Numbers in parentheses represent values in the highest-resolution shell
Used for phasing and initial model building.
Used for structure refinement after molecular replacement with initial model from MAD data.
Figure 4.
Cartoon representation of the AS8 crystal structure. a. The elements of AS8 secondary structure are labeled. Structural elements that are unique to AS8 are colored in orange (helix Aα1), red (helix Aα2) and yellow (AL1, AL2, and AL3). The strands of the N-terminal β-sheet (β1, β2, β3) are colored in purple, cyan, and blue respectively. b. Tertiary structure connecting the subdomain formed by the AS8 insertion peptide and N-terminal domain. Only the N-terminal domain is shown. The side-chains are colored according to their corresponding secondary structure element. Residues Pro28 and Tyr57 are not visible in this orientation.
Although the helix-loop-helix insertion is one of the regions in the structure that has the highest atomic displacement parameters, a network of hydrophobic, aromatic, and polar interactions take place between Aα1, Aα2 and the N-terminal β-sheet. Charged interactions occur between the side chains of Glu60 on helix Aα1 and Tyr26 and Lys95 in the antiparallel β-sheet. Residues in the hydrophobic patch include Phe24, Tyr26, Pro28 on strand β1 and Tyr79, Tyr86 on helix Aα2 (Figure 4b). Interestingly, the residues corresponding to Tyr26 in the known bacterial structures are solvent-exposed polar residues, but a tyrosine is present at this position in the Methanococcus jannaschii structure. The M. jannaschii residues Val24, Tyr26, and Lys62 occupy equivalent positions to Phe24, Tyr26, and Lys95 of AS8. This core contains solvent exposed patches in the M. jannaschii protein while the equivalent patches of AS8 are involved in interactions with the helices of the unique subdomain. This demonstrates that the surface of the N-terminal β-sheet of S8 protein can accommodate structures of similar hydrophobicity in multiple ways.
Two more clusters of interactions are observed between the helix-loop-helix insertion and the conserved core. These include a hydrophobic cluster formed by Pro93 at the N-terminus of β3, Leu52 in AL1, and Tyr57 at the base of Aα1. An additional network of interactions is present between the side-chains of Ser29 on the C-terminal end of β1, Lys53 of AL1, and Tyr92 of AL3.
Conservation of S8
An estimate of the sequence diversity of S8 proteins is presented in Supplementary Figure 1 as position-dependent conformation scores based on a multiple sequence alignment of sixty-two S8 orthologs obtained from the COGS database (www.ncbi.nlm.nih.gov/COG/) (Sequence names are listed in Supplementary Table 1). These scores were calculated for each position based on a combination of the Shannon entropy and a gap penalty resulting in a score of increasing conservation from zero to one 40. The unique sequence of the AS8 insertion was excluded from the alignment to increase accuracy (See Materials and Methods). The scores are presented in Supplementary Figure 1 along with the protein sequence of AS8 and a map of its secondary structure determined from its three dimensional structure. The aligned sequences of S8 proteins with previously-determined three-dimensional structures are also presented.
The greatest region of conservation in these structures include the hydrophobic cores of both the N and C-terminal domains, the core region connecting the two domains, and the C-terminal region of S8 contributing to interactions with the minimal RNA binding site within helix 21 of the 16S central domain. Ser143 (A. aeolicus numbering) in the C-terminal domain is universally conserved within S8 sequences. An interaction between an equivalent Ser and an unpaired A equivalent to A642 in helix 21 makes the only amino acid side-chain to base contact observed within all the structures of S8-RNA complexes.
In general, the sequences of the N-terminal domain of S8 are far less conserved than those of the C-terminal domain. The longest stretch of variable sequences occurs from the beginning of β2 and continues through the linker that connects β3 of the N-terminal domain to β4 of the C-terminal domain. Within this variable stretch is the unique helix-loop-helix subdomain of AS8 (positions 51–92). Low conservation scores in this region result from the unique AS8 insert and the sequence variability of the strands and loops of the N-terminal β-sheet.
Modeled Interactions Between AS8 and 16S rRNA
To model potential contacts with the rRNA, the structure of AS8 was aligned with other S8 proteins within the known structures of the 30S ribosomal subunit (E. coli, PDB code 2AVY Figure 5a and T. thermophilus, PDB code1J5E). The alignment, conducted with MacPymol 41, results in RMSD values of 0.77 Å and 1.39 Å respectively. The AS8 structure was also compared to the M. Jannaschii S8 in complex with helix 21 (PDB code 1I6U). The proposed protein-RNA interactions are described using the A. aeolicus numbering system for AS8 and the E. coli numbering for 16S rRNA. For protein-base contacts where the sequence is not identical between E. coli and A. aeolicus (see Figure 2a), the A. aeolicus sequence is indicated. The models indicate that AS8 could make extensive contacts with the minimal RNA binding site on helix 21, as well as with helices 25 and 26, and the three-way junction of helices 20, 21, and 22. The model built by superposition of AS8 with E.coli S8 bound to rRNA is shown in Figures 5a–c. Contacts were considered to be similar among the two structures when the interacting amino acid residues belong to the same group in the conservation score calculations (see Materials and Methods). The model shows several AS8-rRNA contacts that are conserved with respect to all the reported S8-RNA bound structures. Other interactions are found only in the bacterial (E. coli and T. thermophilus) or archaeal (M. jannaschii) systems. Additional AS8-specific interactions are described below.
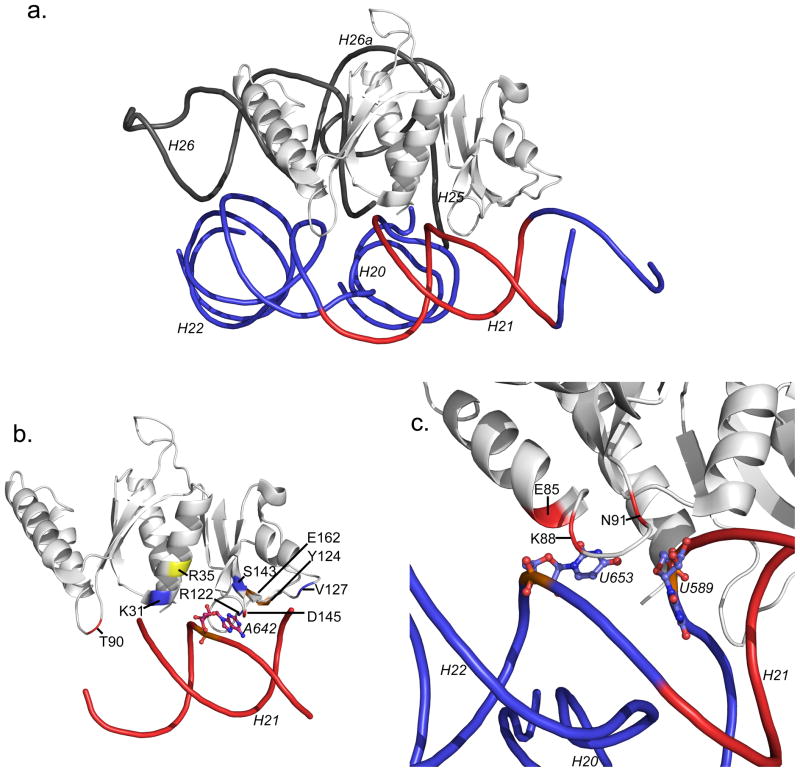
Figure 5.
Modeled rRNA contacts of AS8. a. AS8 is shown after alignment with S8 in the crystal structure of E. coli 30S ribosomal subunit (PDB code 2AVY). The helices of the E. coli RNA are labeled as in Figure 2a. The RNA corresponding to the minimal S8 binding site is colored red; the RNA homologous to the picomolar binding site of A. aeolicus RNA is blue. b. Modeled interactions with the minimal site. The universally conserved A642 is shown in ball and stick representation. The positions of AS8 modeled to contact the RNA are colored on the protein. Contacts similar in all the S8 proteins of known S8-rRNA structures are blue. Contacts similar to only E. coli and T. thermophilus complexes are orange. The contact similar to only the M. jannaschii complex is yellow and contacts unique to AS8 are red. c. Modeled amino acids contributing to the picomolar AS8-rRNA interaction. Glu85, Lys88, and Gln91 are structured to make contact with the platform created by U653 which hydrogen bonds to the ribose of U589. An E85A, K88A, N91A variant of AS8 has an apparent Kd of approximately 500 pM.
Interactions With the Minimal RNA Binding Site
Contacts with the minimal RNA binding site on helix 21 are observed in all the homologous structures and are mediated both by the N- and C-terminal domains of AS8 (Figure 5b). In the C-terminal domain, two loops formed by residues Ser143-Asp145 and residues Val116-Val123 wrap around the bulged bases U641-A642. This interaction is driven both by shape complementarity between the loops and the RNA minor groove surface and base-specific contact between the universally conserved Ser143 and A642. In the RNA-free structure of AS8, the loop formed by Val116-Val123 has high B factors suggesting it may be flexible in the absence of RNA.
Additional contacts between the C-terminal domain of AS8 and the minimal RNA binding site on helix 21 that are conserved in all the RNA-bound S8 structures may include the interaction between the side chain of Arg122 and the ribose of U644 and between the backbone of Val127 and the base of C599 (G in A. aeolicus). Contacts found in the bacterial structures but not in M. jannaschii include the side-chain of Tyr124 and the base of G597 and between the side-chain of Glu162 and the 2′ hydroxyl of C643. Potential AS8-specific interactions found in the C-teminus involve the side-chain of Asp145 and the 2′ hydroxyl of A640.
The N-terminal domain is proposed to interact with helix 21 through a smaller surface. Similar to all of the other structures, the backbone NH group of Lys31 may interact with the phosphate backbone of U590, while its side-chain nitrogen is in close proximity to the phosphate backbones of U590 and U591. The only interaction we propose to be uniquely homologous with the M. jannaschii complex is the contact between the side chain of Arg35 and the phosphate backbone of A642. Potential AS8-specific contacts may occur between Thr90 in the AS8 subdomain and the ribose of U590.
Contacts with the Three-way Junction between helices 20, 21 and 22
Our model suggests that the AL3 loop is deeply inserted into the three-way junction (Figure 5c), which could allow a combination of backbone and side-chain interactions between residues Glu85, Lys88, and Gln91 and the unpaired base of U653 and the ribose of U589. These unique interactions expand the shape complementarity between AS8 and the rRNA beyond the minimal site on helix 21.
Contacts with helices 25 and 26
A cluster of conserved interactions occurs between helix α1 of AS8 and the helix 25 of the 16S rRNA and include side-chain to base contacts between Ser12 to C876 (G in A. aeolicus) and Asn16 to C826. Possible contacts found only within the T. thermopholus structure include the side-chain of Asp9 and the base of G824 (G in A. aeolicus) and the side-chain of Lys15 and the phosphate backbone of C876. AS8-specific interaction include contacts between the backbone and side-chain of Arg20 and the ribose of U827 and the phosphate backbone U828 respectively.
The unique AS8 subdomain packs against the backbone of the minor groove of helix 26. This results in potential additional salt bridges between the phosphate oxygen atoms of the RNA backbone and positively charged protein side chains (Lys75, Lys77, Arg78, Lys81). Possible contacts originating from the subdomain of AS8 include electrostatic interactions between the side-chain of Arg78 and the phosphate backbones of U828 and G829 of helix 25 as well as between the side-chain of Lys77 and the phosphate backbone of U851 of helix 26.
Disruption of High Affinity AS8-rRNA Interactions by Mutation of AL3
The protein-RNA interactions proposed to be AS8-specific occur with the three-way junction of helixes 21, 22, and 23 and with helixes 25 and 26. Our RNA-binding studies indicate that helices 25 and 26 can be removed from the rRNA with a modest two-fold effect on the binding affinity. Based on this observation we focused on the interaction between loop AL3 and U653 and U589 that seem to contribute most significantly to the interaction with the three-way junction. A triple alanine variant of AS8 was made by mutagenesis to positions Glu85, Lys88, and Gln91 to eliminate most of the potential for side-chain interactions with U653 and U589 while retaining some potential for backbone-RNA contacts. The superposition of the 2D [15N, 1H]–HSQC NMR correlation spectra of wild type AS8 and the AS8 triple alanine mutant indicated that the fold is preserved (data not shown). About 90% of the resonances appear at the same position while only a few are shifted due to the change of chemical environment introduced by the mutations. These mutations resulted in a weaker Kd of 520 ± 80 pM, as observed by EMSA using the AFr1 RNA at 65°C (Table 1 and Figure 3b). The reduction in Kd value associated with the triple mutation is consistent with a significant contribution of the AL3 loop to the high-affinity binding of AS8 with the RNA of the central domain.
Discussion
The Unique Subdomain of AS8 Contributes to the Extremely Tight Interaction with 16S rRNA
AS8 binds with an extremely high affinity to the central domain of the 16S rRNA when compared to the homologous interactions in both mesophiles and thermophiles. In a study of both bacterial and archaeal systems, a correlation between affinity of S8 for rRNA and growth temperature was observed 11. Dissociation constants at 40°C ranged from the low nanomolar for the thermophilic T. thermophilus, which has an optimal growth temperature (TOG) of 75°C 11, to the low micromolar for the mesophilic Methanococcus vannielii (TOG =37°C 11). A. aeolicus has a relatively high growth temperature (TOG= 85°C 42) that is consistent with the correlative trend. The low picomolar affinity of AS8 and the ACD RNA is 600 fold tighter than the corresponding T. thermophilus interaction and shows a 200,000 fold increase when compared to M. vanniellii. The differences in Kd values are particularly remarkable considering that the earlier measurements were performed at a temperature 25 degrees below that of the A. aeolicus study.
The thermodynamic and structural data presented in this work confirm that the unique subdomain of AS8 contributes significantly to this picomolar interaction. The insertion subdomain of AS8 clearly has the opportunity for additional interactions between protein positions 85, 88, and 91 and the residues equivalent to U653 and U589 when modeled onto the 30S subunit of E. coli. Mutation of these protein positions significantly reduces the affinity of AS8 for rRNA containing this platform. Similarly, interaction between wildtype AS8 with the minimal site, that does not contain the three-way junction, has only nanomolar affinity. This is tight by comparison to mesophilic organisms, and the affinity is likely modulated by the contacts found similar to other organisms and the contacts unique to AS8. However, the interaction with the minimal site alone is approximately 1,000 fold weaker than the interaction that includes the unique AS8 subdomain and the three-way junction of helices 20, 21 and 22.
The larger surface of the protein provides the opportunity for increased interactions with the rRNA and is consistent with the general observation that thermophilic ribosomal proteins contain long extensions important for the stabilization of the RNA tertiary fold 31. In addition to contacts with the rRNA, the larger structure allows for further contact with other r-proteins. In the T. Thermophilus structure, S8 contacts S2 through the inter-domain loop. Although the corresponding loop in AS8 is not large enough to make a homologous contact, there is the potential for contact between S2 and the apical part of helix Aα1 of the unique subdomain. It is possible that stabilization of the S2 complex is a general feature of enhanced thermostability of ribosomes from thermophilic organisms.
High-affinity protein-RNA interactions may be required throughout the assembly of the entire central domain of the 30S subunit of A. aeolicus. A recent study of the free energy and enthalpy of this assembly assayed the cooperative RNA binding by AS15 and the AS6:AS18 heterodimer and independent binding by AS8 and AS11 12. In that study, the picomolar binding affinity observed for AS8 is the tightest interaction by an order of magnitude. This interaction is followed by the hundreds of picomolar interaction observed for the fully cooperative binding of AS6:AS18 to the preformed complex with AS15. Both AS11 and AS15 have individual Kd values on the orders of nanomolar. Collectively, these interactions are all within the high affinity range when compared to the r-protein-rRNA interactions measured for mesophilic systems. This suggests that the assembly of the entire central domain of A. aeolicus requires more stable interactions than those of mesophiles.
Possible Functions for AS8-RNA Interactions Outside The Tight Binding Interface
For E. coli, the S8 interaction with helices 25 and 26 is linked to progression through transition states along the RNA folding pathway 28. However, this relationship has not been investigated in other organisms. There is extensive sequence diversity for both of these helices when compared to E. coli for both A. Aeolicus and T. Thermopholius 16S rRNA (Figure 2a). Helix 25 of A. aeolicus and T. thermophilus is made entirely of G-C pairs and shares sequence identity at all but one position out of twelve. In contrast, helix 25 of E. coli contains two A-U pairs and shares sequence identity with A. aeolicus at only two positions. Helix 26 is considerably more diverse, with E. coli having four and five more bases than A. aeolicus and T. thermophilus respectively, and with only limited sequence identity between the organisms. It is not clear that the folding transition states for the thermophilic RNAs are the same among these organisms. It is possible that the unique contacts observed between AS8 and helix 26 are necessary for the transition states required by the sequence of A. Aeolicus 16S rRNA.
In conclusion
AS8 contains a unique subdomain that enhances the affinity for the central domain of 16S rRNA through interactions with the unpaired U of the three-way junction of helices 20, 21, and 22. The subdomain is accommodated into the conserved architecture of S8 proteins through extensive interactions with the β-sheet of the N-terminal domain. It is clear that N- and C-terminal additions to highly conserved globular domains of ribosomal proteins have been a frequent feature of structural and functional evolution of the ribosome. In the case of AS8, an insertion sequence has provided an evolutionary advantage to a thermophile by providing additional interactions, which is a novel extension to this general observation. The present investigation into AS8-rRNA interactions provides a structural framework for comparative studies on the evolution of the ribosomal assembly.
Materials and methods
Preparation of A. aeolicus S8
AS8 was overexpressed in E.coli BL21-CodonPlus (DE3)-RIL (Stratagene) and purified as described 12. SeMet substituted AS8 was prepared for multiwavelength anomalous dispersion (MAD) phasing by expression in M9 minimal medium supplemented with 50 mg/L L-selenomethionine. The SeMet-protein was purified using the same protocol used for the wild-type protein, except that the buffers contained 5 mM beta mercaptoethanol to keep the SeMet in a reduced state.
Point mutations were introduced via site-directed mutagenesis. The vector encoding the triple mutant AS8(E85A/K88A/Q91A) was constructed in a single step with PCR primers containing all three mutations.
RNA constructs
The ΔH21 and ΔH23 were prepared by Quickchange mutagenesis of the AFr1 plasmid and sequenced for validation. All RNAs except the minimal helix 21 RNA construct were transcribed as described previously 43. The fluorescein labeled minimal RNA and was purchased from Thermo Scientific. AFr1, ΔH21, and ΔH23 were labeled with 32P as described previously 43.
All RNA sequence and secondary structure displays were adapted from the Comparative RNA Web Site and Project (www.rna.ccbb.utexas.edu).
Electrophoretic mobility shift assay
The EMSA was performed as described previously 43. Prior to protein binding, the 32P-labeled AFrI RNA was heat denatured in water at 90°C for two minutes and cooled on ice for five minutes. The RNA was diluted to 0.2 nM in binding buffer (20 mM K-Hepes pH 7.5, 330 mM KCl, 10 mM MgCl2, 0.1 mM EDTA, 0.1 mg/ml tRNA, 5 mg/ml heparin, 0.01 % IGEPAL) and incubated with an equal volume of protein diluted in the same buffer. The protein concentration in the reconstitution mixture ranged from 50 nM to 0.1 pM. The protein-RNA complexes were incubated for 1 hr at 65°C. The incubation time was optimized as described previously12. 65°C was chosen because volume changes during incubation where both large and variable at higher temperatures. 4 μl of loading buffer (30 % (w/v) glycerol, 0.25 %(w/v) bromophenol blue, 0.25 % (w/v) xylene cyanol) were added to 20μl of reaction mixture and 5 μl of sample were loaded onto 15 %, 29:1 acrylamide:bisacrylamide gel in 0.5 TBE buffer. The gels were run at 500 V in the cold room for 17–19 hours and dried prior to exposure on a Phosphorimager screen. It was previously demonstrated that the temperature during the initial binding and incubation time is the most significant for determining the Kd of the interaction 12. The data were quantified using ImageQuant software (Molecular Dynamics) and analyzed as described with IgorPro software (Wavemetrics).
Competition EMSA experiments were performed by using identical conditions, except that a constant concentration of AS8 was included in each equilibration. The protein concentration was chosen in order to yield a 70%–90% bound complex. Unlabeled competitor RNA was titrated into each reaction at varying concentrations. The observed dissociation constant for each competitor was calculated by fitting the data to a quadratic solution of the Lin Riggs equation as described previously 44.
Fluorescence Anisotropy
The fluorescence anisotropy assay was conducted with 10 nM fluorescein labeled minimal RNA in pH 7.4, 20 mM HEPES, 0.33 M Variable KCl, (0.33 M–0.9 M) 10 mM MgCl2, and 0.1 mg/mL tRNA (High-Purity yeast tRNAphe was purchased from Sigma) at 25°C. The final sample volume was 100 μL per well. Samples were equilibrated in 96-well opaque microtiter plates (Grenier) and the anisotropy determined using a Fusion α-FP plate reader (Packard). The average anisotropy value for each AS8 concentration was calculated from 10 consecutive readings of each plate. These values were averaged over at least three separate titrations and the reported error is one standard deviation calculated from the averaged titrations. Apparent Kd values were determined by nonlinear least squares fitting of the binding data to a model for a single-transition isotherm using Igor (Wavemetrics). The titration data were fit to the following equations:
where Pt is the total concentration of AS8 and Rt is the total concentration of labeled minima-RNA at each step. rr is the anisotropy of free RNA and rpr is the anisotropy of AS8 bound RNA.
Protein crystallization
Prior to crystallization AS8 was dialyzed into 10 mM Tris-HCl pH 7.50, 50 mM KCl and concentrated to 11 mg/ml. AS8 crystals were grown using the vapor-diffusion technique. Large orthorhombic crystals grew in 0.1 M Tris-HCl, 1.8 M ammonium sulfate pH 8.5 but were of poor diffraction quality. Better diffracting crystals were obtained by mixing 2 μl of AS8 with an equal volume of reservoir solution containing 0.1 M sodium acetate pH 4.6, 0.2 M lithium sulfate, 40% PEG400 and equilibrated against 0.8 ml of the same solution at 22 C. Needle clusters appeared in 1–2 days. Despite extensive optimization of the crystallization conditions, no single crystals were obtained. A single needle was broken off from the cluster, harvested from mother liquor and flash cooled in liquid nitrogen for data collection. Se-Met derivatised crystals grew in similar conditions (a slightly lower concentration of PEG400 yielded the best crystals).
Structure determination
X-ray diffraction data were collected at the Stanford Synchrotron Radiation Lightsource (SSRL) beamline 11–1. Two native datasets were collected from the same native crystal at 2.40 and 2.16Å respectively on two different synchrotron trips. The native datasets were merged and processed with HKL2000 45. A three-wavelength MAD dataset was collected to 2.6Å resolution and was processed in space group P4 with HKL2000. Seven selenium sites were identified in P41 21 2 space group with PHENIX AutoSol 45, with a figure of merit of 0.62. An initial model was built with PHENIX Autobuild,. 100 of 168 residues were placed and 50 were residues built without side chains into the SeMet-phased electron-density map. Further model building and fitting were carried out with the program COOT 46 and the model was refined using PHENIX Refine 47. The model was then refined against the higher resolution native dataset and several iterations of building and refinement were performed. No electron density was observed for the first three N-terminal residues. Electron density for residues Ser71 to Lys75 is poor, therefore only the backbone of the amino acids were modeled and the occupancy was set to zero for refinement. The data processing and refinement statistics are summarized in Table 2.
Conservation Score Calculations
A conservation score for each position in the sequence of S8 was determined using the sequences available from the Clusters of Orthologous Groups database (www.ncbi.nlm.nih.gov/COG/). This score was calculated as a combination of the normalized sequence entropy and a gap probability as previously described. 40. To be consistent with other calculations of sequence entropy, the twenty amino acids were grouped by the method of Mirny and Shakhnovivh 48 into the following six groups: (1) Arg, Lys; (2) Asp, Glu; (3) His, Phe, Trp, Tyr,; (4) Asn, Gln, Ser, Thr, (5) Ala, Cys, Ile, Leu, Met, Val; (6) Gly, Pro. Alignments for comparing S8 sequences were calculated using ClustalW (1.8) with the default parameters (European Bioinformatics Institute: http://www.ebi.ac.uk/clustalw/). Alignments where conducted with positions 52–92 edited out of the AS8 sequence to increase accuracy. This sequence was returned for the conservation calculations.
Molecular Modeling
All three dimensional structures and models were rendered using the PyMOL Molecular Graphics System, Version 1.3, Schrödinger, LLC.
Supplementary Material
Conservation of S8 primary structure. The sequence of AS8 is shown with every tenth position indicated by a dot. The sequence was aligned with the 62 known S8 sequences obtained from the COGS database. To increase accuracy, the insert unique to AS8 was edited out of the alignment. For clarity only sequences of S8 proteins of known structure are shown. After returning the unique AS8 sequence, a conservation score was calculated for each position as a combination of the normalized Shannon entropy and a gap penalty. The color coded score is presented from red representing the least conserved sequences to blue representing the most conserved. The secondary structure elements of AS8, as determined from the structure, is mapped above the sequence (rectangles for α-helices and arrows for β-strands). The unique secondary structure of AS8 is labeled in red. The universally conserved serine that contributes to specific binding of the minimal site on the 16S rRNA (S143 in AS8 numbering) is boxed in grey. The least conserved regions of S8 sequence correspond to the loop regions and βstrands of the N-terminus and contain the unique subdomain of AS8.
Acknowledgments
The authors wish to acknowledge Robyn Stanfield for assistance with in house crystal screening, Pavel Afonine for advice on structure refinement and the staff of the Stanford Synchrotron Radiation Lightsource beamline BL11-1. We also wish to thank William Ridgeway for help with installation of crystallography software and critical reading of the manuscript, Pedro Serrano Navarro, Zahra Shajani, and Gabriela Ring for comments on the manuscript. We thank Blair Szymczyna for helpful discussions.
This work was supported by a grant from the NIH (GM-53320 to J.R.W) and an American Heart Association Postdoctoral Fellowship (to E.M.).
Footnotes
Accession number
Coordinates and structure factors have been deposited at the Protein Data Bank with accession number 3RF2.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ratcliff K, Corn J, Marqusee S. Structure, stability, and folding of ribonuclease H1 from the moderately thermophilic Chlorobium tepidum: comparison with thermophilic and mesophilic homologues. Biochemistry. 2009;48:5890–8. doi: 10.1021/bi900305p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Razvi A, Scholtz JM. Lessons in stability from thermophilic proteins. Protein Sci. 2006;15:1569–78. doi: 10.1110/ps.062130306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Razvi A, Scholtz JM. A thermodynamic comparison of HPr proteins from extremophilic organisms. Biochemistry. 2006;45:4084–92. doi: 10.1021/bi060038+. [DOI] [PubMed] [Google Scholar]
- 4.Hollien J, Marqusee S. Comparison of the folding processes of T. thermophilus and E. coli ribonucleases H. J Mol Biol. 2002;316:327–40. doi: 10.1006/jmbi.2001.5346. [DOI] [PubMed] [Google Scholar]
- 5.Hollien J, Marqusee S. Structural distribution of stability in a thermophilic enzyme. Proc Natl Acad Sci U S A. 1999;96:13674–8. doi: 10.1073/pnas.96.24.13674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hollien J, Marqusee S. A thermodynamic comparison of mesophilic and thermophilic ribonucleases H. Biochemistry. 1999;38:3831–6. doi: 10.1021/bi982684h. [DOI] [PubMed] [Google Scholar]
- 7.Kohrer C, Mayer C, Neumair O, Grobner P, Piendl W. Interaction of ribosomal L1 proteins from mesophilic and thermophilic Archaea and Bacteria with specific L1-binding sites on 23S rRNA and mRNA. Eur J Biochem. 1998;256:97–105. doi: 10.1046/j.1432-1327.1998.2560097.x. [DOI] [PubMed] [Google Scholar]
- 8.Shcherbakov D, Dontsova M, Tribus M, Garber M, Piendl W. Stability of the ‘L12 stalk’ in ribosomes from mesophilic and (hyper)thermophilic Archaea and Bacteria. Nucleic Acids Res. 2006;34:5800–14. doi: 10.1093/nar/gkl751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerstner RB, Pak Y, Draper DE. Recognition of 16S rRNA by ribosomal protein S4 from Bacillus stearothermophilus. Biochemistry. 2001;40:7165–73. doi: 10.1021/bi010026i. [DOI] [PubMed] [Google Scholar]
- 10.Powers T, Noller HF. A temperature-dependent conformational rearrangement in the ribosomal protein S4.16 S rRNA complex. J Biol Chem. 1995;270:1238–42. doi: 10.1074/jbc.270.3.1238. [DOI] [PubMed] [Google Scholar]
- 11.Gruber T, Kohrer C, Lung B, Shcherbakov D, Piendl W. Affinity of ribosomal protein S8 from mesophilic and (hyper)thermophilic archaea and bacteria for 16S rRNA correlates with the growth temperatures of the organisms. FEBS Lett. 2003;549:123–8. doi: 10.1016/s0014-5793(03)00760-9. [DOI] [PubMed] [Google Scholar]
- 12.Recht MI, Williamson JR. RNA tertiary structure and cooperative assembly of a large ribonucleoprotein complex. J Mol Biol. 2004;344:395–407. doi: 10.1016/j.jmb.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 13.Mougel M, Ehresmann B, Ehresmann C. Binding of Escherichia coli ribosomal protein S8 to 16S rRNA: kinetic and thermodynamic characterization. Biochemistry. 1986;25:2756–65. doi: 10.1021/bi00358a003. [DOI] [PubMed] [Google Scholar]
- 14.Held WA, Ballou B, Mizushima S, Nomura M. Assembly mapping of 30 S ribosomal proteins from Escherichia coli. Further studies. J Biol Chem. 1974;249:3103–11. [PubMed] [Google Scholar]
- 15.Mizushima S, Nomura M. Assembly mapping of 30S ribosomal proteins from E. coli. Nature. 1970;226:1214. doi: 10.1038/2261214a0. [DOI] [PubMed] [Google Scholar]
- 16.Nomura M, Mizushima S, Ozaki M, Traub P, Lowry CV. Structure and function of ribosomes and their molecular components. Cold Spring Harb Symp Quant Biol. 1969;34:49–61. doi: 10.1101/sqb.1969.034.01.009. [DOI] [PubMed] [Google Scholar]
- 17.Gregory RJ, Zimmermann RA. Site-directed mutagenesis of the binding site for ribosomal protein S8 within 16S ribosomal RNA from Escherichia coli. Nucleic Acids Res. 1986;14:5761–76. doi: 10.1093/nar/14.14.5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moine H, Squires CL, Ehresmann B, Ehresmann C. In vivo selection of functional ribosomes with variations in the rRNA-binding site of Escherichia coli ribosomal protein S8: evolutionary implications. Proc Natl Acad Sci U S A. 2000;97:605–10. doi: 10.1073/pnas.97.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schaup HW, Sogin ML, Kurland CG, Woese CR. Localization of a binding site for ribosomal protein S8 within the 16S ribosomal ribonucleic acid of Escherichia coli. J Bacteriol. 1973;115:82–7. doi: 10.1128/jb.115.1.82-87.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ungewickell E, Garrett R, Ehresmann C, Stiegler P, Fellner P. An investigation of the 16-S RNA binding sites of ribosomal proteins S4, S8, S15, and S20 FROM Escherichia coli. Eur J Biochem. 1975;51:165–80. doi: 10.1111/j.1432-1033.1975.tb03917.x. [DOI] [PubMed] [Google Scholar]
- 21.Zimmermann RA, Mackie GA, Muto A, Garrett RA, Ungewickell E, Ehresmann C, Stiegler P, Ebel JP, Fellner P. Location and characteristics of ribosomal protein binding sites in the 16S RNA of Escherichia coli. Nucleic Acids Res. 1975;2:279–302. doi: 10.1093/nar/2.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zimmermann RA, Singh-Bergmann K. Binding sites for ribosomal proteins S8 and S15 in the 16 S RNA of Escherichia coli. Biochim Biophys Acta. 1979;563:422–31. doi: 10.1016/0005-2787(79)90061-3. [DOI] [PubMed] [Google Scholar]
- 23.Wower I, Brimacombe R. The localization of multiple sites on 16S RNA which are cross-linked to proteins S7 and S8 in Escherichia coli 30S ribosomal subunits by treatment with 2-iminothiolane. Nucleic Acids Res. 1983;11:1419–37. doi: 10.1093/nar/11.5.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thurlow DL, Ehresmann C, Ehresmann B. Nucleotides in 16S rRNA that are required in unmodified form for features recognized by ribosomal protein S8. Nucleic Acids Res. 1983;11:6787–802. doi: 10.1093/nar/11.19.6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mougel M, Eyermann F, Westhof E, Romby P, Expert-Bezancon A, Ebel JP, Ehresmann B, Ehresmann C. Binding of Escherichia coli ribosomal protein S8 to 16 S rRNA. A model for the interaction and the tertiary structure of the RNA binding site. J Mol Biol. 1987;198:91–107. doi: 10.1016/0022-2836(87)90460-8. [DOI] [PubMed] [Google Scholar]
- 26.Svensson P, Changchien LM, Craven GR, Noller HF. Interaction of ribosomal proteins, S6, S8, S15 and S18 with the central domain of 16 S ribosomal RNA. J Mol Biol. 1988;200:301–8. doi: 10.1016/0022-2836(88)90242-2. [DOI] [PubMed] [Google Scholar]
- 27.Jagannathan I, Culver GM. Assembly of the central domain of the 30S ribosomal subunit: roles for the primary binding ribosomal proteins S15 and S8. J Mol Biol. 2003;330:373–83. doi: 10.1016/s0022-2836(03)00586-2. [DOI] [PubMed] [Google Scholar]
- 28.Calidas D, Culver GM. Interdependencies govern multidomain architecture in ribosomal small subunit assembly. RNA. 2011;17:263–77. doi: 10.1261/rna.2332511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–34. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 30.Wimberly BT, Brodersen DE, Clemons WM, Jr, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–39. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 31.Brodersen DE, Clemons WM, Jr, Carter AP, Wimberly BT, Ramakrishnan V. Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: structure of the proteins and their interactions with 16 S RNA. J Mol Biol. 2002;316:725–68. doi: 10.1006/jmbi.2001.5359. [DOI] [PubMed] [Google Scholar]
- 32.Wu H, Wower I, Zimmermann RA. Mutagenesis of ribosomal protein S8 from Escherichia coli: expression, stability, and RNA-binding properties of S8 mutants. Biochemistry. 1993;32:4761–8. doi: 10.1021/bi00069a010. [DOI] [PubMed] [Google Scholar]
- 33.Wu H, Jiang L, Zimmermann RA. The binding site for ribosomal protein S8 in 16S rRNA and spc mRNA from Escherichia coli: minimum structural requirements and the effects of single bulged bases on S8-RNA interaction. Nucleic Acids Res. 1994;22:1687–95. doi: 10.1093/nar/22.9.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kalurachchi K, Nikonowicz EP. NMR structure determination of the binding site for ribosomal protein S8 from Escherichia coli 16 S rRNA. J Mol Biol. 1998;280:639–54. doi: 10.1006/jmbi.1998.1915. [DOI] [PubMed] [Google Scholar]
- 35.Kalurachchi K, Uma K, Zimmermann RA, Nikonowicz EP. Structural features of the binding site for ribosomal protein S8 in Escherichia coli 16S rRNA defined using NMR spectroscopy. Proc Natl Acad Sci U S A. 1997;94:2139–44. doi: 10.1073/pnas.94.6.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nevskaya N, Tishchenko S, Nikulin A, al-Karadaghi S, Liljas A, Ehresmann B, Ehresmann C, Garber M, Nikonov S. Crystal structure of ribosomal protein S8 from Thermus thermophilus reveals a high degree of structural conservation of a specific RNA binding site. J Mol Biol. 1998;279:233–44. doi: 10.1006/jmbi.1998.1758. [DOI] [PubMed] [Google Scholar]
- 37.Davies C, Ramakrishnan V, White SW. Structural evidence for specific S8-RNA and S8-protein interactions within the 30S ribosomal subunit: ribosomal protein S8 from Bacillus stearothermophilus at 1.9 A resolution. Structure. 1996;4:1093–104. doi: 10.1016/s0969-2126(96)00115-3. [DOI] [PubMed] [Google Scholar]
- 38.Tishchenko S, Nikulin A, Fomenkova N, Nevskaya N, Nikonov O, Dumas P, Moine H, Ehresmann B, Ehresmann C, Piendl W, Lamzin V, Garber M, Nikonov S. Detailed analysis of RNA-protein interactions within the ribosomal protein S8-rRNA complex from the archaeon Methanococcus jannaschii. J Mol Biol. 2001;311:311–24. doi: 10.1006/jmbi.2001.4877. [DOI] [PubMed] [Google Scholar]
- 39.Merianos HJ, Wang J, Moore PB. The structure of a ribosomal protein S8/spc operon mRNA complex. RNA. 2004;10:954–64. doi: 10.1261/rna.7030704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Edgcomb SP, Aschrafi A, Kompfner E, Williamson JR, Gerace L, Hennig M. Protein structure and oligomerization are important for the formation of export-competent HIV-1 Rev-RRE complexes. Protein Sci. 2008;17:420–30. doi: 10.1110/ps.073246608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DeLano WL. The PyMOL Molecular Graphic System. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]
- 42.Deckert G, Warren PV, Gaasterland T, Young WG, Lenox AL, Graham DE, Overbeek R, Snead MA, Keller M, Aujay M, Huber R, Feldman RA, Short JM, Olsen GJ, Swanson RV. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature. 1998;392:353–8. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- 43.Recht MI, Williamson JR. Central domain assembly: thermodynamics and kinetics of S6 and S18 binding to an S15-RNA complex. J Mol Biol. 2001;313:35–48. doi: 10.1006/jmbi.2001.5018. [DOI] [PubMed] [Google Scholar]
- 44.Ryder SP, Williamson JR. Specificity of the STAR/GSG domain protein Qk1: implications for the regulation of myelination. RNA. 2004;10:1449–58. doi: 10.1261/rna.7780504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Macromolecular Crystallography, Pt A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 46.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallographica Section D-Biological Crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 47.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–21. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mirny LA, Shakhnovich EI. Universally conserved positions in protein folds: reading evolutionary signals about stability, folding kinetics and function. J Mol Biol. 1999;291:177–96. doi: 10.1006/jmbi.1999.2911. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Conservation of S8 primary structure. The sequence of AS8 is shown with every tenth position indicated by a dot. The sequence was aligned with the 62 known S8 sequences obtained from the COGS database. To increase accuracy, the insert unique to AS8 was edited out of the alignment. For clarity only sequences of S8 proteins of known structure are shown. After returning the unique AS8 sequence, a conservation score was calculated for each position as a combination of the normalized Shannon entropy and a gap penalty. The color coded score is presented from red representing the least conserved sequences to blue representing the most conserved. The secondary structure elements of AS8, as determined from the structure, is mapped above the sequence (rectangles for α-helices and arrows for β-strands). The unique secondary structure of AS8 is labeled in red. The universally conserved serine that contributes to specific binding of the minimal site on the 16S rRNA (S143 in AS8 numbering) is boxed in grey. The least conserved regions of S8 sequence correspond to the loop regions and βstrands of the N-terminus and contain the unique subdomain of AS8.