Viruses infecting higher plants are among the smallest viruses known and typically have four to ten protein encoding genes. By contrast, many viruses that infect algae (classified in the virus family Phycodnaviridae) are among the largest viruses found to date and have up to 600 protein encoding genes. This brief review focuses on one group of plaque-forming phycodnaviruses that infect unicellular chlorella-like green algae. The prototype chlorovirus PBCV-1 has more than 400 protein-encoding genes and 11 tRNA genes. About 40% of the PBCV-1 encoded proteins resemble proteins of known function including many that are completely unexpected for a virus. In many respects, chlorovirus infection resembles bacterial infection by tailed bacteriophages.
Algal viruses
Viruses infecting higher plants are typically small RNA viruses that encode only a few genes [1]. Although small viruses have recently been discovered that infect algae [e.g., 2], many viruses infecting eukaryotic algae are huge dsDNA viruses with genomes ranging from 160 to 560 kb with up to 600 protein encoding genes (CDSs) and are the subject of this review. These large viruses (family Phycodnaviridae), are found in aqueous environments throughout the world and play dynamic, albeit largely undocumented, roles in regulating algal communities such as the termination of massive algal blooms commonly referred to as red and brown tides (e.g., [3-5]). This review focuses on one genus in the Phycodnaviridae, the chloroviruses, which are large, icosahedral, plaque-forming (Figure 1a), dsDNA-containing viruses that replicate in certain unicellular, chlorella-like green algae [6-9]. As noted below, their structure, their initial stages of infection, and many of their genes resemble bacteriophage more than viruses that infect eukaryotes, i.e., they are not your everyday plant virus.
Figure 1.
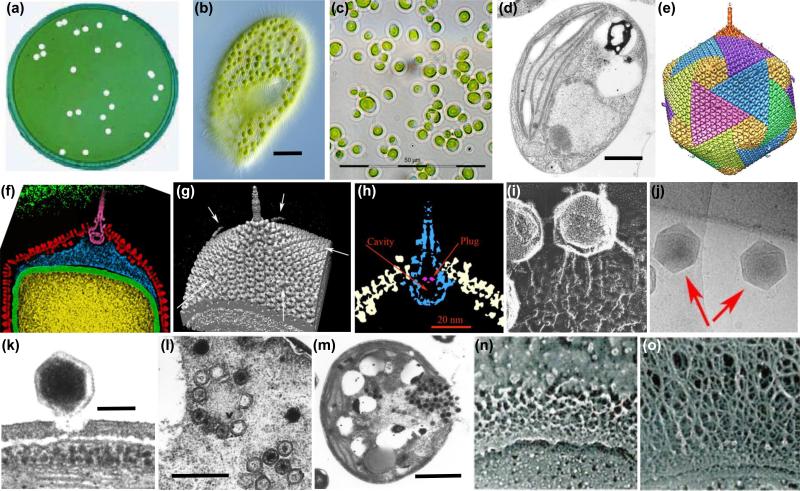
Chlorella cells and chlorovirus PBCV-1. (a) Plaques formed by PBCV-1 on a lawn of C. variabilis. (b) Paramecium bursaria and its symbiotic chlorella cells (size bar: 20 μm). (c) C. variabilis cells cultured in the laboratory free of both Paramecium and virus (size bar: 50 μm). (d) Thin section of a healthy C. variabilis cell (size bar: 1 μm). (e) Five-fold averaged cryo-electron micrograph of PBCV-1 reveals a long narrow cylindrical spike structure at one vertex (orange) and fibers extending from one unique capsomer per trisymmetron. The virion has a diameter varying from 1,650 Å, measured along the 2-fold and 3-fold axes, and 1,900 Å, measured along the 5-fold axes. The glycoprotein shell is composed of 20 triangular units, or “trisymmetrons,” and 12 pentagonal caps, or “pentasymmetrons,” at the 5-fold vertices (yellow). (f) Central cross section of (e). Note the gap between the unique vertex and the membrane enclosing the DNA. (g) Surface view of the PBCV-1 spike structure and fibers. (h) A magnified image of the spike structure (in blue). Also note a plug structure (in pink) inside the spike (size bar: 20 nm). (i) PBCV-1 attached to the cell wall as viewed by the quick-freeze, deep etch procedure. Note the virions attached to the wall by fibers. (j) Initial attachment of PBCV-1 to C. variabilis cell walls. The orientation of the unique vertex (recognized by the pocket under the vertex) can be seen for the two virus particles marked with red arrows. These virus particles have their unique vertex facing the cell wall indicating that these particles recognize the cell surface. (k) Attachment of PBCV-1 to the algal wall and digestion of the wall at the point of attachment. This occurs within 1-3 min p.i. (size bar: 100 nm) (l) Virion particles assemble in defined areas in the cytoplasm named virus assembly centers. Note both DNA containing (dark centers) and empty capsids (size bar: 500 nm). (m) Localized lysis of cell plasma membrane and cell wall and release of progeny viruses at ~8 hr p.i. (size bar: 1 μm). (n/o) C. variabilis cell wall surface of uninfected (n) and 4 hr after PBCV-1 infection (o), as viewed by quick-freeze, deep-etch electron microscopy. Note the accumulation of a dense, fibrous hyaluronan network on the surface of the infected cells. Figure b was kindly provided by Wim van Egmond; Figure e from [13]; Figure f was kindly provided by Xinzheng Zhang and Michael Rossmann; Figures g, h and j from [14]; Figure i from [26]; Figure k from [83], Figures l and m from [84], and Figures n and o from [56] - all published with permission.
Chloroviruses
Chloroviruses exist in freshwater throughout the world with titers as high as 100,000 plaque-forming units (PFU) per ml of indigenous water although titers are typically 1-100 PFU/ml. Titers fluctuate during the year with the highest titers occurring in the spring [7,9]. Chlorovirus hosts, which are normally symbionts and are often referred to as zoochlorellae, are associated with the protozoan Paramecium bursaria (Figure 1b), the coelenterate Hydra viridis or the heliozoon Acanthocystis turfacea [8]. Zoochlorellae are resistant to viruses in their symbiotic state. Fortunately, some zoochlorellae grow independently of their partners, permitting plaque assay of the viruses and synchronous infection of their hosts, which allows one to study the virus life cycle in detail. Three such zoochlorellae are Chlorella NC64A (recently renamed Chlorella variabilis [10] and its viruses are called NC64A viruses), Chlorella SAG 3.83 (renamed Chlorella heliozoae and its viruses are called SAG viruses), and Chlorella Pbi (renamed Micratinium conductrix and its viruses are called Pbi viruses). However, little is known about the natural history of the chloroviruses and we suspect that many more chlorovirus hosts and viruses exist in nature.
Paramecium bursaria chlorella virus (PBCV-1), which infects C. variabilis (Figure 1c,d), is the type member of the genus Chlorovirus [7]. The C. variabilis 46.2 Mb genome was sequenced recently [11] and the availability of both host and virus sequences makes chloroviruses an attractive model system. Given the coding capacity of the chloroviruses, it is not surprising that they encode many unusual proteins. However, with the exception of homologs solely in other chlorovirus members, the majority of their CDSs do not match anything in the databases. Some of these viruses also have introns and inteins, which are rare in viruses.
PBCV-1 structure
Cryo-electron microscopy and 5-fold symmetry averaging 3-dimensional reconstruction of PBCV-1 indicate the outer capsid is an icosahedron and covers a single lipid bilayered membrane, which is required for infection (Figure 1e,f) [12-14]. One of the PBCV-1 vertices has a 560 Å long spike structure; 340 Å protrudes from the surface of the virus. The part of the spike structure that is outside the capsid has an external diameter of 35 Å at the tip expanding to 70 Å at the base (Figure. 1g,h). The spike structure widens to 160 Å inside the capsid and forms a closed cavity inside a large pocket between the capsid and the membrane enclosing the virus DNA (Figure 1h). Therefore, the internal virus membrane departs from icosahedral symmetry adjacent to the unique vertex (Figure 1f). Consequently the virus DNA located inside the envelope is packaged non-uniformly in the particle.
The capsid shell consists of 1,680 donut-shaped trimeric capsomers plus 11 pentameric capsomers, one at each icosahedral vertex except for the spike-containing vertex. The trimeric capsomers are arranged into 20 triangular facets (trisymmetrons, each containing 66 trimers) and 11 pentagonal facets (pentasymmetrons, each containing 30 trimers and one pentamer at the icosahedral vertices) (Figure 1e). External fibers extend from some of the trisymmetron capsomers (probably one per trisymmetron) and presumably aid in virus attachment to the host (Figure 1i).
The PBCV-1 major capsid protein (MCP) is a glycoprotein and consists of two eight-stranded, antiparallel β-barrel jelly-roll domains. The monomeric MCP forms a trimeric capsomer, which has pseudo-sixfold symmetry [15]. Identities of the other virus surface proteins are unknown. A proteomic analysis of highly purified PBCV-1 virions indicates that the particle has 148 virus-encoded proteins (Dunigan et al., unpublished results).
PBCV-1 life cycle
Three-dimensional reconstruction of PBCV-1 in the presence of C. variabilis cell walls established that the spike first contacts the cell wall (Figure 1j) and that the fibers then aid in holding the virus to the wall (Figure 1i) [14]. PBCV-1 attachment to its host receptor is specific and attachment is a major factor in limiting its host range. The spike is too narrow to deliver DNA and likely serves to puncture the wall and is then jettisoned. Following host cell wall degradation by a virus-associated enzyme(s) (Figure 1k), the viral internal membrane presumably fuses with the host membrane, facilitating entry of the viral DNA and virion-associated proteins into the cell, leaving an empty capsid attached to the surface [16]. This fusion process triggers rapid depolarization of the host membrane, possibly by a virus encoded K+ channel (named Kcv for K+ channel from chlorella virus) predicted to be in the internal membrane of the virus releasing K+ from the cell. The rapid loss of K+ and associated water fluxes from the host reduce its turgor pressure, which may aid ejection of viral DNA and virion-associated proteins into the host [16]. Host membrane depolarization also rapidly inhibits many host secondary transporters [17] and likely prevents infection by a second virus [18].
PBCV-1 lacks a recognizable RNA polymerase gene, and so circumstantial evidence suggests PBCV-1 DNA and viral-associated proteins quickly move to the nucleus where early transcription begins 5 to 10 min post infection (p.i.) [7,9]. The rapid initiation of virus transcription suggests that some component(s) facilitates transport of virus DNA to the nucleus. This component could be a product of the PBCV-1 a443r gene. A443R is packaged in the virion and may be a SUMOylated protein; the predicted A443R structure resembles mammalian SUMOylated proteins involved in nuclear trafficking (Van Etten et al., unpublished results).
In this immediate-early phase of infection, host transcription rates decrease [19] and the host transcription machinery is reprogrammed to transcribe viral DNA. Details of reprogramming are unknown but host chromatin remodeling is probably involved. PBCV-1 encodes a SET domain containing protein (referred to as vSET) that methylates Lys-27 in histone 3. vSET is packaged in the PBCV-1 virion, and circumstantial evidence indicates vSET represses host transcription following PBCV-1 infection [20]. In addition, host chromosomal DNA degradation begins within minutes after infection, presumably by PBCV-1 encoded and packaged DNA restriction endonuclease(s) [19]. This degradation also inhibits host transcription. The net result is that by 20 min p.i., 33% to 50% of the polyadenylated mRNAs in the infected cell are of viral origin (Van Etten, unpublished results). This rapid increase in viral mRNAs probably involves the selective degradation of host mRNAs, by an unknown mechanism(s), as well as viral transcription.
Viral DNA replication begins 60 to 90 min p.i. and is followed by transcription of late genes [7]. About two to three hr p.i., assembly of virus capsids begins in localized regions in the cytoplasm, which become prominent three to four hr p.i. (Figure 1l). DNA packaging of empty virus particles may involve a virus encoded DNA packaging ATPase [21]. However, the putative DNA packaging ATPase is part of a larger CDS (A392R), which consists of two predicted fused proteins separated by a caspase cleavage site. We hypothesize that virus DNA packaging begins after a host/virus-encoded enzyme with caspase-like activity cleaves A392R to produce a functional enzyme (Van Etten et al., unpublished results). [Note: Earlier studies on the phycodnavirus Emiliana huxleyi virus established that a caspase-like cleavage activity of one or more virus-encoded proteins was required for its replication [22,23].] Five to six hr after PBCV-1 infection the cytoplasm fills with infectious progeny virus particles and localized lysis of the host cell releases progeny at six to eight hr p.i. (Fig. 1M) Each infected cell releases ~1000 particles, of which ~30% form plaques. A schematic diagram of the PBCV-1 replication cycle is reported in Figure 2.
Figure 2.
Proposed replication cycle of PBCV-1. The virus uncoats at the surface of the alga and the viral DNA, possibly with associated proteins, is assumed to move to the nucleus where early gene transcription begins with 5 to 10 min p.i. The early mRNAs are transported to the cytoplasm for translation, and at least some early proteins presumably return to the nucleus to initiate viral DNA replication, which begins 60 to 90 min p.i., followed by late gene transcription. Late mRNAs are transported to the cytoplasm for translation and many of these late proteins are targeted to the virus assembly centers, where virus capsids are formed and DNA is packaged. The chlorella cell membrane and wall lyses, and infectious PBCV-1 progeny viruses are released at 6 to 8 hr p.i. ( →) Known events; (---->) hypothesized events.
Global transcription of PBCV-1 genes during virus replication was examined by microarrays [24] and the results can be summarized as follows: (i) 98% of the PBCV-1 protein encoding genes are expressed in laboratory conditions; (ii) 63% of the genes are expressed before 60 min p.i. (early genes); (iii) 37% of the genes are expressed after 60 min p.i. (late genes); and (iv) 43% of the early gene transcripts are also detected at late times following infection (early/late genes). Many of the late and early/late genes encode virion-associated proteins, consistent with particle assembly and maturation.
Chlorovirus genomes
The PBCV-1 genome is a linear ~331-kb, nonpermuted dsDNA molecule with covalently closed hairpin termini. Identical ~2.2-kb inverted repeats flank each hairpin end. The remainder of the PBCV-1 genome contains primarily single-copy DNA [7]. The GC content of the PBCV-1 genome is ~40%; by contrast, its host nuclear genome is ~67% GC. The PBCV-1 genome was recently re-sequenced to correct mistakes in the original sequence. Using 40 codons as the minimum CDS size, PBCV-1 contains 416 predicted CDSs (Figure 3) (Dunigan et al., unpublished results, GenBank JF411744). Of the predicted CDSs, ~40% resemble proteins of known function, including many that are novel for a virus. The CDSs are evenly distributed on both DNA strands with minimal intergenic spaces. One exception is a 1,788-nucleotide sequence in the middle of the PBCV-1 genome encoding 11 tRNAs, which is co-transcribed as a large precursor and then processed to mature tRNAs [9].
Figure 3.
Map of the re-sequenced and annotated PBCV-1 genome (GenBank accession number JF411744.1) presented as a circle. However the genome is a linear molecule and the ends are depicted at the top (12 o'clock) of the figure as a crooked black line. Sequences to the right of the black line are numbered from 1 to 330611 base pairs. Open reading frames are indicated by the block arrows. Those pointing clockwise (red) are annotated with a “R”, those pointing counter-clockwise (blue) are annotated with a “L”. The gene tags are annotated with the prefix “a” indicating the ORF is unlikely to be expressed, or with “A” indicating that it is likely to be expressed, and in most cases shown to be expressed at the RNA level and in many cases as proteins. The polycistronic tRNA gene is located near “6 o'clock”. The two brackets on the top-right of the figure depict two classes of deletion mutants; (a) deletion of map position 4.9 to 42.2, (b) deletion of map position 16 to 42.2. The figure was composed using CGView [85]
Not all PBCV-1 genes are required for virus replication. Four spontaneously derived PBCV-1 mutants were isolated that contain 27- to 37-kb deletions at the left end of the genome [25]. Taken together, ~40 kb of single-copy DNA encoding 31 CDSs, or 12% of the PBCV-1 genome, is not essential for PBCV-1 replication in the laboratory, although its replication is attenuated. Deleted CDSs include two putative capsid-like proteins, a putative glycerophosphoryl diesterase, a putative D-lactate dehydrogenase, a glycosyltransferase and a pyrimidine dimer-specific glycosylase.
The chlorella virus genomes contain methylated bases. Genomes from 37 chloroviruses have 5-methylcytosine (5mC) in amounts ranging from 0.12 to 47.5% of the total cytosines. In addition, 24 of the 37 viral DNAs contain N6-methyladenine (6mA) in amounts ranging from 1.5 to 37% of the total adenines [26]. The methylated bases occur in specific DNA sequences, which led to the discovery that chloroviruses encode multiple 5mC and 6mA DNA methyltransferases. About 25% of the virus-encoded DNA methyltransferases have companion DNA site-specific (restriction) endonucleases, including some with unique cleavage specificities that are sold commercially (e.g., [26-28]. The purpose of the other virus-encoded DNA methyltransferases is unknown.
Five additional chloroviruses have been sequenced and annotated including two viruses (NY-2A and AR158) that infect the same host as PBCV-1, C. variabilis [29], two viruses (MT325 and FR483) that infect M. conductrix [30], and one virus (ATCV-1) that infects C. heliozoae [31]. Approximately 80% of the genes are common to all six viruses, suggesting they are important for virus replication. However, the sum total of chlorella virus encoded genes is much larger than those present in any one virus. Not surprisingly, homologs from viruses infecting the same host are the most similar; the average amino acid identity between homologs from PBCV-1 and NY-2A or AR158 is ~73%. PBCV-1 and MT325 or FR483 orthologs have ~50% amino acid identity and PBCV-1 and ATCV-1 orthologs have ~49% amino acid identity. Using PBCV-1 as a model, there is good synteny among the three viruses that infect C. variabilis. By contrast, PBCV-1 has only slight synteny with the two Pbi viruses and the SAG virus [32].
Chlorovirus genes
Many PBCV-1 encoded enzymes are either the smallest or among the smallest proteins in their family including the smallest eukaryotic ATP-dependent DNA ligase [33], the smallest type II DNA topoisomerase [34], the smallest prolyl-4-hydroxylase [35], the smallest histone methyltransferase [36] and the smallest protein to form a functional K+ channel named Kcv [37]. While K+ channels from prokaryotes and eukaryotes often consist of several hundred amino acids, Kcv from PBCV-1 consists of 94 amino acids [37] and virus ATCV-1 Kcv consists of 82 amino acids [38]. In spite of their small sizes, both viral proteins form functional channels that have many of the properties associated with larger K+ channels such as selectivity, gating and sensitivity to inhibitors [39-42]. Kcv proteins readily assemble into tetramers and sort in cells to distinct target membranes [43]. This means that all of the structural requirements for correct assembly of the channel as well as for the basic functional properties of a K+ channel exist in the viral Kcvs [44].
Phylogenetic analyses indicate that some of the viral minimalist proteins are evolutionary precursors of more complex cellular proteins. Despite their small sizes, the virus proteins typically have all of the catalytic properties of larger enzymes. Their small size and the fact that they are often “laboratory friendly” have made them excellent models for mechanistic and structural studies (e.g., see [41,42].
Chloroviruses encode many unexpected proteins, a few examples are listed in Table 1. Some PBCV-1-encoded proteins assemble into metabolic pathways, e.g., hyaluronan synthesis, fucose synthesis and polyamine synthesis. Space only allows us to comment on a few unusual proteins. For example, in addition to the aforementioned K+ channel Kcv, the viruses encode other channel/transporter proteins including an aquaglyceroporin [45], a Ca++ transporting-ATPase [46] and a HAK/KUP/KT-like K+ transporter [47].
Table 1.
Selected unusual gene products encoded by the chlorella viruses
| Polyamine metabolic enzymes | Sugar metabolic enzymes | Polysaccharide degrading enzymes | Channel/transporter proteins | Others |
|---|---|---|---|---|
| *Arginine/ornithine decarboxylase | *GDP-D-mannose dehydratase | *Alkaline alginate lyase | *Aquaglyceroporin | *Pyrimidine dimer-specific glycosylase |
| *Agmatine iminohydrolase | *Mannose-6-phosphate isomerase | *Chitinases (2)a | *Potassium ion channel protein (Kcv) | *Aspartate transcarbamylase |
| *N-carbamoylputrescine amidohydrolase | *UDP-glucose 4,6 dehydratase | *Chitosanase | *Potassium ion transporter | *Thymidylate synthase X |
| *Homospermindine synthase | *Fucose synthase | *β-1,3-glucanase | *Calcium transporting ATPase | *Histone 3, Lys 27 methylase (vSET) |
| *Polyamine acetyltransferase | *UDP-glucose dehydrogenase | *Chitin deacetylase | *Prolyl-4-hydroxylase | |
| dTDP glucose pyrophosphorylase | *Cu/Zn-superoxide dismutase | |||
| *Glucosamine synthetase | Cytosine deaminase | |||
| *Hyaluronan synthase | Histidine decarboxylase | |||
| Chitin synthase | Glycosyltransferases (9) | |||
| #Adenine DNA methyltransferases (11) | ||||
| #Cytosine DNA methyltransferases (8) | ||||
| #DNA restriction endonucleases (6) |
The number in brackets refers to the number of different enzymes with that activity.
Recombinant protein is functional.
Some of the recombinant proteins are known to be functional.
Presumably, polyamines play an essential role in the PBCV-1 life cycle and need to be tightly regulated because PBCV-1 encodes four functional polyamine biosynthetic enzymes, arginine/ornithine decarboxylase, agmatine iminohydrolase, N-carbamoylputrescine amidohydrolase, and homospermidine synthase [48-50]. In addition, PBCV-1 encodes a functional polyamine acetyltransferase, which is presumably involved in polyamine degradation (Charlop-Power et al., manuscript in preparation). These five polyamine metabolic enzymes are highly conserved among all of the chloroviruses; however, their functions are unknown.
Chloroviruses are also unusual because they often encode enzymes involved in sugar metabolism [51]. For example, two PBCV-1 encoded enzymes synthesize either GDP-L-fucose or GDP-D-rhamnose from GDP-D-mannose [52,53]. Other chloroviruses encode additional sugar metabolizing enzymes including a UDP-glucose 4,6 dehydratase [54] and a putative mannose-6-phosphate isomerase. Three PBCV-1 encoded enzymes are involved in the synthesis of the extracellular matrix polysaccharide hyaluronan, including hyaluronan synthase (HAS) [55,56]. Hyaluronan, a ubiquitous constituent of the extracellular matrix in vertebrates, consists of ~20,000 alternating β-1,4-glucuronic acid and β-1,3-N-acetylglucosamine residues [57]. Until the has gene was discovered in PBCV-1, hyaluronan was thought to occur only in vertebrates and a few pathogenic bacteria [57,58]. PBCV-1 also encodes two enzymes involved in the biosynthesis of hyaluronan precursors, glutamine:fructose-6-phosphate amidotransferase and UDP-glucose dehydrogenase [59]. All three genes are expressed early during PBCV-1 infection. These results led to the discovery that hyaluronan lyase-sensitive hair-like fibers accumulate on the surface of PBCV-1 infected host cells beginning at 15 min p.i. By four hr p.i., the infected cells are covered with a dense fibrous hyaluronan network (compare Figure 1n and 1o) [56].
The has gene is present in many, but not all, chloroviruses [56]. Surprisingly, many chloroviruses lacking a has gene have a gene encoding a functional chitin synthase (CHS). Furthermore, cells infected with these viruses produce chitin fibers on their external surface [60]. Chitin, an insoluble linear homopolymer of β-1,4-linked N-acetyl-glucosamine residues, is a common component of insect exoskeletons, shells of crustaceans and fungal cell walls.
A few chloroviruses encode both has and chs genes and form both hyaluronan and chitin on the surface of their infected cells [60,61]. Finally, some chloroviruses probably lack both genes because cells infected with these viruses produce no extracellular polysaccharides [56]. The fact that many chloroviruses encode enzymes involved in extracellular polysaccharide biosynthesis suggests that the polysaccharides, which require a huge amount of ATP for their synthesis, are important in the viral life cycles. However, this function is unknown.
The chloroviruses also encode functional chitinases and chitosanases [62]. The discovery that members of the Chlorella genus have chitin in their walls [63,64] was surprising because chitin is normally absent in green algae. One hypothesis is that components of Chlorella chitin metabolism were acquired horizontally from the chloroviruses [11].
PBCV-1 also encodes at least five putative glycosyltransferases, which likely participate in glycosylating the virus MCP [51,65]. In contrast to all other viruses that use the host machinery located in the endoplasmic reticulum (ER) and Golgi to glycosylate their glycoproteins, chloroviruses encode most, if not all, of the components required to glycosylate their MCPs. Furthermore, all experimental results indicate that glycosylation occurs independent of the ER and Golgi [51,65]. For example, unlike viruses that acquire their glycoprotein(s) by budding through a plasma membrane (which become infectious only during their release from the host), intact infectious PBCV-1 particles accumulate inside its host 30-40 min before virus release [66]. The identification of the glycan-linked Asn residues in the PBCV-1 MCP provided additional evidence that MCP glycosylation does not involve host ER glycosyltransferases [15]. None of the MCP glycan linked Asn residues reside in a NX(T/S) sequence commonly recognized by ER- and Golgi-located glycosyltransferases [67]. Finally, the MCP, like the PBCV-1 glycosyltransferases, lacks an ER or Golgi signal peptide. We hypothesize that PBCV-1 encodes some, if not all, of the machinery involved in glycosylating its MCP and that the process occurs independent of the ER and Golgi.
Evolutionary history
Phycodnaviruses are believed to have an ancient, common evolutionary ancestry with some other large dsDNA viruses such as the poxviruses (e.g., smallpox virus), asfarviruses, iridoviruses, ascoviruses and mimiviruses [68-70]. Collectively these viruses are referred to as nucleocytoplasmic large dsDNA viruses (NCLDVs). Comparative analysis of 45 NCLDVs identified five genes common to all the viruses and 177 additional genes exist in at least two of the virus families [71].
Although a common ancestry of NCLDVs is generally accepted, there is lively discussion on the role these viruses played in the evolution of eukaryotes. These discussions, which are likely to continue for some time, include: (i) Should NCLDVs be included in the tree of life (e.g., see [72-75]; comments by seven groups in Nat. Rev. Microbiol. 2009, vol 7:614-627)? (ii) Did the NCLDV genes arise from the original gene pool that led to prokaryotes and eukaryotes [69]? (iii) Did primitive NCLDVs give rise to the eukaryotic nucleus or vice versa (e.g., [76,77]? (iv) Does the structure of chlorovirus MCP, which resembles MCPs from smaller dsDNA viruses with hosts in all three domains of life (human adenovirus, bacteriophage PRD1, and Archaea virus STIV), indicate these three viruses have a common evolutionary ancestor with the NCLDVs, despite the lack of amino acid sequence similarity among their MCPs (e.g., [78,79]?
Concluding remarks
With the increasing interest in using algae for producing biofuels, it is obvious that pathogens, including viruses, will affect yields [80]. Although this review focuses on certain large dsDNA viruses that infect algae, ssRNA, dsRNA, and ssDNA viruses have recently been discovered that infect algae [e.g., 2]. Algal viruses are potentially a greater problem than plant viruses are to higher plants because plant viruses are usually transmitted by biological vectors. Thus two components are required for a virus disease outbreak in plants, whereas there is no evidence of biological vectoring of algal viruses; thus they only require one component for infection.
Chloroviruses are also a largely unexplored source of genetic elements for engineering algae and higher plants. For example, promoter elements from chloroviruses function well in both monocots and dicots of higher plants, as well as bacteria [81]. Also a translational enhancer element from a chlorella virus functions well in Arabidopsis [82].
The hosts for some algal viruses have been recently sequenced or are in the process of being sequenced. Annotation of these sequences will certainly contribute to studies on these viruses. However, one obstacle to studying the phycodnaviruses is that currently their hosts cannot be transformed. The development of successful and reproducible host transformation procedures will allow molecular genetic analysis of these viruses, which will be a major development.
Acknowledgments
We thank Dr. Leslie Lane for critical reading of the manuscript, and Dr. Alexander Tchourbanov for composing the gene map, and Wim van Egmond for the micrograph of zoochlorella-paramecium. Research in the Van Etten laboratory was supported in part by NSF-EPSCoR EPS-1004094, DOE DE-FG36-08GO88055, DOE DE-EE0003142 and NIH/NCRR 1P20RR15635 from the COBRE program of the National Center for Research Resources.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure statement
The authors are not aware of any affiliations, memberships, funding or financial holdings that might be perceived as affecting the objectivity of this review.
References
- 1.Hull R. Comparative Plant Virology. Elsevier/Academic Press; Amsterdam: 2009. [Google Scholar]
- 2.Nagasaki K. Dinoflagellates, diatoms, and their viruses. J. Microbiol. 2008;46:235–243. doi: 10.1007/s12275-008-0098-y. [DOI] [PubMed] [Google Scholar]
- 3.Fuhrman JA. Marine viruses and their biogeochemical and ecological effects. Nature. 1999;399:541–548. doi: 10.1038/21119. [DOI] [PubMed] [Google Scholar]
- 4.Suttle CA. Marine viruses - major players in the global ecosystem. Nat. Rev. Microbiol. 2005;5:801–812. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- 5.Wommack KE, Colwell RR. Viroplankton: viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 2000;64:69–114. doi: 10.1128/mmbr.64.1.69-114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunigan DD, et al. Phycodnaviruses: a peek at genetic diversity. Virus Res. 2006;117:119–132. doi: 10.1016/j.virusres.2006.01.024. [DOI] [PubMed] [Google Scholar]
- 7.Van Etten JL. Unusual life style of giant chloroviruses. Annu. Rev. Genetics. 2003;37:153–195. doi: 10.1146/annurev.genet.37.110801.143915. [DOI] [PubMed] [Google Scholar]
- 8.Wilson WH, et al. The Phycodnaviridae: the story of how tiny giants rule the world. In: Etten J Van., editor. Lesser Known Large dsDNA Viruses. Springer; Berlin: 2009. pp. 1–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamada T, et al. Chloroviruses. In: Maramorosch K, Shatkin AJ, editors. Advances in Virus Research. Vol. 66. Elsevier Inc.; 2006. pp. 293–366. [Google Scholar]
- 10.Proschold T, et al. The systematics of zoochlorella revisited employing an integrative approach. Environ. Microbiol. 2011;13:350–364. doi: 10.1111/j.1462-2920.2010.02333.x. [DOI] [PubMed] [Google Scholar]
- 11.Blanc G, et al. The Chlorella variabilis NC64A genome reveals adaptation to photosymbiosis, coevolution with viruses, and cryptic sex. Plant Cell. 2010;22:2943–2955. doi: 10.1105/tpc.110.076406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan X, et al. Structure and assembly of large lipid-containing dsDNA viruses. Nature Struct. Biol. 2000;7:101–103. doi: 10.1038/72360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cherrier MV, et al. An icosahedral algal virus has a complex unique vertex decorated by a spike. Proc. Natl. Acad. Sci. USA. 2009;106:11085–11089. doi: 10.1073/pnas.0904716106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X, et al. Three-dimensional structure and function of the Paramecium bursaria chlorella virus capsid. Proc. Natl. Acad. Sci. USA. 2011;108:14837–14842. doi: 10.1073/pnas.1107847108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nandhagopal N, et al. The structure and evolution of the major capsid protein of a large, lipid-containing, DNA virus. Proc. Natl. Acad. Sci. USA. 2002;99:14758–14763. doi: 10.1073/pnas.232580699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thiel G, et al. Initial events associated with virus PBCV-1 infection of Chlorella NC64A. In: Lüttge U, Beyschlag W, Büdel B, editors. Progress in Botany. Vol. 71. Springer -Verlag; Berlin: 2010. pp. 169–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Agarkova IV, et al. Chlorovirus-mediated membrane depolarization of chlorella alters secondary active transport of solutes. J. Virol. 2008;82:12181–12190. doi: 10.1128/JVI.01687-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greiner T, et al. Chlorella viruses prevent multiple infections by depolarizing the host membrane. J. Gen. Virol. 2009;90:2033–2039. doi: 10.1099/vir.0.010629-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Agarkova IV, et al. Virion-associated restriction endonucleases of chloroviruses. J. Virol. 2006;80:8114–8123. doi: 10.1128/JVI.00486-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mujtaba S, et al. Epigenetic transcription repression of cellular genes by a viral SET protein. Nature Cell Biol. 2008;10:1114–1122. doi: 10.1038/ncb1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iyer LM, et al. Comparative genomics of the FtsK-HerA superfamily of pumping ATPases: implications for the origins of chromosome segregation, cell division and viral capsid packaging. Nucleic Acids Res. 2004;32:5260–5279. doi: 10.1093/nar/gkh828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bidle KD, et al. Viral activation and recruitment of metacaspases in the unicellular coccolithophore, Emiliania huxleyi. Proc. Natl. Acad. Sci. USA. 2007;104:6049–6054. doi: 10.1073/pnas.0701240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bidle KD, Vardi A. A chemical arms race at sea mediates algal host-virus interactions. Curr. Opin. Microbiol. 2011;14:449–457. doi: 10.1016/j.mib.2011.07.013. [DOI] [PubMed] [Google Scholar]
- 24.Yanai-Balser GM, et al. Microarray analysis of Paramecium bursaria chlorella virus 1 transcription. J. Virol. 2010;84:532–542. doi: 10.1128/JVI.01698-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Landstein D, et al. Large deletions in antigenic variants of the chlorella virus PBCV-1. Virology. 1995;214:413–420. doi: 10.1006/viro.1995.0051. [DOI] [PubMed] [Google Scholar]
- 26.Van Etten JL, et al. Viruses and virus-like particles of eukaryotic algae. Microbiol. Rev. 1991;55:586–620. doi: 10.1128/mr.55.4.586-620.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nelson M, et al. Chlorella viruses encode multiple DNA methyltransferases. Biol. Chem. 1998;379:423–428. doi: 10.1515/bchm.1998.379.4-5.423. [DOI] [PubMed] [Google Scholar]
- 28.Chan SH, et al. Cloning of Nt.CviQII nicking endonucleases and its cognate methyltransferase: M.CviQII methylates AG sequences. Prot. Expr. Purif. 2006;49:138–150. doi: 10.1016/j.pep.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 29.Fitzgerald LA, et al. Sequence and annotation of the 369-kb NY-2A and 345-kb AR158 viruses that infect Chlorella NC64A. Virology. 2007;358:472–484. doi: 10.1016/j.virol.2006.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fitzgerald LA, et al. Sequence and annotation of the 314-kb MT325 and the 321-kb FR483 viruses that infect Chlorella Pbi. Virology. 2007;358:459–471. doi: 10.1016/j.virol.2006.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fitzgerald LA, et al. Sequence and annotation of the 288-kb ATCV-1 virus that infects an endosymbiotic chlorella strain of the heliozoon Acanthocystis turfacea. Virology. 2007;362:350–361. doi: 10.1016/j.virol.2006.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fitzgerald LA, et al. Putative gene promoter sequences in the chloroviruses. Virology. 2008;380:388–393. doi: 10.1016/j.virol.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ho CK, et al. Characterization of an ATP dependent DNA ligase encoded by chlorella virus PBCV-1. J. Virol. 1997;71:1931–1937. doi: 10.1128/jvi.71.3.1931-1937.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lavrukhin OV, et al. Topoisomerase II from clorella virus PBCV-1. Characterization of the smallest known type II topoisomerase. J. Biol. Chem. 2000;275:6915–6921. doi: 10.1074/jbc.275.10.6915. [DOI] [PubMed] [Google Scholar]
- 35.Eriksson M, et al. Evidence for 4-hydroxproline in viral proteins: characterization of a viral prolyl 4 hydroxylase and its peptide substrates. J. Biol. Chem. 1999;274:22131–22134. doi: 10.1074/jbc.274.32.22131. [DOI] [PubMed] [Google Scholar]
- 36.Manzur KL, et al. A dimeric viral SET domain methyltransferase specific to Lys27 of histone H3. Nature Struct. Biol. 2003;10:187–196. doi: 10.1038/nsb898. [DOI] [PubMed] [Google Scholar]
- 37.Plugge B, et al. A potassium channel protein encoded by chlorella virus PBCV-1. Science. 2000;287:1641–1644. doi: 10.1126/science.287.5458.1641. [DOI] [PubMed] [Google Scholar]
- 38.Gazzarrini S, et al. Eighty-two amino acids are sufficient for making a potassium selective, voltage-dependent channel. Biochem. J. 2009;420:295–303. doi: 10.1042/BJ20090095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pagliuca C, et al. Molecular properties of Kcv, a virus encoded K+ channel. Biochemistry. 2007;46:1079–90. doi: 10.1021/bi061530w. [DOI] [PubMed] [Google Scholar]
- 40.Shim JW, et al. In vitro synthesis, tetramerization and single channel characterization of virus-encoded potassium channel Kcv. FEBS Lett. 2007;581:1027–1034. doi: 10.1016/j.febslet.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 41.Chatelain FC, et al. Selection of inhibitor-resistant viral potassium channels identifies a selectivity filter site that affects barium and amantadine block. PLoS One. 2009;4:e7496. doi: 10.1371/journal.pone.0007496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hertel B, et al. Salt bridges in the miniature viral channel Kcv are important for function. Eur. Biophys. J. 2010;39:1057–1068. doi: 10.1007/s00249-009-0451-z. [DOI] [PubMed] [Google Scholar]
- 43.Balss J, et al. Transmembrane domain length of viral K+ channels is a signal for mitochondria targeting. Proc. Natl. Acad. Sci. USA. 2008;105:12313–12318. doi: 10.1073/pnas.0805709105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thiel G, et al. Minimal art: or why small viral K+ channels are good tools for understanding basic structure and function relations. Biochem. Biophys. Acta. 2011;1808:580–588. doi: 10.1016/j.bbamem.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 45.Gazzarrini S, et al. Chlorella virus MT325 encodes water and potassium channels that interact synergistically. Proc. Natl. Acad. Sci. USA. 2006;103:5355–5360. doi: 10.1073/pnas.0600848103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bonza MC, et al. A functional calcium transporting ATPase encoded by chlorella viruses. J. Gen. Virol. 2010;91:2620–2629. doi: 10.1099/vir.0.021873-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Greiner T, et al. A functional HAK/KUP/KT-like potassium transporter encoded by chlorella viruses. Plant Journal. 2011 doi: 10.1111/j.1365-313X.2011.04748.x. in press. [DOI] [PubMed] [Google Scholar]
- 48.Kaiser A, et al. Chlorella virus PBCV-1 encodes a functional homospermidine synthase. Virology. 1999;263:254–262. doi: 10.1006/viro.1999.9972. [DOI] [PubMed] [Google Scholar]
- 49.Shah R, et al. Chlorella virus PBCV-1 encodes an unusual arginine decarboxylase that is a close homolog of eukaryotic ornithine decarboxylases. J. Biol. Chem. 2004;279:35760–35767. doi: 10.1074/jbc.M405366200. [DOI] [PubMed] [Google Scholar]
- 50.Baumann S, et al. Chlorella viruses contain genes encoding a complete polyamine biosynthesis pathway. Virology. 2007;360:209–217. doi: 10.1016/j.virol.2006.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Van Etten JL, et al. Chloroviruses encode most, if not all, of the machinery to glycosylate their glycoproteins independent of the endoplasmic reticulum and Golgi. Biochim. Biophys. Acta. 2010;1800:152–159. doi: 10.1016/j.bbagen.2009.07.024. [DOI] [PubMed] [Google Scholar]
- 52.Fruscione F, et al. Differential role of NADP+ and NADPH in the activity and structure of GDP-D-mannose 4,6-dehydratase from two chlorella viruses. J. Biol. Chem. 2008;283:184–193. doi: 10.1074/jbc.M706614200. [DOI] [PubMed] [Google Scholar]
- 53.Tonetti M, et al. Chlorella virus PBCV-1 encodes two enzymes involved in the biosynthesis of GDP-L-fucose and GDP-D-rhamnose. J. Biol. Chem. 2003;278:21672–21677. doi: 10.1074/jbc.M301543200. [DOI] [PubMed] [Google Scholar]
- 54.Parakkottil CM, et al. Identification of an L-rhamnose synthetic pathway in two nucleocytoplasmic large DNA viruses. J. Virol. 2010;84:8829–8838. doi: 10.1128/JVI.00770-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.DeAngelis PL, et al. Hyaluronan synthase of chlorella virus PBCV-1. Science. 1997;278:1800–1803. doi: 10.1126/science.278.5344.1800. [DOI] [PubMed] [Google Scholar]
- 56.Graves MV, et al. Hyaluronan synthesis in virus PBCV-1 infected chlorella-like green algae. Virology. 1999;257:15–23. doi: 10.1006/viro.1999.9628. [DOI] [PubMed] [Google Scholar]
- 57.DeAngelis PL. Hyaluronan synthases: fascinating glycosyltransferases from vertebrates, bacterial pthogens, and algal viruses. Cell Mol. Life Sci. 1999;56:670–682. doi: 10.1007/s000180050461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.DeAngelis PL. Evolution of glycosaminoglycans and their glycosyltransferases: implication for the extracellular matrices of animals and the capsules of pathogenic bacteria. Anat. Rec. 2002;268:317–326. doi: 10.1002/ar.10163. [DOI] [PubMed] [Google Scholar]
- 59.Landstein D, et al. Chlorella virus PBCV-1 encodes functional glutamine: fructose-6-phosphate amidotransferase and UDP-glucose dehydrogenase enzymes. Virology. 1998;250:388–396. doi: 10.1006/viro.1998.9388. [DOI] [PubMed] [Google Scholar]
- 60.Kawasaki T, et al. Chitin synthesis in chlorovirus CVK2-infected chlorella cells. Virology. 2002;302:123–131. doi: 10.1006/viro.2002.1572. [DOI] [PubMed] [Google Scholar]
- 61.Yamada T, Kawasaki T. Microbial synthesis of hyaluronan and chitin: new approaches. J. Biosci. Bioeng. 2005;99:521–528. doi: 10.1263/jbb.99.521. [DOI] [PubMed] [Google Scholar]
- 62.Sun L, et al. Characterization of two chitinase genes and one chitosanase gene encoded by chlorella virus PBCV-1. Virology. 1999;263:376–387. doi: 10.1006/viro.1999.9958. [DOI] [PubMed] [Google Scholar]
- 63.Takeda H. Sugar composition of the cell wall and the taxonomy of Chlorella (Chlorophyceae). J. Phycol. 1991;27:224–232. [Google Scholar]
- 64.Kapaun E, et al. Cell wall composition of virus-sensitive symbiotic Chlorella species. Phytochemistry. 1992;31:3103–3104. [Google Scholar]
- 65.Wang IN, et al. Evidence for virus-encoded glycosylation specificity. Proc. Natl. Acad. Sci. USA. 1993;90:3840–3844. doi: 10.1073/pnas.90.9.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Van Etten JL, et al. Growth cycle of a virus, PBCV-1, that infects chlorella-like algae. Virology. 1983;126:117–125. doi: 10.1016/0042-6822(83)90466-x. [DOI] [PubMed] [Google Scholar]
- 67.Reuter G, Gabius HJ. Eukaryotic glycosylation: whim of nature or multipurpose tool. Cell Mol. Life Sci. 1999;55:368–422. doi: 10.1007/s000180050298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Iyer LM, et al. Common origin of four diverse families of large eukaryotic DNA viruses. J Virol. 2001;75:11720–11734. doi: 10.1128/JVI.75.23.11720-11734.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Iyer LM, et al. Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res. 2006;117:156–184. doi: 10.1016/j.virusres.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 70.Koonin EV, Yutin N. Origin and evolution of eukaryotic large nucleo-cytoplasmic DNA viruses. Intervirology. 2010;53:284–292. doi: 10.1159/000312913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yutin N, et al. Eukaryotic large nucleo-cytoplasmic DNA viruses: clusters of orthologous genes and reconstruction of viral genome evolution. Virol. J. 2009;6:223. doi: 10.1186/1743-422X-6-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Moreira D, López-García P. Ten reasons to exclude viruses from the tree of life. Nature Rev. Microbiol. 2009;7:306–311. doi: 10.1038/nrmicro2108. [DOI] [PubMed] [Google Scholar]
- 73.Boyer M, et al. Phylogenetic and phyletic studies of informational genes in genomes highlight existence of a 4th domain of life including giant viruses. Plos One. 2010;5:e15530. doi: 10.1371/journal.pone.0015530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Villarreal LP, Witzany G. Viruses are essential agents within the roots and stem of the tree of life. J. Theor. Biol. 2010;262:698–700. doi: 10.1016/j.jtbi.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 75.Van Etten JL. Giant viruses. American Sci. 2011;99:304–311. [Google Scholar]
- 76.Bell PJL. Viral eukaryogenesis: was the ancestor of the nucleus a complex DNA virus? J. Mol. Evol. 2001;53:251–256. doi: 10.1007/s002390010215. [DOI] [PubMed] [Google Scholar]
- 77.Takemura M. Poxviruses and the origin of the eukaryotic nucleus. J. Mol. Evol. 2001;52:419–425. doi: 10.1007/s002390010171. [DOI] [PubMed] [Google Scholar]
- 78.Krupovic M, Bamford D. Virus evolution: how far does the double b-barrel viral lineage extend? Nat Rev Microbiol. 2008;6:941–948. doi: 10.1038/nrmicro2033. [DOI] [PubMed] [Google Scholar]
- 79.Holmes EC. What does virus evolution tell us about virus origins? J. Virol. 2011;85:5247–5251. doi: 10.1128/JVI.02203-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gachon CMM, et al. Algal diseases: spotlight on a black box. Trends Plant Sci. 2010;15:633–640. doi: 10.1016/j.tplants.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 81.Mitra A, et al. A chlorella virus gene promoter functions as a strong promoter both in plants and bacteria. Biochem. Biophys. Res. Comm. 1994;204:187–194. doi: 10.1006/bbrc.1994.2443. [DOI] [PubMed] [Google Scholar]
- 82.Nguyen PS, et al. The A312L 5'-UTR of chlorella virus PBCV-1 is a translational enhancer in Arabidopsis thaliana. Virus Res. 2009;140:138–146. doi: 10.1016/j.virusres.2008.11.017. [DOI] [PubMed] [Google Scholar]
- 83.Meints RH, et al. Infection of a Chlorella-like alga with the virus, PBCV-1: ultrastructure studies. Virology. 1984;138:341–346. doi: 10.1016/0042-6822(84)90358-1. [DOI] [PubMed] [Google Scholar]
- 84.Meints RH, et al. Assembly site of the virus PBCV-1 in a Chlorella-like green alga: ultrastructural studies. Virology. 1986;154:240–245. doi: 10.1016/0042-6822(86)90448-4. [DOI] [PubMed] [Google Scholar]
- 85.Stothard P, Wishart DS. Circular genome visualization and exploration using CGView. Bioinformatics. 2005;21:537–539. doi: 10.1093/bioinformatics/bti054. [DOI] [PubMed] [Google Scholar]