Abstract
Serum N-glycan profiling for use as clinical biomarkers of disease(s) is of increasing scientific interest. Promising profiles have already been identified in several diseases including cancer, Alzheimer’s, and diabetes. Venipuncture is routinely performed to collect the blood necessary for this type of analysis, but blood from a fingerstick placed on filter paper known as dried blood spots (DBS) is more advantageous. This sampling method is less invasive than ‘classical’ blood drawing, can be performed conveniently at home and avoids cumbersome shipping and storage procedures. Here we present a procedure for N-glycan profiling of DBS samples consisting of reconstitution of DBS in N-glycan release buffer, protein denaturation, enzymatic N-glycan release and PGC Solid phase extraction (SPE) for purification. Samples are then analyzed using nHPLC-PGC-chip-TOF-MS to generate N-glycan profiles. Using this method, around 150 N-glycan structures can be monitored, originating from 44 N-glycan compositions that can be analyzed with good repeatability (%CV around 20%). To assess the stability of the N-glycans during storage, DBS samples were stored at RT and −80°C. No major differences in N-glycan composition could be observed. Moreover, upon comparison of the N-glycan profile of DBS with profiles obtained from serum, which is a classical matrix for N-glycan profiling, similar patterns were observed. The method will facilitate large population studies for N-glycan profiling, and will be especially advantageous for children and the elderly, who have limited blood supplies, as well as animal studies in small mammals.
INTRODUCTION
Over the last five years the development of high-throughput N-glycan profiling methods has enabled researchers to pursue the role of glycomics as potential biomarkers of disease.1–4 Alterations in N-glycan profiles from blood have been observed in cancer (e.g. breast,5, 6 ovarian,7, 8 pancreatic,9, 10 and lung,11), liver cirrhosis and fibrosis.12–14 In addition, large-scale population studies have evaluated the role of aging, sex, smoking behavior and other environmental factors on blood protein glycosylation.15–17 Although collections of blood samples are associated with minimal risk to subjects, inconveniences including the requirement of trained phlebotomists, time and temperature sensitive blood processing, restricted shipping protocols and the need for secure refrigerated storage facilities exist. Alternative approaches such as the analysis of dried blood spots (DBS) –drops of whole blood collected and dried on filter paper, is highly attractive. DBS was first developed in the 1960s, to detect phenylketonuria in newborns.18 Today, DBS specimens are collected from neonates during their first few days of life to allow screening for a series of genetic inborn errors.19 While the use of dried blood spots is widely applied in neonatal screening programs, the use of this technique in population-level research is relatively new as a result of improvement in mass spectrometric assays. For example, the application of DBS in Therapeutic Drug Monitoring is gaining popularity (e.g.20–22).
There are several advantages of DBS:23 1. A finger prick is much less invasive than a regular blood drawing, and can be performed at home, without the need for a medically trained individual. 2. Only a small amount of blood is necessary, which is advantageous for research involving small mammals, children and the elderly. 3. Storage and shipping of DBS specimens is relatively easy. As the analytes are stabilized on the matrix, and dry, no cold transportation is necessary. Moreover, samples have been reported to remain stable at −80°C for long periods of time. For these reasons DBS specimens are increasingly being utilized in biomarker studies. Thus, a method for the analysis of N-glycan patterns from DBS would be valuable.
Current assays for glycosylation profiling comprise of LC or CE based separations, coupled to mass spectrometric or fluorometric analyte detection. The use of HILIC-HPLC with fluorescence detection for the analysis of derivatized glycans is widely accepted,11, 16, 17, 24 but several studies using CGE-LIF have also been conducted.14, 15, 25 We recently developed a strategy to analyze native N-glycans using PGC-LC-MS, resulting in the unambiguous identification of over 100 N-glycan compositions and around 300 N-glycan species from serum.26 Below we describe how we applied this methodology to DBS to achieve a comprehensive profiling of N-glycosylation patterns. The procedure involved reconstitution of DBS in N-glycan release buffer, protein denaturation, enzymatic N-glycan release and PGC Solid phase extraction (SPE) for purification (Figure 1). Samples are then analyzed using nHPLC-PGC-chip-TOF-MS to generate N-glycan profiles. The repeatability of the method is assessed, as well as the stability of the N-glycans during storage. Moreover, the N-glycan profile of DBS is compared to profiles obtained from serum, which is a classical matrix for N-glycan profiling.
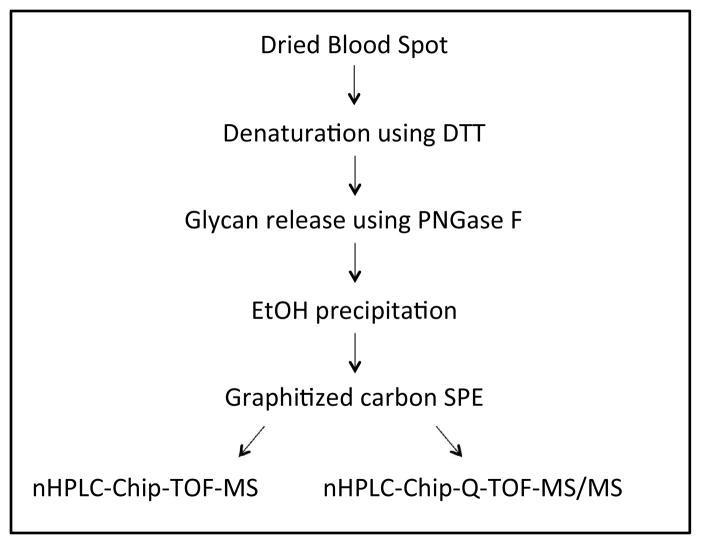
Figure 1.
Schematic overview of the procedure for N-glycan analysis of dried blood spots.
EXPERIMENTAL SECTION
Dried Blood Spots
Filter paper was obtained from Whatman/GE Healthcare (903 Protein saver card, Piscataway, NJ). Blood spots from a healthy volunteer were obtained according to standard procedures.23 In short, a finger was cleaned with 70% isopropyl alcohol (Fisher Scientific, Waltham, MA) and subsequently pricked with a lancet, similar to the ones used for monitoring blood glucose levels. The first drop of blood was removed using a sterile wipe, and subsequent drops were collected on the filter paper without the finger touching the paper. Upon air-drying overnight, the sample cards were stored at −80°C in a closed plastic bag with desiccant.
N-glycan release from a Dried Blood Spot
A 3.1 mm disk was punched out of the middle of a dried blood spot using a manual puncher (Whatman/GE healthcare) and transferred to an Eppendorf tube. Upon addition of 100 μL of 100mM ammonium bicarbonate (Sigma-Aldrich, St. Louis, MO) with 5 mM dithiothreitol (DTT, Promega, Madison, WI) in water, the samples were vortexed and subsequently sonicated for 5 minutes. Proteins in the samples were denatured using six cycles alternating between 100°C and room temperature (RT) for 10 seconds each. 2 μL of PNGaseF (New England Biolabs, Ipswich, MA) was added to the samples, and enzymatic glycan release was performed in a CEM (Matthews, NC) microwave at 20W for 10 min.27 Deglycosylated proteins were precipitated using 400 μL of ice-cold ethanol, and the samples were chilled at −80°C for 1 hour. Upon centrifugation, the supernatant was transferred to new eppendorf tubes, and brought to dryness in vacuo.
N-glycan release from a serum sample
50 μL of human standard serum (Sigma-Aldrich) was mixed together with 50 μL of 200mM ammonium bicarbonate with 10 mM DTT in water. Upon vortexing, the proteins were denatured and the glycans were released. The proteins were precipitated as described for dried blood spots.
N-glycan purification using graphitized carbon SPE
Oligosaccharides released by PNGaseF were purified using graphitized carbon SPE (Grace, Deerfield, IL).2, 26, 28 Briefly, cartridges were conditioned using 4 mL of 80% ACN containing 0.05% TFA. (EMD chemicals, Gibbstown, NJ), followed by 4 mL of water containing 0.05% TFA. Oligosaccharide samples were reconstituted in 500 mL of water and subsequently loaded onto the cartridges. Cartridges were washed using 3 × 4 mL of water and N-glycans were eluted using 4 mL of 40% ACN containing 0.05% TFA. Samples were dried in vacuo prior to analysis.
nHPLC-chip-TOF-MS analysis
N-glycans were analyzed using an Agilent (Santa Clara, CA) 6200 series nanoHPLC-chip-TOF-MS, consisting of an autosampler, which was maintained at 8°C, a capillary loading pump, a nanopump, HPLC-chip-MS interface and an Agilent 6210 Time Of Flight mass spectrometer. The chip (glycan chip II, Agilent) contained a 9 × 0.075 mm i.d. enrichment column coupled to a 43 × 0.075 mm i.d. analytical column; both packed with 5 μm porous graphitized carbon (PGC). N-glycan samples were reconstituted in 45 μL of water and diluted 1:5 with water prior to analysis; 1 μL of sample was used for injection. Upon injection, the sample was loaded onto the enrichment column using 3% ACN containing 0.1% Formic acid (FA, Fluka, St. Louis, MO). After the analytical column was switched in-line, the nano-pump delivered a gradient of 3%ACN with 0.1% FA (solvent A) and 90% ACN with 0.1% FA (solvent B).
Data analysis
Data analysis was performed using Masshunter® qualitative analysis (version B.03.01, Agilent) and Microsoft® Excel® for Mac 2011 (version 14.1.3, Microsoft), according to Hua et al.26 with modifications. Data was loaded into Masshunter qualitative analysis, and glycan features were identified and integrated using the Molecular Feature Extractor algorithm. First, signals above a signal to noise threshold of 5.0 were considered. Then, signals are deconvoluted using a tolerance of 0.0025 m/z ± 10 ppm. The resulting deconvoluted masses are subsequently annotated using a retrosynthetic theoretical glycan library29, where a 15 ppm mass error was allowed. Glycan compositions, retention times and volume were exported to csv-format for further evaluation.
nHPLC-chip-Q-TOF-MS/MS analysis
To allow unambiguous identification of N-glycans, samples were analyzed using an Agilent 6200 series nanoHPLC-chip-Q-TOF-MS/MS, consisting of an autosampler, which was maintained at 8°C, a capillary loading pump, a nanopump, HPLC-chip-MS interface and an Agilent 6520 Quadrupole-Time Of Flight mass spectrometer. The chip and gradient were the same as for the nHPLC-chip-TOF-MS analysis. N-glycans were subjected to collision-induced fragmentation with nitrogen as the collision gas using a series of collision energies that were dependent on the m/z values of the N-glycans. The collision energies correspond to voltages (Vcollision) that were based on the equation: Vcollision = m/z (1.8/100 Da) Volts − 2.4 V, where the slope and offset of the voltages were set at 1.8/100 Da and −2.4, respectively.
RESULTS
A procedure (see Figure 1) is developed for the analysis of N-glycan profiles from dried blood spots using enzymatic N-glycan release followed by protein precipitation using ethanol and graphitized carbon SPE purification. Oligosaccharide samples are then analyzed using nLC-PGC-chip-TOF-MS to obtain isomer specific N-glycan profiles. No derivatization (either oligosaccharide reduction, permethylation or labeling) was performed, as such modifications may result in un-wanted side products and loss of resolving power during PGC separation. All profiles were obtained in the positive ionization mode.
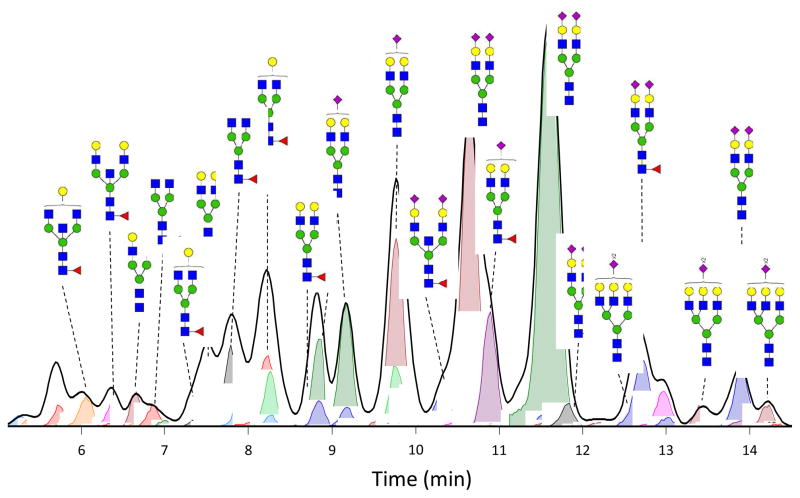
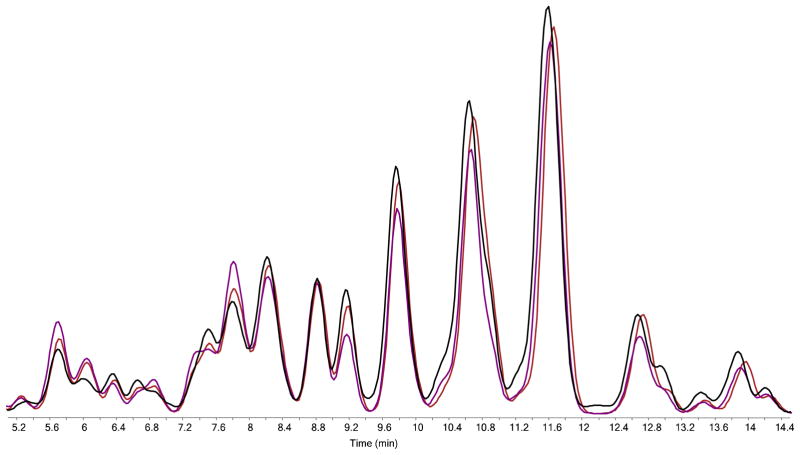
In Figure 2, an nHPLC-PGC-chip-TOF-MS Extracted compound chromatogram from N-glycans derived from DBS is depicted. Approximately 150 glycan compounds were identified and integrated from each DBS sample, relative to around 330 compounds usually obtained from 50 μL of serum. Fewer compounds are detected from DBS specimens, as the amount of serum in the small (3mm) disk is much less (around 2 μL) than the 50 μL used in serum analysis. Here it has to be noted that the analysis is performed on non-reduced glycans, resulting in separation of the α and β anomers. The actual number of linkage isomers detected may, therefore, have to be less. From the 150 N-glycan compounds detected in the DBS samples, 44 glycan compositions could be identified and integrated reproducibly using Masshunter®, as shown in Table 1. Of these compositions, 7 are high mannose type and 37 are hybrid or complex type; 15 are fucosylated and 19 are sialylated.
Figure 2. Extracted N-glycan chromatogram and extracted Compound Chromatograms of N-glycans derived from dried blood spots.
Extracted N-glycan chromatogram (thick black line) as obtained using Masshunter® software, depicts the total ion intensity for all N-glycan compounds observed. Extracted compound chromatograms for each of the 150 N-glycan compositions are automatically extracted (colored lines with integrals, each peak is a different compound) and the most intense signals are annotated with their respective N-glycan composition. Key: red triangle: fucose, blue square: N-acetylglucosamine, green circle: mannose, yellow circle: galactose, purple diamond: sialic acid (NeuAc).
Table 1. Glycan composition from DBS.
Compositions and observed masses of the glycans identified and integrated by Masshunter software are listed. Since Masshunter automatically deconvolutes the mass spectra, the observed monoisotopic molecular masses are given. Relative abundance percentages were calculated in Excel from the volumes obtained from Masshunter.
| No. | Compositiona | Accurate mass | Observed mass | Relative abundance in DBS (%) |
|---|---|---|---|---|
| 1 | H3N2 | 910.328 | 910.326 | 0.13 |
| 2 | H3N3 | 1113.407 | 1113.408 | 0.32 |
| 3 | H3N3F1 | 1259.465 | 1259.464 | 0.23 |
| 4 | H3N4 | 1316.487 | 1316.488 | 0.12 |
| 5 | H3N4F1 | 1462.544 | 1462.546 | 3.10 |
| 6 | H3N5 | 1519.566 | 1519.572 | 0.04 |
| 7 | H3N5F1 | 1665.624 | 1665.631 | 0.92 |
| 8 | H4N2 | 1072.381 | 1072.383 | 0.05 |
| 9 | H4N3 | 1275.460 | 1275.460 | 0.97 |
| 10 | H4N3S1 | 1566.555 | 1566.555 | 1.73 |
| 11 | H4N3F1 | 1421.518 | 1421.517 | 0.09 |
| 12 | H4N4 | 1478.539 | 1478.538 | 0.29 |
| 13 | H4N4S1 | 1769.635 | 1769.634 | 0.38 |
| 14 | H4N4F1 | 1624.597 | 1624.606 | 4.49 |
| 15 | H4N4F1S1 | 1915.693 | 1915.700 | 0.05 |
| 16 | H4N5 | 1681.619 | 1681.621 | 0.13 |
| 17 | H4N5S1 | 1972.714 | 1972.756 | 0.13 |
| 18 | H4N5F1 | 1827.677 | 1827.69 | 1.42 |
| 19 | H5N2 | 1234.433 | 1234.434 | 0.34 |
| 20 | H5N3 | 1437.513 | 1437.509 | 0.09 |
| 21 | H5N3S1 | 1728.608 | 1728.615 | 0.20 |
| 22 | H5N4 | 1640.592 | 1640.599 | 4.03 |
| 23 | H5N4S1 | 1931.688 | 1931.685 | 18.24 |
| 24 | H5N4S2 | 2222.783 | 2222.790 | 34.64 |
| 25 | H5N4F1 | 1786.650 | 1786.652 | 3.01 |
| 26 | H5N4F1S1 | 2077.746 | 2077.756 | 5.73 |
| 27 | H5N4F1S2 | 2368.841 | 2368.843 | 3.68 |
| 28 | H5N5 | 1843.672 | 1843.682 | 0.33 |
| 29 | H5N5S1 | 2134.767 | 2134.766 | 0.69 |
| 30 | H5N5S2 | 2425.862 | 2425.845 | 0.02 |
| 31 | H5N5F1 | 1989.729 | 1989.740 | 1.07 |
| 32 | H5N5F1S1 | 2280.825 | 2280.835 | 2.96 |
| 33 | H5N5F1S2 | 2571.920 | 2571.932 | 1.79 |
| 34 | H6N2 | 1396.486 | 1396.485 | 0.52 |
| 35 | H6N3 | 1599.566 | 1599.566 | 0.08 |
| 36 | H6N5S1 | 2296.820 | 2296.822 | 1.11 |
| 37 | H6N5S2 | 2587.915 | 2587.927 | 3.49 |
| 38 | H6N5F1S2 | 2733.973 | 2733.986 | 0.84 |
| 39 | H6N5F1S3 | 3025.069 | 3025.076 | 0.71 |
| 40 | H7N6S2 | 2953.047 | 2953.052 | 0.15 |
| 41 | H7N6S4 | 3535.238 | 3535.248 | 0.18 |
| 42 | H7N2 | 1558.539 | 1558.543 | 0.35 |
| 43 | H8N2 | 1720.592 | 1720.602 | 0.56 |
| 44 | H9N2 | 1882.645 | 1882.655 | 0.62 |
Key: H = hexose, N = N-acetylhexosamine, F = fucose, S = sialic acid (NeuAc).
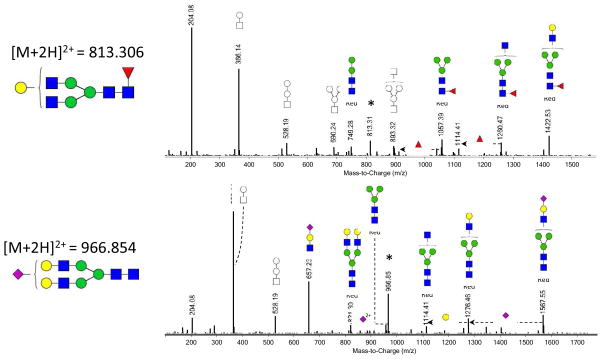
To confirm the glycan compositions determined by accurate m/z signals, fragmentation data was obtained from one DBS sample using nLC-PGC-chip-Q-TOF MS/MS. Fragmentation spectra of two randomly chosen N-glycans (H3N4F1 at m/z 813.306 and H5N4S1 at m/z 966.854) are depicted in Figure 3. Clear characteristic N-glycan fragments were observed (m/z 366, 528 and 657), confirming the N-glycan annotation.
Figure 3. Fragmentation spectra from N-glycans derived from dried blood spots.
CID fragmentation of N-glycans from DBS obtained from nHPLC-PGC-chip-Q-TOF-MS. * indicates the parent ion. Key: square: N-acetylhexosamine, circle: hexose, red triangle: fucose, blue square: N-acetylglucosamine, green circle: mannose, yellow circle: galactose, purple diamond: sialic acid, Red: reducing end. The structures represented in color have the reducing end present, facilitating their annotation. When the reducing end is not present, it is not possible to distinguish between galactose and mannose residues, therefore, these structures are represented with symbols only.
Effects of collection matrix on N-glycosylation profile
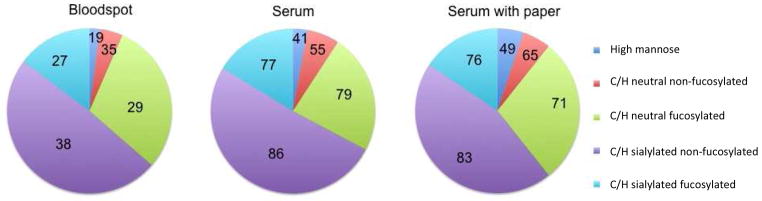
To evaluate the effects of the collection matrix -cellulose-based neonatal screening cards- N-glycosylation profiles obtained from 50 μL of standard serum with and without the addition of a full disc (10 mm diameter) of collection paper. Figure 4 shows that no significant changes were obtained between the samples with and without the collection paper, both in numbers of compounds determined (344 vs 338, respectively) and compositions.
Figure 4. Comparison of N-glycan pattern from serum with and without collection material and dried blood spots.
Pie charts from a dried blood spot and serum samples with and without collection material. Chart areas represent the relative abundance of N-glycan classes, while the numbers represent the number of glycan compounds identified and integrated for each class. Class information is given in terms of high mannose type glycans (high mannose), complex/hybrid neutral non-fucosylated glycans, complex/hybrid neutral fucosylated glycans, complex/hybrid sialylated non-fucosylated glycans and complex/hybrid sialylated and fucosylated glycans.
Comparison between DBS and serum
Current research on glycan-based biomarker discovery is primarily conducted on serum. To determine if our DBS profile compares favorably with serum, we analyzed commercial serum. Pie charts of the compositional analysis of DBS and serum are depicted in Figure 4. Overall, the N-glycan pattern is very similar between the two samples, but a slightly lower amount of high-mannose glycans was observed in the DBS. The DBS and serum, are, however, not obtained from the same individual, and as serum N-glycosylation patterns show high inter-individual variation,30 the difference observed here could well be caused by inter-individual differences.
Repeatability
Measurement variability was evaluated using one DBS that was prepared in triplicate according to the procedure described. All three samples were analyzed using nLC-PGC-chip-TOF-MS on 3 consecutive days. Data analysis was performed as described. The average %CV of the measurement variability was obtained by comparing the 3 daily analyses. Glycan intensities of the 44 glycan compounds consistently observed in all samples (see Table 1) were used to calculate % CV and all individual glycan CVs were averaged to generate the average %CV of the measurement variability, which was 18.8%.
To evaluate the repeatability of the proposed procedure, 3 dried blood spots were collected from the same individual on 3 consecutive days. Blood spots were allowed to dry overnight and N-glycan release was performed the next day. Samples were analyzed using nLC-PGC-chip-TOF-MS and data analysis was performed as previously described. An overlay of the extracted glycan chromatograms obtained from the three analyses on day 1 is depicted in Figure 5. The average within-day %CV was obtained by comparing the analysis of the 3 samples within one day and was 20.5%; the average inter-day %CV was obtained by comparing differences in the analysis of the 3 × 3 samples in-between days and was 20.8%.
Figure 5. Intra-day variability of the analysis of N-glycans from dried blood spots.
Three DBS collected from the same individual on the same day were prepared and analyzed on the same day. Extracted N-glycan chromatograms of the three analyses are overlaid to depict the intra-day variability of the analysis
Storage of Dried Blood Specimens
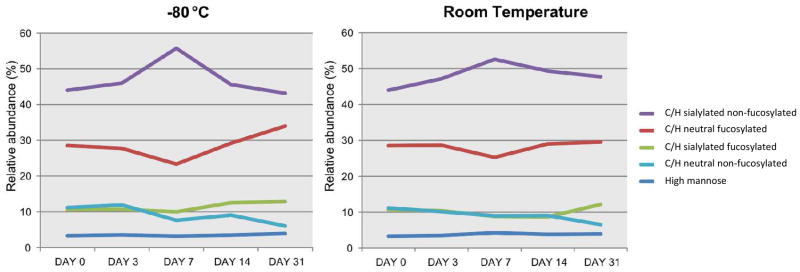
It is generally acknowledged that DBS specimens are a good replacement for serum specimens, with the additional advantage of less degradation at ambient temperatures. To evaluate the degradation of N-glycans during storage, 18 dried blood spots were collected from the same individual at the same time point. Samples were allowed to dry overnight and N-glycans were released the next day (Day 0) or stored in a plastic bag with desiccant at RT (25°C, 8 specimens) or −80°C (8 specimens). N-glycan profiles (n=2) were obtained after 3, 7, 14 and 31 days of storage. Figure 6 reveals that no significant difference was observed between storage at RT and storage at −80°C. Also no difference was observed between day 0 and day 31.
Figure 6. Stability of N-glycans in Dried blood spots during storage.
Dried blood spots were stored either at room temperature or at −80°C. N-glycan profiles (n=2) were obtained on days 0, 3, 7, 14 and 31 after sample collection. Average relative percentage is depicted for each glycan type. For key, see Figure 4.
CONCLUSIONS
This study describes a new procedure for the analysis of N-glycosylation patterns from dried blood spots. Around 150 N-glycan compounds can be identified from DBS samples using this approach, originating from 44 N-glycan compositions. No interference from the collection matrix could be observed. Relative quantitation of the 44 compositions resulted in a %CV of 20.8%, which is similar to serum. Furthermore we showed that storage of DBS at RT or at −80°C for at least one month does not affect N-glycosylation patterns.
The use of DBS technology for the collection of blood-derived specimens has many advantages compared to venous blood drawing, which is the current clinical standard. First, sample collection is less invasive and more convenient for the patient. Only a finger prick is required, which is routinely used by diabetic patients, and the sample can be obtained at home on the patients time schedule. Second, lower volumes are needed. Serum is usually collected in containers of at least 4mL; a drop of blood is approximately 50 μL and is sufficient for a high quality DBS specimen. Third, there is no need for immediate processing of the sample. Serum must be allowed to clot for a minimum of 60 minutes, then centrifuged, separated, aliquoted and frozen. DBS specimens are air-dried prior to RT storage.
These advantages make the use of DBS specimens especially suitable in specific cases: It will be easier to collect DBS specimens from children and elderly, as venipuncture is frequently problematic in these vulnerable patients. Application of DBS specimens in larger scale population studies will most likely result in better participation numbers. A last group of studies that will benefit of the use of DBS are small mammal studies; currently larger amounts of blood are needed, which is not required for DBS analysis.
N-glycans are complex structures, comprising several different monosaccharides that can be attached in several different ways. Not only can monosaccharides be linked to different sugars (topoisomers, e.g. triantennary GlcNAc or bisecting GlcNAc), but the monosaccharides may also be linked at different positions (linkage isomers, e.g. α2–3 sialic acid versus α2–6 sialic acid). Over the years several strategies have been employed for glycan analysis, comprising chromatographic or electromigrative separations as well as mass spectrometry. Recently, especially PGC has been recognized as a good stationary phase for HPLC separation of glycans, due to its efficient separation of both topoisomers as well as linkage isomers.2, 31, 32 Due to the different isomers, the most complete glycan profiles can only be obtained using a combination of separation (here PGC) and mass spectrometry.24, 31
With the recent interest for the use of DBS specimens in therapeutic drug monitoring, research is initiated towards optimization of sampling and processing procedures. New, coated collection cards have been developed, which precipitate proteins (http://www.whatman.com/DMPK.aspx). The use of these cards for DBS glycomics analysis might be beneficial, however it is currently unclear whether these cards would be compatible with mass spectrometry based studies. Moreover, efforts have been made to automate DBS punching, or even omit punching of DBS. Some manufacturers have developed instruments for controlled DBS punching or immediate extraction (e.g.33). Such techniques might also be adapted for automated sample preparation for N-glycan analysis from DBS.
Recently, the use of paper spray ionization for direct analysis of metabolites from dried blood spots has been introduced.34 Several molecules, including epinephrine, serine, cocaine and imatinib (a small molecule tyrosine kinase used for the treatment of cancer), have been analyzed by paper spray mass spectrometry using blood spiked with drugs or metabolites.35, 36 Even though N-glycans need to be released from their proteins, and chromatographic separation is necessary to separate isomeric N-glycans, application of such an approach may be a fast way to obtain global N-glycan profiles.
In conclusion, we showed that the analysis of N-glycan profiles from Dried blood spots is feasible with good repeatability. It is expected that application of the described technology will facilitate further research towards N-glycan biology in children, elderly, large population studies as well as animal models. Our next steps are to evaluate a large number of serum and DSBs collected from the healthy volunteers and subjects with cancer to confirm our findings.
Acknowledgments
Funds provided by the NIH (R21CA135240, HD061923 and HD059127) and DOD (W81XWH1010635) are gratefully acknowledged.
References
- 1.Packer NH, von der Lieth CW, Aoki-Kinoshita KF, Lebrilla CB, Paulson JC, Raman R, Rudd P, Sasisekharan R, Taniguchi N, York WS. Proteomics. 2008;8:8–20. doi: 10.1002/pmic.200700917. [DOI] [PubMed] [Google Scholar]
- 2.Chu CS, Ninonuevo MR, Clowers BH, Perkins PD, An HJ, Yin H, Killeen K, Miyamoto S, Grimm R, Lebrilla CB. Proteomics. 2009;9:1939–1951. doi: 10.1002/pmic.200800249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Royle L, Campbell MP, Radcliffe CM, White DM, Harvey DJ, Abrahams JL, Kim YG, Henry GW, Shadick NA, Weinblatt ME, Lee DM, Rudd PM, Dwek RA. Anal Biochem. 2008;376:1–12. doi: 10.1016/j.ab.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 4.Ruhaak LR, Hennig R, Huhn C, Borowiak M, Dolhain RJ, Deelder AM, Rapp E, Wuhrer M. J Proteome Res. 2010;9:6655–6664. doi: 10.1021/pr100802f. [DOI] [PubMed] [Google Scholar]
- 5.Kirmiz C, Li B, An HJ, Clowers BH, Chew HK, Lam KS, Ferrige A, Alecio R, Borowsky AD, Sulaimon S, Lebrilla CB, Miyamoto S. Mol Cell Proteomics. 2007;6:43–55. doi: 10.1074/mcp.M600171-MCP200. [DOI] [PubMed] [Google Scholar]
- 6.Kyselova Z, Mechref Y, Kang P, Goetz JA, Dobrolecki LE, Sledge GW, Schnaper L, Hickey RJ, Malkas LH, Novotny MV. Clin Chem. 2008;54:1166–1175. doi: 10.1373/clinchem.2007.087148. [DOI] [PubMed] [Google Scholar]
- 7.An HJ, Miyamoto S, Lancaster KS, Kirmiz C, Li B, Lam KS, Leiserowitz GS, Lebrilla CB. J Proteome Res. 2006;5:1626–1635. doi: 10.1021/pr060010k. [DOI] [PubMed] [Google Scholar]
- 8.Saldova R, Royle L, Radcliffe CM, Abd Hamid UM, Evans R, Arnold JN, Banks RE, Hutson R, Harvey DJ, Antrobus R, Petrescu SM, Dwek RA, Rudd PM. Glycobiology. 2007;17:1344–1356. doi: 10.1093/glycob/cwm100. [DOI] [PubMed] [Google Scholar]
- 9.Saldova R, Fan Y, Fitzpatrick JM, Watson RW, Rudd PM. Glycobiology. 2011;21:195–205. doi: 10.1093/glycob/cwq147. [DOI] [PubMed] [Google Scholar]
- 10.Kyselova Z, Mechref Y, Al Bataineh MM, Dobrolecki LE, Hickey RJ, Vinson J, Sweeney CJ, Novotny MV. J Proteome Res. 2007;6:1822–1832. doi: 10.1021/pr060664t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arnold JN, Saldova R, Galligan MC, Murphy TB, Mimura-Kimura Y, Telford JE, Godwin AK, Rudd PM. J Proteome Res. 2011;10:1755–1764. doi: 10.1021/pr101034t. [DOI] [PubMed] [Google Scholar]
- 12.Callewaert N, Van Vlierberghe H, Van Hecke A, Laroy W, Delanghe J, Contreras R. Nat Med. 2004;10:429–434. doi: 10.1038/nm1006. [DOI] [PubMed] [Google Scholar]
- 13.Liu XE, Desmyter L, Gao CF, Laroy W, Dewaele S, Vanhooren V, Wang L, Zhuang H, Callewaert N, Libert C, Contreras R, Chen C. Hepatology. 2007;46:1426–1435. doi: 10.1002/hep.21855. [DOI] [PubMed] [Google Scholar]
- 14.Vanderschaeghe D, Laroy W, Sablon E, Halfon P, Van Hecke A, Delanghe J, Callewaert N. Mol Cell Proteomics. 2009;5:986–949. doi: 10.1074/mcp.M800470-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vanhooren V, Desmyter L, Liu XE, Cardelli M, Franceschi C, Federico A, Libert C, Laroy W, Dewaele S, Contreras R, Chen C. Rejuvenation Res. 2007;10:521–531a. doi: 10.1089/rej.2007.0556. [DOI] [PubMed] [Google Scholar]
- 16.Ruhaak LR, Uh HW, Beekman M, Hokke CH, Westendorp RG, Houwing-Duistermaat J, Wuhrer M, Deelder AM, Slagboom PE. J Proteome Res. 2011;10:1667–1674. doi: 10.1021/pr1009959. [DOI] [PubMed] [Google Scholar]
- 17.Knezevic A, Gornik O, Polasek O, Pucic M, Redzic I, Novokmet M, Rudd PM, Wright AF, Campbell H, Rudan I, Lauc G. Glycobiology. 2010;20:959–969. doi: 10.1093/glycob/cwq051. [DOI] [PubMed] [Google Scholar]
- 18.Guthrie R, Susi A. Pediatrics. 1963;32:338–343. [PubMed] [Google Scholar]
- 19.Levy PA. J Dev Behav Pediatr. 2010;31:622–631. doi: 10.1097/DBP.0b013e3181eedf01. [DOI] [PubMed] [Google Scholar]
- 20.Keevil BG. Clin Biochem. 2011;44:110–118. doi: 10.1016/j.clinbiochem.2010.06.014. [DOI] [PubMed] [Google Scholar]
- 21.Edelbroek PM, van der Heijden J, Stolk LM. Ther Drug Monit. 2009;31:327–336. doi: 10.1097/FTD.0b013e31819e91ce. [DOI] [PubMed] [Google Scholar]
- 22.Millership JS. Paediatr Anaesth. 2011;21:197–205. doi: 10.1111/j.1460-9592.2011.03535.x. [DOI] [PubMed] [Google Scholar]
- 23.McDade TW, Williams S, Snodgrass JJ. Demography. 2007;44:899–925. doi: 10.1353/dem.2007.0038. [DOI] [PubMed] [Google Scholar]
- 24.Ruhaak LR, Zauner G, Huhn C, Bruggink C, Deelder AM, Wuhrer M. Anal Bioanal Chem. 2010;397:3457–3481. doi: 10.1007/s00216-010-3532-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gennaro LA, Salas-Solano O. Anal Chem. 2008;80:3838–3845. doi: 10.1021/ac800152h. [DOI] [PubMed] [Google Scholar]
- 26.Hua S, An HJ, Ozcan S, Ro GS, Soares S, DeVere-White R, Lebrilla CB. Analyst. 2011;136:3663–3671. doi: 10.1039/c1an15093f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kronewitter SR, de Leoz ML, Peacock KS, McBride KR, An HJ, Miyamoto S, Leiserowitz GS, Lebrilla CB. J Proteome Res. 2010;9:4952–4959. doi: 10.1021/pr100202a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Packer NH, Lawson MA, Jardine DR, Redmond JW. Glycoconj J. 1998;15:737–747. doi: 10.1023/a:1006983125913. [DOI] [PubMed] [Google Scholar]
- 29.Kronewitter SR, An HJ, de Leoz ML, Lebrilla CB, Miyamoto S, Leiserowitz GS. Proteomics. 2009;9:2986–2994. doi: 10.1002/pmic.200800760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knezevic A, Polasek O, Gornik O, Rudan I, Campbell H, Hayward C, Wright A, Kolcic I, O’Donoghue N, Bones J, Rudd PM, Lauc G. J Proteome Res. 2009;8:694–701. doi: 10.1021/pr800737u. [DOI] [PubMed] [Google Scholar]
- 31.Pabst M, Altmann F. Proteomics. 2011;11:631–643. doi: 10.1002/pmic.201000517. [DOI] [PubMed] [Google Scholar]
- 32.Ruhaak LR, Deelder AM, Wuhrer M. Anal Bioanal Chem. 2009;394:163–174. doi: 10.1007/s00216-009-2664-5. [DOI] [PubMed] [Google Scholar]
- 33.Kertesz V, Van Berkel GJ. J Mass Spectrom. 2010;45:252–260. doi: 10.1002/jms.1709. [DOI] [PubMed] [Google Scholar]
- 34.Cooks RG, Manicke NE, Dill AL, Ifa DR, Eberlin LS, Costa AB, Wang H, Huang G, Ouyang Z. Faraday Discuss. 2011;149:247–267. doi: 10.1039/c005327a. discussion 333-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu J, Wang H, Manicke NE, Lin JM, Cooks RG, Ouyang Z. Anal Chem. 2010;82:2463–2471. doi: 10.1021/ac902854g. [DOI] [PubMed] [Google Scholar]
- 36.Wang H, Liu J, Cooks RG, Ouyang Z. Angew Chem Int Ed Engl. 2010;49:877–880. doi: 10.1002/anie.200906314. [DOI] [PubMed] [Google Scholar]