Abstract
Mediator is a general coactivator of RNA polymerase II (RNA pol II) bridging enhancer-bound transcriptional factors with RNA pol II. Mediator is organized in three distinct subcomplexes: head, middle, and tail modules. The head and middle modules interact with RNA pol II and the tail module interacts with transcriptional activators. Deletion of one of the tail subunits SIN4 results in derepression of subset of genes, including FLR1, by a largely unknown mechanism. Here we show that derepression of FLR1 transcription in sin4Δ cells occurs by enhanced recruitment of the mediator, as well as Swi/Snf and SAGA complexes. The tail and head/middle modules of the mediator behave as separate complexes at the induced FLR1 promoter. While the tail module remains anchored to the promoter, the head/middle modules are also found in the coding region. The separation of the tail and head/middle modules in sin4Δ cells is also supported by the altered stoichiometry of the tail and head/middle modules at several tested promoters. Deletion of another subunit of the tail module MED2 in sin4Δ cells results in significantly decreased transcription of FLR1, pointing to the importance of the integrity of the separated tail module in derepression. All tested genes exhibited increased recruitment of the tail domain; however, only genes with increased occupancy of the head/middle modules displayed also increased transcription. The separated tail module thus represents a promiscuous transcriptional factor that binds to many different promoters and is necessary for derepression of FLR1 in sin4Δ cells.
Keywords: SIN4, FLR1 expression, transcriptional derepression, Gal11p and Srb4p recruitment, Swi/Snf and SAGA complexes
Introduction
Mediator is a highly conserved complex that plays an important role in transcription by RNA polymerase II (RNA pol II). The mediator subunits were identified by genetic screens and independently by biochemical approaches. Several genes encoding mediator subunits were identified as dominant and recessive suppressors of the cold sensitive phenotype associated with truncations in the carboxyl terminal domain (CTD) of RNA pol II.1–4 Additional genetic screens identified other mediator subunits5,6 and complemented a biochemical approach that resulted in purification of the mediator and identification of its subunits.7 Three-dimensional reconstructions of electron micrographs combined with biochemical and genetic data suggest that mediator is roughly organized in three distinct subcomplexes: head module, middle module, and the tail module.8–10 The head and middle modules interact with the CTD of RNA pol II11 and the tail module interacts with transcriptional activators.12–15 In addition, Srb8p, Srb9p, Srb10p, and Srb11p form a distinct subcomplex that contains the cyclin-kinase pair Cdk8 and CycC that phosphorylates the CTD at the serine 5 residue in the heptapetide repeat.16 This subcomplex is involved in negative regulation of small subset of genes.17
The three-dimensional structure of the mediator suggests that in the presence of RNA pol II the mediator assumes an elongated shape and makes multiple contacts with RNA pol II through the head and middle domains.8 The mediator appears to have a general role in transcription of majority of yeast genes, since inactivation of Med17p or Med22p resulted in almost complete loss of transcription, similar to inactivation of Rpb1p of RNA pol II.17
Mediator plays both positive and negative roles in transcription.6,18–20 The positive role consists in activator-mediated recruitment of mediator and its interaction with RNA polymerase II. Mediator appears to function as a link between the gene-specific activator proteins and the basal RNA pol II transcription machinery. The gene-specific activators interact with subset of mediator subunits, typically involving the tail region. For example, Gal4p interacts directly with the tail subunit Gal11 and the head subunit Srb4,12,13 and Gcn4p interacts with tail subunits Gal11, Pgd1, and Med2 and head subunit Srb2.15
Mediator can also function as a scaffold for repeated rounds of reinitiation by RNA pol II.21,22 Mediator occupancy does not always parallel RNA pol II occupancy and mediator was found also upstream of inactive genes23–25 and, conversely, some highly active promoters did not detectably recruit mediator.23,26 Genome-wide location analysis also found mediator associated with coding region of many highly active genes.24,25
We have found previously that deletion of mediator subunit SIN4 suppresses defect in transcription of the FLR1 gene in plc1Δ cells.27 Plc1p is a budding yeast phospholipase C that is required for the initial step of inositol polyphosphates (InsPs) synthesis. InsPs regulate activity of chromatin remodeling complexes in vivo and in vitro28,29 and are required for induction of the phosphate-responsive PHO5 gene. In this study, we elucidate the molecular mechanism by which sin4Δ mutation derepresses FLR1 gene. Our results are consistent with the model in which sin4Δ mutation increases binding of the tail as well as head/middle modules of the mediator to the FLR1 and other promoters.15 The tail and head/middle modules of the mediator in sin4Δ cells facilitate recruitment of the Swi/Snf and SAGA complexes and assembly of the PIC complex. Interestingly, in induced state, the tail module remains bound to the FLR1 promoter, while the head/middle module is found also in the FLR1 coding region, suggesting that the tail and head/middle modules function as separate complexes in sin4Δ cells.
Results
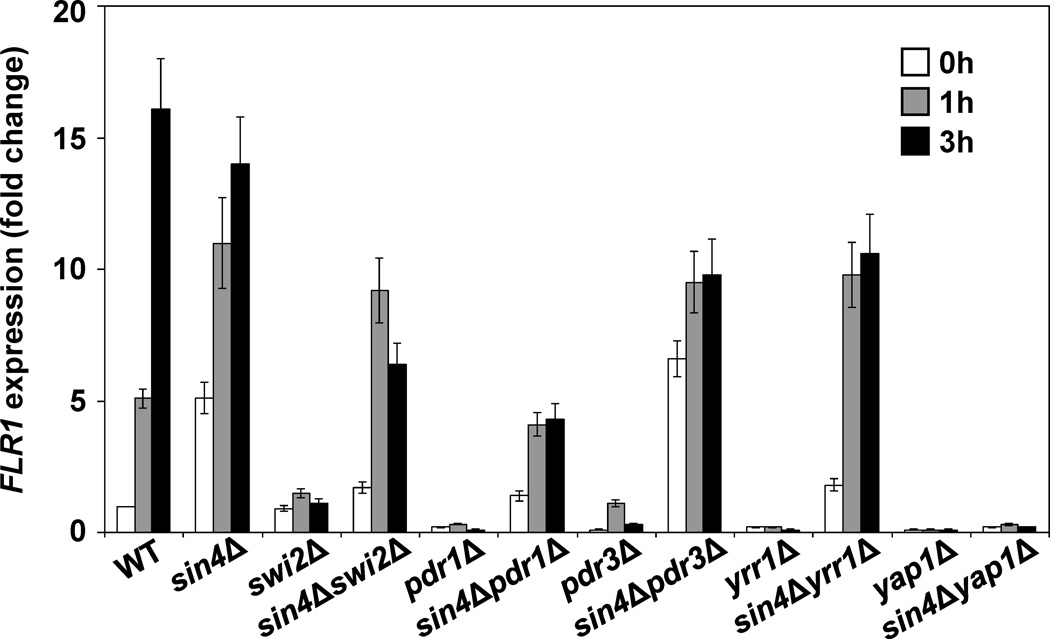
FLR1 expression defect in swi2Δ cells is suppressed by sin4Δ mutation
We have described previously that expression of FLR1 gene requires phospholipase C (Plc1p) – dependent pathway for synthesis of inositol polyphosphates (InsPs) and that the failure of plc1Δ cells to express FLR1 is suppressed by sin4Δ mutation.27 Since plc1Δ cells that are completely devoid of any InsPs30 are not able to recruit Swi/Snf complex and to remodel chromatin of the PHO5 promoter28, we wanted to test whether the induction of the FLR1 gene requires Swi2p, the catalytic subunit of the Swi/Snf chromatin remodeling complex.31,32 Our results show that swi2Δ cells, similarly to plc1Δ cells, are not able to induce FLR1, and this defect is partially suppressed by sin4Δ mutation (Fig. 1). Cells with SIN4 deletion express FLR1 even without induction with benomyl and when induced, the induction occurs faster than in the wild-type cells. However, the maximal expression at 3 h is higher in the wild-type cells than in the sin4Δ cells.
Figure 1.
Defective expression of FLR1 in swi2Δ, pdr1Δ, pdr3Δ, and yrr1Δ cells is partially suppressed by sin4Δ mutation. Indicated trains were grown in YPD medium at 30°C to an A600 of 1.0. Benomyl was added to a final concentration of 5 µg/ml and samples were collected after 0, 1, and 3 h. Total RNA was isolated and assayed for ACT1 and FLR1 transcripts by real-time RT-PCR. The results were normalized to ACT1 RNA and expressed relative to the value for the WT strain at 0 h. The experiment was repeated three times, and the results are shown as means ± SD.
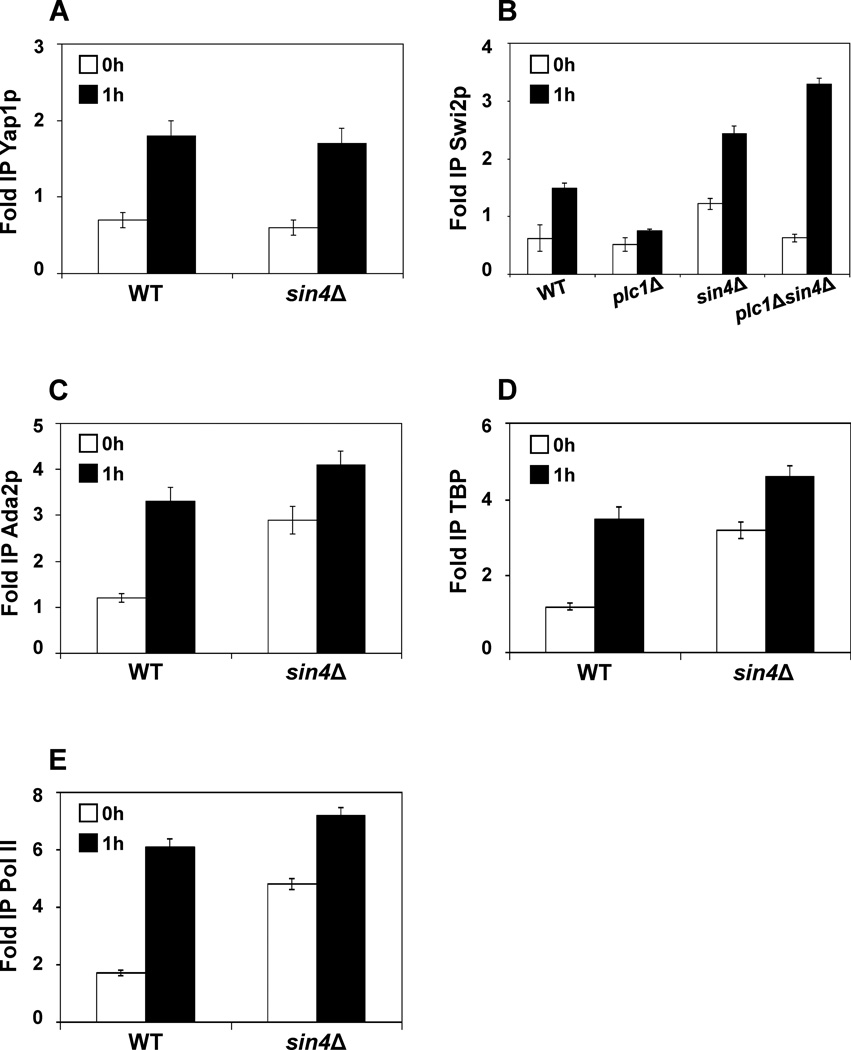
Expression of FLR1 is regulated by several transcriptional factors, including Pdr1p, Pdr3p, Yrr1p, and Yap1p.33–41 Correspondingly, pdr1Δ, pdr3Δ, yrr1Δ, and yap1Δ mutants are not able to express FLR1; however, the expression in pdr1Δ, pdr3Δ, and yrr1Δ mutants can be partially suppressed by sin4Δ mutation (Fig.1). It appears that Yap1p has a unique position in benomyl-induced activation of FLR1 gene, since yap1Δ mutation is not suppressed by sin4Δ mutation (Fig. 1). This result suggests that the sin4Δ mutation does not act by increasing the recruitment of Pdr1p, Pdr3p, or Yrr1p to the FLR1 promoter. However, increased expression of FLR1 in sin4Δ cells before induction could be explained by increased recruitment of Yap1p to the FLR1 promoter. To test this possibility, we determined by chromatin immunoprecipitation (ChIP) the occupancy of Yap1p at the FLR1 promoter before and after induction. The results show that the sin4Δ mutation does not affect Yap1p occupancy at the FLR1 promoter (Fig. 2A).
Figure 2. sin4Δ mutation facilitates PIC assembly.
(A) Recruitment of Yap1p to the FLR1 promoter is not altered in sin4Δ cells. Yap1p occupancy was determined with anti-Yap1p polyclonal antibody. (B) plc1Δ cells fail to recruit Swi2p to the FLR1 promoter and the defect is suppressed by sin4Δ mutation. Swi2p occupancy was determined in strains expressing Swi2p-myc18 using anti-myc antibody. (C) sin4Δ cells display increased recruitment of Ada2p to the FLR1 promoter. Ada2p occupancy was determined in strains expressing Ada2p-myc18 using anti-myc antibody. (D) sin4Δ cells display increased recruitment of TBP to the FLR1 promoter. TBP occupancy was determined in strains expressing Spt15p-3HA using anti-HA antibody. (E) sin4Δ cells display increased recruitment of RNA Pol II to the FLR1 promoter. RNA Pol II occupancy was determined by immunoprecipitation of Rpb1p (the largest subunit of RNA polymerase II) with 8WG16 mAb. (A–E) ChIP was performed using chromatin from the corresponding cells grown in YPD medium (0 h) and treated with benomyl (5 µg/ml) for 1 h. Each immunoprecipitation was performed at least three times using different chromatin samples. The occupancy was calculated with POL1 coding sequence as a negative control. The data are presented as fold occupancy over the POL1 coding sequence control and represent means ± SD.
To better understand the mechanism by which sin4Δ mutation restores the expression of the FLR1 gene in plc1Δ cells, we determined recruitment of Swi2p to the FLR1 promoter in wild-type, plc1Δ, sin4Δ, and plc1Δsin4Δ mutants during FLR1 induction with benomyl. In wild-type and sin4Δ cells, Swi2p is recruited to the FLR1 promoter in response to benomyl induction (Fig. 2B). This recruitment is almost completely eliminated in plc1Δ cells, however, introducing sin4Δ mutation in plc1Δ cells suppresses this defect and allows Swi2p recruitment (Fig. 2B). This result is thus consistent with the notion that InsPs play a role in recruitment of chromatin remodeling complexes28 and suggest that the reduced FLR1 expression in plc1Δ cells is caused by the defect in Swi2p recruitment. In addition, the recruitment of Swi2p in sin4Δ cells before induction and during induction is higher than in wild-type cells. To test the possibility that the assembly of the preinitiation complex (PIC) at the FLR1 promoter is facilitated by sin4Δ mutation, we determined the occupancy of Ada2p, Spt15p, and RNA pol II at the FLR1 promoter before and after induction (Fig. 2C, D, E). The results show an increased occupancy of Ada2p, Spt15p, and RNA Pol II, and suggest that sin4Δ mutation facilitates the PIC assembly at the FLR1 promoter even under non-inducing conditions.
sin4Δ mutation derepresses FLR1 transcription by allowing recruitment of the tail and head/middle modules of the mediator
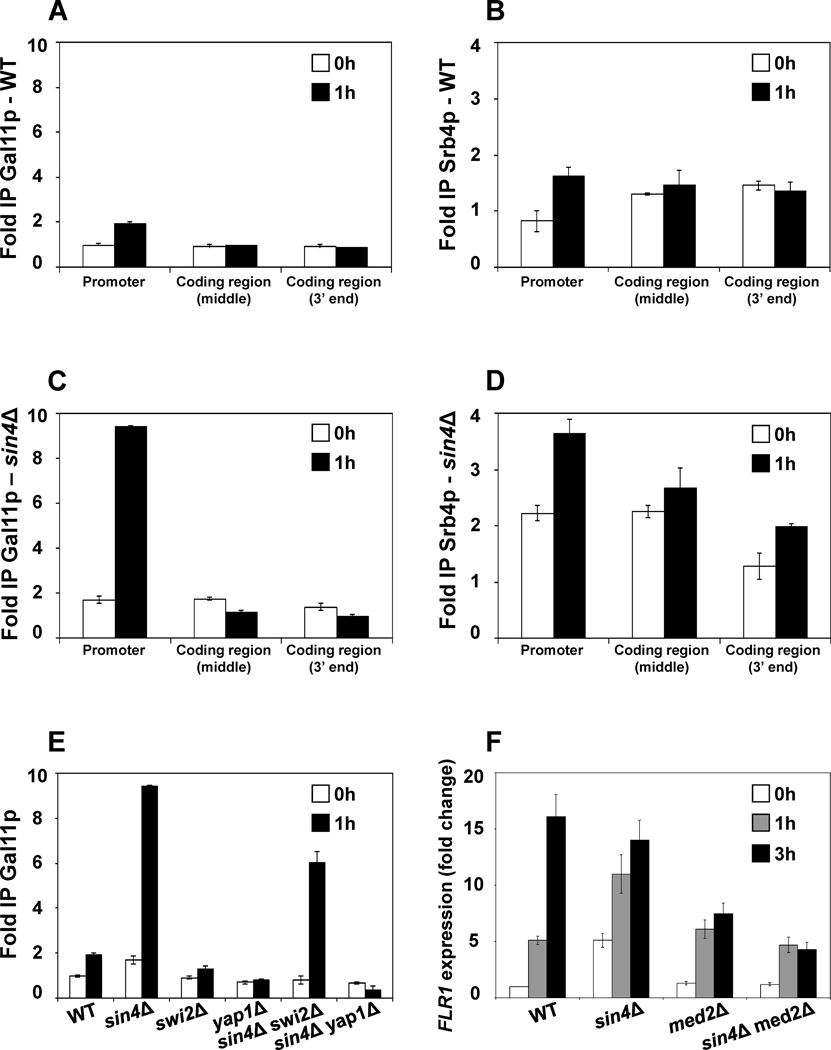
To explain the mechanism of derepression of FLR1 transcription in sin4Δ cells, we considered a possibility that sin4Δ mutation causes separation of the tail domain of the mediator from the middle and head domains15. The tail module is anchored to the middle module of the mediator by Sin4p and deletion of SIN4 results in separation of the tail module from the rest of the mediator composed of the middle and head domains. In sin4Δ mutant, the separated tail domain, composed of Med2p, Pgd1p, and Gal11p is recruited by Gcn4p to the ARG1 promoter, where it stimulates transcription to the same level as intact mediator15. We hypothesized that in sin4Δ mutant even in uninduced state the separated tail domain would be recruited to the FLR1 promoter, facilitate recruitment of the Swi/Snf complex and assembly of the PIC complex. To examine this possibility, we determined the recruitment of Gal11p and Srb4p to the FLR1 promoter and coding region before and after induction. Gal11p and Srb4p are components of the tail and head modules of the mediator, respectively. The occupancy of both Gal11p and Srb4p at the FLR1 promoter in the wild-type cells is at the background level before induction and is elevated about 2 fold after induction (Fig. 3A,B). The occupancy of both proteins in sin4Δ cells before induction is increased approximately 2 fold in comparison to wild-type cells before induction (Fig. 3C, D). Interestingly, while the induction in sin4Δ cells results in increased Gal11p occupancy only at the promoter, the Srb4p occupancy is increased in both the promoter and the coding region (Fig. 3C, D).
Figure 3. The tail and head/middle modules behave as separate complexes in sin4Δ cells.
(A,B) Recruitment of Gal11p and Srb4p to the FLR1 promoter and coding region in wild-type cells. (C,D) Recruitment of Gal11p and Srb4p to the FLR1 promoter and coding region in sin4Δ cells. (E) Recruitment of Gal11p to the FLR1 promoter in wild-type, sin4Δ, swi2Δ, yap1Δ, sin4Δ swi2Δ, and sin4Δ yap1Δ cells. Gal11p and Srb4p occupancies were determined in strains expressing Gal11p-3HA and Srb4p-myc9 using anti-HA and anti-myc antibodies, respectively. ChIP was performed using chromatin from the corresponding cells grown in YPD medium (0 h) and treated with benomyl (5 µg/ml) for 1 h. Each immunoprecipitation was performed at least three times using different chromatin samples. The occupancy was calculated with POL1 coding sequence as a negative control. The data are presented as fold occupancy over the POL1 coding sequence control and represent means ± SD. (F) FLR1 expression in sin4Δ cells requires MED2. Indicated trains were grown in YPD medium at 30°C to an A600 of 1.0. Benomyl was added to a final concentration of 5 µg/ml and samples were collected after 0, 1, and 3 h. Total RNA was isolated and assayed for ACT1 and FLR1 transcripts by real-time RT-PCR. The results were normalized to ACT1 RNA and expressed relative to the value for the WT strain at 0 h. The experiment was repeated three times, and the results are shown as means ± SD.
These results suggest that upon induction, the mediator is recruited as a single complex in the wild-type cells and the occupancy at the FLR1 promoter is increased about 2 fold above the background level. On the other hand, the occupancies of Gal11p and Srb4p suggest that in sin4Δ cells the tail and head/middle modules behave as separate complexes. While the tail remains at the FLR1 promoter, the head/middle module is found also within the coding region. These results are consistent with the physical interactions of the mediator with components of the transcription machinery. The tail module interacts with transcriptional activators that are recruited to the promoter,14 and the head and middle module interact with the CTD of the RNA Pol II.11
The induction of FLR1 expression in sin4Δ cells results in a dramatic recruitment of the tail domain to the promoter. While in the wild-type cells Gal11p is not recruited to FLR1 above background level before induction and the occupancy is increased 2 fold after induction, sin4Δ mutation causes about 2 fold increase in Gal11p recruitment before induction and about 9 fold increase after induction in comparison to wild-type cells before induction (Fig. 3A, C). To determine, whether this recruitment requires Swi/Snf complex or Yap1p transcriptional factor, we determined occupancy of Gal11p also in sin4Δ swi2Δ and sin4Δ yap1Δ strains. Deletion of SWI2 in sin4Δ cells eliminates Gal11p recruitment before induction and reduces the Gal11p occupancy to a level 6 fold higher than in the wild type cells before induction. Deletion of YAP1 completely eliminates Gal11p recruitment (Fig. 3E). These results suggest that Swi/Snf complex is not essential for recruitment of the tail module in sin4Δ cells and has only supportive role; however, Yap1p is essential for recruitment of the tail module.
The above results are consistent with the previous finding that sin4Δ mutation dissociates the tail module from the rest of the mediator15. Our results show that the tail, as well as the head/middle modules are recruited to the FLR1 promoter in sin4Δ cells, and the increased occupancy is associated with elevated FLR1 transcription. The finding that sin4Δ mutation results in elevated expression of HO and several other genes42–47 lead to suggestion that the tail module represents an inhibitory module of the mediator. In this model, the sin4Δ mutation would remove this inhibitory domain and allow the tailless mediator to facilitate the FLR1 transcription. To address this possibility, we determined FLR1 expression in wild-type, sin4Δ, med2Δ, and sin4Δmed2Δ cells (Fig. 3F). Med2p is a subunit of the tail module and in the sin4Δ cells resides in separated tail module.15 Since introducing the med2Δ mutation in sin4Δ cells reduces the FLR1 expression, we conclude that the separated tail module is required for the FLR1 expression in sin4Δ cells and does not represent the inhibitory domain, at least in the context of the FLR1 promoter.
The tail module in sin4Δ cells is a promiscuous transcriptional activator
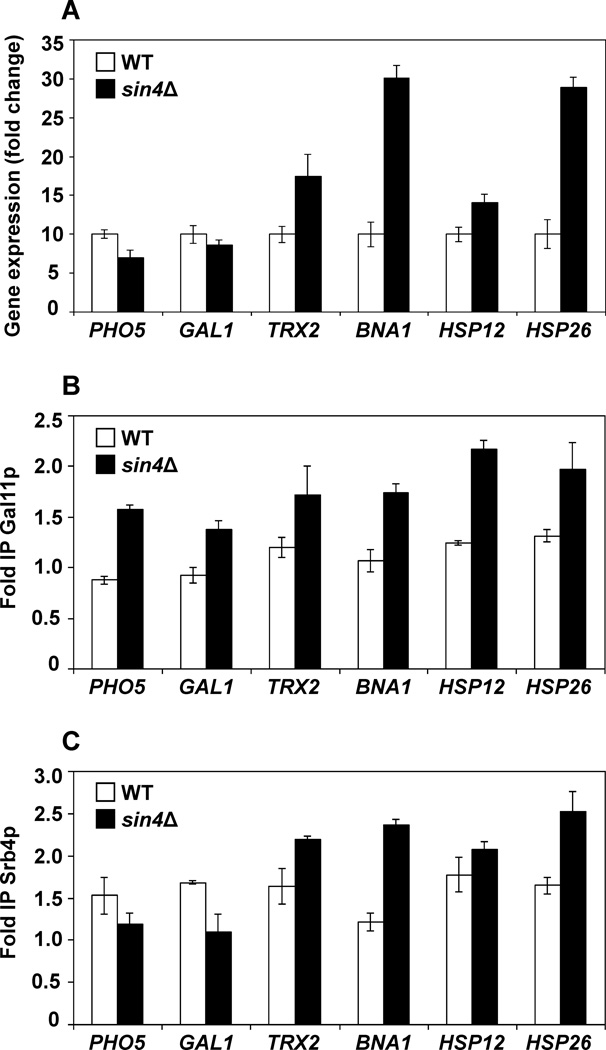
Deletion of SIN4 gene affects transcription of relatively small fraction of the genome.48 To determine, whether the genes with increased expression in sin4Δ cells display a particular pattern of recruitment of the tail and head/middle modules, we determined recruitment of Gal11p and Srb4p to several genes that display higher expression in sin4Δ cells than in the wild-type cells. Gene expression and recruitment of Gal11p and Srb4p were measured in cells grown in YPD medium at 30°C. Our results agreed with the genome-wide expression analysis48 and showed higher expression of TRX2, BNA1, HSP12, and HSP26 in sin4Δ cells than in the wild-type cells (Fig. 4A). For comparison, we also included PHO5 and GAL1 genes that do not differ significantly in expression in wild-type and sin4Δ cells48. Again, our data (Fig. 4A) are in agreement with the genome-wide expression analysis48.
Figure 4. The tail module is recruited to various promoters in sin4Δ cells.
(A) sin4Δ mutation affects expression of several genes. Indicated trains were grown in YPD medium at 30°C to an A600 of 1.0. Total RNA was isolated and assayed for ACT1, PHO5, GAL1, TRX2, BNA1, HSP12, and HSP26 transcripts by real-time RT-PCR. The results were normalized to ACT1 RNA and expressed relative to the value for the WT strain. The experiment was repeated three times, and the results are shown as means ± SD. (B, C) Gal11p and Srb4p occupancies at selected promoters. Gal11p and Srb4p occupancies were determined in strains expressing Gal11p-3HA and Srb4p-myc9 using anti-HA and anti-myc antibodies, respectively. ChIP was performed using chromatin from the corresponding cells grown in YPD medium. Each immunoprecipitation was performed at least three times using different chromatin samples. The occupancy was calculated with POL1 coding sequence as a negative control. The data are presented as fold occupancy over the POL1 coding sequence control and represent means ± SD.
The expression of Gcn4p-regulated ARG1 gene is elevated in sin4Δ cells and the tail module is recruited independently of the rest of the mediator to the ARG1 promoter.15 We found that the expression of Yap1p-regulated FLR1 gene is elevated and the tail module is dramatically recruited to the FLR1 promoter in sin4Δ cells. Since Gcn4p and Yap1p play a major role in transcriptional regulation of ARG1 and FLR1, respectively, we tested other genes that are regulated by these transcriptional factors. Recruitment of Gal11p to TRX2, another Yap1p-regulated gene,40 was significantly elevated in sin4Δ cells (Fig. 4B). In addition, recruitment of Gal11p to Gcn4p-regulated BNA1 gene was also elevated in sin4Δ cells. Surprisingly, however, the tail module in sin4Δ cells was also recruited to PHO5, GAL1, HSP12, and HSP26 promoters that are not regulated by Gcn4p or Yap1p. While the expression of TRX2, BNA1, HSP12, and HSP26 was higher in sin4Δ cells than in the wild-type cells, the expression of PHO5 and GAL1 was not significantly different in wild-type and sin4Δ cells (Fig. 4A). These results suggest that the tail module functions as a promiscuous transcriptional factor that does not require Gcn4p or Yap1p and is recruited to diverse genes that are regulated by different transcriptional factors. Surprisingly, all tested genes displayed increased recruitment of Gal11p in sin4Δ cells than in the wild-type cells; however, the recruitment of Srb4p in sin4Δ cells was elevated only at genes that display increased expression in sin4Δ cells (Fig. 4C). These results suggest that the tail module exhibits an increased recruitment to many different genes in sin4Δ cells and that it’s increased recruitment in sin4Δ cells is not sufficient for the increased expression of the corresponding genes. On the other hand, genes that are upregulated in sin4Δ cells also display increased recruitment of Srb4p. These results suggest that both the tail and head/middle modules are recruited to the upregulated genes.
Discussion
The repressive role of the mediator at many genes was associated with the tail domain. The initial finding that mediator can repress transcription was based on mutation in SIN4 that allowed Swi5-independent expression of HO.42 Deletion of SIN4 results in increased transcription of host of other genes.43–47 These results suggest that the integrity of the tail module is required for proper transcriptional regulation of many genes and disruption of the tail module alleviates the repression. It appears that the tail domain plays specifically a role in repression of genes that are inactive because of low activity of the corresponding transcriptional activator or because they lack the UAS sequence.42,47,49–53
The mechanism for the repressive role of the tail domain was suggested by the finding that in the sin4Δ mutant the tail domain dissociates from the rest of the mediator and is recruited independently to the ARG1 promoter by Gcn4p.15 The isolated tail domain then facilitates recruitment of TBP and RNA pol II. Removal of the tail domain from the rest of the mediator in sin4Δ mutant resulted in significantly decreased recruitment of the tailless mediator, composed of the middle and head modules.15
Our results show that the recruitment of the tail module, measured as Gal11p occupancy, and the recruitment of the head/middle modules, measured as Srb4p occupancy, are significantly increased in sin4Δ cells in comparison to recruitment of the intact mediator in wild-type cells (Fig. 3). This can be explained by a structural change in the mediator in sin4Δ cells that either results in a complete separation of the tail and head/middle modules, or at least repositions the tail module with respect to the head/middle modules. We speculate that this structural change exposes more of the surface of the tail and head/middle modules and allows for increased interaction with the transcriptional factors, RNA pol II, or components of the basal transcriptional machinery.
The expression of FLR1 and recruitment of the tail module to the FLR1 promoter in sin4Δ cells depend on Yap1p. This finding suggests that the isolated tail domain in sin4Δ mutant is not only target of Gcn4p as shown previously for the ARG1 promoter,15 but also target of other transcriptional factors, such as Yap1p. Indeed, we observed recruitment of the tail module also to PHO5, GAL1, HSP12, and HSP26, promoters that are not regulated by Gcn4p or Yap1p.
Our results with the FLR1 promoter differ from the study of the ARG1 promoter in the fact that both the tail and head/middle modules are recruited to the FLR1 promoter, while only the tail module is recruited to the ARG1 promoter. This difference can be explained by two possible mechanisms. The first mechanism assumes that individual promoters differ in their ability to recruit the separated tail module and the head/middle module; ARG1 promoter recruits only the separated tail domain but both tail and the head/middle module are recruited to the FLR1, TRX2, BNA1, and HSP26 promoters (Fig. 3,4). The second possibility is that in highly transcribed genes in sin4Δ cells the tail module remains bound to the promoter while the head/middle module binds to the elongating RNA pol II and its occupancy is highest within the coding region. This mechanism could explain that the head/middle module was not found at the ARG1 promoter under inducing conditions in sin4Δ cells.15
The recruitment of the tail and head/middle modules to the FLR1 promoter can occur by two mechanisms. Either the isolated tail module and head/middle modules are recruited as two separate complexes, or the tail module still remains associated with the middle/head module in sin4Δ cells and the mediator is recruited as a single complex in sin4Δ cells. The first possibility seems to be more likely, since after induction, the tail module remains associated only with the promoter, while the head/middle module is found also in the coding region. We do not know whether the tail and middle/head modules are recruited simultaneously or whether the tail module is recruited first by the transcriptional factor and subsequently recruits the basal machinery and the middle/head module. Regardless of the sequence of the recruiting steps, these results suggest a model in which the tail module is anchored to the promoter by interacting with the transcriptional factors and never leaves the promoter, while the head/middle module moves into the coding region, likely because of interaction with elongating RNA pol II. While our results do not allow conclusion whether the mediator in sin4Δ cells is recruited as a single complex or as two separate complexes, the data show that during active transcription of the FLR1 gene, the tail and the head/middle modules function as two separate complexes. The tail module is recruited by interaction with the transcriptional factors that bind to the FLR1 promoter (Pdr1p, Pdr3p, Yrr1p, and Yap1p). The head/middle module is likely recruited by the interaction with the RNA pol II or by residual interactions between the separated tail module and the head/middle module that might still exist in sin4Δ cells. Alternatively, the head/middle module might be recruited by the interaction with the basal transcription machinery. In this context it is tempting to speculate that the ability of the sin4Δ mutation to activate transcription when the upstream activation sequence (UAS) is far from the TATA box54 is caused by the separation of the tail and head/middle modules of the mediator.
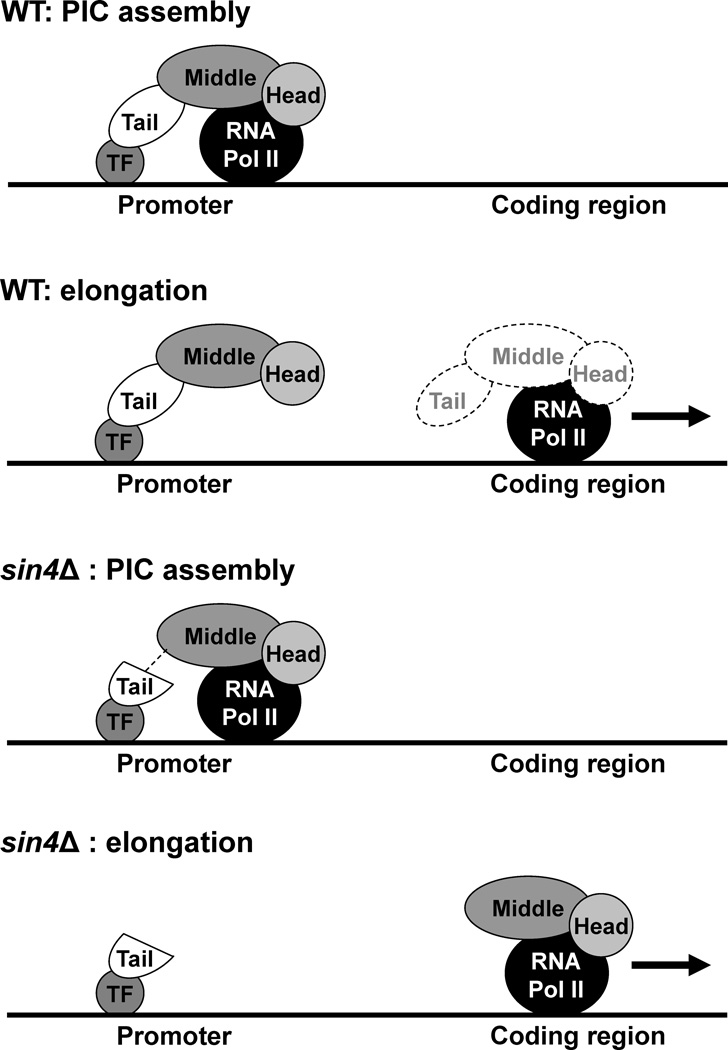
What do our results tell us about the function of the mediator? Mediator is involved in PIC assembly and typically binds to regulatory regions.19 However, we found that in sin4Δ cells during active transcription of the FLR1 gene, the tail module stays anchored to the promoter region while the head/middle module is found also in the coding region. Interestingly, the genome-wide location studies found intact mediator within coding regions of highly transcribed genes.24,25 It is possible that what we observed in sin4Δ cells is a reflection of the role of intact mediator in transcription of highly expressed genes. It is tempting to speculate that the intact mediator, bound to the transcription factor(s) in the promoter through its tail module, can be dislodged and dissociated from the promoter by its interaction with RNA pol II, and carried into the coding region (Fig. 5). This event presumes that the mediator is exposed to a “pulling” force exerted by RNA pol II when it leaves the promoter and enters the coding region. It is possible that the intact Mediator is thus partitioned between the transcription factor(s) in the promoter and RNA pol II in the coding region. If and how the mediator is translocated from the promoter to the coding region and the function of mediator within the coding region requires additional investigation.
Figure 5.
Model depicting the comparison of transcription in wild-type cells expressing intact mediator and transcription in sin4Δ cells. In sin4Δ cells, we found the tail module anchored to the promoter of the FLR1 gene, while the head/middle module was also found in the coding region. Interestingly, intact mediator was found not only in the promoters but also in the coding regions of number of strongly transcribed genes.24 We speculate that in wild-type cells, the mediator partitions between the promoter and coding region, depending on the architecture of the promoter and interactions of the mediator with the DNA-binding transcriptional factors (TF), RNA Pol II, and other components of the transcriptional machinery.
Materials and Methods
Strains and media
All yeast strains are listed in Table 1. All the strains used in this study are isogenic to W303. Standard genetic techniques were used to manipulate yeast strains and to introduce mutations from non-W303 strains into the W303 background.63 Cells were grown in rich medium (YPD; 1% yeast extract, 2% Bacto-peptone, 2% glucose) or under selection in synthetic complete medium containing 2% glucose and, when appropriate, lacking specific nutrients in order to select for a strain with a particular genotype. Meiosis was induced in diploid cells by incubation in 1% potassium acetate.
Table 1.
Yeast strains used in this study
| Strain | Genotype | Source/reference |
|---|---|---|
| W303-1a | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 ssd1-d2 can1-100 | R. Rothstein |
| W303-1α | MATαade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 ssd1-d2 can1-100 | R. Rothstein |
| W303 | MATa/MATαade2-1/ade2-1 his3-11,15/his3-11,15 leu2-3,112/leu2-3,112 trp1-1/ trp1-1 ura3-1/ ura3-1 can1-100/ can1-100 | R. Rothstein |
| HL1-1 | W303-1αplc1::URA3 | Ref. 55 |
| HL1-3 | W303-1a plc1::URA3 | Ref. 56 |
| DY1699 | W303-1a sin4::LEU2 | Ref. 57 |
| PD169 | W303-1a sin4::LEU2 | This study |
| DY2348 | W303-1a swi2::HIS3 | Ref. 57 |
| PD185 | W303-1 pdr1::kanMX | This study |
| PD187 | W303-1 pdr3::kanMX | This study |
| PD183 | W303-1 yrr1::kanMX | This study |
| PD181 | W303-1 yap1::kanMX | This study |
| DY5696 | W303-1a med2::TRP1 | Ref. 14 |
| ND96 | W303-1αsin4::TRP1 plc1::URA3 | Ref. 27 |
| PD176 | W303-1a sin4::LEU2 swi2::HIS3 | This study |
| PD184 | W303-1a sin4::LEU2 pdr1::kanMX | This study |
| PD186 | W303-1a sin4::LEU2 pdr3::kanMX | This study |
| PD182 | W303-1a sin4::LEU2 yrr1::kanMX | This study |
| PD180 | W303-1a sin4::LEU2 yap1::kanMX | This study |
| LG065 | W303-1a sin4::LEU2 med2::TRP1 | This study |
| K8135 | W303-1a ADA2-myc18::TRP1 | Ref. 58 |
| PD171 | W303-1a sin4::LEU2 ADA2-myc18::TRP1 | This study |
| AD066 | W303-1a SPT15-HA3::URA3 | Ref. 59 |
| PD152 | W303-1a sin4::TRP1 SPT15-HA3::URA3 | This study |
| DY6260 | W303-1a SRB4-myc9::TRP1 | Ref. 14 |
| NG034 | W303-1a SRB4-myc9::TRP1 | This study |
| PD149 | W303-1a sin4::LEU2 SRB4-myc9::TRP1 | This study |
| DY6361 | W303-1αSWI2-myc18::TRP1 | Ref. 60 |
| NG022 | W303-1a SWI2-myc18::TRP1 | Ref. 61 |
| NG019 | W303-1a plc1::URA3 SWI2-myc18::TRP1 | Ref. 61 |
| LG181 | W303-1a sin4::LEU2 SWI2-myc18::TRP1 | This study |
| YNYC108 | MATa ura3-52, his3Δ200 leu2Δ1 trp1Δ63 lys2Δ385 gal4Δ11 GAL11-3HA::HIS5 | Ref. 62 |
| LG102 | W303-1a GAL11-3HA::HIS5 | This study |
| LG070 | W303-1a sin4::TRP1 GAL11-3HA::HIS5 | This study |
| LG173 | W303-1a plc1::URA3 sin4::LEU2 SWI2-myc18::TRP1 | This study |
| LG136 | W303-1a sin4::TRP1 swi2::kanMX GAL11-HA3::HIS5 | This study |
| LG076 | W303-1a sin4::LEU2 yap1::kanMX GAL11-HA3::HIS5 | This study |
Chromatin immunoprecipitation and quantitative real-time PCR analysis
In vivo chromatin crosslinking and immunoprecipitation were performed as described previously59,61,64 with the following antibodies: anti-myc polyclonal antibody A-14, anti-HA monoclonal antibody F7, anti-Yap1p polyclonal antibody (all Santa Cruz Biotechnology), and anti-RNA polymerase II monoclonal antibody 8WG16 (Covance). Primers used for real-time PCR analysis are as follows: POL1 (5’-TCCTGACAAAGAAGGCAATAGAAG-3’and 5’-TAAAACACCC TGATCCACCTCTG -3’), FLR1 promoter (5’-AATGGGCGGGATAATTAGTCAG-3’ and 5’-GTGTGTCTGTACGTT GAAGTGTATACC-3’), FLR1, middle of the coding region (5’-GTTGGTTGTGCTACTGT GCATAAC-3’ and 5’-AAACGAGAGGAACCATTTCTGG-3’), FLR1, 3’ end of the coding region (5’-TACCCAAAGTATGTTGCATCCG-3’ and 5’-CCATGCCACAGGATAGTTCTT AGT-3’), PHO5 (5’-CCATTTGGGATAAGGGTAAACATC-3’ and 5’-AGAGATGAAGCCATACTAACCTCG -3’), GAL1 (5’-CGCTTAACTGCTCATTGCTATATTG-3’ and 5’-TTGTTCGGAGCAGTGCGG-3’), TRX2 (5’-ATACGACAGTGCTTTAGCATCTGG-3’ and 5’-GTTCTGCAAACTTTTCAATCATTG-3’), BNA1 (5’-GCGTCAGGATAAATTGTGAGTCAC-3’ and CCTTCGTTCTCCTTCAACCATTTG-3’), HSP12 (5’-AAGCACTCTAGACGGAGAGTAACTAG-3’ and 5’-GAATCCTTTTCTACCTGCGTCAG-3’), HSP26 (5’-CGGATCTATGCACGTTCTTGAGTG-3’ and 5’-CCTTCACCCAGCAATCTGTTAAAG-3’).
Real time RT-PCR analysis
Total RNA was isolated from cultures grown in YPD medium to optical density A600nm = 1.0 by the hot phenol method, treated with RNase-free DNase (Qiagen) and purified with an RNeasy Mini Kit (Qiagen). Reverse transcription and real time PCR amplification were performed with iScript kit (BioRad) using 100 ng of RNA and the following primers: ACT1 (5’-TATGTGTAAAGCCGGTTTTGC-3’ and 5’-GACAATACCGTGTTCAATTGGG-3’), FLR1 (5’-ACAAGAAAACCCGAGAATACCG-3’ and 5’-TGGGTTCTCAGGATCACTGG-3’), PHO5 (5’-CAATTTTAGCCGCTTCTTTGG-3’ and 5’-AAAGAGTAGTATGGTCCGGCAC-3’), GAL1 (5’-CCTGAGTTCAATTCTAGCGCAA-3’ and 5’-GCAACAAAATCCGGTTTAGCA-3’), TRX2 (5’-ATACGACAGTGCTTTAGCATCTGG-3’ and 5’-GTTCTGCAAACTTTTCAATCATTG-3’), BNA1 (5’-TGGGCCTAATGAAAGAACCG-3’ and 5’-GAACAGGACTGTGAGGAACATTTC-3’), HSP12 (5’-AGTCATACGCTGAACAAGGTAAGG-3’ and 5’-AGTCGTGGACACCTTGGAAGAC-3’), HSP26 (5’-AAGACGTCAGTTAGCAAACACACC-3’ and 5’-CATTGTCGAACCAATCATCTAAGG-3’).
Derepression of FLR1 transcription in sin4 cells depends on Yap1p.
Assembly of the PIC is facilitated in sin4 cells.
The tail and head/middle modules of the mediator are separated in sin4 cells.
In sin4 cells the tail is at the promoter and the head/middle module is at the orf.
Acknowledgements
We thank Drs. Nasmyth, Ptashne, and Stillman for strains and plasmids, Dr. Holstege for communicating unpublished results, and members of Vancura lab for helpful comments. This work was supported by grant from the National Institutes of Health (GM087674) to A.V.
Abbreviations
- RNA pol II
RNA polymerase II
- CTD
carboxyl terminal domain of RNA pol II
- InsPs
inositol polyphosphates
- PIC
preinitiation complex
- ChIP
chromatin immunoprecipitation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nonet ML, Young RA. Intragenic and extragenic supressors of mutations in the heptapeptide repeat domain of Saccharomyces cerevisiae RNA polymerase II. Genetics. 1989;123:715–724. doi: 10.1093/genetics/123.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thompson CM, Koleske AJ, Chao DM, Young RA. A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell. 1993;73:1361–1375. doi: 10.1016/0092-8674(93)90362-t. [DOI] [PubMed] [Google Scholar]
- 3.Hengartner CJ, Thompson CM, Zhang J, Chao DM, Liao SM, Koleske AJ, Okamura S, Young RA. Association of an activator with an RNA polymerase II holoenzyme. Genes & Dev. 1995;9:897–910. doi: 10.1101/gad.9.8.897. [DOI] [PubMed] [Google Scholar]
- 4.Liao SM, Zhang J, Jeffery DA, Koleske AJ, Thompson CM, Chao DM, Viljoen M, van Vuuren HJ, Young RA. A kinase-cyclin pair in the RNA polymerase II holoenzyme. Nature. 1995;374:193–196. doi: 10.1038/374193a0. [DOI] [PubMed] [Google Scholar]
- 5.Carlson M. Genetics of transcriptional regulation in yeast: connections to the RNA polymerase II CTD. Annu. Rev. Cell Dev. Biol. 1997;13:1–23. doi: 10.1146/annurev.cellbio.13.1.1. [DOI] [PubMed] [Google Scholar]
- 6.Biddick R, Young ET. Yeast mediator and its role in transcriptional regulation. C.R. Biologies. 2005;328:773–782. doi: 10.1016/j.crvi.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 7.Kim YJ, Bjorklund S, Li Y, Sayre MH, Kornberg RD. A multiprotein mediator of transcriptional activation and its inteteraction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 8.Asturias FJ, Jiang YW, Myers LC, Gustafsson CM, Kornberg RD. Conserved structures of mediator and RNA polymerase II holoenzyme. Science. 1999;283:985–987. doi: 10.1126/science.283.5404.985. [DOI] [PubMed] [Google Scholar]
- 9.Dotson MR, Yuan CX, Roeder RG, Myers LC, Gustafsson CM, Jiang YW, Li Y, Kornberg RD, Asturias FJ. Structural organization of yeast and mammalian mediator complexes. Proc. Natl. Acad. Sci. USA. 2000;97:14307–14310. doi: 10.1073/pnas.260489497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guglielmi B, van Berkum NL, Klapholz B, Bijma T, Boube M, Boschiero C, Bourbon HM, Holstege FC, Werner M. A high-resolution protein interaction map of the yeast Mediator complex. Nucleic Acids Res. 2004;32:5379–5391. doi: 10.1093/nar/gkh878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee YC, Kim YJ. Requirement for a functionalo interaction between mediator components Med6 and Srb4 in RNA polymerase II transcription. Mol. Cell. Biol. 1998;18:5364–5370. doi: 10.1128/mcb.18.9.5364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koh SS, Ansari AZ, Ptashne M, Young RA. An activator target in the RNA polymerase II holoenzyme. Mol. Cell. 1998;1:895–904. doi: 10.1016/s1097-2765(00)80088-x. [DOI] [PubMed] [Google Scholar]
- 13.Park JM, Kim HS, Han SJ, Hwang MS, Lee YC, Kim YJ. In vivo requirement of activator-specific binding targets of mediator. Mol. Cell Biol. 2000;20:8709–8719. doi: 10.1128/mcb.20.23.8709-8719.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhoite LT, Yu Y, Stillman DJ. The Swi5 activator recruits the Mediator complex to the HO promoter without RNA polymerase II. Genes & Dev. 2001;15:2457–2469. doi: 10.1101/gad.921601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang F, Sumibcay L, Hinnebusch AG, Swanson M. A triad of subunits from the Gal11/tail domain of Srb Mediator is an vivo target of transcriptional activator Gcn4p. Mol. Cell Biol. 2004;24:6871–6886. doi: 10.1128/MCB.24.15.6871-6886.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hengartner CJ, Myer VE, Liao SM, Wilson CJ, Koh SS, Young RA. Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol. Cell. 1998;2:43–53. doi: 10.1016/s1097-2765(00)80112-4. [DOI] [PubMed] [Google Scholar]
- 17.Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 18.Bjorklund S, Gustafsson CM. The yeast Mediator complex and its regulation. Trends Biochem. Sci. 2005;30:240–244. doi: 10.1016/j.tibs.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 19.Kornberg RD. Mediator and the mechanism of transcriptional activation. Trends Biochem. Sci. 2005;30:235–239. doi: 10.1016/j.tibs.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 20.Myers LC, Kornberg RD. Mediator of transcriptional regulation. Annu. Rev. Biochem. 2000;69:729–749. doi: 10.1146/annurev.biochem.69.1.729. [DOI] [PubMed] [Google Scholar]
- 21.Yudkovsky N, Ranish JA, Hahn S. A transcriptional reinitiation intermediate that is stabilized by activator. Nature. 2000;408:225–229. doi: 10.1038/35041603. [DOI] [PubMed] [Google Scholar]
- 22.Reeves WM, Hahn S. Activator-independent functions of the yeast mediator Sin4 complex in preinitiation complex formation and transcription reinitiation. Mol. Cell. Biol. 2003;23:349–358. doi: 10.1128/MCB.23.1.349-358.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan X, Chou DM, Struhl K. Activator-specific recruitment of Mediator in vivo. Nat. Struct. Mol. Biol. 2006;13:117–120. doi: 10.1038/nsmb1049. [DOI] [PubMed] [Google Scholar]
- 24.Andrau J-C, van de Pasch L, Lijnzaad P, Bijma T, Koerkamp MG, van de Peppel J, Werner M, Holstege FCP. Genome-wide location of the coactivator mediator: binding without activation and transient Cdk8 interaction on DNA. Mol. Cell. 2006;22:179–192. doi: 10.1016/j.molcel.2006.03.023. [DOI] [PubMed] [Google Scholar]
- 25.Zhu X, Wiren M, Sinha I, Rasmussen NN, Linder T, Holmberg S, Ekwal K, Gustafsson CM. Genome-wide occupancy profile of mediator and Srb8-11 module reveals interaction with coding regions. Mol. Cell. 2006;22:169–178. doi: 10.1016/j.molcel.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 26.Kuras L, Borggrefe T, Kornberg RD. Association of the mediator complex with enhancers of active genes. Proc. Natl. Acad. Sci. USA. 2003;100:13887–13891. doi: 10.1073/pnas.2036346100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Romero C, Desai P, DeLillo N, Vancura A. Expression of FLR1 Transporter Requires Phospholipase C and Is Repressed by Mediator. J. Biol. Chem. 2006;281:5677–5685. doi: 10.1074/jbc.M506728200. [DOI] [PubMed] [Google Scholar]
- 28.Steger DJ, Haswell ES, Miller AL, Wente SR, O'Shea EK. Regulation of chromatin remodeling by inositol polyphosphates. Science. 2003;299:114–116. doi: 10.1126/science.1078062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen X, Xiao H, Ranallo R, Wu W-H, Wu C. Modulation of ATP-dependent chromatin-remodeling complexes by inositol polyphosphates. Science. 2003;299:112–114. doi: 10.1126/science.1078068. [DOI] [PubMed] [Google Scholar]
- 30.York JD, Odom AR, Murphy R, Ives EB, Wente SR. A phospholipase C-dependent inositol polyphosphate kinase pathway required for efficient messenger RNA export. Science. 1999;285:96–100. doi: 10.1126/science.285.5424.96. [DOI] [PubMed] [Google Scholar]
- 31.Laurent BC, Treich I, Carlson M. The yeast SNF2/SWI2 protein has DNA-stimulated ATPase activity required for transcriptional activation. Genes & Dev. 1993;7:583–591. doi: 10.1101/gad.7.4.583. [DOI] [PubMed] [Google Scholar]
- 32.Vignali M, Hassan AH, Neely KE, Workman JL. ATP-dependent chromatin-remodeling complexes. Mol. Cell. Biol. 2000;20:1899–1910. doi: 10.1128/mcb.20.6.1899-1910.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alarco A-M, Balan I, Talibi D, Mainville N, Raymond M. AP1-mediated multidrug resistance in Saccharomyces cerevisiae requires FLR1 encoding a transporter of the major facilitator superfamily. J. Biol. Chem. 1997;272:19304–19313. doi: 10.1074/jbc.272.31.19304. [DOI] [PubMed] [Google Scholar]
- 34.Broco N, Tenreiro S, Viegas CA, Sa-Correia I. FLR1 gene (ORF YBR008c) is required for benomyl and methotrexate resistance in Saccharomyces cerevisiae and its benomyl-induced expression is dependent on Pdr3 transcriptional regulator. Yeast. 1999;15:1595–1608. doi: 10.1002/(SICI)1097-0061(199911)15:15<1595::AID-YEA484>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 35.Tenreiro S, Fernandes AF, Sa-Correia I. Transcriptional activation of FLR1 gene in Saccharomyces cerevisiae adaptation to growth with benomyl: role of Yap1p and Pdr1p. Biochem. Biophys. Res. Commun. 2001;280:216–222. doi: 10.1006/bbrc.2000.4100. [DOI] [PubMed] [Google Scholar]
- 36.Nguyen D-T, Alarco A-M, Raymond M. Multiple Yap1p-binding sites mediate induction of the yeast major facilitator FLR1 gene in response to drugs, oxidants, and alkylating agents. J. Biol. Chem. 2001;276:1138–1145. doi: 10.1074/jbc.M008377200. [DOI] [PubMed] [Google Scholar]
- 37.Jungwirth H, Wendler F, Platzer B, Bergler H, Hogenauer G. Diazaborine resistance in yeast involves the efflux pumps Ycf1p and Flr1p and is enhanced by a gain-of-function allele of gene YAP1. Eur. J. Biochem. 2000;267:4809–4816. doi: 10.1046/j.1432-1327.2000.01537.x. [DOI] [PubMed] [Google Scholar]
- 38.Le Crom S, Devaux F, Marc P, Zhang X, Moye-Rowley WS, Jacq C. New insights into the pleiotropic drug resistance network from genome-wide characterization of the YRR1 transcription factor regulation system. Mol. Cell. Biol. 2002;22:2642–2649. doi: 10.1128/MCB.22.8.2642-2649.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wehrschutz-Sigl E, Jungwirth H, Bergler H, Hogenauer G. The transporters Pdr5p and Snq2p mediate diazaborine resistance and are under the control of the gain-of-function allele PDR1-12. Eur. J. Biochem. 2004;271:1145–1152. doi: 10.1111/j.1432-1033.2004.04018.x. [DOI] [PubMed] [Google Scholar]
- 40.Lucau-Danila A, Lelandais G, Kozovska Z, Tanty V, Delaveau T, Devaux F, Jacq C. Early expression of yeast genes affected by chemical stress. Mol. Cell. Biol. 2005;25:1860–1868. doi: 10.1128/MCB.25.5.1860-1868.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Teixeira MC, Dias PJ, Simoes T, Sa-Correia I. Yeast adaptation to mancozeb involves the up-regulation of FLR1 under the coordinate control of Yap1, Rpn4, Pdr3, and Yrr1. Biochem. Biophys. Res. Commun. 2008;367:249–255. doi: 10.1016/j.bbrc.2007.12.056. [DOI] [PubMed] [Google Scholar]
- 42.Sternberg PW, Stren MJ, Clark I, Herskowitz I. Activation of the yeast HO gene by release from multiple negative controls. Cell. 1987;48:567–577. doi: 10.1016/0092-8674(87)90235-2. [DOI] [PubMed] [Google Scholar]
- 43.Nishizawa M. Negative regulation of transcription by the yeast global transcription factors, Gal11 and Sin4. Yeast. 2001;18:1099–1110. doi: 10.1002/yea.754. [DOI] [PubMed] [Google Scholar]
- 44.Jiang YW, Stillman DJ. Regulation of HIS4 expression by the Saccharomyces cerevisiae SIN4 transcriptional regulator. Genetics. 1995;140:103–114. doi: 10.1093/genetics/140.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang Y-W, Howard SC, Budovskaya YV, Rine J, Herman PK. The rye mutants identify a role for Ssn/Srb proteins of the RNA polymerase II holoenzyme during stationary phase entry in Saccharomyces cerevisiae. Genetics. 2001;157:17–26. doi: 10.1093/genetics/157.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Howard SC, Chang YW, Budovskaya YV, Herman PK. The Ras/PKA signaling pathway of Saccharomyces cerevisiae exhibits a functional interaction with the Sin4p complex of the RNA polymerase II holoenzyme. Genetics. 2001;159:77–89. doi: 10.1093/genetics/159.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang X, Michels CA. Mutations in SIN4 and RGR1 cause constitutive expression of MAL structural genes in Saccharomyces cerevisiae. Genetics. 2004;168:747–757. doi: 10.1534/genetics.104.029611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van de Peppel J, Kettelarij N, van Bakel, Kockelkorn TTJP, van Leenen D, Holstege FCP. Mediator expression profiling epistasis reveals a signal transduction pathway with antagonistic submodules and highly specific downstream targets. Mol. Cell. 2005;19:511–522. doi: 10.1016/j.molcel.2005.06.033. [DOI] [PubMed] [Google Scholar]
- 49.Mizuno T, Harashima S. Activation of basal transcription by a mutation in SIN4, a yeast global repressor, occurs through a mechanism different from activator-mediated transcriptional enhancement. Mol. Gen. Genet. 2000;263:48–59. doi: 10.1007/pl00008675. [DOI] [PubMed] [Google Scholar]
- 50.Piruat JI, Chavez S, Aguilera A. The yeast HRS1 gene is involved in positive and negative regulation of transcription and shows genetic characteristics similar to SIN4 and GAL11. Genetics. 1997;147:1585–1594. doi: 10.1093/genetics/147.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiang YW, Dohrmann PR, Stillman DJ. Genetic and physical interactions between yeast RGR1 and SIN4 in chromatin organization and transcriptional regulation. Genetics. 1995;140:47–54. doi: 10.1093/genetics/140.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singh H, Erkine AM, Kremer SB, Duttweiler HM, Davis DA, Iqbal J, Gross RR, Gross DS. A functional module of yeast mediator tjhat governs the dynamic range of heat-shock gene expression. Genetics. 2006;172:2169–2184. doi: 10.1534/genetics.105.052738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Swanson MJ, Qiu H, Sumibcay L, Krueger A, Kim S-J, Natarajan K, Yoon S, Hinnebusch AG. A multiplicity of coactivators is required by Gcn4p at individual promoters in vivo. Mol. Cell. Biol. 2003;23:2800–2820. doi: 10.1128/MCB.23.8.2800-2820.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dobi KC, Winston F. Analysis of transcriptional activation at a distance in Saccharomyces cerevisiae. Mol. Cell Biol. 2007;27:5575–5586. doi: 10.1128/MCB.00459-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin H, Choi JH, Hasek J, DeLillo N, Lou W, Vancura A. Phospholipase C is involved in kinetochore function in Saccharomyces cerevisiae. Mol. Cell Biol. 2000;20:3597–3607. doi: 10.1128/mcb.20.10.3597-3607.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.DeLillo N, Romero C, Lin H, Vancura A. Genetic evidence for a role of phospholipase C at the budding yeast kinetochore. Mol. Genet. Genomics. 2003;269:261–270. doi: 10.1007/s00438-003-0832-4. [DOI] [PubMed] [Google Scholar]
- 57.Yu Y, Eriksson P, Stillman DJ. Architectural transcription factors and the SAGA complex function in parallel pathways to activate transcription. Mol. Cell. Biol. 2000;20:2350–2357. doi: 10.1128/mcb.20.7.2350-2357.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cosma MP, Tanaka T, Nasmyth K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell. 1999;97:299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- 59.Demczuk A, Guha N, Nguyen PH, Desai P, Chang J, Guzinska K, Rollins J, Ghosh C, Goodwin L, Vancura A. Saccharomyces cerevisiae phospholipase C regulates transcription of Msn2p-dependent stress-responsive genes. Eukaryot. Cell. 2008;7:967–979. doi: 10.1128/EC.00438-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu Y, Eriksson P, Bhoite LT, Stillman DJ. Regulation of TATA-binding protein binding by the SAGA complex and the Nhp6 high-mobility group protein. Mol. Cell. Biol. 2003;23:1910–1921. doi: 10.1128/MCB.23.6.1910-1921.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guha N, Desai P, Vancura A. Plc1p is required for SAGA recruitment and derepression of Sko1p-regulated genes. Mol. Biol. Cell. 2007;18:2419–2428. doi: 10.1091/mbc.E06-10-0946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bryant GO, Ptashne M. Independent recruitment in vivo by Gal4 of two complexes required for transcription. Mol. Cell. 2003;11:1301–1309. doi: 10.1016/s1097-2765(03)00144-8. [DOI] [PubMed] [Google Scholar]
- 63.Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 64.Desai P, Guha N, Galdieri L, Hadi S, Vancura A. Plc1p is required for proper chromatin structure and activity of the kinetochore in Saccharomyces cerevisiae by facilitating recruitment of the RSC complex. Mol. Genet. Genomics. 2009;281:511–523. doi: 10.1007/s00438-009-0427-9. [DOI] [PubMed] [Google Scholar]