Abstract
Cbln1 is the prototype of a family of secreted neuronal glycoproteins (Cbln1-4) and its genetic elimination results in synaptic alterations in cerebellum and striatum. In cerebellum, Cbln1 acts as a bi-functional ligand bridging pre-synaptic β-neurexins on granule cells to post-synaptic Grid2 on Purkinje neurons. Although much is known concerning the action of Cbln1, little is known of the function of its other family members. Here we show that Cbln1 and Cbln2 have similar binding activities to β-neurexins and Grid2 and the targeted ectopic expression of Cbln2 to Purkinje cells in transgenic mice rescues the cerebellar deficits in Cbln1-null animals: suggesting the two proteins have redundant function mediated by their common receptor binding properties. Cbln1 and Cbln2 are also co-expressed in the endolysosomal compartment of the thalamic neurons responsible for the synaptic alterations in striatum of Cbln1-null mice. Therefore, to determine whether the two family members have similar functions we generated Cbln2-null mice. Cbln2-null mice do not show the synaptic alterations evident in striatum of Cbln1-null mice. Thus Cbln2 can exhibit functional redundancy with Cbln1 in cerebellum but it does not have the same properties as Cbln1 in thalamic neurons, implying one or both utilize different receptors/mechanisms in this brain region.
Keywords: cerebellum, striatum, synapse, transgenic, knockout
Cerebellin precursor protein 1 (Cbln1) is a secreted glycoprotein (Urade et al. 1991; Bao et al. 2005) essential for normal synaptic structure and function in the cerebellum (Hirai et al. 2005). In cerebellum Cbln1 acts as a bi-functional ligand bridging pre-synaptic β-neurexins on granule neurons to post-synaptic Grid2 in Purkinje cells (Uemura et al. 2010; Matsuda et al. 2010). However, this cannot be the sole mechanism of action of Cbln1 as it is expressed in neurons in many brain regions (Mugnaini et al. 1987; Miura et al. 2006; Wei et al. 2007) whereas Grid2 expression is largely confined to cerebellum (Araki et al. 1993). In particular, Cbln1 deficiency in glutamatergic neurons of the parafascicular nucleus (PF) of the thalamus results in increased synaptic spine density on dendrites of striatal medium spiny neurons (MSN), the recipients of axonal projections from the Cbln1-positive PF neurons (Kusnoor et al. 2010). As MSNs and PF neurons lack Grid2 this implies that additional membrane proteins mediate the function of Cbln1 in the thalamo-striatal projection.
The dissection of the mechanisms underlying Cbln1 function is further complicated by the fact that Cbln1 is also the prototype of a family of structurally related proteins (Cbln1-4) of largely unknown function (Bao et al. 2005). These proteins have broad, sometimes overlapping, distributions in brain (Miura et al. 2006; Wei et al. 2007) and notably in situ hybridization indicates that Cbln1 and Cbln2 are both expressed in PF neurons (Miura et al. 2006). Besides having the potential to assemble into homomeric complexes all Cbln1 family members are capable of assembling into heteromeric complexes with each other (Bao et al. 2005). This might provide a mechanism to regulate or modify receptor binding specificities, as well as protein turnover, trafficking or function of Cbln1 family members in vivo. For example, Cbln1 and Cbln3 are co-expressed in cerebellar granule cells and are found in heteromeric complexes in vivo (Bao et al. 2006). Moreover, genetic elimination of either Cbln1 or Cbln3 has profound effects on the turnover of the other family member (Bao et al. 2006). Therefore, it is important to determine whether family members have redundant function and underpin similar aspects of neurobiology.
As Cbln1 and Cbln2 have broad expression in the adult nervous system, are able to associate with each other in heteromeric complexes, and are co-expressed in PF neurons we have used transgenic and knockout mouse strategies to determine whether they have identical or independent functions in vivo.
Material and Methods
Animals
Wild type FVB/NJ and C57BL/6J mice and Bax-null mice (Baxtm1Sjk/J) were obtained from Jackson Laboratory (Bar Harbor, ME). All other genetically modified strains of mice were generated at St. Jude Children’s Research Hospital (see below for details). For all experiments male and female mice of the indicated ages and genotypes were used. For behavioral analysis gender and age balanced groups of mice were used. All mice including Cbln1-null mice (Hirai et al. 2005) were maintained at St. Jude Children’s Research Hospital and had free access to food and water. Investigational procedures conformed to all applicable federal rules and guidelines and were approved by the Institutional Animal Care and Use Committee.
Generation of L7-Cbln1 and L7-Cbln2 transgenic mice
The entire coding regions of either Cbln1 or Cbln2 were inserted into the unique BamHI site of the pL7-ΔAUG cloning vector (Oberdick et al. 1990). The resulting plasmids were digested with HindIII and EcoRI to remove the vector backbone. The DNA fragments containing either L7-Cbln1 or L7-Cbln2 were purified by gel electrophoresis and injected into fertilized oocytes of wild type FVB mice using standard techniques (Gordon J. W., 1993). Mice were genotyped by PCR to determine the presence or absence of the transgene using the following primers: L7-Cbln1 or L7-Cbln2 (F, common) 5′-TGAGCCTCATGTTGAACGGG-3′, L7-Cbln1 (R) 5′-GGAAGCTGAGTGCAGCAGGATC-3′, L7-Cbln2 (R) 5′-TGTCGTTCTGAGCCCTTACG-3′. PCR conditions were: denaturation at 94°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for 30 s (35 cycles). Relative expression of the respective transgene in cerebellum was determined by Northern blot. Mice from the strain with the highest expression level of the respective transgene were mated with Cbln1+/− mice to generate appropriate genotypes for analysis.
Generation of Cbln2-null mice
To generate Cbln2-null mice we deleted the region encompassing exon1 through exon3 of Cbln2 using the recombineering approach described by Liu et al. (2003). Briefly, from a BAC clone containing the Cbln2 gene (Wellcome Trust Sanger Institute, Hinxton, Cambridge, UK), we cloned a 13.7 kb DNA fragment spanning the region including a 6 kb 5′arm, 3.7 kb exon 1 to exon 3 of the Cbln2 gene, and a 4 kb 3′arm into the PL253 vector (NCI-Frederick, Frederick, MD). The entire region encompassing exon1 through exon3 was subsequently deleted by inserting the neomycin resistance gene cassette as detailed in Figure 7A. The construct was then linearized using NotI, and electroporated into W9.5 embryonic stem cells derived from the 129S1/SvImJ strain (kindly provided by Dr. P. McKinnon, St. Jude Children’s Research Hospital). ES cells containing the construct were selected using G418 (Cellgro, Herndon, VA) and FIAU (Movarek Biochemicals, Brea, CA). ES clones were tested for deletion of Cbln2 as follows: genomic DNA from ES cells was digested by AseI and analyzed by Southern blotting using a 0.25 kb external probe and a neo probe. Several independently targeted clones were used to generate chimeric mice and two different ES clones underwent germ line transmission and generated founder strains.
Fig. 7.
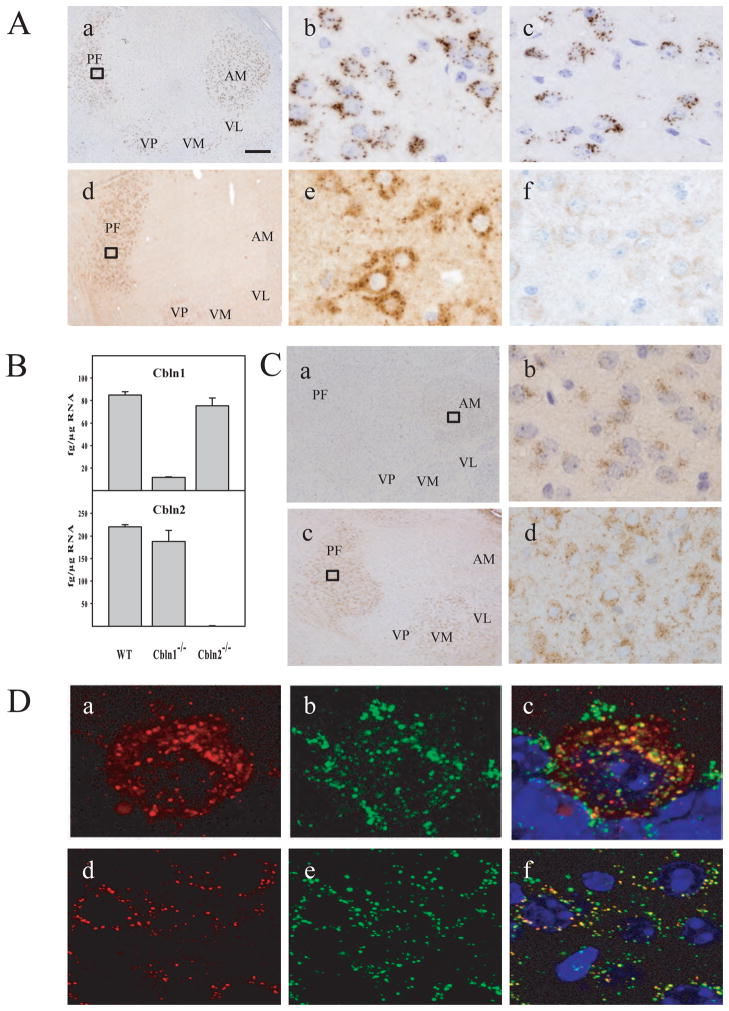
(A) Expression of Cbln1-like immunoreactivity and Cbln2-like immunoreactivity in thalamic neurons. In the adult brain, immunoreactivity to both Cbln1 (a and b) and Cbln2 (d and e) exhibited a punctate pattern in the cytoplasm of thalamic neurons. (b) and (e) show enlarged views of the boxed regions from (a) and (d), respectively. Compared with Cbln1 (a), the localization of Cbln2 (d) is found only in the PF of the thalamus, whereas Cbln1- immunoreactions is more broadly expressed in ventral and anterior thalamic nuclei. The expression pattern of Cbln1 is not markedly changed in thalamic neurons of Cbln2-null mice (c). As the Cbln2 antiserum cross reacts with Cbln1, we determined overall specificity of the anti-Cbln2 antiserum in thalamic neurons from Cbln1-/Cbln2- double null mice (f). AM, anteromedial nucleus; VL, ventrolateral nucleus; VM, ventromedial nucleus; VP, ventroposterior nucleus. Scale bar: a, d, 200 μm; b, c, e, f, 15 μm. (B) No compensatory changes in Cbln1 or Cbln2 mRNA levels in thalamus from Cbln1- and Cbln2-null mice. qRT-PCR was used to assess levels of Cbln1 and Cbln2 mRNA in microdissected thalami from adult wild type (WT), Cbln1−/− and Cbln2−/− mice. Note mRNA levels of Cbln2 are somewhat higher than those for Cbln1 in thalamus from wild type animals. (C) Expression of Cbln1- and Cbln2-like immunoreactivities in thalamus of P7 mice. At P7, Cbln1-like immunoreactivity is weak and largely confined to the anteromedial nucleus, with little or no staining in PF neurons (a, b). In contrast, Cbln2-like immunoreactivity is prominently expressed in the PF and ventromedial nuclei (c, d) at P7. Both Cbln1- and Cbln2-like immunoreactivities are also largely punctate in thalamic neurons of P7 mice. (D) Subcellular localization of Cbln2-like immunoreactivity. Immunofluorescent labeling was performed on sections from cerebellum of Cbln1−/−/L7-Cbln2 mice (a–c) or the thalamus from Cbln1−/− (d–f) mice using the Cbln2 antiserum (red, a,c,d,f), the lysosome marker cathepsin D (green, b,c,e,f) and DAPI to visualize nuclei (c, f). Merged confocal images (c and f) revealed that the Cbln2 vesicle-like immunostaining almost always localizes (92%) with cathepsin D (yellow staining) in both Purkinje cells (c) and PF neurons (f). Scale bar: 10 μm.
Animal genotyping was performed by Southern blotting and PCR. PCR primers were Cbln2 wild type (WT) 5′ primer: 5′-GACCACCTCAGCCTCACCTTCT-3′ and Cbln2 WT 3′ primer: 5′-CATTGGCAACCACTTTGACCTT -3′, as well as Cbln2 KO 5′ primer: 5′-CCCAAGCCATCCTATCCAGACA –3′, and Cbln2 KO 3′ primer: 5′-CAGAAAGCGAAGGAGCCAAAGC -3′.
cDNA constructs
Nrxn1β, Nrxn1β(−S4) and Nrxn2β CDS were synthesized by Genscript (Piscataway, NJ) and cloned into p3XFLAG-CMV-14 from Sigma (St Louis, MO). Nrxn3β CDS was cloned by PCR from C57BL/6J mouse cerebellum using the following primers: Nrxn3β F 5′-CCCAAGCTTACCACCATGCACCTGAGAATCCACCCAAG-3′ and Nrxn3β R 5′-GGGATCCCACATAATACTCCTTGTCCT-3′. The PCR product was then cloned into p3XFLAG-CMV-14 using HindIII and XbaI to obtain Nrxn3β-FLAG. Cloning of Cbln1 and Cbln2 was as described previously (Bao et al. 2005). The cDNA encoding a hemagglutinin (HA) tag was added to the 5′ end of Cbln1 and Cbln2 between glutamine (amino acid 22 for Cbln1, Accession: NP_062600; amino acid 52 for Cbln2, Accession: NP_766221) and asparagine (amino acid 23 for Cbln1 and 53 for Cbln2) amino acids. A mouse Grid2 cDNA was cloned by reverse transcriptase-polymerase chain reaction (RT-PCR) from mouse cerebellum total RNA. Grid2, HA-Cbln1, HA-Cbln2 were inserted into pcDNA3.1 (Invitrogen, Carlsbad, CA) according to manufacturer instructions.
In vitro binding assay
HEK-293T were maintained in DMEM-complete (DMEM containing 10% fetal calf serum, 100 μ/ml penicillin, 100 μg/ml streptomycin and 292 μg/ml glutamine). DMEM was purchased from Lonza (Basel, Switzerland), fetal calf serum from Millipore (Billerica, MA) and penicillin/streptomycin/L-glutamine from Invitrogen.
To study the binding of HA-Cbln1 and HA-Cbln2 to Grid2 and β-neurexins, HEK-293T cells were plated in 6-well plates (300,000 cells/well) and transfected 24 h later with one of empty vector (Mock), HA-Cbln1 or HA-Cbln2 (ligands) and Grid2, Nrxn1β, Nrxn1β(−S4), Nrxn2β and Nrxn3β (receptors). Cells were used 48 h after transfection. Medium containing the HA-Cbln1 or HA-Cbln2 (average concentration 5μg/ml determined using purified HA-Cbln1/2 standards) was collected 48 h after transfection and added to 6-well plates (2 ml/well) containing receptor (Grid2, Nrxn1β, Nrxn1β(−S4), Nrxn2β, Nrxn3β) or mock transfected cells whose media had been removed. Plates were incubated for 4 h. At the end of the experiment, cells were washed three times with ice-cold medium and lysed with 1X XT-running buffer (BioRad, Hercules, CA) containing 10 μM DTT. Samples were run on Criterion 12% Bis-Tris gels (BioRad), and binding of HA-Cbln1or HA-Cbln2 was detected using mouse anti-HA antibody (Covance, Princeton, NJ).
RNA isolation, cDNA synthesis, and qRT-PCR
Total RNA was isolated from different brain regions of mice using Trizol (Invitrogen) and cDNA synthesized using random hexamer and the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Carlsbad, CA) according to manufacturer instructions. Primers and probes for Cbln1 and Cbln2 were designed using Primer Express 3.0 (Applied Biosystems), analyzed for homology to other known sequences using the BLAST (NCBI web address), and prepared by Applied Biosystems. The following primers were used for quantitative real time PCR (qRT-PCR): Cbln1 (NM_019626) forward 5′-GGTGAAAGTCTACAACAGACAGACCAT-3′, probe 5′-FAM- CAGGTGAGCCTCATGTTGAACGGGT-BHQ1-3′, reverse 5′-CGGCGAAGGCTGAAATCA-3′; Cbln2 (NM_172633) forward 5′-GTGGTCAAAGTGTACAACAGACAAACT-3′, probe 5′-FAM- TCCAGGTCAGCTTAATGCAGAATGGCTACC-BHQ1-3′, reverse 5′-CCGGCAAATGCAGAGATCA-3′.
qRT-PCR was performed using 2 X TaqMan® Fast Reagents Starter Kit (Applied Biosystems) according to manufacturer instructions, using the ABI 7900 Fast Real-Time PCR system (Applied Biosystems). PCR conditions were as follows: uracil-N-glycosylase incubation, 50°C for 2 min, AmpliTaq Gold activation, 95°C for 10 min, denaturation step 95°C for 15 s, annealing step 60°C for 1 min; 40 cycles were performed. Standards for absolute quantification were obtained by cloning Cbln1 and Cbln2 into pcDNA3.1. PCR results were normalized for beta-actin (Actb).
Northern blotting
Total RNA (20 μg) from mouse cerebellum was resolved by electrophoresis on 1 % agarose gel containing formaldehyde and transferred onto nylon membrane by vacuum blotting. A digoxigenin (DIG)-labeled probe was generated from mouse L7 cDNA as previously described (Wang et al. 2006). Hybridization was carried out in EasyHyb (Roche) according to manufacturer instructions.
Rota-Rod test
All mice used in this experiment were generated from appropriate heterozygous crosses to obtain animals of the desired genotypes with the same mixed genetic backgrounds. To further minimize any variations due to background strain effects, gender and age matched littermate mice were used. Wild type, Cbln1−/−, Cbln2−/− and Cbln1−/− with L7 transgene strains were tested on an accelerating rota-rod (San Diego Instruments, San Diego, CA) to assess motor coordination, balance, and motor learning. The rota-rod was programmed to accelerate from 0 to 40 rpm in 4 min and then hold constant speed for an additional minute. The amount of time that elapsed before the mouse fell off was recorded. The maximum observation time was 5 minutes. Animals were given 2 trials per day with a 20-min inter-trial interval. Animals were tested for 5 consecutive days. The latency of the mice to fall from the rod was scored as an index of their motor coordination. Improvement in performance across training days indicates motor learning (Buitrago et al. 2004; Wei et al. 2011). The test data were expressed as mean ± SEM and were analyzed for statistical significance using one-way analysis of variance (ANOVA) followed by Bonferroni’s multiple comparison test or Student’s t-test depending on the case. The level of significance was set at p < 0.05.
Histological methods
One-month-old mice were anesthetized, perfused transcardially with buffered 4% paraformaldehyde (PFA), and their brains removed and post-fixed as described (Wei et al. 2009). Paraffin sections (5 μm) were stained using hematoxylin and eosin for standard histological analysis. Cbln2 antiserum was raised to an epitope lying N-terminal to the C1q motif in Cbln2. The synthetic peptide SGSAKVAFSATRSTNHE corresponding to amino acids 88–104 of mouse Cbln2 (NP_766221) was coupled to keyhole limpet hemocyanin (KLH). Rabbits were immunized with KLH-peptide conjugate (Rockland, Gilbertsville, PA). Antisera were further purified with an affinity column comprising Sepharose coupled to the peptide antigen. The specificity of the anti-Cbln2 antiserum was established by immunoblotting of lysates of HEK 293 cells transfected with recombinant vectors expressing Cbln1, 2, 3 or 4 fused to a C-terminus V5 epitope tag. The anti-Cbln2 antiserum detected proteins in both Cbln1-V5 and Cbln2-V5 transfected cells (data not shown). For immunohistochemistry, all sections were heat retrieved in 0.01 M sodium citrate buffer (pH 6.0) containing 0.05% Tween-20 (Sigma) as described previously (Wei et al. 2009). Affinity purified rabbit polyclonal antiserum to Cbln2 (1:1000) or Cbln1 (E3 epitope) antibody (1:1000, Bao et al. 2005) was used to detect Cbln2-ir or Cbln1-ir as previously described (Wei et al. 2007). Rabbit anti-activated caspase-3 (1: 100; BD, Franklin Lakes, NJ) was used to detect the cleaved active form of caspase-3. After immunostaining, the sections were counterstained with hematoxylin (Vector Labs, Burlingame, CA).
To establish the subcellular localization of Cbln2, heat-mediated antigen retrieval treated paraffin sections were incubated overnight with a mixture of rabbit anti-Cbln2 (1:1000) antibody and the lysosome marker, goat anti-cathepsin D (1:300, Santa Cruz Biotechnology, Santa Cruz, CA) (Erickson and Blobel, 1979). Sections were incubated for 1 h with Alexa 488- or Alexa 594- labeled species-specific secondary antibodies (1:200, Invitrogen) and images acquired using confocal laser microscopy as described previously (Wei et al. 2007). Co-localization of Cbln2 with cathepsin D-positive vesicle-like structures was quantitatively assessed by counting puncta positive for Cbln2-immunoreactivity alone or co-labeled with cathepsin D using confocal reconstruction from 6 sections from 2 separate mice.
In situ labeling of fragmented DNA (Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL))
Paraffin embedded tissue sections were prepared as described above. A Diaminobenzidine (DAB) TUNEL-based apoptosis detection assay was performed using the TdT In Situ Apoptosis Detection Kit (R&D systems, Minneapolis, MN) according to the manufacturer’s specifications. After immunostaining, the sections were counterstained with hematoxylin.
Golgi impregnation
Cbln2-null mutant mice and their wild type littermates were perfused with 0.1 M PBS followed by a solution containing 2% PFA/2.5% glutaraldehyde in 0.1 M PBS. Coronal sections (150 μm) through the precommissural striatum were cut on a vibrating microtome. The sections were then incubated in 1% osmium tetroxide for 30 min and kept in 3.5% potassium dichromate overnight. The next day the sections were developed in 1% silver nitrate for 4–6 hours before being washed extensively, mounted, and cover slipped.
Dendritic analyses of MSNs
Golgi-stained striatal MSNs in the lateral striatum were reconstructed using Neurolucida (MicroBrightField, Colchester, VT). Dendritic spine density was determined on segments of secondary or tertiary dendrites at distances between 70 and 90 μm from the soma, on second or third order dendrites. Segments from three different primary basal dendrites per neuron and 5–6 neurons per animal were analyzed to compute an average dendritic spine density for a given animal; these spine density/animal values were then compared by means of two tailed t-tests.
Dendritic spine length was measured in > 50 spines/animal of Cbln2-null and wild type mice. We also determined the shape of dendritic spines in Cbln2−/− mice following the classification scheme of Peters and Kaiserman-Abramof, (1970).
Statistical analysis
The rota-rod data were analyzed for statistical significance using repeated-measures analysis of variance (ANOVA), and where appropriate multiple-comparison tests were performed with Bonferroni’s adjustment. For other experiments, t-test was carried out for comparing two independent samples. Significance was set at a p of < 0.05. All statistical analyses were performed using SAS version 9.2 (SAS Institute Inc., Cary, NC).
Results
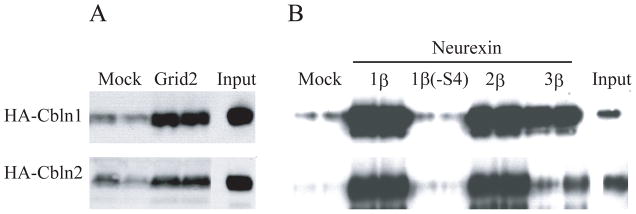
To establish whether Cbln1 and Cbln2 have similar receptor specificities an in vitro binding assay was performed. HEK293 cells were transfected with empty vector (Mock) or plasmids expressing Grid2 (Fig. 1A), the S4-containing splice variants of neurexins-1β, -2β, -3β, or neurexin-1β lacking the S4 insert (Fig. 1B). Subsequently the cells were incubated with conditioned medium (Input in Fig. 1A, B) from cells transfected with either HA-Cbln1 or HA-Cbln2 and bound ligand determined 4 hours later using western blotting (see Materials and Methods for details). Both HA-Cbln1 and HA-Cbln2 bound to Grid2, although in our hands binding of HA-Cbln1 was more robust than the binding of HA-Cbln2 (Fig. 1A). HA-Cbln1 and HA-Cbln2 both bound to the S4-containing variants of all three β-neurexins with similar specificities (Nrxn1β = Nrxn2β ≫ Nrxn3β) but they did not bind to the S4-lacking variant of neurexin-1β (Fig. 1B). Binding of both ligands to neurexin-3β was consistently much lower than to neurexins-1β and -2β (Fig. 1B). These data support other studies (Uemura et al. 2010; Joo et al. 2011) and indicate that Cbln1 and Cbln2 have similar receptor binding specificities.
Fig. 1.
Comparison of binding of HA-Cbln1 and HA-Cbln2 to Grid2 and β-neurexins. (A) HEK293T cells were transfected with Grid2 or empty vector (Mock). Cells were subsequently exposed to conditioned medium (Input) containing HA-Cbln1 or HA-Cbln2 for 4 h. After washing with medium, cells were lysed with sample buffer and subjected to SDS-PAGE and immunoblotted with anti- HA antiserum. HA-Cbln1 and HA-Cbln2 show elevated binding to Grid2 compared with mock transfection. (B) HEK293T cells were transfected with Nrxn1β, Nrxn1β(−S4), Nrxn2β, Nrxn3β or empty vector (Mock). Then cells were incubated with conditioned medium (Input) containing HA-Cbln1 or HA-Cbln2 for 4 h. After washing with medium, cells were lysed with sample buffer and subjected to SDS-PAGE and immunoblotted with anti- HA. Compared with mock transfection, both HA-Cbln1 and HA-Cbln2 bound to Nrxn1β, Nrxn2β and Nrxn3β, but not to Nrxn1β lacking the S4 splice variant sequence.
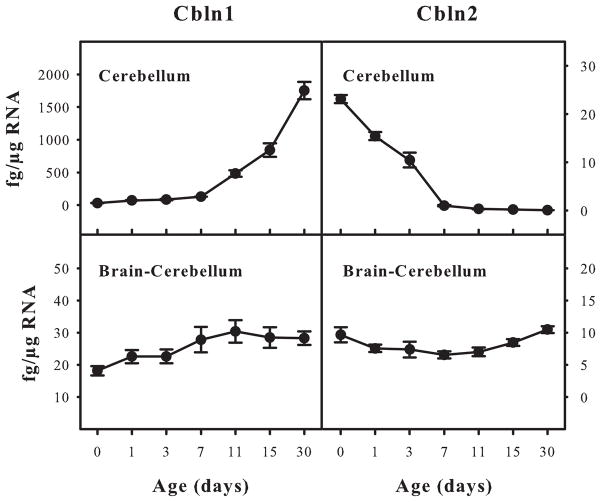
As Cbln1 and Cbln2 have qualitatively the same receptor binding properties we next determined whether their patterns of expression were the same in the adult and developing nervous system of mice using qRT-PCR. Whereas Cbln1 mRNA levels rise in the developing cerebellum from postnatal day 0 onwards, the levels of Cbln2 mRNA decline over the same period and are barely detectable at postnatal day 30 (Fig. 2). In contrast, Cbln1 and Cbln2 were expressed at roughly equivalent levels throughout development in the extra-cerebellar regions of brain (Fig. 2).
Fig. 2.
Quantitative assessment of Cbln1 and Cbln2 mRNA levels in developing and adult mouse brain. Cbln1 and Cbln2 mRNA levels from wild type FVB mice of various ages were evaluated by qRT-PCR (described in Materials and Methods). Samples were prepared from cerebellum and whole brain less cerebellum. Data are presented as mean ± SEM of 3 animals. Whereas Cbln1 and Cbln2 mRNA levels change in cerebellum during development they are relatively constant over the same period in the rest of brain.
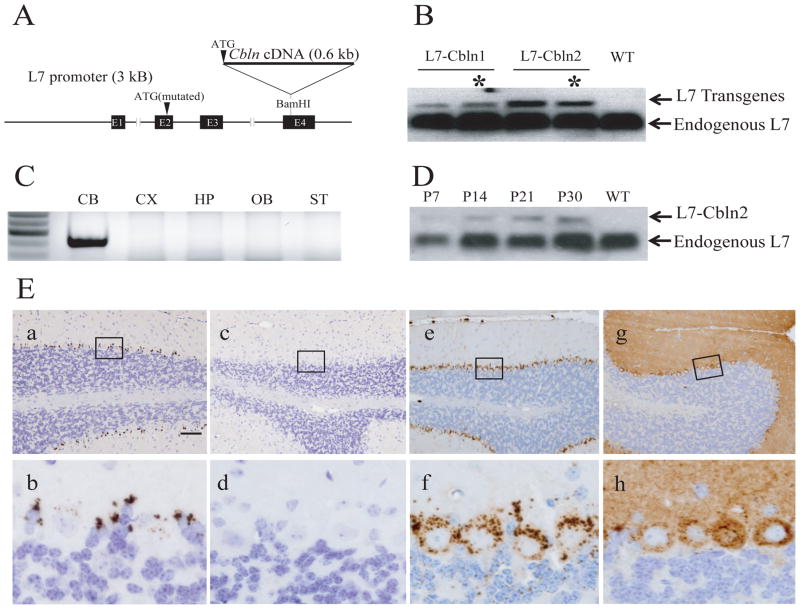
We showed previously that ectopic expression of Cbln1 in Purkinje cells rescued the ataxic phenotype of Cbln1-null mice (Wei et al. 2009). Therefore, the absence of Cbln2 in adult cerebellum afforded the possibility of using the same strategy to test whether over-expression of Cbln2 rescues the cerebellar deficits of Cbln1-null mice. Cbln1 or Cbln2 were expressed from the L7/Pcp2 Purkinje cell-specific promoter (Oberdick et al. 1990) in transgenic mice generated using the strategy depicted in Figure 3A. The transgenic mRNA is a chimera between Cbln1 or Cbln2 coding sequences and L7/Pcp2 non-coding sequences. Therefore, individual founder lines were screened by northern blot using a probe to the L7 component of the fusion mRNA to identify lines that expressed each family member to approximately the same level using endogenous L7 mRNA as an internal standard (see Fig. 3B for representative examples and data not shown). Individual lines from amongst those with relatively high levels of expression of the respective mRNA were then chosen based upon selective expression of the transgene in cerebellum (CB in Fig. 3C for a L7-Cbln2 line) and appropriate expression profile in the developing cerebellum (Fig. 3D). The presence of the respective protein in Purkinje cells was then confirmed by immunohistochemistry (Fig. 3Ee–h). The selected transgenic lines (asterisks in Fig. 3B) were subsequently crossed onto a Cbln1-null background.
Fig. 3.
L7-Cbln1 and L7-Cbln2 constructs and their expression in transgenic mice. (A) Cbln1 or Cbln2 cDNA was inserted into a unique BamHI site in the fourth exon of the L7 gene to make the L7-Cbln1 or L7-Cbln2 transgene construct. (B) Northern blot using a probe to L7 in cerebellum from wild type (WT), L7-Cbln1 and L7-Cbln2 transgenic mouse lines. The fusion RNAs (L7 Transgenes) have higher molecular weights than endogenous L7 mRNA that acts as an internal standard. Asterisks denote the transgenic lines selected for further investigation. (C) RT-PCR of different brain regions in an L7-Cbln2 transgenic mouse line shows that L7-Cbln2 recombinant RNA is only detected in the cerebellum (CB). CX, cerebellar cortex; HP, hippocampus; OB, olfactory bulb; ST, striatum. (D) Northern blot using a probe to L7 in cerebellum from a developing L7-Cbln2 transgenic mouse line indicates that L7-Cbln2 recombinant mRNA is expressed at P7 although at lower levels than at later time points. (E) Expression of transgenic Cbln1- and Cbln2-immunoreactivity in Purkinje cells of Cbln1−/−/L7-Cbln1 and Cbln1−/−/L7-Cbln2 mice. In the cerebellum from adult wild-type mice, Cbln1-immunoreactivity is found in a punctate pattern (a and b) in Purkinje neurons and Bergmann glia. Specificity of anti-Cbln1 antiserum is shown by a lack of immunoprecipitate in cerebellum from a Cbln1-null mouse (c and d). In cerebellum of adult Cbln1−/−/L7-Cbln1 (e and f) and Cbln1−/−/L7-Cbln2 mice (g and h), the immunoreactivities of Cbln1 and Cbln2 is detected in Purkinje neurons. (b), (d), (f), (h) show enlarged views of the boxed regions from (a), (c), (e), (g), respectively. Immunostaining for Cbln1 is predominantly punctate and confined to the soma and proximal dendrites of Purkinje cells whereas Cbln2 immunoreactivity is seen in the soma and the entire dendritic tree. Scale bar: a, c, e, g, 100 μm; b, d, f, h, 10 μm.
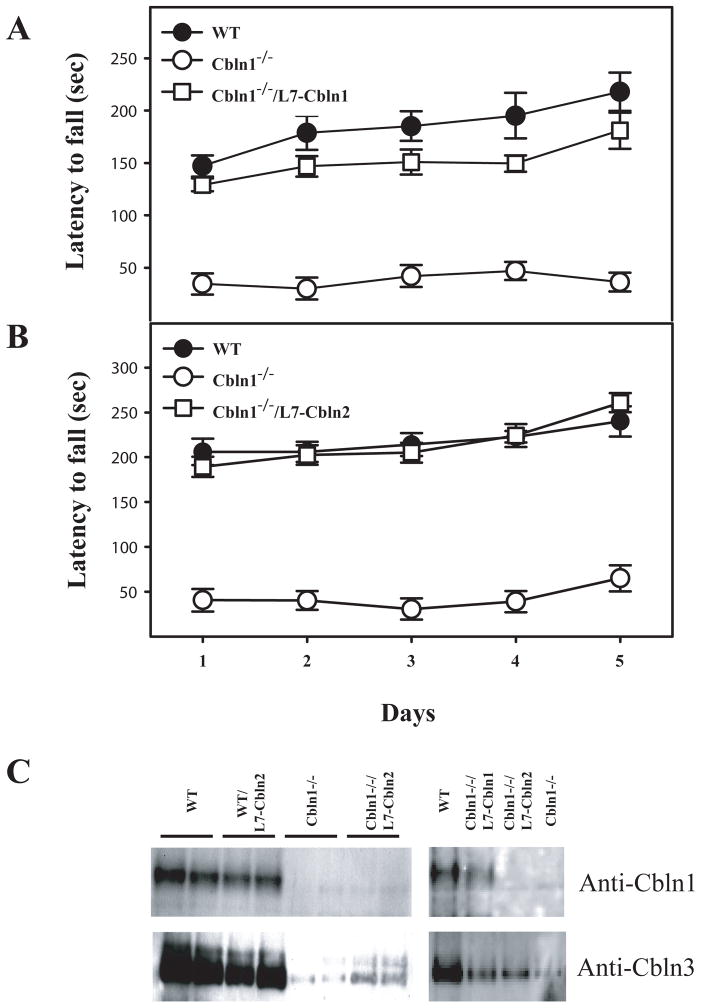
As Cbln1 knockout mice are ataxic and perform poorly on the accelerating rota-rod (Hirai et al. 2005; Wei et al. 2009) this test was used to determine the efficacy of the respective transgene in rescuing the Cbln1-null phenotype. Ectopic expression of either Cbln1 or Cbln2 markedly improved the rota-rod performance of Cbln1-null mice (Fig. 4A, B) (also see Wei et al. 2009 for Cbln1). Indeed, the L7-Cbln2/Cbln1-null mice were indistinguishable from wild type littermates. Therefore, we asked whether Cbln2 corrected other alterations in the cerebellum of Cbln1-null mice.
Fig. 4.
Ectopic expression of Cbln1 (A) or Cbln2 (B) in Purkinje cells of Cbln1-null mice improves Rota-rod performance. Gender balanced littermates of each genotype (n = 6–8/genotype) between 45 and 55 days old were tested on a standardized accelerating Rota-rod. Wild type (WT) mice, Cbln1-null (Cbln1−/−) mice and Cbln1-null animals carrying a transgene (Cbln1−/−/L7-Cbln1; Cbln1−/−/L7-Cbln2) were tested for 5 consecutive days. The latency to fall in second for all animals of a given genotype was recorded, and data presented as mean ± SEM. Multiple comparisons between groups using the Bonferroni post-hoc test revealed that the presence of L7-Cbln1 (p = 0.0015) and L7-Cbln2 (p = 0.0003) significantly improved the performance of Cbln1-null animals. One-way ANOVA showed that only the Cbln1-null group differed significantly from all other groups. (C) Adult mouse cerebellum from the indicated genotypes was subjected to SDS–PAGE and immunoblotting with anti-Cbln1and anti-Cbln3 antisera. Both Cbln1 and Cbln3 levels were greatly reduced in the Cbln1-null cerebellum (Cbln1−/−) compared to wild type (WT) littermates. The reductions in Cbln3 levels in Cbln1-null mice were not restored to wild type values by ectopic expression of either Cbln1 (Cbln1−/−/L7-Cbln1) or Cbln2 (Cbln1−/−/L7-Cbln2). However, the Cbln1-null mice harboring a transgene had slightly higher levels of Cbln3 than Cbln1-null mice without a transgene.
A second aspect of the Cbln1-null phenotype is the near absence of Cbln3 protein in cerebellum (Bao et al. 2006). This is attributable to Cbln1 being required for the intracellular trafficking and secretion of Cbln3 (Bao et al. 2006). Therefore, we asked whether expression of Cbln1 or Cbln2 in Purkinje cells rescued the loss of Cbln3 in granule neurons of Cbln1-null mice using western blotting. As shown in Fig. 4C, Cbln3 levels are greatly reduced in the Cbln1-null cerebellum but there is little if any rescue of this deficit by either Cbln1 or Cbln2.
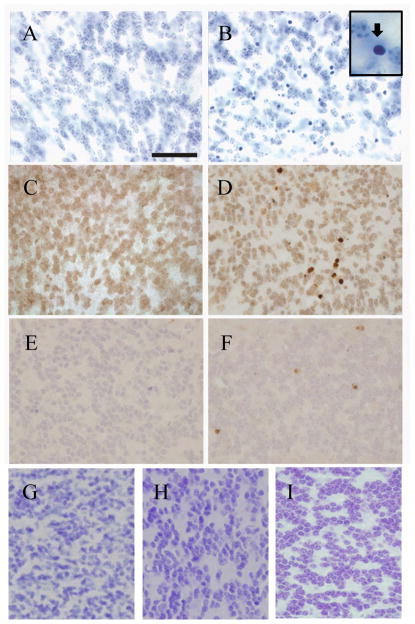
A third phenotype in the cerebellum of Cbln1-null mice is the presence of sporadic pyknotic cells in the internal granule cell layer between postpartum day 21 and approximately 2-months of age that ultimately results in an approximately 10–15% depletion of granule cells (Hirai et al. 2005; Fig. 5B and insert and data not shown). Although we do not see nuclear fragmentation, these degenerating cells appear to be neurons undergoing apoptosis as they are positive for TUNEL (Fig. 5D) and activated caspase-3 (Fig. 5F) staining. Several mutant strains of mice that exhibit Purkinje cell degeneration have a secondary loss of granule cells (Sotelo et al. 1974; Wetts et al. 1983; Triarhou, 1998) and we showed previously that elimination of Bax function rescued granule neuron death in Purkinje cell degeneration (pcd) mice without sparing Purkinje cells (Wang and Morgan, 2007). Therefore, we crossed the Cbln1-null animals onto a Bax-null background. Cbln1/Bax-double knockout mice do not exhibit pyknotic granule cells (Fig. 5G). However, the mice were still grossly ataxic (data not shown) indicating that the delayed loss of granule neurons does not cause the locomotor defects in Cbln1-null mice. We next assessed whether ectopic expression of Cbln1 or Cbln2 in Purkinje cells also rescued granule neurons loss in Cbln1-null mice. There were no pyknotic neurons evident in Cbln1-null mice harboring either the L7-Cbln1 (Fig. 5H) or L7-Cbln2 (Fig. 5I) transgenes. Therefore, both Cbln1 and Cbln2 rescue this secondary phenotype of Cbln1-null mice.
Fig. 5.
Cbln1-null mice exhibit granule cell degeneration that is rescued by elimination of Bax or ectopic expression of Cbln1 or Cbln2. Compared to wild type P25 mouse cerebellum (A), cerebellum from a littermate Cbln1−/− mouse (B) has many pyknotic nuclei. Boxed area in B shows morphology of a pyknotic neuron. TUNEL staining of P25 wild type (C) and cbln1−/−mice (D) showed TUNEL-positive cells in the knockout strain. Activated caspase-3 staining of P25 wild type (E) and Cbln1−/−mice (F) showed activated caspase-3-positive cells only in the knockout mouse cerebellum. When Cbln1−/−mice are crossed onto a Bax-null background there are no pyknotic nuclei evident in cerebellum (G). When Cbln1−/− mice are crossed onto either the L7-Cbln1 (H) or the L7-Cbln2 (I) strain there are no pyknotic nuclei evident in cerebellum. Scale bar = 50 μm for panels A–I.
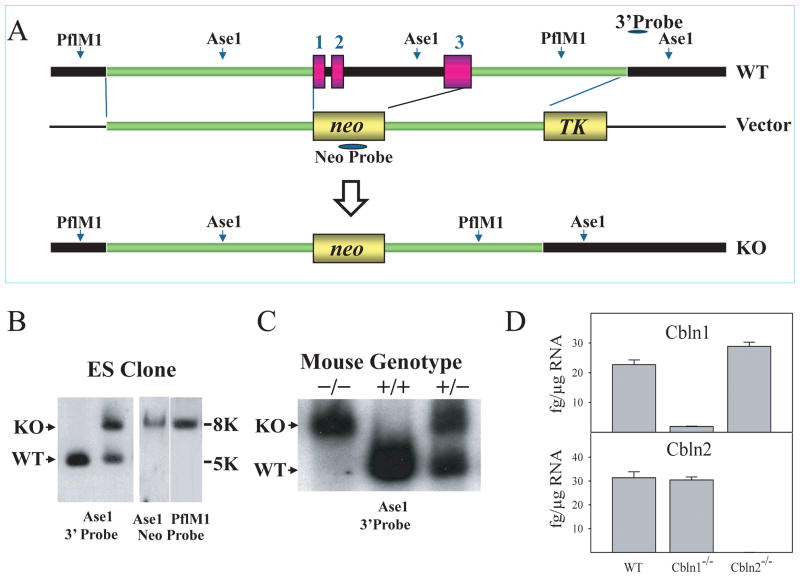
As ectopic expression of Cbln2 in cerebellum rescues Cbln1 deficiency we generated Cbln2-null mice to assess whether they had Cbln1-null-like phenotypes in either cerebellum or thalamus. Cbln2-null mice were produced using standard techniques and the strategy summarized in Fig. 6A. The elimination of the gene and the absence of the cognate mRNA were confirmed by Southern blotting (Fig. 6B, C) and qRT-PCR (Fig. 6D), respectively. Using qRT-PCR we also found no transcriptional compensation amongst family members in the brains of Cbln1- or Cbln2-null mice (Fig. 6D), precluding this potential confounder.
Fig. 6.
Generation of Cbln2-null mice. (A) Schematic representations of Cbln2 genomic DNA (WT), targeting vector (Vector) and targeted genome (KO). Shaded bars indicate the probes (neo and 3′) for Southern blot analysis. All 3 exons and introns of Cbln2 were replaced with a neo cassette. Southern blot analysis of genomic DNA was prepared from ES cells (B) and tissues of wild-type (+/+), heterozygote (+/−), and knockout (−/−) mice (C). DNA from ES cells and mouse tissues was digested with AseI (for 3′probe and neo probe) and PflMI (for neo probe), then analyzed using a DIG-labeled 0.25 kb 3′ external probe and a neo probe, respectively. Bands with the indicated sizes denote correct targeting in both ES cells and tissues. (D) qRT-PCR showing loss of Cbln1 and Cbln2 mRNA in Cbln1-null and Cbln2-null mouse brain, respectively. There is no compensatory change of Cbln1 or Cbln2 mRNA levels in Cbln2- or Cbln1-null mice, respectively.
Prior studies indicated that Cbln1 is expressed in extra-cerebellar neurons, notably within neurons of the parafascicular nucleus of the thalamus (Miura et al. 2006; Wei et al. 2007; Kusnoor et al. 2010). Cbln2 is also expressed outside of cerebellum (Fig. 2) including the thalamus (Miura et al. 2006). As the Cbln1-positive neurons of the PF are responsible for the synaptic alterations in the striatum of Cbln1-null mice (Kusnoor et al. 2010) we examined the localization of Cbln2-ir in thalamus using immunohistochemistry. Like Cbln1-ir (Fig. 7Aa, b), Cbln2-ir is present in neurons in the PF nucleus (Fig. 7A, d, e). However, Cbln1-ir is more widespread in thalamus than that of Cbln2-ir, being observed in neurons in both ventral and anterior thalamic nuclei (compare Fig. 7Aa with d). Nevertheless, within the PF nucleus, Cbln2-ir is present in the large majority of neurons. As our anti-Cbln1 family antisera are all raised in rabbits, co-localization is difficult. However, Cbln1 is expressed in essentially all neurons in this brain region (Kusnoor et al. 2010) and therefore given the present data on Cbln2-ir it must be co-expressed with Cbln2 in a large proportion of the PF neurons. As loss of Cbln1 can affect the level of Cbln3 (Bao et al. 2006) we also wanted to assess whether loss of Cbln2 influenced the expression profile of Cbln1 in thalamus. Cbln1-ir staining intensity and subcellular distribution did not appear to be markedly altered in Cbln2-null animals (Fig. 7Ac) compared to wild type (Fig. 7Ab). As the anti-Cbln2 antiserum cross-reacts with Cbln1 (although it is specific to these two proteins as there is no immunostaining in Cbln1/Cbln2 double null mice, Fig. 7Af) we cannot perform the reverse experiment. To circumvent this limitation, we performed qRT-PCR on dissected thalamus from wild, Cbln1-null and Cbln2-null mice. In thalamus from adult mice, Cbln2 mRNA levels are higher than those of Cbln1 (Fig. 7B). Furthermore, loss of Cbln1 does not affect the level of Cbln2 mRNA and likewise loss of Cbln2 does not lead to a change in Cbln1 mRNA levels, supporting the findings using immunohistochemistry.
As the temporal emergence of the synaptic spine alterations in Cbln1-null animals has not been characterized, we assessed the expression of Cbln1-ir and Cbln2-ir in thalamus of P7 mice (Fig. 7C). In contrast to its broad distribution in adult thalamus (Fig 7Aa) at P7, Cbln1-ir is weak and limited to the AM (Fig. 7Ca, b). Notably, there are no Cbln1-ir-positive neurons in the PF. Cbln2-ir on the other hand is robustly expressed in PF neurons at postnatal day 7 (Fig. 7Cc, d) and is also evident in ventral tier neurons that do not prominently express Cbln2 at later stages (compare Fig 7Ad with Fig 7Cc). These data indicate that expression of Cbln2 starts earlier in PF thalamic neurons compared to Cbln1, which may have implications for the interpretation of the cellular basis of synaptic alterations in Cbln1-null mice.
Notably, both Cbln1-ir and Cbln2-ir exhibited punctate patterns of staining in both Purkinje neurons ectopically expressing the proteins (Fig. 3Ee–h) and in neurons of the parafascicular nucleus of the thalamus in non-transgenic mice (Fig. 7Ab, e). We showed previously that Cbln1-ir was localized to the endolysosomal system of neurons (Wei et al. 2009) and these data indicate that Cbln2 may be subject to the same mode of intracellular trafficking. To confirm localization, we performed a confocal double immunofluorescence analysis using an anti-Cbln2 antiserum (red) and an antibody to the lysosome marker, cathepsin D (green) (Fig. 7D). In Purkinje cells ectopically expressing Cbln2 (Fig. 7Da–c) Cbln2-ir is present in both the cytoplasm and vesicle-like structures (Fig. 7Da) that in merged images are frequently doubled labeled for cathepsin D (Fig. 7Dc). Within thalamic neurons (Fig. 7Dd–f) Cbln2-ir is also punctate and almost always (92% +/− 1.5%) co-localized with cathepsin D (Fig. 7Df). The presence of Cbln2-ir in cytoplasm as well as punctate structures in Purkinje cells of L7-Cbln2 mice is also seen using immunohistochemistry (Fig. 3Eg, h). In comparison, Cbln1-ir is almost exclusively punctate with little or no staining of cytoplasm in either wild type (Fig. 3Eb) or L7-Cbln1 transgenic animals (Fig. 3Ef). Whether this reflects differences in the trafficking of the two family members is uncertain. The level of Cbln2 mRNA in the L7-Cbln2 strain was slightly higher than in the L7-Cbln1 transgenic strain so this may be a detection limit effect or an effect of over-expression of the protein that does not normally occur. However, in PF thalamic neurons of non-transgenic mice, Cbln2-ir also had some cytoplasmic staining as well as punctate staining (Fig. 7Ae) that was not evident with Cbln1-ir (Fig. 7Ab), suggesting that Cbln2 may indeed be present in two compartments. However, the antibodies used here recognize totally distinct regions of the two proteins, and so differences in apparent subcellular distributions may be a product of the epitope targeted. Cbln1 undergoes extensive stereotyped proteolytic processing in vivo (Bao et al. 2005; Hirai et al. 2005) and it is unknown if Cbln2 is subject to similar processing. Therefore, it is possible that Cbln1 also has both a cytoplasmic and vesicular distribution, but the epitope recognized by the antiserum is only prevalent in the endolysosomal compartment whereas the one recognized by the Cbln2 antiserum is in both.
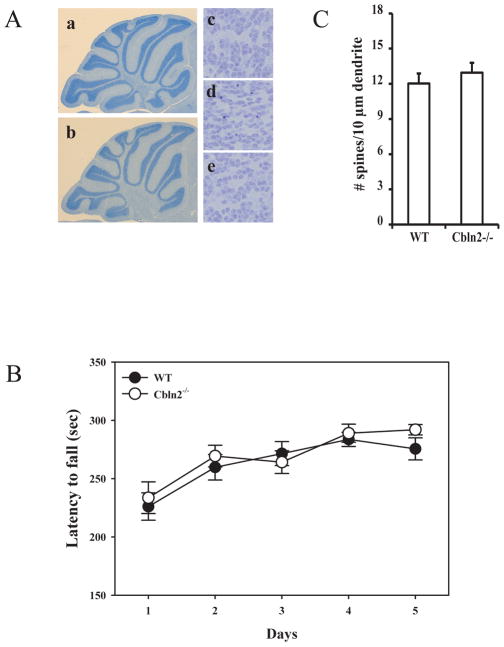
Homozygous Cbln2-null mice are fertile, have normal life spans and have no overt anatomical or neuroanatomical defects (Fig. 8A). As Cbln2 is expressed in the developing cerebellum (Fig. 2) and corrects the cerebellar deficits of Cbln1-null mice (Fig. 4, 5) we first assessed their locomotor behavior using the accelerating rota-rod test. Cbln2-null mice (empty circles) were indistinguishable from wild type (filled circles) littermates on the first trial day, confirming they are not ataxic. Thereafter both strains improved their performance to the same extent over the subsequent test days indicating intact locomotor learning in the Cbln2-null animals (Fig. 8B; p > 0.34, 7 mice/genotype). Finally, as Cbln2 is co-expressed with Cbln1 in the PF neurons (Fig. 7Aa, b, d, e) that underlie the synaptic alterations in the striatum of Cbln1-null mice we determined whether there were any changes in striatal MSN synaptic spine density in Cbln2-null mice. Unlike Cbln1-null mice (Kusnoor et al. 2010), synaptic spine densities in Cbln2-null mice were not significantly different from wild type littermates (Fig. 8C; p > 0.1, 4 mice/genotype).
Fig. 8.
Cbln2-null mice do not exhibit the same deficits as Cbln1-null mice. (A) Compared to wild type P30 mouse cerebellum (a, c), cerebellum from a littermate Cbln2−/− mouse (b, e) did not show any anatomic defects, whereas P30 cerebellum from a Cbln1−/− mouse (d) has many pyknotic nuclei. (B) Gender balanced littermates of each genotype (n = 7/genotype) between 45 and 55 days old were tested on a standardized accelerating rota-rod. Cbln2-null mice (open circles) are indistinguishable from wild type mice (filled circles) (p > 0.34). Data is mean ± SEM. (C) There is no difference in dendritic spine densities on striatal medium spiny neurons of Cbln2-null (Cbln2−/−) mice compared to wild type (WT) littermates (p > 0.1, n = 4/genotype).
Discussion
Although Cbln2 is biologically redundant with Cbln1 in cerebellum (Fig. 4, 5), its functional loss in vivo does not mimic the Cbln1-null phenotype in either the cerebellum or striatum (Fig. 8). This result is unsurprising in the adult cerebellum as Cbln2 is expressed at levels that are orders of magnitude lower than Cbln1 (Fig. 2) and so would likely be insufficient to support the maintenance of the numerous granule cell-Purkinje cell synapses. Thus the temporal-spatial pattern of Cbln2 expression can explain this dichotomy. In addition, the absence of overt changes in cerebellar morphology or function in Cbln2-null mice also indicates that the higher levels of Cbln2 detected in cerebellum during the perinatal period (Fig. 2) are not critical for cerebellar development. These findings are consistent with Cbln2 being able to interact with the same receptors as Cbln1 in cerebellum in a manner that is functionally redundant. Our in vitro binding data as well as prior studies from other groups (Uemura et al. 2010; Joo et al. 2011) also indicate that Cbln1 and Cbln2 have similar binding profiles to neurexins and Grid2 (Fig. 1), further supporting this conclusion.
Cbln2 also rescues another feature of the Cbln1-null phenotype, namely the degeneration of a proportion (~10–15%) of cerebellar granule cells that occurs over a period of approximately 8 weeks starting in the fourth week after birth. The degenerating neurons show several features typical of cells undergoing apoptosis, such as activation of caspase-3 and DNA fragmentation, although they do not have the characteristic fragmented nuclear morphology of apoptotic cells but rather appear pyknotic. A number of mutant strains of mice that have degeneration of Purkinje neurons as their primary phenotype, such as staggerer, pcd and lurcher, exhibit a secondary degeneration of granule neurons (Sotelo et al. 1974; Wetts et al. 1983; Triarhou, 1998). Furthermore, whereas Bax deficiency does not influence Purkinje cell survival it does rescue the degeneration of granule cells in Lurcher (Doughty et al. 2000) and pcd mice (Wang and Morgan, 2007). As granule cell loss is also Bax-dependent in Cbln1-null mice, this implies a common degenerative mechanism. The secondary loss of granule neurons in the mutant strains is thought to result from the disruption of trans-synaptic signaling and the possible loss of target-derived trophic support (Sotelo et at. 1974; Wetts et al. 1983; Vogel et al. 1989; Triarhou, 1998). Therefore, granule cell degeneration in Cbln1-null mice might result from the loss of synaptic contacts with Purkinje cells. However, as Cbln1 is a secreted ligand that can bind to pre-synaptic neurexins, one cannot exclude the possibility that it might also have an autocrine trophic effect upon granule cells, and Cbln2 might share this activity. Interestingly, the Grid2-null mouse, which also does not loose Purkinje cells and mimics the cerebellar synaptic phenotype of Cbln1-null mice (Kashiwabuchi et al. 1995: Hirai et al. 2005) was also reported to have less granule neurons than wild type littermates (Kurihara et al. 1997) although the basis of this difference was not investigated. This provides not only another phenotypic parallel between Grid2- and Cbln1-null mice, but it also suggests that Cbln1 is not a survival factor for granule cells (as it is present in Grid2-null animals that lack only its post-synaptic receptor) and that neuronal death is a consequence of a loss of synaptic contact.
Cbln3 protein, but not mRNA, levels are markedly reduced in Cbln1-null mice, making these animals effectively Cbln1/Cbln3-double null and thereby complicating the interpretation of the basis of the phenotype (Bao et al. 2006). This is because Cbln3 is unique amongst the Cbln1 family in that it cannot form homotrimers and has an obligatory requirement to heterotrimerize with Cbln1 (or potentially other family members) in order to exit the ER and undergo trafficking and secretion (Bao et al. 2006). In the absence of Cbln1, Cbln3 undergoes ER retention and subsequent degradation (Bao et al. 2006). In L7-Cbln1/Cbln1-null and L7-Cbln2/Cbln1-null mice Cbln3 levels are still greatly reduced. This indicates that Cbln3 protein is not required for Cbln2 (or Cbln1) to rescue the Cbln1-null phenotype and implies that loss of Cbln3 does not contribute to the Cbln1-null phenotype. In addition, as Purkinje cells do not express any of the Cbln1 family members the findings in L7-Cbln2/Cbln1-null mice also establish that Cbln2 (or Cbln1) homomeric complexes are sufficient to support proper cerebellar synapse development and maintenance in vivo.
Unlike in the cerebellum where Cbln1 is vastly more abundant than Cbln2, these two family members are expressed in the same population of PF thalamic neurons at roughly equivalent levels. Indeed, Cbln2 mRNA levels in thalamus are even higher than those for Cbln1, suggesting it may be the more predominant protein in PF neurons. Therefore, the lack of concordance in the phenotypes of Cbln1- and Cbln2-null mice in this brain region cannot simply be attributed to the abundance of the proteins, but rather must reflect some other aspect of biology. There is no obvious change in Cbln1-ir intensity in PF neurons of Cbln2-null mice and Cbln1 mRNA levels are unaltered in thalamus of Cbln2-null mice, indicating the lack of phenotype in striatum cannot be attributed to a marked increase or decrease in Cbln1 levels. Furthermore, the proteins appear to be located in the same subcellular compartment, the endolysosomal system, and loss of Cbln2 does not affect the subcellular localization of Cbln1. Thus functional differences between the two knockout strains cannot readily be attributed to dichotomous routes of intracellular trafficking or turnover of Cbln1 and Cbln2. Therefore, the lack of a Cbln1-null like phenotype in the striatum of Cbln2-null mice suggests that these proteins may have distinct functions. Indeed, it would be difficult to understand the evolutionary advantage of having two genes with identical function that are only distinguished by their temporal-spatial expression patterns.
One distinction between Cbln1 and Cbln2 in thalamus is that Cbln2 is already prominently expressed in PF neurons at P7 whereas Cbln1-ir cannot be detected. This contrasts with cerebellum, where Cbln1 is detected even in immature granule cell precursors in the external granule cell layer whereas Cbln3 is only detected in terminally differentiated neurons of the internal granule cell layer (Pang et al. 2000; Miura et al. 2006; Wei et al. 2007). Therefore, whatever its function, Cbln2 is likely involved in processes already operating in the neonatal thalamus, whereas Cbln1’s role is associated with neurobiological processes that emerge or mature later. Another confounder is that Cbln2 (but not Cbln1) is expressed in cortical neurons that may also project to striatal MSNs (Miura et al. 2006). Thus, elimination of Cbln2 may affect two inputs to MSNs that may offset each other.
Cbln1 and Cbln2 bind to all three β-neurexins and Grid2, although Cbln1 binds more avidly than Cbln2 to Grid2. Whereas β-neurexins are ubiquitously expressed in brain, Grid2 is not detectable in striatum, and thus Cbln1 and Cbln2 presumptively interact either with neurexins exclusively in the thalamo-striatal tract or in combination with additional as yet uncharacterized proteins. Given that Cbln1 and Cbln2 have similar binding to neurexins, it would be difficult to explain the lack of a striatal synaptic spine phenotype in the Cbln2-null mice if only neurexins were involved. This is the more so since neurexins are generally considered to be presynaptic proteins (Ushkaryov et al. 1992; Graf et al. 2004; Ko et al. 2009). Therefore, even though there are numerous splice variants of the neurexins, some of which might conceivably discriminate between Cbln1 and Cbln2, a more parsimonious explanation is that additional post-synaptic proteins with selectivity for Cbln1 or Cbln2 mediate the function of these two proteins. The identification of such proteins will provide greater insight into the biology of the Cbln1 family of proteins as well as the signaling pathways in which they function.
Whereas Cbln1 is critical for synaptic integrity in cerebellum, it may not fulfill the same function in other brain regions. In the cerebellum of Cbln1-null mice Purkinje neuron synaptic spine numbers are unaltered but these spines lack their pre-synaptic component (Hirai et al. 2005). In marked contrast in the striatum of Cbln1-null mice there is an increase in synaptic spines on MSNs but these spines are occupied by pre-synaptic terminals (Kusnoor et al. 2010); i.e. there is no detectable loss of synaptic integrity. As the binding of Cbln1 and Cbln2 to neurexins is so robust and Grid2 is present at such low levels outside of cerebellum, the role of the Cbln1 family may be to regulate the properties of presynaptic neurexins rather than serving as a bi-functional ligand bridging pre- and postsynaptic membrane proteins. A number of scenarios can be envisioned. First, neurexins are known to be important for synaptic transmission and exocytosis (Missler et al. 2003). Therefore, binding of Cblns to neurexins could potentially influence their membrane recycling, with secondary consequences on phenomena such as neurotransmitter vesicle release. Second, neurexins and their known ligands, the neuroligins (Ichtchenko et al. 1995) and leucine-rich repeat transmembrane proteins (Laurén et al. 2003), have been implicated in regulating the development/determination of glutamatergic and GABAergic synapses (Ko et al. 2009; Baudouin et al. 2010; Zhang et al. 2010). Therefore, the binding of Cbln1 family members to pre-synaptic neurexins may potentiate or inhibit their interactions with one or more of their postsynaptic ligands, thereby influencing synapse determination rather than synapse stability. The generation of the L7-Cbln2 transgenic and Cbln2 knockout mice reported here will enable studies aimed at addressing such hypotheses.
Acknowledgments
This work was supported in part by the NCI Cancer Center Support Grant CA 21765, National Institutes of Health grants NS040361 and NS042828, and ALSAC (American Lebanese Syrian Associated Charities) to J. I. M. and NS44282 and MH-077298 and National Parkinson Foundation Center of Excellence at Vanderbilt to A. Y. D.
We thank Dr. R. J. Smeyne, Department of Developmental Neurobiology, St. Jude Children’s Research Hospital, for advice with neuroanatomical analysis. We thank the Hartwell Center for Bioinformatics and Biotechnology at St Jude Children’s Research Hospital for all DNA sequencing and synthesis.
Abbreviations used
- AM
anteromedial nucleus
- ANOVA
one-way analysis of variance
- Cbln
cerebellin precursor protein
- DAB
Diaminobenzidine
- DIG
digoxigenin
- HA
hemagglutinin
- LH
lateral thalamic nucleus
- MSN
medium spiny neurons
- Nrxn
neurexin
- PF
parafascicular nucleus
- PFA
paraformaldehyde
- qRT-PCR
quantitative real time PCR
- TUNEL
terminal deoxynucleotidyl transferase dUTP nick end labeling
- VL, VM, and VP
ventrolateral, ventromedial, and ventroposterior nucleus
- WT
wild type
Footnotes
The authors declare no conflicts of interest.
References
- Araki K, Meguro H, Kushiya E, Takayama C, Inoue Y, Mishina M. Selective expression of the glutamate receptor channel delta 2 subunit in cerebellar Purkinje cells. Biochem Biophys Res Commun. 1993;197:1267–1276. doi: 10.1006/bbrc.1993.2614. [DOI] [PubMed] [Google Scholar]
- Bao D, Pang Z, Morgan JI. The structure and proteolytic processing of Cbln1 complexes. J Neurochem. 2005;95:618–29. doi: 10.1111/j.1471-4159.2005.03385.x. [DOI] [PubMed] [Google Scholar]
- Bao D, Pang Z, Morgan MA, Parris J, Rong Y, Li L, Morgan JI. Cbln1 is essential for interaction-dependent secretion of Cbln3. Mol Cell Biol. 2006;26:9327–37. doi: 10.1128/MCB.01161-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudouin S, Scheiffele P. SnapShot: Neuroligin-neurexin complexes. Cell. 2010;141:908–908. doi: 10.1016/j.cell.2010.05.024. [DOI] [PubMed] [Google Scholar]
- Buitrago MM, Schulz JB, Dichgans J, Luft AR. Short and long-term motor skill learning in an accelerated rotarod training paradigm. Neurobiol Learn Mem. 2004;81:211–216. doi: 10.1016/j.nlm.2004.01.001. [DOI] [PubMed] [Google Scholar]
- Doughty ML, De Jager PL, Korsmeyer SJ, Heintz N. Neurodegeneration in Lurcher mice occurs via multiple cell death pathways. J Neurosci. 2000;20:3687–94. doi: 10.1523/JNEUROSCI.20-10-03687.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson AH, Blobel G. Early events in the biosynthesis of the lysosomal enzyme cathepsin D. J Biol Chem. 1979;254:11771–4. [PubMed] [Google Scholar]
- Graf ER, Zhang X, Jin SX, Linhoff MW, Craig AM. Neurexins induce differentiation of GABA and glutamate postsynaptic specializations via neuroligins. Cell. 2004;119:1013–1026. doi: 10.1016/j.cell.2004.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai H, Pang Z, Bao D, Miyazaki T, Li L, Miura E, Parris J, Rong Y, Watanabe M, Yuzaki M, Morgan JI. Cbln1 is essential for synaptic integrity and plasticity in the cerebellum. Nat Neurosci. 2005;8:1534–41. doi: 10.1038/nn1576. [DOI] [PubMed] [Google Scholar]
- Gordon JW. Production of transgenic mice. Guide to techniques in mouse development. In: Wassarman P, DePamphilis ML, editors. Method in enzymology. Vol. 225. 1993. pp. 747–770. [Google Scholar]
- Ichtchenko K, Hata Y, Nguyen T, Ullrich B, Missler M, Moomaw C, Südhof TC. Neuroligin 1: a splice site-specific ligand for beta-neurexins. Cell. 1995;81:435–43. doi: 10.1016/0092-8674(95)90396-8. [DOI] [PubMed] [Google Scholar]
- Joo JY, Lee SJ, Uemura T, Yoshida T, Yasumura M, Watanabe M, Mishina M. Differential interactions of cerebellin precursor protein (Cbln) subtypes and neurexin variants for synapse formation of cortical neurons. Biochem Biophys Res Commun. 2011;406:627–32. doi: 10.1016/j.bbrc.2011.02.108. [DOI] [PubMed] [Google Scholar]
- Kashiwabuchi N, Ikeda K, Araki K, Hirano T, Shibuki K, Takayama C, Inoue Y, Kutsuwada T, Yagi T, Kang Y, Aizawa S, Mishina M. Impairment of motor coordination, Purkinje cell synapse formation, and cerebellar long-term depression in GluRδ2 mutant mice. Cell. 1995;81:245–252. doi: 10.1016/0092-8674(95)90334-8. [DOI] [PubMed] [Google Scholar]
- Ko J, Fuccillo MV, Malenka RC, Südhof TC. LRRTM2 Functions as a Neurexin Ligand in Promoting Excitatory Synapse Formation. Neuron. 2009;64:791–798. doi: 10.1016/j.neuron.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara H, Hashimoto K, Kano M, Takayama C, Sakimura K, Mishina M, Inoue Y, Watanabe M. Impaired Parallel Fiber→Purkinje Cell Synapse Stabilization during Cerebellar Development of Mutant Mice Lacking the Glutamate Receptor δ2 Subunit. J Neurosci. 1997;17:9613–23. doi: 10.1523/JNEUROSCI.17-24-09613.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusnoor SV, Parris J, Muly EC, Morgan JI, Deutch AY. Extracerebellar role for Cerebellin1: modulation of dendritic spine density and synapses in striatal medium spiny neurons. J Comp Neurol. 2010;518:2525–37. doi: 10.1002/cne.22350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurén J, Airaksinen MS, Saarma M, Timmusk T. A novel gene family encoding leucine-rich repeat transmembrane proteins differentially expressed in the nervous system. Genomics. 2003;81:411–421. doi: 10.1016/s0888-7543(03)00030-2. [DOI] [PubMed] [Google Scholar]
- Liu P, Jenkins NA, Copeland NG. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003;13:476–484. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda K, Miura E, Miyazaki T, Kakegawa W, Emi K, Narumi S, Fukazawa Y, Ito-Ishida A, Kondo T, Shigemoto R, Watanabe M, Yuzaki M. Cbln1 is a ligand for an orphan glutamate receptor delta2, a bidirectional synapse organizer. Science. 2010;328:363–8. doi: 10.1126/science.1185152. [DOI] [PubMed] [Google Scholar]
- Missler M, Zhang W, Rohlmann A, Kattenstroth G, Hammer RE, Gottmann K, Sudhof TC. Alpha-neurexins couple Ca2+ channels to synaptic vesicle exocytosis. Nature. 2003;423:939–948. doi: 10.1038/nature01755. [DOI] [PubMed] [Google Scholar]
- Miura E, Iijima T, Yuzaki M, Watanabe M. Distinct expression of Cbln family mRNAs in developing and adult mouse brains. Eur J Neurosci. 2006;24:750–756. doi: 10.1111/j.1460-9568.2006.04950.x. [DOI] [PubMed] [Google Scholar]
- Mugnaini E, Morgan JI. The neuropeptide cerebellin is a marker for two similar neuronal circuits in rat brain. Proc Natl Acad Sci USA. 1987;84:8692–8696. doi: 10.1073/pnas.84.23.8692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberdick J, Smeyne RJ, Mann JR, Zackson S, Morgan JI. A promoter that drives transgene expression in cerebellar Purkinje and retinal bipolar neurons. Science. 1990;248:223–6. doi: 10.1126/science.2109351. [DOI] [PubMed] [Google Scholar]
- Pang Z, Zuo J, Morgan JI. Cbln3, a novel member of the precerebellin family that binds specifically to Cbln1. J Neurosci. 2000;20:6333–9. doi: 10.1523/JNEUROSCI.20-17-06333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters A, Kaiserman-Abramof IR. The small pyramidal neuron of the rat cerebral cortex. The perikaryon, dendrites and spines. Am J Anat. 1970;127:321–355. doi: 10.1002/aja.1001270402. [DOI] [PubMed] [Google Scholar]
- Sotelo C, Changeux JP. Transsynaptic degeneration ‘en cascade’ in the cerebellar cortex of staggerer mutant mice. Brain Res. 1974;67:519–526. doi: 10.1016/0006-8993(74)90499-5. [DOI] [PubMed] [Google Scholar]
- Triarhou LC. Rate of neuronal fallout in a transsynaptic cerebellar model. Brain Res Bull. 1998;47:219–222. doi: 10.1016/s0361-9230(98)00076-8. [DOI] [PubMed] [Google Scholar]
- Uemura T, Lee SJ, Yasumura M, Takeuchi T, Yoshida T, Ra M, Taguchi R, Sakimura K, Mishina M. Trans-synaptic interaction of GluRdelta2 and Neurexin through Cbln1 mediates synapse formation in the cerebellum. Cell. 2010;141:1068–79. doi: 10.1016/j.cell.2010.04.035. [DOI] [PubMed] [Google Scholar]
- Urade Y, Oberdick J, Molinar-Rode R, Morgan JI. Precerebellin is a cerebellum-specific protein with similarity to the globular domain of complement C1q B chain. Proc Natl Acad Sci USA. 1991;88:1069–73. doi: 10.1073/pnas.88.3.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushkaryov YA, Petrenko AG, Geppert M, Sudhof TC. Neurexins: synaptic cell surface proteins related to the alpha-latrotoxin receptor and laminin. Science. 1992;257:50–56. doi: 10.1126/science.1621094. [DOI] [PubMed] [Google Scholar]
- Vogel MW, Sunter K, Herrup K. Numerical matching between granule and Purkinje cells in lurcher chimeric mice: a hypothesis for the trophic rescue of granule cells from target related cell death. J Neurosci. 1989;9:3454–3462. doi: 10.1523/JNEUROSCI.09-10-03454.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Parris J, Li L, Morgan JI. The carboxypeptidase-like substrate-binding site in Nna1 is essential for the rescue of the Purkinje cell degeneration (pcd) phenotype. Mol Cell Neurosci. 2006;33:200–13. doi: 10.1016/j.mcn.2006.07.009. [DOI] [PubMed] [Google Scholar]
- Wang T, Morgan JI. The Purkinje cell degeneration (pcd) mouse: an unexpected molecular link between neuronal degeneration and regeneration. Brain Res. 2007;1140:26–40. doi: 10.1016/j.brainres.2006.07.065. [DOI] [PubMed] [Google Scholar]
- Wei P, Blundon JA, Rong Y, Zakharenko SS, Morgan JI. Impaired Locomotor Learning and Altered Cerebellar Synaptic Plasticity in pep-19/pcp4-Null Mice. Mol Cell Biol. 2011;31:2838–2844. doi: 10.1128/MCB.05208-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei P, Rong Y, Li L, Bao D, Morgan JI. Characterization of trans-neuronal trafficking of Cbln1. Mol. Cell Neurosci. 2009;41:258–73. doi: 10.1016/j.mcn.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei P, Smeyne RJ, Bao D, Parris J, Morgan JI. Mapping of Cbln1-like immunoreactivity in adult and developing mouse brain and its localization to the endolysosomal compartment of neurons. Eur J Neurosci. 2007;26:2962–78. doi: 10.1111/j.1460-9568.2007.05913.x. [DOI] [PubMed] [Google Scholar]
- Wetts R, Herrup K. Direct correlation between Purkinje and granule cell number in the cerebella of lurcher chimeras and wild-type mice. Brain Res. 1983;312:41–47. doi: 10.1016/0165-3806(83)90119-0. [DOI] [PubMed] [Google Scholar]
- Zhang C, Atasoy D, Araç D, Yang X, Fucillo MV, Robison AJ, Ko J, Brunger AT, Südhof TC. Neurexins physically and functionally interact with GABA(A) receptors. Neuron. 2010;66:403–416. doi: 10.1016/j.neuron.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]