Abstract
The hTau mouse model of tauopathy was utilized to assess gene expression changes in vulnerable hippocampal CA1 neurons. CA1 pyramidal neurons were microaspirated via laser capture microdissection followed by RNA amplification in combination with custom-designed microarray analysis and qPCR validation in hTau mice and nontransgenic (ntg) littermates aged 11-14 months. Statistical analysis revealed ∼8% of all the genes on the array platform were dysregulated, with notable downregulation of several synaptic-related markers including synaptophysin (Syp), synaptojanin, and synaptobrevin, among others. Downregulation was also observed for select glutamate receptors (GluRs), Psd-95, TrkB, and several protein phosphatase subunits. In contrast, upregulation of tau isoforms and a calpain subunit were found. Microarray assessment of synaptic-related markers in a separate cohort of hTau mice at 7-8 months of age indicated only a few alterations compared to the 11-14 month cohort, suggesting progressive synaptic dysfunction occurs as tau accumulates in CA1 pyramidal neurons. An assessment of SYP and PSD-95 expression was performed in the hippocampal CA1 sector of hTau and ntg mice via confocal laser scanning microscopy along with hippocampal immunoblot analysis for protein-based validation of selected microarray observations. Results indicate significant decreases in SYP-immunoreactive and PSD-95-immunoreactive puncta as well as downregulation of SYP-immunoreactive and PSD-95-immunoreactive band intensity in hTau mice compared to age-matched ntg littermates. In summary, the high prevalence of downregulation of synaptic-related genes indicates that the moderately aged hTau mouse may be a model of tau-induced synaptodegeneration, and has profound effects on how we perceive progressive tau pathology affecting synaptic transmission in AD.
Keywords: Alzheimer's disease, hippocampus, laser capture microdissection, PSD-95, RNA amplification, synaptophysin, TrkB
Introduction
Alzheimer's disease (AD) is a progressive neurodegenerative disorder affecting >5.4 million people and is the 6th leading cause of death in the USA and AD-associated costs exceed $183 billion dollars annually (Thies and Bleiler, 2011). AD is characterized by pathological hallmarks including extracellular accumulation of amyloid-beta (Aβ) peptides in senile plaques, and intracellular accumulation of the microtubule-associated protein tau in neurofibrillary tangles (NFTs). In addition, hippocampal disconnection is an invariant pathological hallmark of AD along with regionally selective synaptic loss (Hyman et al., 1984; Kordower et al., 2001). Tau protein is principally found within axons in normal brain, and is believed to promote microtubule assembly and stabilization (Ballatore et al., 2007; Duff, 2006). Tau localization to dendrites has also been identified, and is impacted by Aβ oligomers (Zempel et al., 2010). In AD, tau is redistributed into somatodendritic compartments, which include intracellular aggregates of NFTs formed by hyperphosphorylation and conformational alterations of tau (Binder et al., 2005; Cripps et al., 2006; Fischer et al., 2009; Jicha et al., 1997; Mondragon-Rodriguez et al., 2008). Tau hyperphosphorylation and intracellular accumulation may be directly related to synaptic loss, neuronal loss, and cognitive deficits in tauopathies (Binder et al., 2005; Duff, 2006; Hyman et al., 1984, 1990; Mufson et al., 2006).
A goal is to understand alterations in gene expression levels, and ultimately their protein products, in a mouse model of tauopathy, within a vulnerable cell type seen in AD. We are employing the hTau mouse model for single population expression profiling studies (Ginsberg, 2008; Ginsberg et al., 2006a). hTau mice are a mouse model of progressive tauopathy generated by overexpressing six human tau isoforms on a mouse null tau background (Andorfer et al., 2003; Duff et al., 2000). hTau mice develop pathological alterations including age-related phosphorylated intracellular tau accumulation within hippocampal and neocortical neurons (Andorfer et al., 2003, 2005). Delivery of minocycline, a derivative of tetracycline, reduces tau phosphorylation and aggregation (Noble et al., 2009), making this an attractive model to assess potential therapeutic interventions. hTau mice also develop moderate age-related dendritic alterations and cognitive deficits (Dickstein et al., 2010; Polydoro et al., 2009). Specifically, neuropathology progresses in the forebrain, and learning and memory deficits associated with cortical and hippocampal function manifest by 12 months of age (Polydoro et al., 2009).
The present study was undertaken to delineate cellular and molecular events that occur in hTau mice at two timepoints to help understand mechanisms that underlie neurodegeneration in human tauopathies, including AD. We employed custom-designed microarray analysis to evaluate multiple classes of transcripts relevant to neuroscience and neurodegeneration similar to our approach using postmortem human brains (Alldred et al., 2008, 2009; Ginsberg et al., 2006a, 2006b, 2010). Confirmation studies were performed using hTau and nontransgenic (ntg) littermates for select genes that demonstrated differential expression on the microarray platform via real-time quantitative PCR (qPCR). Furthermore, immunocytochemical evaluation of several synaptic-related marker protein products found to be dysregulated by microarray and qPCR assessments was performed via confocal laser scanning microscopy and immunoblot analysis.
Materials and Methods
Tissue preparation for microarray analysis
Animal protocols were approved by the Institutional Animal Care and Use Committee (IACUC) of the Nathan Kline Institute/NYU Langone Medical Center and were in full accordance with NIH guidelines. A cohort of hTau (n = 15; 8M/7F) and ntg (n = 11; 6M/5F) mice 11-14 months of age and a cohort of hTau (n = 7; 4M/3F) and ntg (n = 7; 3M/4F) mice 7-8 months of age were given an overdose of ketamine and xylazine and perfused transcardially with ice-cold 4% paraformaldehyde buffered in 0.15 M phosphate buffer. Tissue blocks containing the dorsal hippocampus were paraffin embedded and 6 μm-thick tissue sections were cut in the coronal plane on a rotary microtome for immunocytochemistry as described previously (Ginsberg, 2005a, 2010). RNase-free precautions were used throughout the experimental procedures, and solutions were made with 18.2 mega Ohm RNase-free water (Nanopure Diamond, Barnstead, Dubuque, IA).
Immunocytochemistry to identify CA1 neurons for subsequent microdissection was performed as described previously (Alldred et al., 2008; Ginsberg, 2005a; Ginsberg et al., 2010). Briefly, deparaffinized tissue sections were blocked in an RNase-free 0.1 M Tris (pH 7.6) solution containing 2% donor goat serum (DGS) and 0.01% Triton X-100 for 1 hour and then incubated with a primary monoclonal antibody directed against neurofilaments (RMdO20) which recognizes an epitope on the poorly phosphorylated NF-M/NF-H subunits (Lee et al., 1987) at a 1:200 dilution in a 0.1 M Tris/2% DGS solution overnight at 4 °C in a humidified chamber. An adjacent series was incubated with a primary monoclonal antibody directed against hyperphosphorylated tau (PHF1; a gift of Dr. Peter Davies; Jicha et al., 1997) at a 1:100 dilution. Sections were processed with the ABC kit (Vector Labs, Burlingame, CA) and developed with 0.05% diaminobenzidine, 0.03% hydrogen peroxide, and 0.01 M imidazole in Tris buffer for 10 minutes. Neurofilament-immunostained tissue sections were not coverslipped or counterstained, and were immersed in RNase-free 0.1 M Tris for microaspiration and subsequent TC RNA amplification.
Single cell microaspiration and Terminal Continuation (TC) RNA amplification
LCM and TC RNA amplification procedures have been described in detail previously by our group (Alldred et al., 2008, 2009; Che and Ginsberg, 2004; Ginsberg, 2010; Ginsberg et al., 2010). Individual CA1 pyramidal neurons were microaspirated via LCM (Arcturus PixCell IIe, Applied Biosystems, Foster City, CA; Fig. 1A-D). Fifty cells were captured per reaction for population cell analysis (Alldred et al., 2008; Ginsberg, 2010; Ginsberg et al., 2010). A total of 2-6 microarrays (containing 50 LCM-captured CA1 neurons each) were performed per mouse brain. Reproducibility and linearity of the TC RNA amplification procedure has been published previously, including the use of CA1 neurons as input sources of RNA (Alldred et al., 2008; Che and Ginsberg, 2004; Ginsberg, 2008). The TC RNA amplification protocol is available at http://cdr.rfmh.org/pages/ginsberglabpage.html. This method entails synthesizing first strand cDNA complementary to the RNA template, re-annealing the primers to the cDNA, and finally in vitro transcription using the synthesized cDNA as a template (Fig. 1E). Microaspirated CA1 neurons were homogenized in 500 μl of Trizol reagent (Invitrogen, Carlsbad, CA), extracted with chloroform, and precipitated utilizing isopropanol (Alldred et al., 2009). RNAs were reverse transcribed in a solution containing poly d(T) primer (100 ng/μl) and TC primer (100 ng/μl) in 1× first strand buffer (Invitrogen), 2 μg of linear acrylamide (Applied Biosystems), 0.5 mM dNTPs, 5 μM DTT, 20 U of SuperRNase Inhibitor (Applied Biosystems), and 200 U of reverse transcriptase (Superscript III, Invitrogen). Single-stranded cDNAs were then subjected to RNase H digestion and re-annealing of the primers to generate cDNAs with double-stranded regions at the primer interfaces. Single stranded cDNAs were digested by adding the following and then placed in a thermal cycler: 10 mM Tris (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, and 10 U RNase H (Invitrogen) in a final volume of 100 μl. RNase H digestion step at 37 °C, 30 minutes; denaturation step 95 °C, 3 minutes; primer re-annealing step 60 °C, 5 minutes (Che and Ginsberg, 2004). Samples were purified by column filtration (Montage PCR filters; Millipore, Billerica, MA). Column reservoirs were filled with 300 μl of 18.2 mega Ohm RNase-free water and the cDNA reaction was then added to the reservoir. The columns were then spun at 1000 × g for 15 minutes. To recover the cDNA, 20 μl of 18.2 mega Ohm RNase-free water was added to the columns, and the columns were inverted into clean microfuge tubes and spun at 1000 × g for 2 minutes (Alldred et al., 2008, 2009). Hybridization probes were synthesized by in vitro transcription using 33P incorporation in 40 mM Tris (pH 7.5), 6 mM MgCl2, 10 mM NaCl, 2 mM spermidine, 10 mM DTT, 2.5 mM ATP, GTP and CTP, 100 μM of cold UTP, 20 U of SuperRNase Inhibitor, 2 KU of T7 RNA polymerase (Epicentre, Madison, WI), and 120 μCi of 33P -UTP (Perkin-Elmer, Boston, MA) (Ginsberg, 2005b, 2008). The reaction was performed at 37 °C for 4 hours. Radiolabeled TC RNA probes were hybridized to custom-designed cDNA arrays without further purification.
Figure 1.
LCM of CA1 neurons and TC RNA amplification.
A. LCM was performed on 6 μm thick tissue sections of hTau and ntg mice. The CA1 region of hippocampus was immunostained with antibodies directed against neurofilaments for hippocampal pyramidal neuron detection and subsequent microaspiration. Scale bar: 25 μm.
B. CA1 neurons were isolated by applying laser pulses (black circles) over desired neurons. Note that tissue sections were dehydrated and not coverslipped to enable proper execution of the LCM process. Scale bar: 20 μm.
C. CA1 region of hippocampus seen after microaspiration, with cells captured by LCM removed and adjacent immunostained CA1 neurons remaining in the tissue section. Scale bar: 40 μm.
D. Captured CA1 neurons were visualized on a clean slide for contrast. Scale bar: 25 μm.
E. Schematic overview of the TC RNA amplification procedure.
F. Representative photomicrograph illustrating intensely labeled PHF1-immunoreactive CA1 neurons within the hippocampus of an aged hTau mouse. The inset depicts profuse intracellular phospho-tau immunoreactivity within the cell body of CA1 neurons.
G. PHF1 immunoreactivity was localized principally to neuropil surrounding the CA1 pyramidal layer in age-matched ntg littermates compared to the intracellular labeling in hTau mice. Scale bar F, G: 100 μm. Inset scale bar F, G: 50 μm.
Custom-designed cDNA array platforms and array hybridization
Array platforms consist of 1 μg of linearized cDNA purified from plasmid preparations adhered to high-density nitrocellulose (Hybond XL, GE Healthcare, Piscataway, NJ) using an arrayer robot (VersArray, Bio-Rad, Hercules, CA) (Ginsberg, 2005b; Ginsberg, 2008). Each cDNA and/or expressed sequence-tagged cDNA (EST) was verified by sequence analysis and restriction digestion. Mouse and human clones were employed on the custom-designed array. Notably, all of the tau isoforms were derived from human sequences. Approximately 576 cDNAs/ESTs were utilized on the current array platform, organized into 19 gene ontology groups (Table I). The majority of genes are represented by one transcript on the array platform, although the neurotrophin receptors TrkA, TrkB, and TrkC are represented by ESTs that contain the extracellular domain (ECD) as well as the tyrosine kinase domain (TK) (Ginsberg et al., 2010; Ginsberg et al., 2006b).
Table I. Classes of transcripts.
| Gene Category | # of genes | Gene Category | # of genes |
|---|---|---|---|
| AD-related genes | 42 | GluRs, transporters, & interacting | 42 |
| calcium, potassium, & sodium channels | 20 | immediate early genes | 12 |
| catecholamine synthesis, receptors, & transporters | 28 | neurotrophins & neurotrophin receptors | 20 |
| cell death-related genes | 28 | protease-related markers | 60 |
| cytoskeletal elements | 24 | protein phosphatases & kinases | 40 |
| development-related markers | 24 | steroid synthesis & receptors | 34 |
| G-protein subunits | 42 | synaptic-related markers | 24 |
| GABA synthesis, receptors, & transporters | 20 | transcription factors & regulators | 42 |
| glial-associated markers | 10 | others | 49 |
| glucose utilization | 15 | TOTAL | 576 |
Arrays were prehybridized (4 hours) and hybridized (16 hours) in a solution consisting of 6× saline–sodium phosphate–ethylenediaminetetraacetic acid (SSPE), 5× Denhardt's solution, 50% formamide, 0.1% sodium dodecyl sulfate (SDS), and denatured salmon sperm DNA (200 μg/ml) at 42 °C in a rotisserie oven (Che and Ginsberg, 2004; Ginsberg, 2008). Following hybridization, arrays were washed sequentially in 2× SSC/0.1% SDS, 1× SSC/0.1% SDS and 0.5× SSC/0.1% SDS for 15 min each at 37 °C. Arrays were placed in a phosphor screen for 24 hours and developed on a phosphor imager (GE Healthcare). All array phosphor images were adjusted to the same brightness and contrast levels for data acquisition and analysis.
Data collection and statistical analysis for custom-designed microarrays
Hybridization signal intensity was determined by utilizing ImageQuant TL (GE Healthcare). This array analysis program quantifies signal intensity, subtracts background by utilizing a spot edge average for each clone, and normalizes hybridization signal intensity. Statistical procedures for custom-designed microarray analysis have been described in detail previously (Ginsberg, 2007, 2009; Ginsberg and Mirnics, 2006). Briefly, expression of TC amplified RNA bound to each linearized cDNA (576 cDNAs/ESTs on the array platform) minus background was expressed as a ratio of the total hybridization signal intensity of the array (a global normalization approach). Global normalization effectively minimizes variation due to differences in the specific activity of the synthesized probe and absolute probe quantity (Eberwine et al., 2001; Ginsberg, 2008). These data do not allow the absolute quantitation of transcript levels. Rather, an expression profile of relative changes in mRNA levels was generated. Relative changes in total hybridization signal intensity were analyzed by one-way analysis of variance (ANOVA) with post-hoc analysis (Neumann-Keuls test) and correction for multiple observations for individual comparisons. The level of statistical significance was set at the more conservative (p < 0.01, two-sided; p-values (p < 0.02 – p < 0.05 were considered as trends) to account for the large number of analyses performed (Ginsberg, 2010; Ginsberg et al., 2010). p-values were also adjusted using a false discovery rate controlling procedure (Reiner et al., 2003) to reduce sampling (Type I error) due to the large number of genes being analyzed simultaneously. Expression levels were graphed using a bioinformatics software package (GeneLinker Gold, Predictive Patterns, Kingston, ON).
qPCR
qPCR was performed on microdissected paraffin-embedded tissue sections {adjacent to the tissue sections used for microarray analysis (Alldred et al., 2008; Ginsberg, 2008)} containing the hippocampal CA1 region from 11-14 month hTau (n = 9; 5M/4F) and ntg (n = 7; 3M/4F) mice. Taqman qPCR primers (Applied Biosystems, Foster City, CA) were utilized for the following genes: Arc (mM00479619_g1), synaptophysin (Syt; mM00436854_m1), TrkB (mM01341760_m1), Gria1 (mM01342711_m1), Gria2 (mM01220174_m1), Gria3 (mM01322408_m1), and the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (Gapdh; mM99999915_g1). Samples were assayed on a real-time qPCR cycler (7900HT, Applied Biosystems) in 96-well optical plates as described previously (Alldred et al., 2008; Ginsberg et al., 2006b, 2010; Jiang et al., 2010). Standard curves and cycle threshold (Ct) were generated using standards obtained from total mouse brain RNA. The ddCT method was employed to determine relative gene level differences between hTau and ntg mice with Gapdh qPCR products used as a control (ABI, 2004; Alldred et al., 2008; Ginsberg et al., 2010; Jiang et al., 2010). A total of 3-5 independent samples per subject were run in triplicate for the qPCR assessments. Negative controls consisted of the reaction mixture without input RNA. Alterations in PCR product synthesis were analyzed by one-way ANOVA with post-hoc analysis (Neumann-Keuls test; level of statistical significance was set at p < 0.05).
Confocal laser scanning microscopy
hTau (n = 8) and ntg (n = 6) mice 12-14 months of age were perfused transcardially with ice-cold 4% paraformaldehyde and post fixed for 2 hours in 4% paraformaldehyde followed by immersion in sucrose solutions (12%, 18%, and 30% in PBS for 24 hours each) prior to frozen sectioning in a cryostat. Tissue sections were cut at 20 μm for immunohistochemical labeling and mounted onto Superfrost plus glass slides (Thermo Fisher Scientific, Waltham, MA). Double label immunofluorescence for subsequent confocal microscopy was performed as described previously (Alldred et al., 2005; Jiang et al., 2010). Briefly, tissue sections were permeabilized with 0.3% Triton X-100, 10% normal horse serum (NHS) in PBS for 1 hour and then incubated with primary antibodies directed against PSD-95 (MA-0146; monoclonal; Thermo Fisher Scientific, Waltham, MA; 1:400 dilution) and synaptophysin (101-002; rabbit polyclonal; Synaptic Systems, Goettingen, Germany; 1:200 dilution) overnight in 0.05% Triton X-100, 10% NHS in PBS at 4 °C in a humidified chamber. Sections were washed three times in PBS containing 0.05% Triton X-100 and incubated with secondary antibodies coupled to Alexa 488 and Alexa 594 dyes, respectively (Molecular Probes Invitrogen, Eugene, OR). Sections were washed in PBS and coverslipped with buffered glycerol. Control experiments included omission of primary antisera and swapping Alexa dyes for the respective primary antibodies (Alldred et al., 2005; Ginsberg et al., 1995; Jiang et al., 2010).
Fluorescent images were captured with a confocal laser scanning microscope (LSM 510 Meta, Carl Zeiss, Thornwood, NY) with a Plan-Apochromat 100× (N.A. 1.4) objective. PSD-95-immunoreactive and synaptophysin-immunoreactive puncta were assessed using an unbiased estimation approach within stacks of optical z-sections acquired by confocal microscopy. Image acquisition parameters were adjusted for each section (1024 × 1024), with one slice analyzed within each of two z-stacks (10 optical slices per 10 μm) taken per section. Three sections were analyzed per subject (a total of 6 slices per subject were employed). AxioVision Automeasure software (Carl Zeiss) was utilized for stereologic assessments, measuring area (μm2), total puncta number, and mean intensity of the puncta. Area, number and intensity of puncta were quantified if they measured within the target area between 0.04 μm2 and 3.0 μm2. The mean signal intensity was compared to background measurements taken for each image (Alldred et al., 2005; Jiang et al., 2010). Alterations in immunoreactive puncta intensity, size, and number were analyzed by one-way ANOVA with post-hoc analysis (Neumann-Keuls test; level of statistical significance was set at p < 0.05).
Immunoblot analysis
Frozen microdissected CA1 sector hippocampal tissue samples obtained from hTau and ntg mice aged 3-5 months (hTau; n = 10; 5M/5F) (ntg; n = 5; 3M/2F), 7-10 months (hTau; n = 8; 5M/3F) (ntg; n = 6; 3M/3F), and 12-16 months (hTau; n = 11; 7M/4F) (ntg; n = 9; 5M/4F). Hippocampal tissues were homogenized in a 20 mM Tris-HCl (pH 7.4) buffer containing 10% (w/v) sucrose, 1 mM ethylenediaminetetraacetic acid (EDTA), 5 mM ethylene glycol-bis (β-aminoethylether)-N,N,N′,N′-tetra-acetic acid (EGTA), 2 mg/ml of the following: (aprotinin, leupeptin, and chymostatin), 1 mg/ml of the following: {pepstatin A, antipain, benzamidine, and phenylmethylsulfonyl fluoride (PMSF)}, 100 μg/ml of the following: {soybean trypsin inhibitor, Na-p-tosyl-L-lysine chloromethyl ketone (TLCK), and N-tosyl-L-phenylalanine chloromethyl ketone (TPCK)}, 1 mM of the following: (sodium fluoride and sodium orthovanadate) and centrifuged as described previously (Counts et al., 2004; Ginsberg, 2005a, 2010). All protease inhibitors were purchased from Sigma (St. Louis MO). Identical amounts of homogenates (10 μg) were loaded into a gel electrophoresis apparatus, subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; 4-15% gradient acrylamide gels; Bio-Rad), and transferred to nitrocellulose by electroblotting (Mini Transblot, Bio-Rad). Nitrocellulose membranes were blocked in blocking buffer (LiCor, Lincoln, NE) for 1 hour at 4 °C prior to being incubated with antibodies directed against GRIA1 (101-002; rabbit polyclonal; Synaptic Systems; 1:1,000 dilution), GRIA2/3 (09-307; rabbit polyclonal; Millipore; 1:1,000 dilution), GRIN1 (NMDA R1; sc-1467; goat polyclonal, Santa Cruz Biotechnology, Santa Cruz, CA; 1:1,000 dilution), PSD-95 (MA-0146; monoclonal; Thermo Fisher Scientific; 1:1,000 dilution), SYP (S-5768; monoclonal; Sigma; 1:1,000 dilution), tau (CP27; monoclonal; gift of Dr. Peter Davies; 1:2,500 dilution; (Cripps et al., 2006; Jicha et al., 1997), or β-tubulin (TUBB; monoclonal antibody; T-5293; Sigma, 1:1,000 dilution) in blocking buffer overnight at 4 °C. Membranes were developed with affinity–purified secondary antibodies conjugated to IRDye 800 (Rockland, Gilbertsville, PA) and visualized using an infrared detection system (Odyssey, LiCor). Immunoblots were quantified by densitometric software supplied with the instrument. Signal intensity of immunoreactive bands was normalized to TUBB immunoreactivity for each assay. Alterations in immunoreactive band intensity were analyzed by one-way ANOVA with post-hoc analysis (Neumann-Keuls test; level of statistical significance was set at p < 0.05).
Results
Microarray analysis identifies selectively altered genes including synaptic-related markers and glutamate receptors within CA1 neurons in aged hTau mice
CA1 pyramidal neurons in hTau mice displayed profuse intracellular accumulation of phosphorylated tau as compared to ntg littermates (Fig. 1F, G). Expression profiling was performed on a total of 90 custom-designed microarrays for hTau and ntg mice 11-14 months of age (50 CA1 neurons were analyzed per microarray) following the TC RNA amplification protocol. Quantitative analysis revealed differential regulation of 48 genes out of a total 576 (8.3%) on the custom-designed array (Table II). Notably, expression profiling results show upregulation of all 6 human isoforms of tau in the hTau mouse model compared to ntg littermates (Fig 2A). This observation effectively serves as a positive control, and is consistent with previous results from the hTau mouse that demonstrated an increase in tau protein expression levels (Andorfer et al., 2003, 2005). No significant effect of gender was detected in the cohorts of hTau and ntg mice.
Table II. Genes altered in moderately aged hTau mice (11-14 months) compared to ntg littermates.
| Unigene Annotation | Gene | Direction of Change |
|---|---|---|
| ADRA1B | (adrenergic alpha 1B receptor) | − |
| AQP1 | (aquaporin 1) | + |
| ARC | (activity regulated cytoskeletal-associated protein) | − |
| CAPN1 | (calpain 1, mu/I large subunit) | + |
| CCND2 | (cyclin D2) | +* |
| CCNE1 | (cyclin E1) | + |
| CNR1 | (CB1 cannabinoid receptor) | +* |
| ELAVL1 | (embryonic lethal abnormal vision like 1; Hu antigen R) | + |
| ELAVL4 | (embryonic lethal abnormal vision like 4; Hu antigen D) | + |
| FMR1 | (fragile × mental retardation 1) | − |
| FOSB | (FBJ osteosarcoma oncogene B) | + |
| GABARAP-L1 | (GABA-A receptor-associated protein like 1) | − |
| GAD67 | (glutamate decarboxylase isoform 67) | − |
| GALR1 | (galanin receptor 1) | − |
| GNAQ | (G protein, q polypeptide) | − |
| GNB2 | (G protein, beta polypeptide 2) | −* |
| GRIA1 | (GluR1 AMPA1) | − |
| GRIA2 | (GluR2 AMPA2) | − |
| GRIK4 | (KA receptor 1) | − |
| HMGCR | (3-hydroxy-3-methylglutaryl-coenzyme A reductase) | + |
| IGF1 | (insulin-like growth factor 1) | −* |
| IGF2 | (insulin-like growth factor 2) | −* |
| IP3KA | (inositol-(1,4,5) trisphosphate 3-kinase A subunit) | − |
| MAOB | (monoamine oxidase B) | −* |
| MAPT1 | (microtubule-associated protein tau 3Rtau 0 insert) | + |
| MAPT2 | (microtubule-associated protein tau 4Rtau 0 insert) | + |
| MAPT3 | (microtubule-associated protein tau 3Rtau 1 insert) | + |
| MAPT4 | (microtubule-associated protein tau 4Rtau 1 insert) | + |
| MAPT5 | (microtubule-associated protein tau 3Rtau 2 inserts) | +* |
| MAPT6 | (microtubule-associated protein tau 4Rtau 2 inserts) | + |
| MENTHO | (STARD3; N-terminal like MLN64 homolog) | + |
| NCSTN | (nicastrin) | +* |
| NR3C1 | (glucocorticoid receptor Grl) | −* |
| NRF1 | (nuclear respiratory factor 1) | + |
| NTRK2ECD | (BDNF receptor TrkB, extracellular domain) | − |
| NTRK2TK | (BDNF receptor TrkB, tyrosine kinase domain) | − |
| PPP1CC | (protein phosphatase 1 catalytic subunit, γ isoform) | − |
| PPP2CA | (protein phosphatase 2 catalytic subunit, α isoform) | −* |
| PPP2R1A | (protein phosphatase 2 regulatory subunit A, α isoform) | − |
| PRKCB1 | (protein kinase C, β1 subunit) | − |
| PSD-95 | (postsynaptic density protein 95; disc homolog 4) | − |
| RGS2 | (regulator of G-protein signalling 2) | +* |
| RGS13 | (regulator of G-protein signalling 13) | −* |
| SNAP-29 | (synaptosomal-associated protein, 29 Kd) | − |
| STX4A | (syntaxin 4A) | − |
| STX7 | (syntaxin 7) | − |
| SYB | (synaptobrevin 1; vesicle-associated membrane protein 1) | − |
| SYN3 | (synapsin 3) | − |
| SYNJ | (synaptojanin) | − |
| SYP | (synaptophysin) | − |
Data represents significant (p < 0.01 or greater) gene changes for hTau mice compared to ntg littermates.
+, up regulation; −, down regulation. Asterisk indicates a trend where (p < 0.02 – p < 0.05).
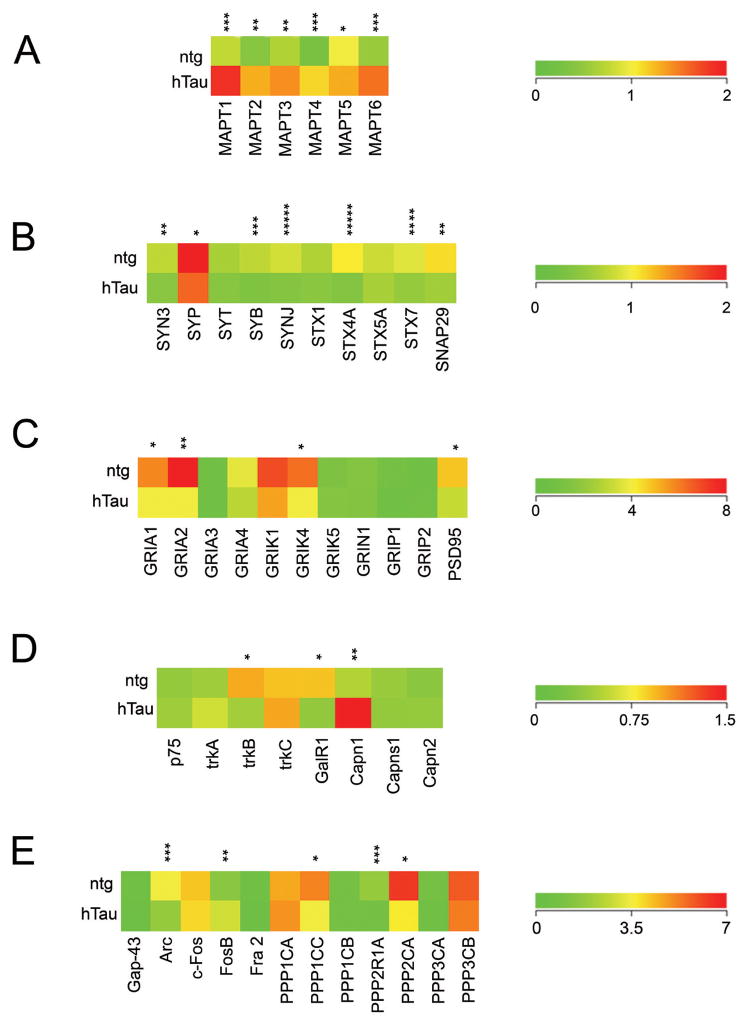
Figure 2.
Color-coded heatmaps depicting differential regulation of select transcripts in 11-14 month old hTau mice compared to age-matched ntg littermates.
A. Upregulation of the 6 tau isoforms were found in hTau mice. Asterisk denotes (p < 0.05), double asterisk denotes (p < 0.005), and triple asterisk denotes (p < 0.001).
B. Downregulation of a high percentage of individual synaptic-related markers was observed (e.g., Syn3, Syb, Syp, Synj, Stx4a, Stx7, and Snap29), making this class of transcripts the most affected overall within the aged hTau mouse. Asterisk denotes (p < 0.01), double asterisk denotes (p < 0.004), triple asterisk denotes (p < 0.003), quadruple asterisk denotes (p < 0.002), and quintuple asterisk denotes (p < 0.001).
C. Downregulation of select AMPA receptor subunits (Gria1 and Gria2), a KA receptor (Grik4), and Psd95 is demonstrated. No differential regulation of NMDA receptors was found. Asterisk denotes (p < 0.006) and double asterisk denotes (p < 0.001).
D. Downregulation of the BDNF receptor TrkB, but not p75, TrkA, or TrkC was observed along with the galanin receptor Galr1. In contrast, significant upregulation of the large subunit of the protease calpain 1 (Capn1) was found in hTau mice. Asterisk denotes (p < 0.002) and double asterisk denotes (p < 0.001).
E. Selective downregulation of PP1 (Ppp1cc) and PP2 (Ppp2r1a and Ppp2ca) subunits that regulate tau dephosphorylation were observed in hTau mice. Additionally, significant downregulation of the immediate-early gene Arc contrasted the up regulation immediate-early gene Fosb in hTau mice. Asterisk denotes (p < 0.01), double asterisk denotes (p < 0.004), and triple asterisk denotes (p < 0.001).
Classes of transcripts that were selectively downregulated included synaptic-related markers and glutamate receptors (GluRs), indicating deficits in excitatory synaptic transmission. Alterations included genes encoding proteins located within presynaptic and postsynaptic membranes. Specifically, several synaptic-related genes displayed downregulation in aged hTau CA1 neurons including synapsin 3 (Syn3; 1.8 fold; p < 0.004), synaptobrevin (Syb; 2.2 fold; p < 0.003), synaptojanin (Synj; 2.2 fold; p < 0.001), synaptosomal-associated protein 29 kD (Snap29; 1.9 fold; p < 0.004), syntaxin 4A (Stx4a; 2.8 fold; p < 0.001), syntaxin 7 (Stx7; 1.8 fold; p < 0.002), and synaptophysin (Syp; 1.7 fold; p < 0.01; Fig. 2B). In addition, GluRs that were downregulated included α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid (AMPA) receptor subunits Gria1 (1.6 fold; p < 0.006), Gria2 (2 fold; p < 0.001), and the kainate (KA) receptor Grik4 (1.6 fold; p < 0.006; Fig. 2C). There was no differential regulation of AMPA receptors Gria3, Gria4, N-methyl-D-aspartate (NMDA) receptor subunits, and metabotropic GluRs (mGluRs). The GluR-interacting protein/postsynaptic density protein Psd95 was also downregulated (1.6 fold; p < 0.006). No differential regulation of the GluR-interacting proteins Grip1 or Grip2 was observed (Fig. 2C).
Dysregulation of the brain derived neurotrophic factor (BDNF) receptor TrkB was also found within aged hTau CA1 neurons, including significant downregulation of the extracellular domain (TrkBecd; 2.3 fold; p < 0.002) and the tyrosine kinase domain (TrkBtk; 2.2 fold; p < 0.003). No significant alterations were found for gene expression of the pan NGF receptor p75NTR, nerve growth factor receptor TrkA or the neurotrophin-3 receptor TrkC (Fig. 2D). Downregulation of fragile × mental retardation 1 gene (Fmr1; 1.9 fold; p < 0.008), G-protein q polypeptide (Gnaq; 2.7 fold; p < 0.001), and galanin receptor 1 (Galr1; 2.5 fold; p < 0.002) was found along with downregulation of the insulin-like growth factors Igf1 (1.8 fold; p < 0.01), and Igf2 (2 fold; p < 0.01). Additionally, genes within the GABAergic neurotransmission class displayed selective downregulation including Gad67 (2.2 fold; p < 0.001) and GABA A receptor-associated protein like 1 (Gabarap-l1; 2 fold; p < 0.004). Interestingly, downregulation of genes that encode protein phosphatase (PP) PP1 and PP2 subunits that regulate tau dephosphorylation including PP1 catalytic subunit γ (Ppp1cc; 1.5 fold; p < 0.01), PP2 regulatory subunit Aα (Ppp2r1a; 2.4 fold; p < 0.001), and PP2 catalytic subunit α (Ppp2ca; 1.7 fold; p < 0.01; Fig. 2E) were observed along with downregulation of the kinases protein kinase Cβ1 (Prkcb1; 2.5 fold; p < 0.002) and a trend for inositol-(1,4,5) trisphosphate 3-kinase A subunit (Ip3ka; 1.6 fold; p < 0.03).
Transcripts that displayed upregulation within CA1 neurons in hTau mice included the large subunit of the protease calpain 1 (Capn1; 3.1 fold; p < 0.001) and a trend for the intramembrane protease nicastrin (Ncstn; 1.6 fold; p < 0.03) as well as the cell cycle regulatory gene cyclin E1 (Ccne1; 2.2 fold; p < 0.004), aquaporin 1 (Aqp1; 2 fold; p < 0.004), steroid-related gene 3-hydroxy-3-methylglutaryl-coenzyme A reductase (Hmgcr; 2.1 fold; p < 0.005), and the cannabinoid receptor Cnr1 (1.8 fold; p < 0.01). Upregulation of the transcription factors embryonic lethal abnormal vision Drosophila-like (ELAV) Elav1 (2.23 fold; p < 0.001), Elav4 (1.9 fold; p < 0.002), and nuclear respiratory factor 1 (Nrf1; 1.8 fold; p < 0.01) were also observed. Moreover, differential regulation of the immediate-early genes Arc (downregulated; 2.1 fold; p < 0.001) and Fosb (upregulated; 1.9 fold; p < 0.004; Fig. 2E) was identified in CA1 neurons. In summary, the classes of transcripts that displayed the most significant alterations in hTau mice included synaptic-related markers, select GluR subunits, select PP1 and PP2 subunits, and several transcription factors/immediate-early genes among other individual genes that did not represent a single gene ontology grouping.
In contrast to the marked downregulation of select synaptic-related markers and GluRs in CA1 pyramidal neurons microaspirated from 11-14 month old hTau mice (Fig. 2B, C), only a few select genes were downregulated in hTau mice aged 7-8 months compared to ntg littermates via assessment of 37 custom-designed microarrays. Specifically, downregulation was observed for Syp (1.6 fold; p < 0.01) and Psd95 (1.7 fold; p < 0.01) with a trend for downregulation for Synj (1.5 fold; p < 0.03) and Arc (1.5 fold; p < 0.03). No differential regulation was seen for the other identified synaptic-related markers downregulated at 11-14 months, including Syn3, Syb, Snap29, Stx4a, and Stx7 the or GluRs Gria1, Gria2, and Grik4, suggesting the alterations seen in these genes at 11-14 months are due to progressive tau accumulation during aging.
qPCR validation of select gene expression alterations in the CA1 sector of the hippocampus in hTau mice
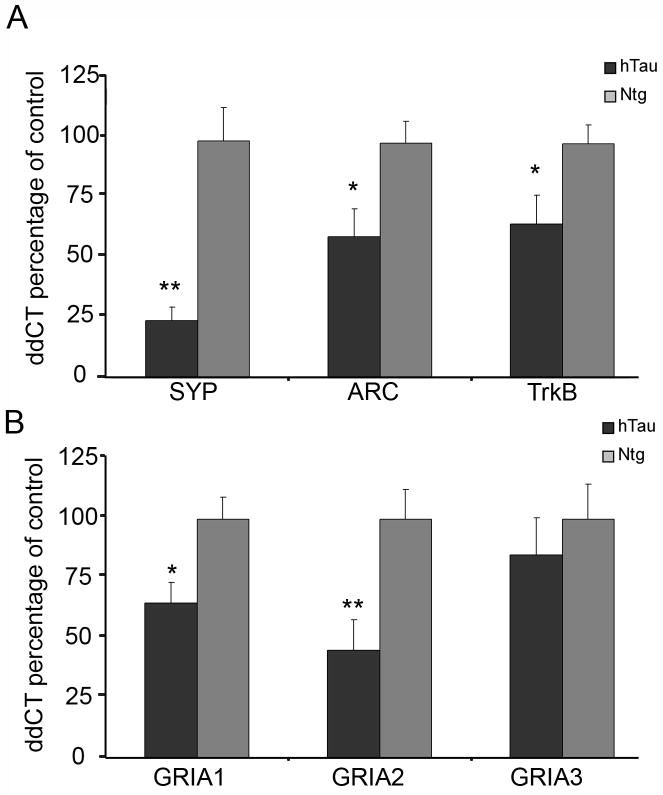
Based upon results gleaned from the custom-designed microarray analysis of CA1 pyramidal neurons in 11-14 month old mice, select gene expression levels were assessed via qPCR using microdissected hippocampal CA1 tissue harvested from hTau and ntg mice. The ddCT method was employed to determine relative gene level differences using Gapdh qPCR products as a control (Alldred et al., 2008, 2009; Ginsberg, 2008; Ginsberg et al., 2010). No differences in Gapdh expression levels were found between hTau and ntg mice, consistent with previously published observations using the CA1 sector of the hippocampus from mouse models of neurodegeneration (Alldred et al., 2008, 2009; Ginsberg, 2005a, 2010). Data was presented as percent of ntg ± standard deviation (SD). qPCR analysis independently validated the microarray observations using a separate cohort of mice, including downregulation of the synaptic-related marker Syp (p < 0.001), immediate early gene Arc (p < 0.04), and the neurotrophin receptor TrkB (p < 0.04) within the hippocampal CA1 region in hTau mice (Fig. 3A). qPCR analysis also demonstrated a statistically significant decrease in expression levels for Gria1 (p < 0.03) and Gria2 (p < 0.02), but not Gria3, consistent with microarray findings in aged mice (Fig. 3B). Two additional genes that did not differ between hTau and ntg mice on the microarray platform, the synaptic-related marker synaptopodin (Synpo) and the neurotrophin receptor TrkA were also examined. However, both transcripts were at the limit of resolution for qPCR assessment, which precluded microarray confirmation.
Figure 3.
qPCR validation of microarray observations in microdissected CA1 pyramidal neurons in 11-14 month old hTau and age-matched ntg littermates. The ddCT method was used for quantitative analysis and data is depicted as percentage of ntg expression ± SD.
A. Syp, Arc, and TrkB PCR products displayed significant downregulation in hTau mice compared to ntg littermates. Asterisk denotes (p < 0.04) and double asterisk denotes (p < 0.01).
B. Downregulation of Gria1 and Gria2, but not Gria3 PCR products was found, confirming the microarray results. Asterisk denotes (p < 0.03) and double asterisk denotes (p < 0.02).
SYP and PSD-95 levels are reduced within puncta in the CA1 hippocampus of aged hTau mice
Following qPCR validation, we analyzed whether downregulation of Syp and/or Psd95 mRNA levels would result in changes in protein levels denoted by loss of punctate synaptic clusters and/or protein localization within postsynaptic sites. Dual fluorescence confocal microscopy was performed within the pyramidal cell layer of the CA1 region in hTau and ntg mice 12-14 months of age to assess SYP-immunoreactive and PSD-95-immunoreactive puncta size, number, and staining intensity. Confocal microscopy demonstrated a colocalized punctate pattern for SYP and PSD-95 (Fig. 4A). Quantitative morphometric analyses identified a significant decrease in the staining intensity for both SYP (p < 0.02; Fig. 4B) and PSD-95 (p < 0.03; Fig. 4C) in hTau mice, indicating that the reduction in gene expression for these two transcripts seen in the microarray and qPCR studies also reflects a loss at the respective protein expression level. Although a significantly lower expression level of SYP and PSD-95 per puncta was observed in hTau mice, no differences were observed in the total number of immunoreactive puncta or the punctal area in hTau mice compared to ntg littermates (Fig. 4B, C).
Figure 4.
Confocal microscopy combined with unbiased quantitative morphometric analysis was performed within the CA1 region of hippocampus for two synaptic proteins (SYP and PSD-95), which were downregulated via microarray and qPCR analyses.
A. Representative confocal images depicting downregulation of SYP (red) puncta and PSD-95 (green) puncta in hTau mice compared to ntg littermates. A merged image indicates the overlapping distribution of SYP and PSD-95 (yellow) puncta within the CA1 pyramidal layer.
B. Morphometric analyses of the confocal images confirmed the expression profiling results, as significantly lower signal intensity levels for SYP were found in hTau mice. No significant differences were found in the number of puncta in hTau and ntg mice. Moreover, no significant differences exist between hTau and ntg mice in terms of punctal area measurements. Asterisk denotes (p < 0.02).
C. Similar to the SYP-immunoreactive results, down regulation of PSD-95-immunoreactivity was found in hTau mice. No significant differences in the number of puncta or punctal area measurements were observed. Asterisk denotes (p < 0.03).
Downregulation of select synaptic and GluR protein levels within the hippocampal CA1 sector in aged hTau mice
Protein-based assessment was performed via immunoblotting using well-established antibodies combined with frozen hippocampal CA1 sector punches to assess whether transcriptional alterations, particularly in aged hTau mice, resulted in protein level changes. Expression level data was normalized to TUBB expression ± SD. No significant changes were observed within hTau mice at 3-5 months of age (data not shown). Consistent with microarray analysis at 7-8 months of age, hTau mice displayed significant downregulation of SYP (p < 0.01) and PSD-95 (p < 0.008) expression in a separate cohort of 7-10 month old hTau mice compared to age-matched ntg littermates, with no significant changes in GluR expression levels (Fig. 5A). Moreover, quantitative analysis of select GluRs and GluR-interacting proteins demonstrated a significant downregulation of GRIA1 (p < 0.01), GRIA2/3 (p < 0.01), and PSD-95 (p < 0.005), but not GRIN1 (NMDA R1) in 12-16 month old hTau mice (Fig. 5B, C), consistent with microarray and qPCR findings in the CA1 region. Significant down regulation of SYP was also observed (p < 0.005; Fig. 5B, C), validating (along with the PSD-95 immunoblot analysis) the tissue-based confocal observations in aged hTau mice (Fig. 4).
Figure 5.
Immunoblotting using hippocampal CA1 sector punches to assess whether selected transcriptional alterations resulted in commensurate protein level changes in hTau mice compared to ntg littermates at 7-10 months of age (A) and 12-16 months of age (B). Expression level data was normalized to TUBB expression ± SD.
A. Downregulation of SYP and PSD-95 expression was observed in hTau mice compared to age matched littermates. Asterisk denotes (p < 0.01) and double asterisk denotes (p < 0.008).
B. Downregulation of GRIA1, GRIA2/3, PSD-95, and SYP was observed within the CA1 sector in 12-16 month old hTau mice, consistent with microarray, qPCR, and confocal analyses. Asterisk indicates (p < 0.01) and double asterisk indicates (p < 0.005).
C. Representative immunoblots obtained from 13-14 month old hTau and ntg mice demonstrating downregulation of GRIA1, GRIA2/3, PSD-95, and SYP in hTau mice, consistent with microarray and qPCR findings in the CA1 region. Upregulation of tau is demonstrated in the hTau mice as a positive control.
Discussion
Postmortem brain and biomarker studies of tau in human cerebrospinal fluid indicate the importance of assessing tau biology during AD pathogenesis. The hTau mouse line was created to model this pathology by expression of the normal human tau gene in the absence of mouse tau. hTau mice display pathological changes including accumulation of phosphorylated tau in the somatodendritic compartment of neurons, age-related increases in tau phosphorylation, and conformational alterations of tau (Andorfer et al., 2003, 2005). NFTs are not likely to occur solely due to tau overexpression, as another mouse model of tauopathy, 8c mice, has human tau and mouse tau overexpression, but does not develop overt tau cytopathology (Andorfer et al., 2003, 2005; Yuan et al., 2008). We are interested in identifying mechanisms of how tau dysfunction affects vulnerable neuronal populations. To this end, we performed microarray analysis, qPCR, and protein-based validation studies on CA1 neurons in hTau mice prior to and following an established timepoint where a behavioral phenotype has been identified (Andorfer et al., 2003, 2005; Polydoro et al., 2009).
The single population expression profiling paradigm employed in this study enabled an extensive, concurrent representation of hundreds of genes relevant to neurodegeneration and neuroscience. Notably, upregulation of the 6 isoforms of tau was seen within CA1 neurons in hTau mice, commensurate with increased tau protein levels in the hippocampus (Andorfer et al., 2003, 2005). Approximately 8% of the genes interrogated on the custom-designed array platform were dysregulated in 11-14 month old hTau mice compared to ntg littermates, including a disproportionately large number of synaptic-related markers and discrete AMPA and KA GluR subunits displaying downregulation which progressed from 7-8 months of age where only a few genes (including Syp and Psd95) were initially observed to be dysregulated. Interestingly, dysfunction of excitatory neurotransmission has been observed in aged hTau mice in behavioral and physiological paradigms (Andorfer et al., 2003; Polydoro et al., 2009), and these deficits coincide with dendritic alterations observed in CA1 neurons of older tauopathic mice (Dickstein et al., 2010; Gotz et al., 2010; Luebke et al., 2010). Notably, alterations in expression profiles and cytoarchitecture may not be as dependent on the biophysics of NFT formation as much as on the accumulation of tau within the somatodendritic domain (Berger et al., 2007; Luebke et al., 2010; Rocher et al., 2010). This has basic, translational, and drug development implications, as our data support the emerging concept that developing methods to regulate progressively accumulating intracellular tau levels may be a key aspect to therapeutic interventions. Further support of this hypothesis comes from in vitro studies whereby transfection of tau in hippocampal neurons causes somatodendritic tau resdistribution with concomitant synaptodegeneration, including spine loss and decrements in presynaptic and postsynaptic markers similar to our in vivo observations (Thies and Mandelkow, 2007). It is noteworthy that several key proteins are translated locally at synaptic sites, including mRNAs present directly within dendritic processes as well as somatic mRNAs that are transported to synaptic zones for local translation (Steward et al., 1998; Swanger and Bassell, 2011). Although our single cell microarray analysis is more sensitive than global or regional microarray analyses (Ginsberg et al., 2011), we cannot formally exclude the possibility of under representing expression level changes at local synaptic sites. Notwithstanding this caveat, the present findings at the genomic and protein levels provide a cellular backdrop for the aforementioned behavioral and morphological measures, implicating a loss of integral synaptic and GluR subunits, among others, in the progressive development of tauopathy.
Although the number of dysregulated genes in the hTau mouse at 11-14 months of age is moderate (and even less so at 7-8 months of age) compared to our previous microarray studies in NFT-bearing CA1 neurons obtained postmortem from AD brains, where we observed alterations in approximately 24%-36% of interrogated genes depending on the array platform (Ginsberg et al., 2000, 2006a, 2010, 2011), there is considerable overlap in the dysregulation of genes encoding integral synaptic proteins and GluR subunits (Table III), indicating the utility of this model to mimic synaptodegeneration found in AD. Notably, a consistent finding in both regional and single cell hippocampal analyses within the AD brain is the downregulation of genes encoding several synaptic-related markers (including Syp, synapsins, and syntaxins, among others) (Callahan et al., 1999; Colangelo et al., 2002; Ginsberg et al., 2000, 2006a, 2011; Gutala and Reddy, 2004; Mufson et al., 2002). Gene expression alterations are also reflected in respective protein level deficits, and importantly, correlation with cognitive decline (Counts et al., 2006; Heffernan et al., 1998; Heinonen et al., 1995; Masliah et al., 1991; Sze et al., 1997). Thus, hippocampal synaptodegeneration (particularly in CA1 neurons) is becoming recognized as one of the cardinal observations of AD pathogenesis, although the relationship(s) of this degeneration to tauopathy and amyloid deposition remain speculative and in need of further characterization in relevant animal and cellular models. Furthermore, down regulation of discrete GluRs and GluR-interacting proteins has been recognized as a feature of AD pathology (Armstrong et al., 1994; Carter et al., 2004; Ginsberg et al., 2004, 2006a; Ikonomovic et al., 1995; Proctor et al., 2010; Yasuda et al., 1995) that is reflected in gene expression and protein level changes in discrete AMPA and KA subunits as well as in PSD-95 within the hTau mouse. Combined with independent physiological and morphological assessments (Andorfer et al., 2003; Dickstein et al., 2010; Noble et al., 2009; Polydoro et al., 2009), the present results underscore the observation that the hTau mouse is emerging as a model of progressive, age-related synaptic dysregulation within vulnerable hippocampal neurons, which can be exploited for the assessment of therapeutic approaches aimed at restoring synaptic integrity, and warrants future high-density microarray analysis in the hTau mouse and parallel studies in AD postmortem brains. Moreover, the hTau mouse is devoid of amyloid-related pathology, which is perceived as a decided advantage, as effects of tau overexpression can be delineated in the absence of potential confounds of amyloidosis, as these two pathological hallmarks may have both distinct, as well as overlapping, effects on hippocampal circuitry and vulnerability (Small and Duff, 2008). Models of synaptodegeneration independent of APP and/or Aβ overproduction are increasingly becoming an area of great interest, since anti-amyloid therapies have not been effective to date in AD patients, as evidenced by the apparent lack of success of high-profile clinical trials of γ-secretase inhibitors and Aβ vaccination therapies (Imbimbo and Giardina, 2011; Sambamurti et al., 2011; Schor, 2011).
Table III. Genes altered within CA1 pyramidal neurons in the hTau mouse and in postmortem AD brain tissues.
| Unigene Annotation/Gene | hTau mice | AD | |
|---|---|---|---|
| ARC | (activity regulated cytoskeletal-associated protein) | − | − |
| FOSB | (FBJ osteosarcoma oncogene B) | + | +* |
| GAD67 | (glutamate decarboxylase isoform 67) | − | NS |
| GAT1 | (GABA A transporter 1) | NS | − |
| GRIA1 | (GluR1 AMPA1) | − | − |
| GRIA2 | (GluR2 AMPA2) | − | − |
| GRIA3 | (GluR3 AMPA3) | NS | NS |
| GRIA4 | (GluR4 AMPA4) | NS | NS |
| GRIK4 | (KA receptor 1) | − | NS |
| GRIK5 | (KA receptor 2) | NS | NS |
| GRIN1 | (NMDA receptor NR1) | NS | NS |
| GRIN2A | (NMDA receptor NR2A) | NS | NS |
| GRIN2B | (NMDA receptor NR2B) | − | − |
| GRIP1 | (glutamate receptor interacting protein 1) | NS | NS |
| GRIP2 | (glutamate receptor interacting protein 2) | − | − |
| NTRK2ECD | (BDNF receptor TrkB, extracellular domain) | − | − |
| NTRK2TK | (BDNF receptor TrkB, tyrosine kinase domain) | − | − |
| NTRK3ECD | (NT-3 receptor TrkC, extracellular domain) | NS | − |
| NTRK3TK | (NT-3 receptor TrkC, tyrosine kinase domain) | NS | − |
| P75NTR | (pan-neurotrophin receptor) | NS | NS |
| PPP1CC | (protein phosphatase 1 catalytic subunit, γ isoform) | − | − |
| PPP2CA | (protein phosphatase 2 catalytic subunit, α isoform) | −* | − |
| PPP2R1A | (protein phosphatase 2 regulatory subunit A, α isoform) | − | − |
| PRKCB1 | (protein kinase C, β1 subunit) | − | NS |
| PSD-95 | (postsynaptic density protein 95; disc homolog 4) | − | − |
| SNAP-29 | (synaptosomal-associated protein, 29 Kd) | − | − |
| SNCA | (α-synuclein) | NS | − |
| STX1 | (syntaxin 1) | NS | −* |
| STX4A | (syntaxin 4A) | − | − |
| STX7 | (syntaxin 7) | − | − |
| SYB | (synaptobrevin 1; VAMP1) | − | − |
| SYN1 | (synapsin 1) | NS | − |
| SYN3 | (synapsin 3) | − | − |
| SYNGR1 | (synaptogyrin 1) | NS | −* |
| SYNJ | (synaptojanin) | − | −* |
| SYNPO | (synaptopodin) | NS | − |
| SYP | (synaptophysin) | − | − |
| SYT1 | (synaptotagmin 1) | NS | − |
Data represents significant (p < 0.01 or greater) gene changes for hTau mice compared to ntg littermates (left column) and AD compared to cognitively normal, age-matched controls (right column) as described in (Ginsberg et al., 2011; Ginsberg et al., 2000).
+, up regulation; −, down regulation. Asterisk indicates a trend where (p < 0.02 – p < 0.05). Note the high similarity in downregulation of several synaptic-related markers, GluRs, and PP subunits in hTau and AD CA1 pyramidal neurons.
CA1 pyramidal neuron expression profiling at 11-14 months of age also demonstrated significant downregulation of select PP subunits, consistent with our previous human CA1 neuron molecular fingerprinting studies in AD (Ginsberg et al., 2000, 2006a; Mufson et al., 2002), and supports the concept that decrements in phosphatase activity are associated with the progression of AD (Goedert et al., 1995; Liu et al., 2005; Pei et al., 1994; Sontag et al., 2004; Vogelsberg-Ragaglia et al., 2001). Moreover, upregulation of Capn1 in aged hTau mice is consistent with observations of dysregulated protease activity in AD and in AD models, particularly within the calpain activation system (Di Rosa et al., 2002; Liang et al., 2010; Nixon, 2003; Rao et al., 2008). Although a description of the relevance of each gene change in the hTau mouse is beyond the scope of this report, it is worth noting that many of the expression level changes are relevant to AD pathology and disrupted synaptic plasticity, including the dysregulation of the neurotrophin receptor TrkB and the immediate-early gene Arc. Interestingly, the single population expression profiling paradigm in CA1 neurons within postmortem AD and mild cognitive impairment brains demonstrated a highly significant downregulation of TrkB (both the TrkBecd and TrkBtk forms; Table III) that correlated with levels of cognitive decline (Ginsberg et al., 2010). TrkB expression is widespread throughout the BDNF-responsive cortical mantle and hippocampal formation (Mufson et al., 2006), and may be a novel site for pharmacotherapeutic interventions via small molecule activators and/or TrkB transactivation (Longo et al., 2007; Rajagopal et al., 2004; Skaper, 2008). Based on the present study, employing the aged hTau mouse, with particular emphasis on hippocampal CA1 pyramidal neurons, is an exciting site of research for translational development of TrkB-based therapeutics for amelioration of neurodegeneration and associated cognitive decline during the progression of AD.
In summary, the coordinated microarray, qPCR, confocal microscopy, and immunoblotting studies presented herein indicate that the moderately aged hTau mouse has relevance towards understanding the source(s) of expression level changes found in human AD and related tauopathies. Specifically, the present results identified progressive gene and protein level changes in a vulnerable hippocampal cell type critical for learning and memory that indicates the hTau mouse is a model of progressive synaptodegeneration associated with tauopathy. Moreover, these findings in the hTau mouse at 11-14 months (which show progressive increases from observations starting at 7-8 months of age) potentially mimic intracellular tau level changes consistent with mild cognitive impairment and/or mild AD levels, and emphasize the importance of studying synaptic-related markers and GluRs in animal models of neurodegenerative disorders, particularly at the single population level.
Research Highlights.
Microarrays, qPCR, and confocal microscopy assessed CA1 neurons in tauopathic mice. Analysis of hTau CA1 pyramidal neurons revealed progressive synaptic dysfunction. Downregulation was observed for select glutamatergic markers at 11-14 months of age. qPCR and confocal analysis validated PSD-95 and TrkB microarray results in hTau mice.
Acknowledgments
Support for this project comes from NIH grants NS050276, AG14449, and the Alzheimer's Association. We thank Dr. Peter Davies for his assistance with the hTau mice and tau antibodies, and Dr. Shaoli Che for critical review of the manuscript. We thank Irina Elarova, Shaona Fang, and Arthur Saltzman for expert technical assistance.
Abbreviations
- AD
Alzheimer's disease
- Aβ
amyloid-β peptide
- AMPA
α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid
- APP
amyloid-β precursor protein
- ANOVA
analysis of variance
- BDNF
brain derived neurotrophic factor
- Ct
cycle threshold
- DGS
donor goat serum
- ESTs
expressed sequence-tagged cDNAs
- ECD
extracellular domain
- ELAV
embryonic lethal abnormal vision Drosophila-like
- GluRs
glutamate receptors
- GAPDH
glyceraldehyde-3 phosphate dehydrogenase
- IACUC
Institutional Animal Care and Use Committee
- KA
kainate
- LCM
laser capture microdissection
- NFTs
neurofibrillary tangles
- ntg
nontransgenic
- NMDA
N-methyl-D-aspartate
- NHS
normal horse serum
- qPCR
real-time quantitative polymerase-chain reaction
- PP
protein phosphatase
- SSPE
6× saline–sodium phosphate–ethylenediaminetetraacetic acid
- SDS
sodium dodecyl sulfate
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- TC
terminal continuation
- TK
tyrosine kinase domain
- TLCK
Na-p-tosyl-L-lysine chloromethyl ketone
- TPCK
N-tosyl-L-phenylalanine chloromethyl ketone
- TUBB
β-tubulin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- ABI. Guide to Performing Relative Quantitation of Gene Expression Using Real-Time Quantitative PCR. Applied Biosystems Product Guide. 2004:1–60. [Google Scholar]
- Alldred MJ, et al. Terminal continuation (TC) RNA amplification enables expression profiling using minute RNA input obtained from mouse brain. Int J Mol Sci. 2008;9:2091–2104. doi: 10.3390/ijms9112091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alldred MJ, et al. Terminal continuation (TC) RNA amplification without second strand synthesis. J Neurosci Methods. 2009;177:381–385. doi: 10.1016/j.jneumeth.2008.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alldred MJ, et al. Distinct gamma2 subunit domains mediate clustering and synaptic function of postsynaptic GABAA receptors and gephyrin. J Neurosci. 2005;25:594–603. doi: 10.1523/JNEUROSCI.4011-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andorfer C, et al. Cell-cycle reentry and cell death in transgenic mice expressing nonmutant human tau isoforms. J Neurosci. 2005;25:5446–5454. doi: 10.1523/JNEUROSCI.4637-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andorfer C, et al. Hyperphosphorylation and aggregation of tau in mice expressing normal human tau isoforms. J Neurochem. 2003;86:582–90. doi: 10.1046/j.1471-4159.2003.01879.x. [DOI] [PubMed] [Google Scholar]
- Armstrong DM, et al. AMPA-selective glutamate receptor subtype immunoreactivity in the entorhinal cortex of non-demented elderly and patients with Alzheimer's disease. Brain Res. 1994;639:207–216. doi: 10.1016/0006-8993(94)91732-9. [DOI] [PubMed] [Google Scholar]
- Ballatore C, et al. Tau-mediated neurodegeneration in Alzheimer's disease and related disorders. Nat Rev Neurosci. 2007;8:663–672. doi: 10.1038/nrn2194. [DOI] [PubMed] [Google Scholar]
- Berger Z, et al. Accumulation of pathological tau species and memory loss in a conditional model of tauopathy. J Neurosci. 2007;27:3650–3662. doi: 10.1523/JNEUROSCI.0587-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder LI, et al. Tau, tangles, and Alzheimer's disease. Biochim Biophys Acta. 2005;1739:216–223. doi: 10.1016/j.bbadis.2004.08.014. [DOI] [PubMed] [Google Scholar]
- Callahan LM, et al. Quantitative decrease in synaptophysin message expression and increase in cathepsin D message expression in Alzheimer disease neurons containing neurofibrillary tangles. J Neuropathol Exp Neurol. 1999;58:275–287. doi: 10.1097/00005072-199903000-00007. [DOI] [PubMed] [Google Scholar]
- Carter TL, et al. Differential preservation of AMPA receptor subunits in the hippocampi of Alzheimer's disease patients according to Braak stage. Exp Neurol. 2004;187:299–309. doi: 10.1016/j.expneurol.2003.12.010. [DOI] [PubMed] [Google Scholar]
- Che S, Ginsberg SD. Amplification of transcripts using terminal continuation. Lab Invest. 2004;84:131–137. doi: 10.1038/labinvest.3700005. [DOI] [PubMed] [Google Scholar]
- Colangelo V, et al. Gene expression profiling of 12633 genes in Alzheimer hippocampal CA1: transcription and neurotrophic factor down-regulation and up-regulation of apoptotic and pro-inflammatory signaling. J Neurosci Res. 2002;70:462–473. doi: 10.1002/jnr.10351. [DOI] [PubMed] [Google Scholar]
- Counts SE, et al. Differential expression of synaptic proteins in the frontal and temporal cortex of elderly subjects with mild cognitive impairment. J Neuropathol Exp Neurol. 2006;65:592–601. doi: 10.1097/00005072-200606000-00007. [DOI] [PubMed] [Google Scholar]
- Counts SE, et al. Reduction of cortical TrkA but not p75(NTR) protein in early-stage Alzheimer's disease. Ann Neurol. 2004;56:520–531. doi: 10.1002/ana.20233. [DOI] [PubMed] [Google Scholar]
- Cripps D, et al. Alzheimer disease-specific conformation of hyperphosphorylated paired helical filament-Tau is polyubiquitinated through Lys-48, Lys-11, and Lys-6 ubiquitin conjugation. J Biol Chem. 2006;281:10825–10838. doi: 10.1074/jbc.M512786200. [DOI] [PubMed] [Google Scholar]
- Di Rosa G, et al. Calpain inhibitors: a treatment for Alzheimer's disease. J Mol Neurosci. 2002;19:135–141. doi: 10.1007/s12031-002-0024-4. [DOI] [PubMed] [Google Scholar]
- Dickstein DL, et al. Changes in dendritic complexity and spine morphology in transgenic mice expressing human wild-type tau. Brain Struct Funct. 2010;214:161–179. doi: 10.1007/s00429-010-0245-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duff K. Normal and abnormal tau neurobiology. Alzheimer Dis Assoc Disord. 2006;20:202–205. doi: 10.1097/01.wad.0000213881.01289.d9. [DOI] [PubMed] [Google Scholar]
- Duff K, et al. Characterization of pathology in transgenic mice over-expressing human genomic and cDNA tau transgenes. Neurobiol Dis. 2000;7:87–98. doi: 10.1006/nbdi.1999.0279. [DOI] [PubMed] [Google Scholar]
- Eberwine J, et al. mRNA expression analysis of tissue sections and single cells. J Neurosci. 2001;21:8310–8314. doi: 10.1523/JNEUROSCI.21-21-08310.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer D, et al. Conformational changes specific for pseudophosphorylation at serine 262 selectively impair binding of tau to microtubules. Biochemistry. 2009;48:10047–10055. doi: 10.1021/bi901090m. [DOI] [PubMed] [Google Scholar]
- Ginsberg SD. Glutamatergic neurotransmission expression profiling in the mouse hippocampus after perforant-path transection. Am J Geriatr Psychiatry. 2005a;13:1052–1061. doi: 10.1176/appi.ajgp.13.12.1052. [DOI] [PubMed] [Google Scholar]
- Ginsberg SD. RNA amplification strategies for small sample populations. Methods. 2005b;37:229–237. doi: 10.1016/j.ymeth.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Ginsberg SD. Expression profile analysis of brain aging. In: Riddle DR, editor. Brain Aging: Models, Methods and Mechanisms. CRC Press; New York: 2007. pp. 159–185. [PubMed] [Google Scholar]
- Ginsberg SD. Transcriptional profiling of small samples in the central nervous system. Methods Mol Biol. 2008;439:147–158. doi: 10.1007/978-1-59745-188-8_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg SD. Microarray use for the analysis of the CNS. In: Squire LR, editor. Encyclopedia of Neuroscience. Vol. 5. Academic Press; Oxford; 2009. pp. 835–841. [Google Scholar]
- Ginsberg SD. Alterations in discrete glutamate receptor subunits in adult mouse dentate gyrus granule cells following perforant path transection. Anal Bioanal Chem. 2010;397:3349–3358. doi: 10.1007/s00216-010-3826-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg SD, et al. Gene expression levels assessed by CA1 pyramidal neuron and regional hippocampal dissections in Alzheimer's disease. Neurobiol Dis. 2011 doi: 10.1016/j.nbd.2011.07.013. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg SD, et al. Microarray analysis of hippocampal CA1 neurons implicates early endosomal dysfunction during Alzheimer's disease progression. Biol Psychiatry. 2010 doi: 10.1016/j.biopsych.2010.05.030. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg SD, et al. Single cell gene expression profiling in Alzheimer's disease. NeuroRx. 2006a;3:302–318. doi: 10.1016/j.nurx.2006.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg SD, et al. Down regulation of trk but not p75 gene expression in single cholinergic basal forebrain neurons mark the progression of Alzheimer's disease. J Neurochem. 2006b;97:475–487. doi: 10.1111/j.1471-4159.2006.03764.x. [DOI] [PubMed] [Google Scholar]
- Ginsberg SD, et al. Single cell gene expression analysis: implications for neurodegenerative and neuropsychiatric disorders. Neurochem Res. 2004;29:1054–1065. doi: 10.1023/b:nere.0000023593.77052.f7. [DOI] [PubMed] [Google Scholar]
- Ginsberg SD, et al. Expression profile of transcripts in Alzheimer's disease tangle-bearing CA1 neurons. Ann Neurol. 2000;48:77–87. [PubMed] [Google Scholar]
- Ginsberg SD, Mirnics K. Functional genomic methodologies. Prog Brain Res. 2006;158:15–40. doi: 10.1016/S0079-6123(06)58002-1. [DOI] [PubMed] [Google Scholar]
- Ginsberg SD, et al. GluR3 is the major AMPA glutamate receptor of oxytocinergic magnocellular neurons and is localized at synapses. Neuroscience. 1995;65:563–575. doi: 10.1016/0306-4522(94)00513-5. [DOI] [PubMed] [Google Scholar]
- Goedert M, et al. Protein phosphatase 2A is the major enzyme in brain that dephosphorylates tau protein phosphorylated by proline-directed protein kinases or cyclic AMP-dependent protein kinase. J Neurochem. 1995;65:2804–2807. doi: 10.1046/j.1471-4159.1995.65062804.x. [DOI] [PubMed] [Google Scholar]
- Gotz J, et al. Animal models reveal role for tau phosphorylation in human disease. Biochim Biophys Acta. 2010;1802:860–871. doi: 10.1016/j.bbadis.2009.09.008. [DOI] [PubMed] [Google Scholar]
- Gutala RV, Reddy PH. The use of real-time PCR analysis in a gene expression study of Alzheimer's disease post-mortem brains. J Neurosci Methods. 2004;132:101–107. doi: 10.1016/j.jneumeth.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Heffernan JM, et al. Temporal cortex synaptophysin mRNA is reduced in Alzheimer's disease and is negatively correlated with the severity of dementia. Exp Neurol. 1998;150:235–239. doi: 10.1006/exnr.1997.6772. [DOI] [PubMed] [Google Scholar]
- Heinonen O, et al. Loss of synaptophysin-like immunoreactivity in the hippocampal formation is an early phenomenon in Alzheimer's disease. Neuroscience. 1995;64:375–384. doi: 10.1016/0306-4522(94)00422-2. [DOI] [PubMed] [Google Scholar]
- Hyman BT, et al. Memory-related neural systems in Alzheimer's disease: an anatomic study. Neurology. 1990;40:1721–1730. doi: 10.1212/wnl.40.11.1721. [DOI] [PubMed] [Google Scholar]
- Hyman BT, et al. Alzheimer's disease: cell-specific pathology isolates the hippocampal formation. Science. 1984;225:1168–1170. doi: 10.1126/science.6474172. [DOI] [PubMed] [Google Scholar]
- Ikonomovic MD, et al. AMPA-selective glutamate receptor subtype immunoreactivity in the hippocampal formation of patients with Alzheimer's disease. Hippocampus. 1995;5:469–86. doi: 10.1002/hipo.450050509. [DOI] [PubMed] [Google Scholar]
- Imbimbo BP, Giardina GA. Gamma-secretase inhibitors and modulators for the treatment of Alzheimer's disease: disappointments and hopes. Curr Top Med Chem. 2011;11:1555–1570. doi: 10.2174/156802611795860942. [DOI] [PubMed] [Google Scholar]
- Jiang Y, et al. Alzheimer's-related endosome dysfunction in Down syndrome is A{beta}-independent but requires APP and is reversed by BACE-1 inhibition. Proc Natl Acad Sci USA. 2010;107:1630–1635. doi: 10.1073/pnas.0908953107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jicha GA, et al. A conformation- and phosphorylation-dependent antibody recognizing the paired helical filaments of Alzheimer's disease. J Neurochem. 1997;69:2087–2095. doi: 10.1046/j.1471-4159.1997.69052087.x. [DOI] [PubMed] [Google Scholar]
- Kordower JH, et al. Loss and atrophy of layer II entorhinal cortex neurons in elderly people with mild cognitive impairment. Ann Neurol. 2001;49:202–213. [PubMed] [Google Scholar]
- Lee VMY, et al. Monoclonal antibodies distinguish several differentially phosphorylated states of the two largest rat neurofilament subunits (NF-H and NF-M) and demonstrate their existence in the normal nervous system of adult rats. J Neurosci. 1987;7:3474–3488. doi: 10.1523/JNEUROSCI.07-11-03474.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang B, et al. Calpain activation promotes BACE1 expression, amyloid precursor protein processing, and amyloid plaque formation in a transgenic mouse model of Alzheimer disease. J Biol Chem. 2010;285:27737–27744. doi: 10.1074/jbc.M110.117960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, et al. Contributions of protein phosphatases PP1, PP2A, PP2B and PP5 to the regulation of tau phosphorylation. Eur J Neurosci. 2005;22:1942–50. doi: 10.1111/j.1460-9568.2005.04391.x. [DOI] [PubMed] [Google Scholar]
- Longo FM, et al. Small molecule neurotrophin receptor ligands: novel strategies for targeting Alzheimer's disease mechanisms. Curr Alzheimer Res. 2007;4:503–506. doi: 10.2174/156720507783018316. [DOI] [PubMed] [Google Scholar]
- Luebke JI, et al. Dendritic vulnerability in neurodegenerative disease: insights from analyses of cortical pyramidal neurons in transgenic mouse models. Brain Struct Funct. 2010;214:181–199. doi: 10.1007/s00429-010-0244-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masliah E, et al. Immunoelectron microscopic study of synaptic pathology in Alzheimer's disease. Acta Neuropathol. 1991;81:428–433. doi: 10.1007/BF00293464. [DOI] [PubMed] [Google Scholar]
- Mondragon-Rodriguez S, et al. Cleavage and conformational changes of tau protein follow phosphorylation during Alzheimer's disease. Int J Exp Pathol. 2008;89:81–90. doi: 10.1111/j.1365-2613.2007.00568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mufson EJ, et al. Neuronal gene expression profiling: uncovering the molecular biology of neurodegenerative disease. Prog Brain Res. 2006;158:197–222. doi: 10.1016/S0079-6123(06)58010-0. [DOI] [PubMed] [Google Scholar]
- Mufson EJ, et al. Single cell gene expression profiles of nucleus basalis cholinergic neurons in Alzheimer's disease. Neurochem Res. 2002;27:1035–1048. doi: 10.1023/a:1020952704398. [DOI] [PubMed] [Google Scholar]
- Nixon RA. The calpains in aging and aging-related diseases. Ageing Res Rev. 2003;2:407–418. doi: 10.1016/s1568-1637(03)00029-1. [DOI] [PubMed] [Google Scholar]
- Noble W, et al. Minocycline reduces the development of abnormal tau species in models of Alzheimer's disease. Faseb J. 2009;23:739–750. doi: 10.1096/fj.08-113795. [DOI] [PubMed] [Google Scholar]
- Pei JJ, et al. Expression of protein phosphatases (PP-1, PP-2A, PP-2B and PTP-1B) and protein kinases (MAP kinase and P34cdc2) in the hippocampus of patients with Alzheimer disease and normal aged individuals. Brain Res. 1994;655:70–6. doi: 10.1016/0006-8993(94)91598-9. [DOI] [PubMed] [Google Scholar]
- Polydoro M, et al. Age-dependent impairment of cognitive and synaptic function in the htau mouse model of tau pathology. J Neurosci. 2009;29:10741–10749. doi: 10.1523/JNEUROSCI.1065-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor DT, et al. Reduction in post-synaptic scaffolding PSD-95 and SAP-102 protein levels in the Alzheimer inferior temporal cortex is correlated with disease pathology. J Alzheimers Dis. 2010;21:795–811. doi: 10.3233/JAD-2010-100090. [DOI] [PubMed] [Google Scholar]
- Rajagopal R, et al. Transactivation of Trk neurotrophin receptors by G-protein-coupled receptor ligands occurs on intracellular membranes. J Neurosci. 2004;24:6650–6658. doi: 10.1523/JNEUROSCI.0010-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao MV, et al. Marked calpastatin (CAST) depletion in Alzheimer's disease accelerates cytoskeleton disruption and neurodegeneration: neuroprotection by CAST overexpression. J Neurosci. 2008;28:12241–12254. doi: 10.1523/JNEUROSCI.4119-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner A, et al. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003;19:368–375. doi: 10.1093/bioinformatics/btf877. [DOI] [PubMed] [Google Scholar]
- Rocher AB, et al. Structural and functional changes in tau mutant mice neurons are not linked to the presence of NFTs. Exp Neurol. 2010;223:385–393. doi: 10.1016/j.expneurol.2009.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambamurti K, et al. Targets for AD treatment: conflicting messages from gamma-secretase inhibitors. J Neurochem. 2011;117:359–374. doi: 10.1111/j.1471-4159.2011.07213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schor NF. What the halted phase III gamma-secretase inhibitor trial may (or may not) be telling us. Ann Neurol. 2011;69:237–239. doi: 10.1002/ana.22365. [DOI] [PubMed] [Google Scholar]
- Skaper SD. The biology of neurotrophins, signalling pathways, and functional peptide mimetics of neurotrophins and their receptors. CNS Neurol Disord Drug Targets. 2008;7:46–62. doi: 10.2174/187152708783885174. [DOI] [PubMed] [Google Scholar]
- Small SA, Duff K. Linking Abeta and tau in late-onset Alzheimer's disease: a dual pathway hypothesis. Neuron. 2008;60:534–542. doi: 10.1016/j.neuron.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontag E, et al. Altered expression levels of the protein phosphatase 2A ABalphaC enzyme are associated with Alzheimer disease pathology. J Neuropathol Exp Neurol. 2004;63:287–301. doi: 10.1093/jnen/63.4.287. [DOI] [PubMed] [Google Scholar]
- Steward O, et al. Synaptic activation causes the mRNA for the IEG Arc to localize selectively near activated postsynaptic sites on dendrites. Neuron. 1998;21:741–751. doi: 10.1016/s0896-6273(00)80591-7. [DOI] [PubMed] [Google Scholar]
- Swanger SA, Bassell GJ. Making and breaking synapses through local mRNA regulation. Curr Opin Genet Dev. 2011 doi: 10.1016/j.gde.2011.04.002. in press Epub 5/3/2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze CI, et al. Loss of the presynaptic vesicle protein synaptophysin in hippocampus correlates with cognitive decline in Alzheimer disease. J Neuropathol Exp Neurol. 1997;56:933–944. doi: 10.1097/00005072-199708000-00011. [DOI] [PubMed] [Google Scholar]
- Thies E, Mandelkow EM. Missorting of tau in neurons causes degeneration of synapses that can be rescued by the kinase MARK2/Par-1. J Neurosci. 2007;27:2896–2907. doi: 10.1523/JNEUROSCI.4674-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thies W, Bleiler L. 2011 Alzheimer's disease facts and figures. Alzheimers Dement. 2011;7:208–244. doi: 10.1016/j.jalz.2011.02.004. [DOI] [PubMed] [Google Scholar]
- Vogelsberg-Ragaglia V, et al. PP2A mRNA expression is quantitatively decreased in Alzheimer's disease hippocampus. Exp Neurol. 2001;168:402–12. doi: 10.1006/exnr.2001.7630. [DOI] [PubMed] [Google Scholar]
- Yasuda RP, et al. Reduction of AMPA-selective glutamate receptor subunits in the entorhinal cortex of patients with Alzheimer's disease pathology: a biochemical study. Brain Res. 1995;678:161–167. doi: 10.1016/0006-8993(95)00178-s. [DOI] [PubMed] [Google Scholar]
- Yuan A, et al. Axonal transport rates in vivo are unaffected by tau deletion or overexpression in mice. J Neurosci. 2008;28:1682–1687. doi: 10.1523/JNEUROSCI.5242-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zempel H, et al. A{beta} oligomers cause localized Ca2+ elevation, missorting of endogenous tau into dendrites, tau phosphorylation, and destruction of microtubules and spines. J Neurosci. 2010;30:11938–11950. doi: 10.1523/JNEUROSCI.2357-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]